Abstract
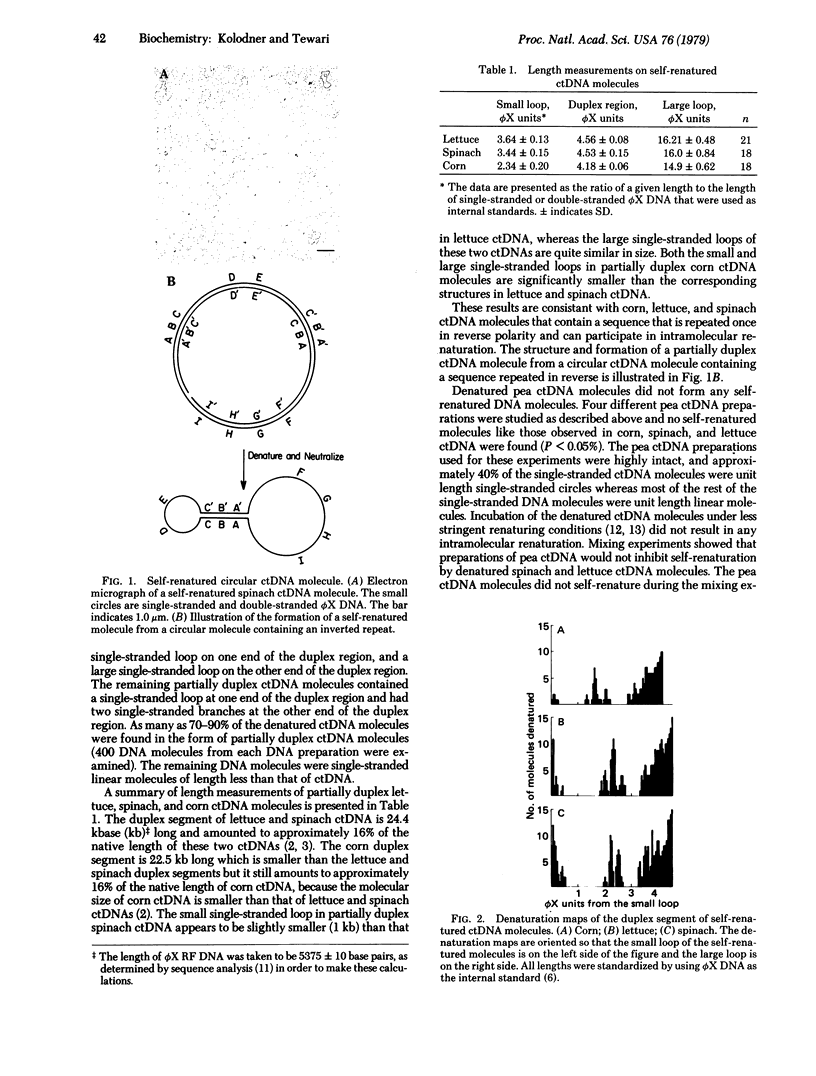
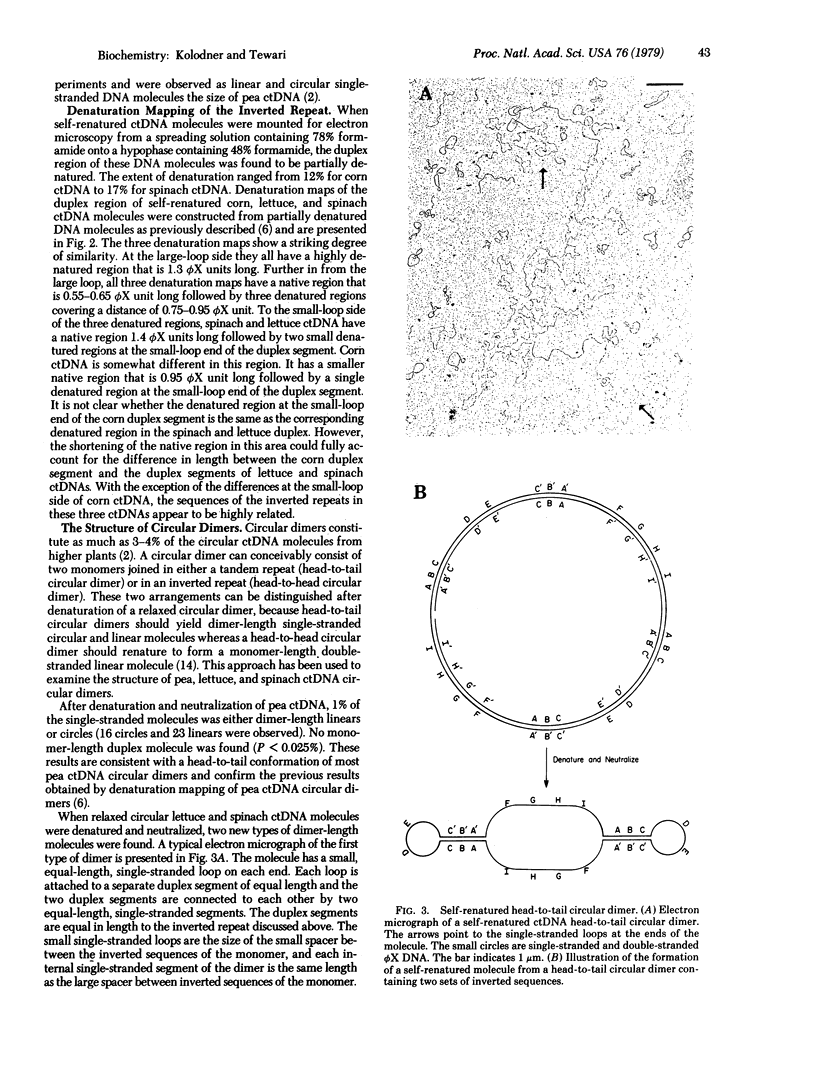
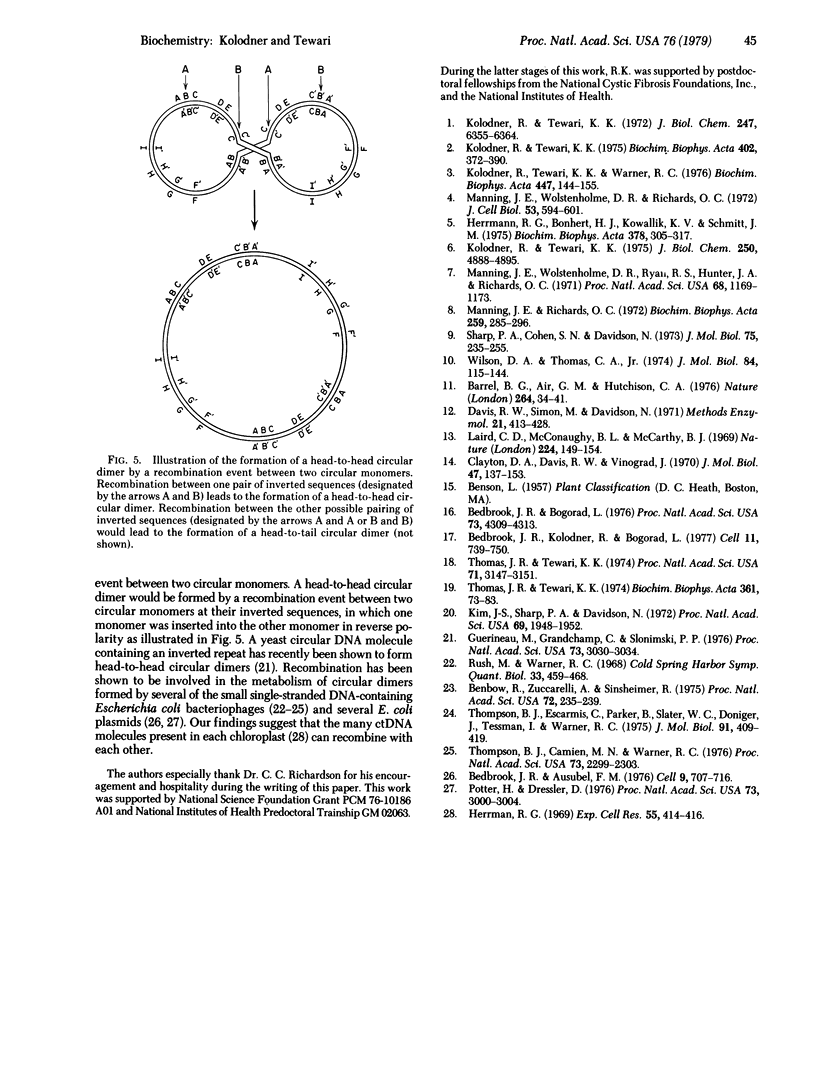
The circular chloroplast DNAs from spinach, lettuce, and corn plants have been examined by electron microscopy and shown to contain a large sequence repeated one time in reverse polarity. The inverted sequence in spinach and lettuce chloroplast DNA has been found to be 24,400 base pairs long. The inverted sequence in the corn chloroplast DNA is 22,500 base pairs long. Denaturation mapping studies have shown that the structure of the inverted sequence is highly conserved in these three plants. Pea chloroplast DNA does not contain an inverted repeat. All of the circular dimers of pea chloroplast DNA are found to be in a head-to-tail confirmation. Circular dimers of spinach and lettuce were also found to have head-to-tail conformation. However, approximately 70-80% of the circular dimers in preparations of lettuce and spinach chloroplast DNA were found to be in a head-to-head conformation. We propose that the head-to-head circular dimers are formed by a recombination event between two circular monomers in the inverted sequence.
Keywords: circular DNA, electron microscopy, denaturation mapping, circular dimers
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrell B. G., Air G. M., Hutchison C. A., 3rd Overlapping genes in bacteriophage phiX174. Nature. 1976 Nov 4;264(5581):34–41. doi: 10.1038/264034a0. [DOI] [PubMed] [Google Scholar]
- Bedbrook J. R., Ausubel F. M. Recombination between bacterial plasmids leading to the formation of plasmid multimers. Cell. 1976 Dec;9(4 Pt 2):707–716. doi: 10.1016/0092-8674(76)90134-3. [DOI] [PubMed] [Google Scholar]
- Bedbrook J. R., Bogorad L. Endonuclease recognition sites mapped on Zea mays chloroplast DNA. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4309–4313. doi: 10.1073/pnas.73.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook J. R., Kolodner R., Bogorad L. Zea mays chloroplast ribosomal RNA genes are part of a 22,000 base pair inverted repeat. Cell. 1977 Aug;11(4):739–749. doi: 10.1016/0092-8674(77)90288-4. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Zuccarelli A. J., Sinsheimer R. L. Recombinant DNA molecules of bacteriophage phi chi174. Proc Natl Acad Sci U S A. 1975 Jan;72(1):235–239. doi: 10.1073/pnas.72.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A., Davis R. W., Vinograd J. Homology and structural relationships between the dimeric and monomeric circular forms of mitochondrial DNA from human leukemic leukocytes. J Mol Biol. 1970 Jan 28;47(2):137–153. doi: 10.1016/0022-2836(70)90335-9. [DOI] [PubMed] [Google Scholar]
- Guerineau M., Grandchamp C., Slonimski P. P. Circular DNA of a yeast episome with two inverted repeats: structural analysis by a restriction enzyme and electron microscopy. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3030–3034. doi: 10.1073/pnas.73.9.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann R. G. Are chloroplasts polyploid? Exp Cell Res. 1969 Jun;55(3):414–416. doi: 10.1016/0014-4827(69)90576-x. [DOI] [PubMed] [Google Scholar]
- Herrmann R. G., Bohnert H. J., Kowallik K. V., Schmitt J. M. Size, conformation and purity of chloroplast DNA of some higher plants. Biochim Biophys Acta. 1975 Jan 20;378(2):305–317. doi: 10.1016/0005-2787(75)90118-5. [DOI] [PubMed] [Google Scholar]
- Kim J., Sharp P. A., Davidson N. Electron microscope studies of heteroduplex DNA from a deletion mutant of bacteriophage phiX-174. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1948–1952. doi: 10.1073/pnas.69.7.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner R. D., Tewari K. K. Denaturation mapping studies on the circular chloroplast deoxyribonucleic acid from pea leaves. J Biol Chem. 1975 Jul 10;250(13):4888–4895. [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. Molecular size and conformation of chloroplast deoxyribonucleic acid from pea leaves. J Biol Chem. 1972 Oct 10;247(19):6355–6364. [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K. The molecular size and conformation of the chloroplast DNA from higher plants. Biochim Biophys Acta. 1975 Sep 1;402(3):372–390. doi: 10.1016/0005-2787(75)90273-7. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Tewari K. K., Warner R. C. Physical studies on the size and structure of the covalently closed circular chloroplast DNA from higher plants. Biochim Biophys Acta. 1976 Oct 4;447(2):144–155. doi: 10.1016/0005-2787(76)90338-5. [DOI] [PubMed] [Google Scholar]
- Laird C. D., McConaughy B. L., McCarthy B. J. Rate of fixation of nucleotide substitutions in evolution. Nature. 1969 Oct 11;224(5215):149–154. doi: 10.1038/224149a0. [DOI] [PubMed] [Google Scholar]
- Manning J. E., Richards O. C. Isolation and molecular weight of circular chloroplast DNA from Euglena gracilis. Biochim Biophys Acta. 1972 Feb 15;259(3):285–296. doi: 10.1016/0005-2787(72)90304-8. [DOI] [PubMed] [Google Scholar]
- Manning J. E., Wolstenholme D. R., Richards O. C. Circular DNA molecules associated with chloroplasts of spinach, Spinacia oleracea. J Cell Biol. 1972 May;53(2):594–601. doi: 10.1083/jcb.53.2.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning J. E., Wolstenholme D. R., Ryan R. S., Hunter J. A., Richards O. C. Circular chloroplast DNA from Euglena gracilis. Proc Natl Acad Sci U S A. 1971 Jun;68(6):1169–1173. doi: 10.1073/pnas.68.6.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter H., Dressler D. On the mechanism of genetic recombination: electron microscopic observation of recombination intermediates. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3000–3004. doi: 10.1073/pnas.73.9.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush M. G., Warner R. C. Molecular recombination in a circular genome-phi X174 and S13. Cold Spring Harb Symp Quant Biol. 1968;33:459–466. doi: 10.1101/sqb.1968.033.01.053. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Thomas J. R., Tewari K. K. Conservation of 70S ribosomal RNA genes in the chloroplast DNAs of higher plants. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3147–3151. doi: 10.1073/pnas.71.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. R., Tewari K. K. Ribosomal-RNA genes in the chloroplast DNA of pea leaves. Biochim Biophys Acta. 1974 Aug 15;361(1):73–83. doi: 10.1016/0005-2787(74)90210-x. [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Escarmis C., Parker B., Slater W. C., Doniger J., Tessman I., Warner R. C. Figure-8 configuration of dimers of S13 and phiX174 replicative form DNA. J Mol Biol. 1975 Feb 5;91(4):409–419. doi: 10.1016/0022-2836(75)90269-7. [DOI] [PubMed] [Google Scholar]
- Wilson D. A., Thomas C. A., Jr Palindromes in chromosomes. J Mol Biol. 1974 Mar 25;84(1):115–138. doi: 10.1016/0022-2836(74)90216-2. [DOI] [PubMed] [Google Scholar]






