Abstract
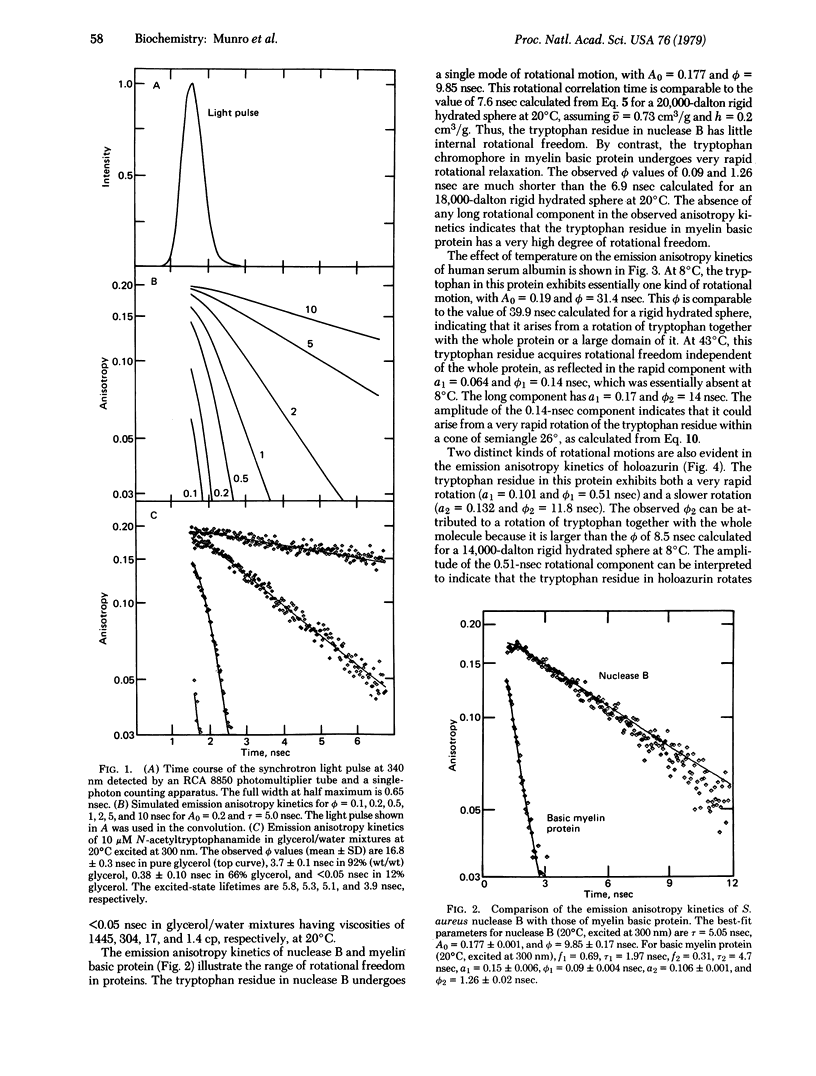
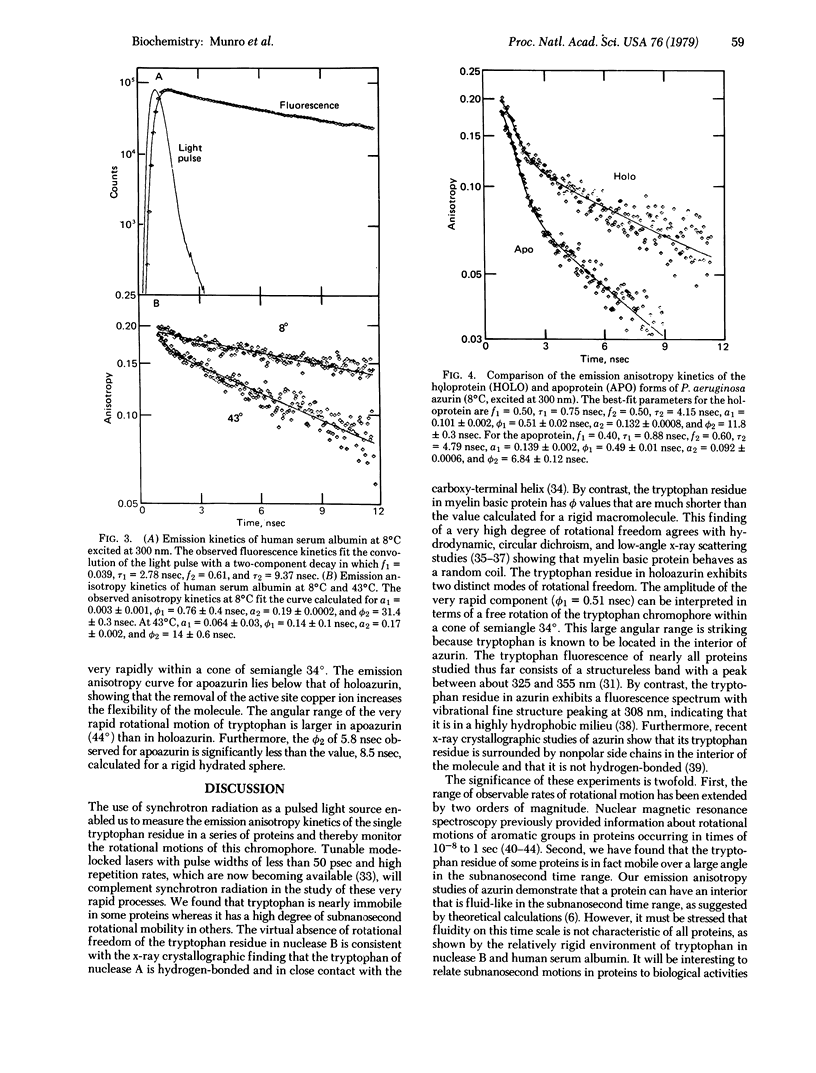
The dynamics of protein molecules in the subnanosecond and nanosecond time range were investigated by time-resolved fluorescence polarization spectroscopy. Synchrotron radiation from a storage ring was used as a pulsed light source to excite the single tryptophan residue in a series of proteins. The full width at half maximum of the detected light pulse was 0.65 nsec, making it feasible to measure emission anisotropy kinetics in the subnanosecond time range and thereby to resolve internal rotational motions. The proteins investigated exhibit different degrees of rotational freedom of their tryptophan residue, ranging from almost no mobility to nearly complete freedom in the subnanosecond time range. The tryptophan residue of Staphylococcus aureus nuclease B (20,000 daltons) has a single rotational correlation time (ϕ) of 9.9 nsec at 20°C, corresponding to a rotation of the whole protein molecule. By contrast, bovine basic A1 myelin protein (18,000 daltons) exhibits ϕ of 0.09 and 1.26 nsec, showing that the tryptophan residue in this protein is highly flexible. The single tryptophan of human serum albumin (69,000 daltons) has almost no rotational freedom at 8°C (ϕ = 31.4 nsec), whereas at 43°C it rotates rapidly (ϕ1 = 0.14 nsec) within a cone of semiangle 26° in addition to rotating together with the whole protein (ϕ2 = 14 nsec). Of particular interest in the large angular range (semiangle, 34°) and fast rate (ϕ1 = 0.51 nsec) of the rotational motion of the tryptophan residue in Pseudomonas aeruginosa azurin (14,000 daltons). This residue is known to be located in the hydrophobic interior of the protein. The observed amplitudes and rates of these internal motions of tryptophan residues suggest that elementary steps in functionally significant conformational changes may take place in the subnanosecond time range.
Keywords: protein dynamics, rotational relaxation, fluorescence polarization, emission anisotropy, synchrotron radiation
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T., Stenkamp R. E., Sieker L. C., Jensen L. H. A crystallographic model for azurin a 3 A resolution. J Mol Biol. 1978 Jul 25;123(1):35–47. doi: 10.1016/0022-2836(78)90375-3. [DOI] [PubMed] [Google Scholar]
- Belford G. G., Belford R. L., Weber G. Dynamics of fluorescence polarization in macromolecules. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1392–1393. doi: 10.1073/pnas.69.6.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R. Structural origins of mammalian albumin. Fed Proc. 1976 Aug;35(10):2141–2144. [PubMed] [Google Scholar]
- Cotton F. A., Bier C. J., Day V. W., Hazen E. E., Jr, Larsen S. Some aspects of the structure of staphylococcal nuclease. I. Crystallographic studies. Cold Spring Harb Symp Quant Biol. 1972;36:243–249. doi: 10.1101/sqb.1972.036.01.032. [DOI] [PubMed] [Google Scholar]
- Davis A., Moore I. B., Parker D. S., Taniuchi H. Nuclease B. A possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1977 Sep 25;252(18):6544–6553. [PubMed] [Google Scholar]
- Eylar E. H., Brostoff S., Hashim G., Caccam J., Burnett P. Basic A1 protein of the myelin membrane. The complete amino acid sequence. J Biol Chem. 1971 Sep 25;246(18):5770–5784. [PubMed] [Google Scholar]
- Eylar E. H., Caccam J., Jackson J. J., Westall F. C., Robinson A. B. Experimental allergic encephalomyelitis: synthesis of disease-inducing site of the basic protein. Science. 1970 Jun 5;168(3936):1220–1223. doi: 10.1126/science.168.3936.1220. [DOI] [PubMed] [Google Scholar]
- Eylar E. H., Thompson M. Allergic encephalomyelitis: the physico-chemical properities of the basic protein encephalitogen from bovine spinal cord. Arch Biochem Biophys. 1969 Feb;129(2):468–479. doi: 10.1016/0003-9861(69)90204-5. [DOI] [PubMed] [Google Scholar]
- Finazzi-Agrò A., Rotilio G., Avigliano L., Guerrieri P., Boffi V., Mondovì B. Environment of copper in Pseudomonas fluorescens azurin: fluorometric approach. Biochemistry. 1970 Apr 28;9(9):2009–2014. doi: 10.1021/bi00811a023. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Schlessinger J., Pecht I., Steinberg I. Z. Homogeneity and variability in the structure of azurin molecules studied by fluorescence decay and circular polarization. Biochemistry. 1975 May 6;14(9):1921–1929. doi: 10.1021/bi00680a018. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. On the analysis of fluorescence decay kinetics by the method of least-squares. Anal Biochem. 1974 Jun;59(2):583–598. doi: 10.1016/0003-2697(74)90312-1. [DOI] [PubMed] [Google Scholar]
- Grinvald A. The use of standards in the analysis of fluorescence decay data. Anal Biochem. 1976 Sep;75(1):260–280. doi: 10.1016/0003-2697(76)90077-4. [DOI] [PubMed] [Google Scholar]
- Hammes G. G., Wu C. W. Kinetics of allosteric enzymes. Annu Rev Biophys Bioeng. 1974;3(0):1–33. doi: 10.1146/annurev.bb.03.060174.000245. [DOI] [PubMed] [Google Scholar]
- Hetzel R., Wüthrich K., Deisenhofer J., Huber R. Dynamics of the aromatic amino acid residues in the globular conformation of the basic pancreatic trypsin inhibitor (BPTI). II. Semi-empirical energy calculations. Biophys Struct Mech. 1976 Aug 23;2(2):159–180. doi: 10.1007/BF00863707. [DOI] [PubMed] [Google Scholar]
- Hirshfeld H., Teitelbaum D., Arnon R., Sela M. Basic encephalitogenic protein: A simplified purification on sulphoethyl-sephadex. FEBS Lett. 1970 May 1;7(4):317–320. doi: 10.1016/0014-5793(70)80193-4. [DOI] [PubMed] [Google Scholar]
- Hull W. E., Sykes B. D. Fluorotyrosine alkaline phosphatase: internal mobility of individual tyrosines and the role of chemical shift anisotropy as a 19F nuclear spin relaxation mechanism in proteins. J Mol Biol. 1975 Oct 15;98(1):121–153. doi: 10.1016/s0022-2836(75)80105-7. [DOI] [PubMed] [Google Scholar]
- Kinosita K., Jr, Kawato S., Ikegami A. A theory of fluorescence polarization decay in membranes. Biophys J. 1977 Dec;20(3):289–305. doi: 10.1016/S0006-3495(77)85550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krigbaum W. R., Hsu T. S. Molecular conformation of bovine A1 basic protein, a coiling macromolecule in aqueous solution. Biochemistry. 1975 Jun 3;14(11):2542–2546. doi: 10.1021/bi00682a038. [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Weber G. Quenching of protein fluorescence by oxygen. Detection of structural fluctuations in proteins on the nanosecond time scale. Biochemistry. 1973 Oct 9;12(21):4171–4179. doi: 10.1021/bi00745a021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy C., Holowka D. A., Cathou R. E. Nanosecond fluorescence spectroscopy of pyrenebutyrate anti-pyrene antibody complexes. Biochemistry. 1977 Aug 9;16(16):3668–3672. doi: 10.1021/bi00635a025. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Dynamics of folded proteins. Nature. 1977 Jun 16;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- Mendelson R. A., Morales M. F., Botts J. Segmental flexibility of the S-1 moiety of myosin. Biochemistry. 1973 Jun 5;12(12):2250–2255. doi: 10.1021/bi00736a011. [DOI] [PubMed] [Google Scholar]
- Rosen P., Pecht I. Conformational equilibria accompanying the electron transfer between cytochrome c (P551) and azurin from Pseudomonas aeruginosa. Biochemistry. 1976 Feb 24;15(4):775–786. doi: 10.1021/bi00649a008. [DOI] [PubMed] [Google Scholar]
- Snyder G. H., Rowan R., 3rd, Karplus S., Sykes B. D. Complete tyrosine assignments in the high field 1H nuclear magnetic resonance spectrum of the bovine pancreatic trypsin inhibitor. Biochemistry. 1975 Aug 26;14(17):3765–3777. doi: 10.1021/bi00688a008. [DOI] [PubMed] [Google Scholar]
- Veatch W. R., Stryer L. Effect of cholesterol on the rotational mobility of diphenylhexatriene in liposomes: a nanosecond fluorescence anisotrophy study. J Mol Biol. 1977 Dec 25;117(4):1109–1113. doi: 10.1016/s0022-2836(77)80017-x. [DOI] [PubMed] [Google Scholar]
- Visscher R. B., Gurd F. R. Rotational motions in myoglobin assessed by carbon 13 relaxation measurements at two magnetic field strengths. J Biol Chem. 1975 Mar 25;250(6):2238–2242. [PubMed] [Google Scholar]
- WEBER G. Fluorescence-polarization spectrum and electronic-energy transfer in proteins. Biochem J. 1960 May;75:345–352. doi: 10.1042/bj0750345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEBER G. Fluorescence-polarization spectrum and electronic-energy transfer in tyrosine, tryptophan and related compounds. Biochem J. 1960 May;75:335–345. doi: 10.1042/bj0750335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber G. Ligand binding and internal equilibria in proteins. Biochemistry. 1972 Feb 29;11(5):864–878. doi: 10.1021/bi00755a028. [DOI] [PubMed] [Google Scholar]
- Wüthrich K., Wagner G. NMR investigations of the dynamics of the aromatic amino acid residues in the basic pancreatic trypsin inhibitor. FEBS Lett. 1975 Feb 1;50(2):265–268. doi: 10.1016/0014-5793(75)80504-7. [DOI] [PubMed] [Google Scholar]
- Yguerabide J., Epstein H. F., Stryer L. Segmental flexibility in an antibody molecule. J Mol Biol. 1970 Aug;51(3):573–590. doi: 10.1016/0022-2836(70)90009-4. [DOI] [PubMed] [Google Scholar]
- Yguerabide J. Nanosecond fluorescence spectroscopy of macromolecules. Methods Enzymol. 1972;26:498–578. doi: 10.1016/s0076-6879(72)26026-8. [DOI] [PubMed] [Google Scholar]


