Abstract
Unlike activated CD4+ T cells, resting CD4+ T cells are highly resistant to productive HIV-1 infection1–8. Early after HIV-1 entry, a major block limits reverse transcription of incoming viral genomes. Here we show that the deoxynucleoside triphosphate triphosphohydrolase SAMHD1 prevents reverse transcription of HIV-1 RNA in resting CD4+ T cells. SAMHD1 is abundantly expressed in resting CD4+ T cells circulating in peripheral blood and residing in lymphoid organs. The early restriction to infection in unstimulated CD4+ T cells is overcome by HIV-1 or HIV-2 virions into which viral Vpx is artificially or naturally packaged, respectively, or by addition of exogenous deoxynucleosides. Vpx-mediated proteasomal degradation of SAMHD1 and elevation of intracellular deoxynucleotide pools precede successful infection by Vpx-carrying HIV. Resting CD4+ T cells from healthy donors following SAMHD1 silencing or from a patient with Aicardi-Goutières syndrome homozygous for a nonsense mutation in SAMHD1 were permissive for HIV-1 infection. Thus, SAMHD1 imposes an effective restriction to HIV-1 infection in the large pool of noncycling CD4+ T cells in vivo. Bypassing SAMHD1 was insufficient for the release of viral progeny, implicating other barriers at later stages of HIV replication. Together, these findings may unveil new ways to interfere with the immune evasion and T cell immunopathology of pandemic HIV-1.
Human CD4+ T lymphocytes are the major target cells of HIV-1. Activated CD4+ T cells are highly permissive to infection, but resting CD4+ T cells circulating in peripheral blood and residing in lymphoid organs resist infection. Blocks to infection in these cells occur at different post-entry steps of the viral life cycle1–10.
The accessory lentiviral gene product protein X (Vpx) is encoded by HIV-2 and its related simian immunodeficiency virus (SIV) strains, but not by HIV-1. It is incorporated into virions via an interaction motif in the p6 domain of the structural polyprotein Gag. Notably, delivery of Vpx by virus-like particles or by virion-packaging allows HIV-1 to overcome the early post-entry restriction at the level of reverse transcription in dendritic cells, monocytes and macrophages11–17. Here we asked whether Vpx would allow HIV-1 to infect resting human CD4+ T cells. To efficiently incorporate Vpx into HIV-1 virions, we added a Vpx-interaction motif into the Gag p6 of the CXCR4-tropic HIV-1 strain NL4-3 to generate a replication-competent HIV-1 GFP reporter virus (HIV-1* GFP). Resting CD4+ T cells generally support efficient entry of CXCR4-tropic HIV.
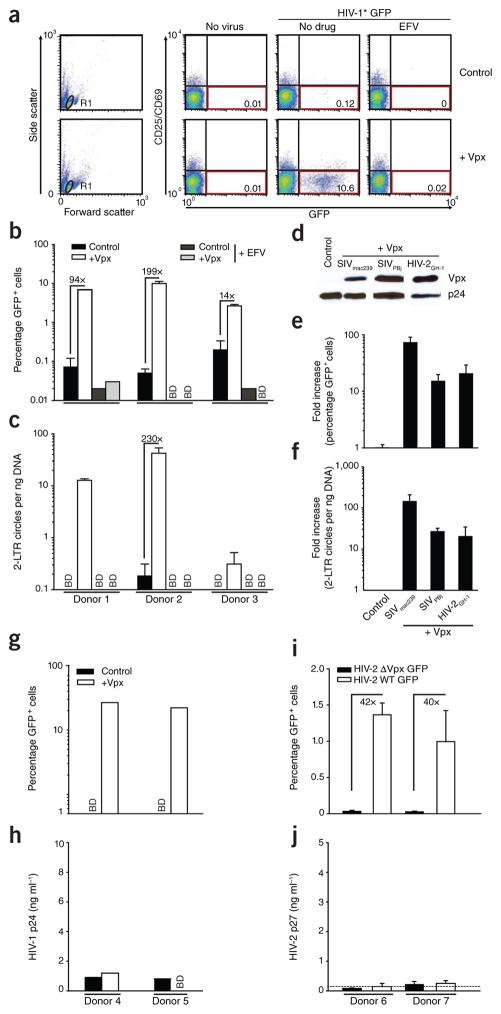
CD4+ T cells purified from peripheral blood of healthy donors were challenged with the engineered HIV-1* GFP virions with or without packaged Vpx. Most CD4+ T cells had a resting phenotype18 with little or no expression of the activation markers CD25 and CD69, and no indicators of cell proliferation (Supplementary Fig. 1) or cell cycle progression (Supplementary Fig. 2). As expected2–6, resting CD4+ T cells were virtually refractory to infection with HIV-1* GFP (Fig. 1a; 0.12% GFP+ cells). In contrast, infection with HIV-1* GFP virions containing Vpx from SIVmac239 (HIV-1* GFP plus Vpx) yielded >10% of CD25−CD69−CD4+ T cells that were GFP+ 3 d after challenge (Fig. 1a) with GFP expression reflecting early viral gene expression from reverse- transcribed, integrated HIV-1 genomes. Vpx enhanced HIV-1 infection of resting CD4+ T cells in a donor-dependent manner by more than 30-fold (mean: 31.3 ± 8.4; range: 2.2–199.4-fold) (Fig. 1b). Vpx also increased levels of late reverse transcription products (RU5/gag) (near full-length HIV-1 cDNA) and 2-LTR circles (HIV-1 cDNA byproduct as a marker of successful nuclear entry) (Fig. 1c and Supplementary Fig. 3), which is consistent with an effect of the accessory lentiviral protein on HIV-1 reverse transcription. Furthermore, the infection rate and accumulation of 2-LTR circles in resting CD4+ T cells challenged with HIV-1* GFP virions that had incorporated Vpx proteins of SIVPBj or HIV-2GH-1 were similar to those observed with HIV-1* GFP virions containing Vpx from SIVmac239 (Fig. 1d–f), indicating that this is a conserved function of lentiviral Vpx proteins.
Figure 1.
Vpx overcomes a restriction to HIV-1 and HIV-2 infection in resting human CD4+ T cells. (a–c) Resting CD4+ T cells were challenged with equivalent infectious units of HIV-1* GFP virions with (+Vpx) or without (Control) incorporated Vpx from SIVmac239 and analyzed on day 3 after infection. The reverse transcriptase inhibitor efavirenz (EFV) served as specificity control. (a) Dot plots of flow cytometric analysis for 1 out of 23 donors. The percentages of viable (gate R1, left graphs) resting (CD25−CD69−) and infected (GFP+) CD4+ T cells are shown in the bottom right quadrants. (b,c) Percentage of GFP+ cells (b) and relative copy numbers of HIV-1 2-LTR circles (c). Bars represent means of triplicates + s.d., with the factors of increase by Vpx indicated. (d) Immunoblotting of HIV-1 virions for incorporated epitope-tagged Vpx proteins from SIVmac239, SIVPBj or HIV-2GH-1. HIV-1 p24 served as a loading control. (e,f) Percentage of GFP+ cells (e) and relative 2-LTR circle copy numbers (f) of resting CD4+ T cells challenged with the Vpx-carrying viruses relative to the Vpx-negative control virus. Bars represent means + s.e.m. of the factors of increase (n = 4). Percentages of GFP+ cells ranged from 0.5% to 3.1% for Vpx of SIVmac239, from 0.2% to 0.7% for Vpx of SIVPBj and from 0.1% to 0.9% for Vpx of HIV-2GH-1. (g–j) Resting CD4+ T cells from the same donors were challenged with equivalent infectious units of HIV-1* GFP virions with (+Vpx) or without (Control) incorporated Vpx (g,h) or HIV-2ROD9 GFP or HIV-2ROD9 ΔVpx GFP (i,j) and analyzed for the percentage of GFP+ cells (g,i) and the amount of HIV-1 p24 or HIV-2 p27, respectively, released into the cell culture supernatant on day 3 after infection (h,j). Bars in i,j represent means of triplicates + s.d. The dotted line in j indicates the cut-off of the p27 ELISA. BD, below detection.
Although Vpx enabled early steps of the HIV-1 life cycle, viral progeny were not released from resting CD4+ T cells (Fig. 1g,h). Virions of the replication-competent HIV-2 strain ROD9 GFP, which naturally encodes Vpx, consistently overcame the early restriction in resting CD4+ T cells in a Vpx-dependent manner, but, similarly to Vpx-carrying HIV-1, they did not lead to the production of new virions (Fig. 1i,j). The susceptibility of resting CD4+ T cells to the Vpx-carrying HIV-1* GFP reached only ~25% of that of activated CD4+ T cells from the same donors challenged with HIV-1* GFP lacking Vpx, and virion-incorporated Vpx did not significantly enhance the HIV-1 susceptibility of activated cells (P = 0.33; Supplementary Fig. 4). Thus, Vpx overcomes a block to the early steps of HIV-1 and HIV-2 replication in resting CD4+ T cells, but additional, Vpx-insensitive blocks probably exist at later stages of the viral life cycle.
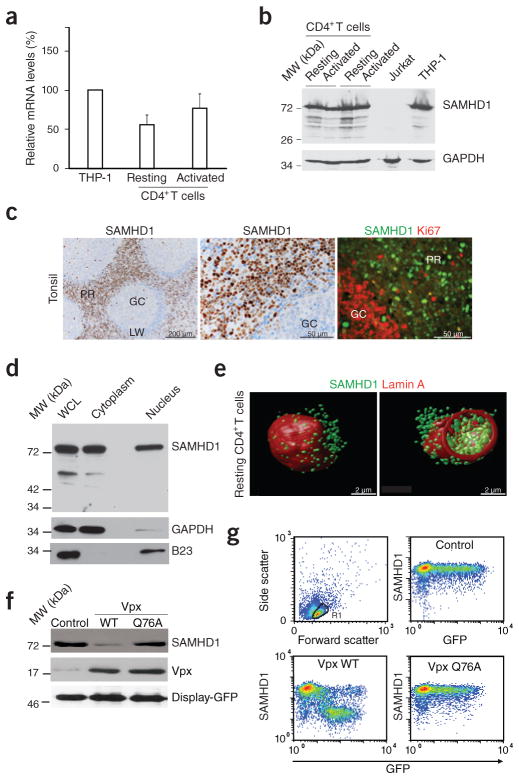
SAMHD1 is expressed in dendritic cells, monocytes and macrophages, but not in T cell lines, and it has been reported to act as a lineage-specific infection barrier for HIV-1 (refs. 19,20). Notably, SAMHD1 is targeted by Vpx for CRL4DCAF1 ubiquitin ligase–dependent proteasomal degradation17,19,20. In spite of this proposed lineage specificity, we detected high levels of endogenous SAMHD1 mRNA and protein in resting CD4+ T cells that were comparable to those in the monocytic THP-1 cell line (Fig. 2a,b). Cell activation with phyto-hemagglutinin (PHA) and interleukin-2 (IL-2) did not affect overall SAMHD1 levels (Fig. 2a,b). SAMHD1 was also abundantly expressed in explants of human tonsil, a lymphoid tissue targeted by HIV-1 in vivo, where the majority of SAMHD1-expressing cells localized to the perifollicular region and were Ki67 negative, that is, nonproliferating (Fig. 2c). Co-staining with cell type–specific markers demonstrated SAMHD1 expression predominantly in CD68+ macrophages, DC-SIGN+ dendritic cells and CD4+ lymphocytes (data not shown). Nuclear-cytoplasmic fractionation and three-dimensional reconstructions of deconvolution confocal images revealed substantial amounts of SAMHD1 in the nucleus and cytoplasm of resting CD4+ T cells (Fig. 2d,e and Supplementary Video 1), activated CD4+ T cells and macrophages (Supplementary Fig. 5). Thus, SAMHD1 might directly deplete levels of dNTPs in the cytoplasm of resting CD4+ T cells where HIV-1 reverse transcription occurs.
Figure 2.
SAMHD1 is expressed in resting CD4+ T cells and depleted by Vpx. (a) RT-PCR–based quantification of SAMHD1 mRNA levels in CD4+ T cells. RNaseP-normalized mRNA levels in resting and PHA- and IL-2-activated CD4+ T cells are presented relative to those from THP-1 cells that were arbitrarily set to 100%. Means + s.e.m. of four donors. (b) Immunoblotting for endogenous expression of SAMHD1 in CD4+ T cells (two donors). GAPDH: loading control. (c) In situ expression of SAMHD1 in explants of human tonsil. Left and center, immunohistochemical detection of SAMHD1. Nuclei were counterstained with hematoxylin. GC, germinal center; LW, lymphocyte wall; PR, perifollicular region. Right, immunofluorescence analysis of SAMHD1 expression (green) relative to the proliferation marker Ki67 (red). (d) SAMHD1 immunoblot analysis of a nuclear-cytoplasmic fractionation of resting CD4+ T cells. WCL, whole cell lysate. GAPDH and B23 served as cytoplasmic and nuclear markers, respectively. (e) Three-dimensional reconstructions of deconvolution confocal microscopy images of a resting CD4+ T cell co-stained for SAMHD1 (green) and the nuclear envelope protein lamin A (red). At right is an optical section across the cell to show nuclear and cytoplasmic SAMHD1. (f) Immunoblots of endogenous SAMHD1 in nucleofected resting CD4+ T cells expressing WT Vpx, Vpx Q76A or an empty vector control. Co-expression of surface-exposed Display-GFP was used to purify a homogenous population of nucleofected cells. Similar results were obtained for two other donors. (g) Quantification of intracellular SAMHD1 amounts in nucleofected resting CD4+ T cells. Cells were nucleofected to co-express GFP together with WT Vpx, Vpx Q76A or an empty control and 16 h later assessed for SAMHD1 expression in relation to GFP by flow cytometry.
To assess the ability of Vpx to degrade endogenous SAMHD1 in resting CD4+ T cells from healthy donors, we co-expressed Myc-tagged SIVmac239 Vpx and a cell surface–exposed GFP variant (Display-GFP; see Online Methods) to facilitate isolation of a homogenous population of viable, nucleofected cells. SAMHD1 pools in both the cytoplasm and the nucleus were depleted after expression of wild-type (WT) Vpx, but not after expression of the CRL4DCAF1 interaction–deficient mutant Q76A16,19 (Fig. 2f and Supplementary Fig. 6), irrespective of the cells’ activation status (Supplementary Fig. 6). We also detected a potent and Q76-dependent reduction of SAMHD1 levels upon Vpx co-expression by intracellular flow cytometry (Fig. 2g and Supplementary Fig. 7). Low levels of co-expressed GFP and WT Vpx were sufficient to maximally degrade SAMHD1 and diminished intracellular SAMHD1 levels by more than 90% (Fig. 2g, left lower panel).
To explore the relationship between Vpx-mediated degradation of SAMHD1 and permissivity to HIV-1 in resting CD4+ T cells, we followed both parameters over time at a single-cell level. Whereas infection with HIV-1* GFP had no effect, SAMHD1 was massively depleted in 20–80% of cells 1 d after challenge with HIV-1* GFP plus SIVmac239 Vpx (Fig. 3a,b and Supplementary Fig. 8b). This degree of depletion was constant until day 3 after infection, when GFP reporter expression indicated proviral integration and early viral gene expression (4.3% of cells in Fig. 3a). Of note, GFP+ cells were detected exclusively within the population of cells containing low amounts of SAMHD1, whereas the residual cell population expressing high amounts of SAMHD1 remained refractory to HIV-1 infection. Virion-incorporated Vpx seemed insufficient to render all resting CD4+ T cells permissive for HIV-1 infection, suggesting either additional Vpx-insensitive limitations for HIV-1 in the post-entry phase2,5,9 or SAMHD1 depletion by Vpx-carrying, but otherwise defective, virions.
Figure 3.
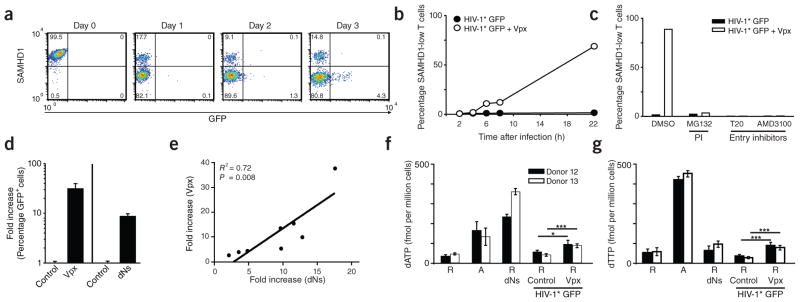
Susceptibility of resting CD4+ T cells to Vpx-carrying HIV-1 is paralleled by proteasomal degradation of SAMHD1 and increased dNTP levels. (a) Time course of SAMHD1 and GFP expression in resting CD4+ T cells after challenge with HIV-1* GFP + Vpx (SIVmac239). The percentages of cells in the respective quadrants are shown. (b) Quantification of SAMHD1 expression in resting CD4+ T cells within the first 22 h after exposure to HIV-1* GFP with (+ Vpx) or without (Control) incorporated Vpx. Data points mark the percentages for cells with low SAMHD1 levels (corresponding to the bottom left quadrants in FACS panels shown in a). (c) Effect of proteasome or HIV-1 entry inhibitors on SAMHD1 levels in resting CD4+ T cells after exposure to HIV-1* GFP ± Vpx. Resting CD4+ T cells were pretreated for 1 h with either the proteasome inhibitor (PI) MG132 (10 μM), the fusion inhibitor T20 (50 μM) or the CXCR4 antagonist AMD3100 (5 μM) before infection with HIV-1* GFP with or without virion-packaged Vpx. Drugs were removed 20 h later. Shown are the percentages of resting CD4+ T cells with low levels of SAMHD1 24 h after infection for one of three donors. (d) Relative factor of increase of infection of resting CD4+ T cells with HIV-1* GFP by virion-packaged Vpx (n = 15) or by incubation with deoxynucleosides (dNs) (n = 8). Means + s.e.m. (e) Correlation between the enhancement of infection of resting CD4+ T cells with HIV-1* GFP either by virion-packaged Vpx or by dN treatment. Depicted is the relative factor of increase in infection for both parameters obtained in parallel experiments for cells from eight different donors. (f,g) Quantification of dATP (f) and dTTP (g) levels in primary CD4+ T cells under different experimental conditions determined by single-nucleotide incorporation assay. R, resting CD4+ T cells; A, activated CD4+ T cells. dNs, cultivation in the presence of dNs (2 mM) for 3 h. Challenge with HIV-1* GFP + Vpx had resulted in depletion of SAMHD1 in 17% (donor 12) or 31% (donor 13) of resting CD4+ T cells 24 h after challenge, respectively, at which time cells were harvested for subsequent dNTP analyses. See also Supplementary Figure 10 for primary data of the single-nucleotide incorporation assay. *P < 0.05; ***P < 0.005.
SAMHD1 degradation in resting CD4+ T cells was specific to HIV-1* GFP virions containing Vpx; it was observed as early as 6 h after virion challenge and correlated in magnitude with the percentage of GFP+ cells (Fig. 3a,b). The peptidic virion fusion inhibitor T20 or the CXCR4 antagonist AMD3100 inhibited Vpx-dependent depletion of SAMHD1 (Fig. 3c), demonstrating that this process strictly depends on virus entry and subsequent release of Vpx from disassembled viral capsids. Furthermore, the depletion of SAMHD1 and the enhancement of resting T cell infection by virion-packaged Vpx was abrogated by treatment of cells with the proteasome inhibitors MG132 (Fig. 3c) or ALLN (data not shown). In line with the targeting of SAMHD1 for proteasomal degradation by Vpx having a central role in HIV-1* GFP infection, incorporation of the Vpx Q76A mutant, which does not deplete cellular pools of SAMHD1, failed to promote infection of resting CD4+ T cells (Supplementary Fig. 8a–c). Similarly, successful HIV-2 GFP infection correlated with Vpx-dependent degradation of SAMHD1 (Fig. 1i and Supplementary Fig. 8d).
In myeloid cells, SAMHD1 hydrolyzes dNTPs, which most likely lowers their concentrations below a threshold required for synthesis of HIV-1 cDNA by reverse transcriptase21–23. To explore this as a mode of restriction for HIV-1 in resting CD4+ T cells, we added pyrimidine and purine deoxynucleosides (dNs) as dNTP precursors to the culture medium. In the absence of Vpx, dNs enhanced the permissivity of resting CD4+ T cells for HIV-1 GFP ninefold (n = 8) over solvent-treated control cells (Fig. 3d) while not affecting the cell cycle or activation status of the cells (Supplementary Fig. 2a and Supplementary Fig. 9). We conducted a side-by-side analysis of the efficacy of virion-packaged Vpx proteins and dN treatment in resting CD4+ T cells from eight donors. The strongly positive correlation of both approaches to augment infection supports a common cellular mechanism that overcomes the HIV-1 restriction (R2 = 0.72; P = 0.008; Fig. 3e). Next, we directly determined the effect of T cell activation or of treatment with either dNs or virion-delivered Vpx on intracellular concentrations of dNTPs in resting CD4+ T cells by the single-nucleotide incorporation assay. PHA- and IL-2-mediated activation of CD4+ T cells increased dATP and dTTP concentrations by 2.9- to 7.8-fold (Fig. 3f,g and Supplementary Fig. 10), as previously reported22,24. Exogenous dN treatment of resting cells resulted, on average, in 4.4-fold higher cellular dNTP levels. Notably, infection of resting CD4+ T cells with HIV-1* GFP also moderately elevated cellular nucleotide concentrations in a Vpx-dependent manner (1.7- to 2.8-fold, P = 0.05–0.005; Fig. 3f,g and Supplementary Fig. 10). This increase was observed despite the relatively low percentage of SAMHD1-depleted cells in this experiment (17% (donor 12) and 31% (donor 13)). Collectively, these results are consistent with the concepts that pools of intracellular dNTPs in resting CD4+ T cells are rate limiting for HIV reverse transcription8 and that SAMHD1 may be a key regulator of this cellular antiviral state.
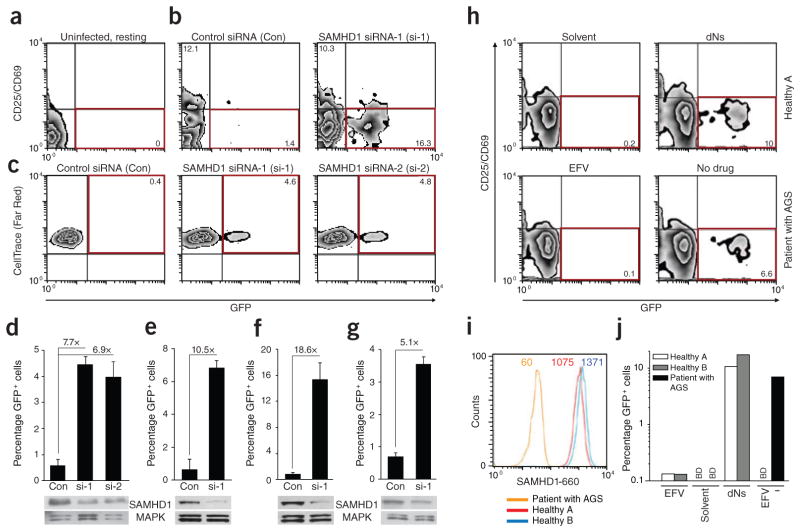
To directly probe the ability of SAMHD1 to restrict HIV-1 infection in resting CD4+ T lymphocytes from healthy donors, we used two RNAi strategies to silence its expression25,26. We activated primary CD4+ T cells to allow effective siRNA delivery by nucleofection or transduction with lentiviral vectors carrying shRNAs. We gradually reduced IL-2 concentrations and analyzed the cells for HIV-1 permissivity once they returned to a quiescent and HIV-1–restrictive state, typically by day 14 after activation (see also Supplementary Figs. 11a and 12a). We classified post-activation CD4+ T cells as resting on the basis of three criteria: (i) their lack of CD25 and CD69 expression, (ii) their lack of proliferation markers and (iii) their resistance to HIV-1 infection after treatment with control siRNA (Con) (Fig. 4a–g and Supplementary Fig. 11b–d). Silencing of SAMHD1 with three independent siRNAs markedly reduced cellular SAMHD1 levels (Fig. 4d–g and Supplementary Fig. 11e) and rendered post-activation resting CD4+ T cells from multiple donors (n = 6) permissive to HIV-1 infection (Fig. 4b–g and Supplementary Fig. 11b–d). The enhancement of HIV-1 infection ranged from 5.1- to 18.6-fold, depending on the donor and knockdown efficiency, corresponding to 3.6–15.3% infected GFP+ cells (Fig. 4d–g and Supplementary Fig. 11d). We observed a similar but slightly less potent increase in HIV-1 infection in post-activation resting CD4+ T cells when SAMHD1 was silenced by shRNA transduction (Supplementary Fig. 12b–d) of two targeting sequences that are distinct from those of the siRNAs used. Thus, depletion of cellular SAMHD1 by RNAi is sufficient to enhance the permissiveness of resting CD4+ T cells for HIV-1 infection. Finally, we wanted to determine whether resting CD4+ T cells with a nonsense mutation in SAMHD1 obtained from a patient with Aicardi-Goutières syndrome (AGS)17,27 were susceptible to HIV-1 infection. By flow cytometric analysis of peripheral blood mononuclear cells (PBMCs), SAMHD1 protein was not detected in CD25−CD69− CD3+CD4+ T cells from this patient. Unlike cells from healthy donors, resting CD4+ T cells from this patient with AGS were intrinsically permissive for HIV-1 GFP infection (Fig. 4h–j).
Figure 4.
Silencing of SAMHD1 by RNA interference or a homozygous nonsense mutation renders resting CD4+ T cells permissive for HIV-1 infection. (a–c) Flow cytometric analysis of freshly isolated, uninfected resting CD4+ T cells (staining reference) (a) or of post-activated resting CD4+ T cells infected with HIV-1 GFP after treatment with the indicated siRNAs (b,c). Red boxes indicate populations of infected cells with relative percentages indicated. Cells’ activation state was assessed by surface expression of CD25 and CD69 (a,b) or CellTrace (Far Red) dilution (c). (d–g) Quantification of the percentage of GFP+ cells and the relative increase of HIV-1 GFP infection in resting CD4+ T cells from four donors (each corresponding to a panel) following silencing with SAMHD1-specific siRNAs (si-1 or si-2) relative to a nontargeting control siRNA (Con). Shown are means + s.d. of triplicate infections. SAMHD1 levels in corresponding cell lysates were determined by immunoblotting. MAPK, loading control. See also Supplementary Figure 11b–e for results with cells from two additional donors, including the use of SAMHD1 si-3. (h) Flow cytometric analyses of PBMCs from healthy donor A (solvent- or dN-treated, top graphs) or PBMCs from a patient with AGS with a homozygous nonsense mutation in the SAMHD1 gene (EFV-treated or untreated, bottom graphs) 3 d after challenge with HIV-1 GFP (multiplicity of infection = 1). The percentages of resting (CD25−CD69−), infected (GFP+) CD3+CD4+ PBMCs are shown in the bottom right quadrants boxed in red, with the percentage of infected cells indicated. (i) Intracellular SAMHD1 expression in resting CD4+ T cells of PBMCs from the patient with AGS and from healthy donors A and B, as determined by flow cytometry. The mean fluorescence intensity of SAMHD1 levels is indicated. (j) Quantification of the percentage of infected (GFP+) resting CD4+ T cells from the patient with AGS and from healthy donors (see panel h for primary data). Shown are arithmetic means of duplicates. BD, below detection.
Our results suggest that HIV-1 reverse transcription is actively suppressed in resting human CD4+ T cells, and we identify SAMHD1 as a long-sought cellular factor that is responsible for this restriction. SAMHD1 thus emerges as a ubiquitous and potent barrier to productive HIV-1 infection in dendritic and myeloid cells17,19,20,22 and also in the large pool of noncycling CD4+ T cells in vivo. Although not excluding additional SAMHD1-dependent mechanisms, the available data are consistent with a scenario in which this restriction is a direct consequence of SAMHD1’s enzymatic activity, which limits the availability of dNTPs required for HIV-1 reverse transcription21–23. Regulation of deoxynucleotide levels in resting cells by SAMHD1 may have evolved to protect T helper cells from retroviral insult without the risk of disrupting the homeostasis of these noncycling cells. Therapeutic manipulation of intracellular dNTP pools thus emerges as a potential antiretroviral strategy. Given the overall potency of virus-encoded SAMHD1 antagonists, such as Vpx, to expand the target cell population of HIV-1 to noncycling cells, it seems surprising that they were lost in lentiviral evolution28,29. This indicates that overcoming SAMHD1 restriction in these types of cells may in fact carry significant adverse consequences for pandemic HIV-1 and that the abortive SAMHD1-restricted infection by HIV-1 contributes to innate sensing-mediated T cell immunopathology1 and immune evasion17,30,31, and thus to AIDS pathogenesis. Further insight into the regulation and consequences of the SAMHD1-imposed restriction in CD4+ T cells will help identify new pathways for interfering with immunodeficiency in HIV-1-infected individuals.
ONLINE METHODS
Ethics statement
Fresh tonsillar tissue was removed during routine tonsillectomy from HIV-, hepatitis B virus– and hepatitis C virus–negative patients with informed consent. The use of anonymous surgical waste was approved by the Ethics Committee of Heidelberg University (approval No. 077/2005). Paraffinized tonsil specimens were provided by the tissue bank of the National Center for Tumor Diseases (Heidelberg, Germany) and approved by the Ethics Committee of Heidelberg University (approval No. 206/2005). Whole blood was obtained from a patient with AGS who signed an informed consent form. The research was approved by the Ethics Committee of the Chamber of Physicians Westfalen-Lippe and the Medical Faculty of the Westfalian Wilhelms University Münster (Reference No 2006-556-f-S) and performed according to the principles expressed in the Declaration of Helsinki.
Cells and reagents
Peripheral blood mononuclear cells (PBMCs) from healthy blood donors were purified by Ficoll-Hypaque gradient centrifugation32. Resting CD4+ T cells were isolated from PBMCs by negative selection with either the CD4+ T Cell Isolation Kit II (Miltenyi Biotec) and separated by the autoMACS Pro Separator or with the RosetteSep Human CD4+ T Cells Enrichment Cocktail (StemCell Technologies). Resting CD4+ T cells were cultured at a density of 2 × 106 cells per ml in RPMI-1640 medium (Gibco) supplemented with 10% heat-inactivated fetal calf serum, glutamine (2 mM) and antibiotics (100 U/ml penicillin, 100 mg/ml streptomycin). For T cell activation, PHA-P (5 μg/ml) or PHA-L (2 μg/ml) (both from Sigma-Aldrich) were added to the culture medium for 2–3 d together with IL-2 (20 or 100 IU/ml) (Biomol). To obtain post-activation resting T cells, the IL-2 concentration was gradually decreased as indicated in Supplementary Figures 11a and 12a. The human cell lines THP-1 and Jurkat were cultivated as reported17,33. Human macrophages were derived from PBMCs and cultivated as previously described32.
The following fluorochrome-conjugated antibodies were used: anti–human CD3 (clone SK7, 1:20), anti-human CD4 (clone RPA-T4, 1:20), anti–human CD8 (clone SK1, 1:20), anti-human CD25 (clone M-A251, 1:40), anti–human CD69 (clone FN50, 1:40) (all from BD Pharmingen). Additional antibodies were: sheep anti–HIV-1 p24 (1:5,000, B. Müller), mouse anti-B23 (1:5,000, Zymed), mouse anti-GAPDH (1:5,000, Santa Cruz Biotechnology), rabbit anti-MAPK (Erk1/2) (1:1,000, Cell Signaling), rabbit anti-SAMHD1 (1:100 for immunofluorescence microscopy, 1:1,000 for western blotting, Proteintech), mouse anti-Ki67 (1:800, GeneTex), mouse anti–lamin A (1:200, Sigma-Aldrich), mouse anti-GFP (1:1,000, Sigma-Aldrich), mouse anti-Myc (1:200, Santa Cruz Biotechnology), mouse anti-HA (1:1,000, Covance) and mouse anti-Flag (1:2,000, Sigma-Aldrich). CellTrace Far Red DDAO-SE was from Invitrogen, and the BrdU Cell Proliferation Assay Kit and 7-AAD were from BD Pharmingen. Pyronin Y was from Sigma-Aldrich.
The following dNs were purchased from Sigma-Aldrich: 2-deoxyadenosine, 2′-deoxyguanosine, thymidine and 2′-deoxycytidine hydrochloride. dNs were dissolved in either RPMI (pH 4.1) or RPMI (pH 7.4; 2′-deoxycytidine hydrochloride). MG132 and ALLN (Sigma-Aldrich) were used as reported34. T20 (enfuvirtide, Roche) and efavirenz (Bristol-Myers Squibb) were used as described35. AMD3100 was a gift from H.-G. Kräusslich. The p24 ELISA (Innotest HIV Antigen mAB, Innogenetics) and p27 ELISA (SIV p27 antigen capture assay, Advanced Bioscience) were performed according to the manufacturer’s protocol.
Plasmids
The infectious HIV-1 GFP proviral clone is based on HIV-1NL4-3 and contains EGFP in the 5′-region of nef and a polypurine track 3′ of EGFP followed by the remaining 3′-LTR sequence (gift from J. Münch). The infectious WT HIV-2 GFP WT and ΔVpx proviral clones, based on pROD9-ΔEnv-GFP and pROD9-ΔEnv-delVpx-GFP from M. Emerman, were generated by reintroduction of Rod9 Env. The Vpx-binding motif DPAVDLL from SIVmac Gag was introduced into p6 of HIV-1 GFP (HIV-1* GFP) by overlapping PCR using the SpeI and SbfI sites of HIV-1 GFP14. Vpx expression constructs used were: pcDNA3.1Vpx SIVmac239-Myc (WT and Q76A)36,37, pcDNA3.1VpxSIVPBj38 and pEF-FVpxHIV-2GH1 (gift from M. Fujita). pDisplay-GFP was from B. Müller.
DNA nucleofection of CD4+ T cells
CD4+ T cells (1 × 107) were nucleofected with either pcDNA3.1Vpx-WT-Myc or pcDNA3.1Vpx-Q76A-Myc (5 μg) together with pDisplay-GFP (3 μg) using the Human T cell Nucleofector Kit (Lonza) (program U-14). Thirty-six hours after nucleofection, cells were incubated with anti-GFP microbeads (Miltenyi Biotec) on ice for 30 min. Display-GFP–expressing cells were isolated by autoMACS Pro Separator selection. Nucleofection of resting CD4+ T cells does not induce T cell activation39.
Virus production
Sucrose cushion–purified HIV-1 virus stocks were produced as previously described40. HIV-1* GFP virus stocks, carrying virion-packaged Vpx, were produced by co-transfection of the proviral HIV-1* GFP DNA and the indicated Vpx expression constructs36. DNase treatment of virus stocks was performed as previously reported41. The infectious titer was determined in a standardized TZM-bl infectivity assay42.
HIV-1 infection
If not stated otherwise, resting primary CD4+ T cells were infected with identical infectious units (105 TZM-bl IU per 106 cells corresponding to a multiplicity of infection (MOI) of 0.1) of HIV-1 GFP or HIV-1*GFP ± Vpx using spin-infection for 90 min at 800 r.p.m. at room temperature. The medium was exchanged 4–6 h after infection. Three days later, cells were stained for CD25 and CD69 expression, fixed in 4% PFA in PBS and analyzed on a FACSCalibur. For the experiment shown in Figure 4h–j, PBMCs (5 × 105) from two healthy donors and one patient with AGS were infected in duplicate with HIV-1 GFP at an MOI of 1 in relation to CD4+ target cells. For dN treatment, PBMCs were preincubated with dNs (2 mM) or solvent for 30 min at 37 °C. On day 3 after infection, cells were washed and stained with PerCP-labeled anti-CD3 (1:20), PE-labeled anti-CD4 (1:20), APC-labeled anti-CD25 and APC-labeled anti-CD69 (both 1:40) monoclonal antibodies (all from BD Pharmingen) for 30 min at 4 °C. After several washing steps with PBS containing 1% BSA, samples were fixed in 4% PFA in PBS for 90 min at room temperature. A BD FACS LSR II SORP instrument and BD FACSDiva software were used for analysis.
Cell fractionation
For nuclear-cytoplasmic fractionation43, resting and activated CD4+ T cells (2 × 107) or macrophages were first washed in cold PBS, followed by incubation in a hypotonic lysis buffer (10 mM HEPES, pH 7.9; 1.5 mM MgCl2; 10 mM KCl; 0.5 mM DDT) containing a protease inhibitor cocktail (Sigma-Aldrich). Next, an aliquot was taken representing the whole cell lysate (WCL). After mechanical disruption of the plasma membrane by douncing, cells were spun twice at 1,700 r.p.m. for 10 min to separate the cytoplasmic and nuclear fraction. Next, an aliquot was taken representing the cytoplasm. The remaining nuclear pellet was resuspended in buffer S1 (0.25 mM sucrose; 10 mM MgCl2) containing protease inhibitors and overlaid with buffer S3 (0.88 mM sucrose; 0.5 mM MgCl2). After centrifugation (3,000 r.p.m., 15 min), the nuclear pellet was resuspended in RIPA buffer and briefly sonicated, and the nuclear fraction was harvested. Aliquots of the respective fractions were mixed with 2× SDS sample buffer and analyzed by immunoblotting.
Immunostaining of tonsil sections
Immunoenzyme staining of paraffinized tonsil tissue was performed as reported44. Briefly, antigen retrieval was achieved by steam-cooking slides in 10 mM citrate buffer (pH 6.1; Dako) for 30 min. A solution of 10% Earle’s balanced salt solution (EBSS, Sigma-Aldrich) supplemented with 1% HEPES, 0.2% BSA and 0.1% saponin (all from Sigma-Aldrich), pH 7.4, was used as a washing, permeabilization and staining buffer. The rabbit anti-SAMHD1 antibody (1:100) was added and incubated overnight. As secondary reagent biotinylated donkey anti–rabbit IgG (1:200; Jackson ImmunoResearch) was applied for 30 min at room temperature, followed by incubation with an avidin-containing ABC-AP Kit (Vectastain; Vector Laboratories). Naphthol AS-biphosphate (Sigma-Aldrich) with New Fuchsin (Merck) was used as the substrate for alkaline phosphatase (AP). Images were acquired on an Olympus BX45 microscope. For double immunofluorescence staining, paraffinized tonsil tissue sections, following antigen retrieval, were incubated overnight with primary antibodies (rabbit anti-SAMHD1 (1:100, Proteintech), mouse anti-Ki67 (1:800, GeneTex)). As secondary reagent, a biotinylated donkey anti–mouse IgM antibody (1:100, Zymed) was applied for 30 min at room temperature, followed by incubation with fluorescently labeled antibodies donkey anti–rabbit IgG Alexa488 (1:2,000, Invitrogen) and streptavidin-Cy3 (1:800, Jackson ImmunoResearch). Images were acquired with an Olympus IX81 microscope.
Immunofluorescence microscopy and three-dimensional reconstruction
Resting and activated CD4+ T cells as well as macrophages were placed on poly-L-lysine–coated cover glasses and fixed for 30 min with 3% paraformaldehyde in PBS. After permeabilization with 0.1% Triton X-100 in PBS, cells were incubated with 2% BSA in PBS for 30 min. Next, cells were stained with rabbit anti-SAMHD1 and mouse anti–lamin A primary antibodies followed by incubation with goat anti-rabbit Alexa568 (1:1,000, Invitrogen) and goat anti-mouse Alexa488 (1:1,000, Invitrogen)-labeled secondary antibodies. Cover glasses were mounted in ProLong Gold antifade reagent (Invitrogen) and analyzed with a six-line spinning-disk confocal microscope (PerkinElmer). The three-dimensional reconstruction of deconvolution images was carried out using the 3D Huygens Deconvolution Software.
Single-nucleotide incorporation assay
The quantification of dATP and dTTP concentrations was performed as reported22,24.
RNA interference
For siRNA-mediated silencing of SAMHD1 (see also schematic in Supplementary Fig. 11a), freshly isolated resting CD4+ T cells (2 ×106 cells per ml) were stimulated overnight with PHA-L (2 μ;g/ml). The following day, the medium was exchanged and cells were cultivated in RPMI medium with 20 IU/ml IL-2. On day 5, the supernatants from each donor culture were collected and filtered, to be subsequently used as conditioned medium. Cells (total of 6 ×107 cells) were subjected to a first nucleofection with SAMHD1-specific siRNA or nontargeting siRNAs (each 2.4 μmol) using the Human T cell Nucleofector Solution and program U-14. The following siRNAs were used: SAMHD1-specific siRNAs: si-1: 5′-GAUUCAUUG UGGCCAUAUA-3′ (Eurofins MWG/ operon); si-2: 5′-CACCUCAAUUUCAACG UCUUCGAUA-3′ (Invitrogen); si-3: 5′-CAACCAGAGCUGCAGAUAA-3′ (Eurofins MWG/operon). Control siRNAs: 5′-AGGUAGUGUAAUCGCCUU GUU-3′; 5′-AGG UAG UGU AAU CGC CUU G-3′ (Eurofins MWG/operon). Nucleofected cells were resuspended in 8 ml culture medium containing 50% conditioned medium and 50% fresh medium with 20 IU/ml IL-2. On day 8, the second siRNA nucleofection was performed analogously to the first. Following nucleofection, cells were resuspended in 6 ml medium containing one-third conditioned medium and two-thirds fresh medium with a final IL-2 concentration of 7 IU/ml. On day 11, 2 ml of medium were replaced with 4 ml of fresh medium without IL-2 to lower the final IL-2 concentration to 3.5 IU/ml. To enhance SAMHD1 silencing, cells from some donors (Fig. 4d–g) underwent a third round of siRNA nucleofection on day 11. On day 12, cells were stained with CellTrace and resuspended in conditioned medium containing 1.75 IU/ml IL-2. On day 14, cells (5×105 per well) were infected with HIV-1 GFP. On day 16, cells were stained with PE-conjugated anti-CD25 and anti-CD69 monoclonal antibodies (both 1:20; BD Pharmingen), fixed and analyzed by flow cytometry.
For shRNA-mediated silencing of SAMHD1 (see schematic in Supplementary Fig. 12a), CD4+ T cells (3 ×106 cells per ml) were stimulated with PHA-P (5 μg/ml) and IL-2 (100 IU/ml). On day 3, activated cells (2 ×107) were transduced by spinoculation with vesicular stomatitis virus G protein–pseudotyped lentiviral vectors carrying either pLK0.1-puro-control-shRNA or pLK0.1-puro-SAMHD1-shRNA2 (ref. 17) in medium containing 50 IU/ml IL-2. On day 5, transduced cells were pooled and cultivated at a concentration of 2 ×107 cells per 6 ml in a 25-cm2 flask in medium containing 30 IU/ml IL-2 and puromycin (1.5 μ;g/ml). On day 7, 5 ml of fresh medium were added with final concentrations of 2 μ;g/ml puromycin and 20 IU/ml IL-2. On day 8, puromycin-resistant, viable cells were purified by Ficoll gradient centrifugation and resuspended in 5 ml fresh medium containing 20 IU/ ml IL-2. On day 10, 50% of the culture medium was replaced with fresh medium to lower the final IL-2 concentration to 10 IU/ml. On day 12, the IL-2 concentration was adjusted to 5 IU/ml. On day 13, cells were stained with CellTrace and cultured in medium containing 2.5 IU/ml IL-2. On day 14, cells (5 ×105 per well) were infected with HIV-1 GFP. On day 16, cells were stained with PE-conjugated anti-CD25 and anti-CD69 mAbs, fixed and analyzed by flow cytometry.
Intracellular SAMHD1 staining by flow cytometry
For fixation and antibody staining, CD4+ T cells were seeded in a 96-well V-bottom plate at 5 ×105 cells per 100 μl per well. Cells were fixed for 90 min at room temperature with BD Cytofix Fixation Buffer and permeabilized in BD Phosflow Perm Buffer III for 2 min at 4 °C in the dark. After two washes in PBS containing 1% FCS and 0.09% sodium azide, cells were stained with the rabbit anti-SAMHD1 antiserum followed by a goat anti-rabbit Alexa660 antibody (1:200, Invitrogen). A FACSCalibur flow cytometer and either BD CellQuest Pro 4.0.2 software or FlowJo software (TreeStar) were used for analyses. For the experiment shown in Figure 4h–j, to detect intracellular SAMHD1 in resting CD4+ T cells, the cells were co-stained with a surface stain antibody mixture of FITC-labeled anti-CD3, PE-labeled anti-CD4, BD Horizon V450–labeled anti-CD25 and BD Horizon V450–labeled anti-CD69 (all 1:20, from BD Bioscience) at room temperature for 1 h. After two washing steps, the samples were analyzed on a BD FACS LSR II SORP instrument.
PCR analyses
TaqMan-based quantifications of HIV-1 2-LTR circles (HIV-1 cDNA byproduct as a marker of successful nuclear entry) in DNA extracts from infected resting CD4+ T cells were performed as described35,45. The TaqMan-based quantification of RU5/gag (near full-length HIV-1 cDNA) was established and validated based on the primer regions chosen by Zack et al.6. The PCR conditions for RU5/gag were: 50 °C for 2 min, 95 °C for 10 min with a denaturation step at 95 °C for 15 s, an annealing step at 55 °C for 30 s, and elongation step at 72 °C for 1 min for 40 cycles. Data acquisition was performed at the elongation step. RNA extraction and TaqMan-based mRNA quantification of SAMHD1 (Applied Biosystems: assay no. Hs00210019_m1) and RNaseP (Applied Biosystems: TaqMan RNase P Control Reagents Kit (4316844), endogenous reference control) were performed as reported46. Relative mRNA levels were calculated as 1/ ΔCt and are presented relative to results obtained for THP-1 cells.
Cell cycle analysis
Cell cycle analysis was performed by staining DNA and RNA of resting and activated CD4+ T cells with 7-amino-actinomycin-D (7-AAD) and pyronin Y, respectively, as previously described47.
Supplementary Material
Acknowledgments
We thank M. Emerman (Fred Hutchinson Cancer Research Center, Seattle, USA; for pROD9, pROD9-ΔEnv-GFP, pROD9-ΔEnv-delVpx-GFP), M. Fujita (Research Institute for Drug Discovery, Kumamoto University, Japan; for pEF-FVpxHIV-2GH1), H.-G. Kräusslich (Department of Infectious Diseases, Virology, University of Heidelberg, Germany; for AMD3100), B. Müller (Department of Infectious Diseases, Virology, University of Heidelberg, Germany; for pDisplay GFP and sheep anti–HIV-1 p24 antibody) and J. Münch (Institute of Molecular Virology, University Hospital Ulm, Germany; for HIV-1 GFP) for reagents. We thank T. Adam, A. Imle, P. Klein, S. Kutscheidt, A. Ruggieri and the Nikon Imaging Center at University of Heidelberg for technical assistance, and G. Howard for editorial assistance. This work was in part funded by the Deutsche Forschungsgemeinschaft (O.T.K., grant KE742/4-1), SFB 938/Z2 (F.L.) and the US National Institutes of Health (GM1041981 and AI049781 to B.K.; and F31 GM095190 to W.D.). O.T.F. and O.T.K. are members of the CellNetworks Cluster of Excellence EXC81 and the German Centre for Infection Research, University of Heidelberg. H.-M.B. is recipient of a fellowship of the Medical Faculty of the University of Heidelberg.
Footnotes
AUTHOR CONTRIBUTIONS
O.T.K. and O.T.F. conceived of the study, designed experiments and wrote the manuscript. H-M.B., X.P., E.E., S. Schmidt, W.D., M.B., K.S., B.K., S.P. and R.K. designed and conducted experiments and discussed and interpreted the data with O.T.F. and O.T.K. I.A., E.F., G.W., T.G. and N.R.L. provided reagents and expertise. S. Sertel, F.R. and F.L. provided tissue samples and expertise and interpreted data.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Note: Supplementary information is available in the online version of the paper.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Doitsh G, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganesh L, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 3.Korin YD, Zack JA. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierson TC, et al. Molecular characterization of preintegration latency in human immunodeficiency virus type 1 infection. J Virol. 2002;76:8518–8531. doi: 10.1128/JVI.76.17.8518-8531.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stevenson M, Stanwick TL, Dempsey MP, Lamonica CA. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zack JA, et al. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 7.Dai J, et al. Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J Virol. 2009;83:4528–4537. doi: 10.1128/JVI.01910-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plesa G, et al. Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. J Virol. 2007;81:13938–13942. doi: 10.1128/JVI.01745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoder A, et al. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swingler S, et al. HIV-1 Nef intersects the macrophage CD40L signalling pathway to promote resting-cell infection. Nature. 2003;424:213–219. doi: 10.1038/nature01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergamaschi A, et al. The human immunodeficiency virus type 2 Vpx protein usurps the CUL4A-DDB1 DCAF1 ubiquitin ligase to overcome a postentry block in macrophage infection. J Virol. 2009;83:4854–4860. doi: 10.1128/JVI.00187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berger G, Goujon C, Darlix JL, Cimarelli A. SIVMAC Vpx improves the transduction of dendritic cells with nonintegrative HIV-1–derived vectors. Gene Ther. 2009;16:159–163. doi: 10.1038/gt.2008.128. [DOI] [PubMed] [Google Scholar]
- 13.Goujon C, et al. Characterization of simian immunodeficiency virus SIVSM/human immunodeficiency virus type 2 Vpx function in human myeloid cells. J Virol. 2008;82:12335–12345. doi: 10.1128/JVI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pertel T, Reinhard C, Luban J. Vpx rescues HIV-1 transduction of dendritic cells from the antiviral state established by type 1 interferon. Retrovirology. 2011;8:49. doi: 10.1186/1742-4690-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharova N, et al. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 2008;4:e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava S, et al. Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate adaptor for cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. 2008;4:e1000059. doi: 10.1371/journal.ppat.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger A, et al. SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 2011;7:e1002425. doi: 10.1371/journal.ppat.1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckstein DA, et al. HIV-1 actively replicates in naive CD4+ T cells residing within human lymphoid tissues. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 19.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell–specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 22.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286:43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diamond TL, et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wabnitz GH, et al. Sustained LFA-1 cluster formation in the immune synapse requires the combined activities of L-plastin and calmodulin. Eur J Immunol. 2010;40:2437–2449. doi: 10.1002/eji.201040345. [DOI] [PubMed] [Google Scholar]
- 26.Santoni de Sio FR, Trono D. APOBEC3G-depleted resting CD4+ T cells remain refractory to HIV1 infection. PLoS ONE. 2009;4:e6571. doi: 10.1371/journal.pone.0006571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiele H, et al. Cerebral arterial stenoses and stroke: novel features of Aicardi-Goutieres syndrome caused by the Arg164X mutation in SAMHD1 are associated with altered cytokine expression. Hum Mutat. 2010;31:E1836–E1850. doi: 10.1002/humu.21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim ES, et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11:194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laguette N, et al. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe. 2012;11:205–217. doi: 10.1016/j.chom.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manel N, et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010;467:214–217. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manel N, Littman DR. Hiding in plain sight: how HIV evades innate immune responses. Cell. 2011;147:271–274. doi: 10.1016/j.cell.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keppler OT, et al. Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J Virol. 2001;75:8063–8073. doi: 10.1128/JVI.75.17.8063-8073.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haller C, et al. The HIV-1 pathogenicity factor Nef interferes with maturation of stimulatory T-lymphocyte contacts by modulation of N-Wasp activity. J Biol Chem. 2006;281:19618–19630. doi: 10.1074/jbc.M513802200. [DOI] [PubMed] [Google Scholar]
- 34.Goffinet C, et al. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe. 2009;5:285–297. doi: 10.1016/j.chom.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Goffinet C, Allespach I, Keppler OT. HIV-susceptible transgenic rats allow rapid preclinical testing of antiviral compounds targeting virus entry or reverse transcription. Proc Natl Acad Sci USA. 2007;104:1015–1020. doi: 10.1073/pnas.0607414104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunseri N, O’Brien M, Bhardwaj N, Landau NR. Human immunodeficiency virus type 1 modified to package simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J Virol. 2011;85:6263–6274. doi: 10.1128/JVI.00346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gramberg T, Sunseri N, Landau NR. Evidence for an activation domain at the amino terminus of simian immunodeficiency virus Vpx. J Virol. 2010;84:1387–1396. doi: 10.1128/JVI.01437-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger A, et al. Interaction of Vpx and apolipoprotein B mRNA-editing catalytic polypeptide 3 family member A (APOBEC3A) correlates with efficient lentivirus infection of monocytes. J Biol Chem. 2010;285:12248–12254. doi: 10.1074/jbc.M109.090977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keppler OT, Tibroni N, Venzke S, Rauch S, Fackler OT. Modulation of specific surface receptors and activation sensitization in primary resting CD4+ T lymphocytes by the Nef protein of HIV-1. J Leukoc Biol. 2006;79:616–627. doi: 10.1189/jlb.0805461. [DOI] [PubMed] [Google Scholar]
- 40.Geuenich S, et al. Aqueous extracts from peppermint, sage and lemon balm leaves display potent anti-HIV-1 activity by increasing the virion density. Retrovirology. 2008;5:27. doi: 10.1186/1742-4690-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tervo HM, Goffinet C, Keppler OT. Mouse T-cells restrict replication of human immunodeficiency virus at the level of integration. Retrovirology. 2008;5:58. doi: 10.1186/1742-4690-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keppler OT, et al. Rodent cells support key functions of the human immunodeficiency virus type 1 pathogenicity factor Nef. J Virol. 2005;79:1655–1665. doi: 10.1128/JVI.79.3.1655-1665.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruggieri A, et al. Human endogenous retrovirus HERV-K(HML-2) encodes a stable signal peptide with biological properties distinct from Rec. Retrovirology. 2009;6:17. doi: 10.1186/1742-4690-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erikson E, et al. In vivo expression profile of the antiviral restriction factor and tumor-targeting antigen CD317/BST-2/HM1.24/tetherin in humans. Proc Natl Acad Sci USA. 2011;108:13688–13693. doi: 10.1073/pnas.1101684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swiggard WJ, et al. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J Virol. 2005;79:14179–14188. doi: 10.1128/JVI.79.22.14179-14188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goffinet C, Schmidt S, Kern C, Oberbremer L, Keppler OT. Endogenous CD317/ Tetherin limits replication of HIV-1 and murine leukemia virus in rodent cells and is resistant to antagonists from primate viruses. J Virol. 2010;84:11374–11384. doi: 10.1128/JVI.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhoeyen E, et al. IL-7 surface-engineered lentiviral vectors promote survival and efficient gene transfer in resting primary T lymphocytes. Blood. 2003;101:2167–2174. doi: 10.1182/blood-2002-07-2224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.