Abstract
Epigenome is an emerging field that demands selective cell-permeable chemical probes to perturb, especially in vivo, the activity of specific enzymes involved in modulating the epigenetic codes. Coactivator Associated Arginine (R) Methyltransferase 1 (CARM1) is a coactivator of estrogen receptor α (ERα), the main target in human breast cancer. We previously showed that overexpression of CARM1 by two-fold in MCF7 breast cancer cells increased the expression of ERα-target genes involved in differentiation and reduced cell proliferation, leading to the hypothesis that activating CARM1 by chemical activators may be therapeutically effective in breast cancer. Selective, potent, cell-permeable CARM1 activators will be essential to test this hypothesis. Here we report the development of a cell-based, time-resolved Förster resonance energy transfer (TR-FRET) assay using poly (A) binding protein 1 (PABP1) methylation to monitor cellular activity of CARM1. The LanthaScreen® TR-FRET assay utilizes MCF7 cells expressing GFP-PABP1 fusion protein via BacMam gene delivery system, methyl-PABP1 specific antibody and terbium-labeled secondary antibody. This assay has been validated to reflect the expression and/or activity of CARM1 and optimized for high throughput screening to identify CARM1 allosteric activators. This TR-FRET platform serves as a generic tool for functional screening of cell-permeable, chemical modulators of CARM1 for elucidation of its in vivo functions.
Keywords: Arginine methylation, CARM1, PABP1, TR-FRET
Introduction
Epigenetic modifications of DNA and histone proteins, including methylation, acetylation and phosphorylation, are widely recognized to have significant effects on gene expression and human health. Changes in the epigenetic marks in cancer cells have stimulated great interest in the enzymes that generate, remove, or recognize the marks on chromatin.[1] A few histone modification enzymes are already strongly implicated in some types of cancer and are targets for “epigenetic therapies”, e.g., inhibitors of histone deacetylases are FDA-approved for treatment of cutaneous T cell lymphoma.[2] A critical barrier in the field is the lack of cell-permeable, potent and selective chemical modulators to facilitate study the roles of the many proteins that are responsible for epigenetic modifications in normal cells and disease and to validate the therapeutic potential of these targets.[3] Most chemical biology and drug discovery efforts have focused on the identification and design of small-molecule inhibitors as loss-of-function probes to perturb enzyme function, yet activators provide a means to better understand the de novo enzyme activation mechanism and the sufficiency of an enzyme to drive a particular cellular phenotype in normal and disease conditions.[4]
Coactivator associated arginine (R) methyltransferase 1 (CARM1) belongs to the type I protein arginine methyltransferase (PRMT) family, which asymmetrically dimethylate protein substrates on arginines.[5] CARM1 functions as a transcriptional coactivator with a series of cancer-relevant transcription factors including NF-kB, p53, E2F1, β-catenin, and steroid receptors, among which coactivation of estrogen receptor alpha (ERα) targets is best characterized.[6] ERα regulates a number of genes that are essential for the etiology and progression of breast cancer. CARM1 has a variety of protein substrates, making it a multifunctional protein engaged in diverse cellular processes. For instance, CARM1 methylates histone H3 at R17 and R26,[7] which correlates with activation of ERα-target genes.[8] In addition, CARM1 methylates a number of non-histone proteins including RNA polymerase II,[9] transcription co-factor CBP/p300,[10] RNA binding proteins and RNA splicing factors,[11] as well as poly (A) binding protein 1 (PABP1).[12] Importantly, loss of CARM1 in the mouse embryo leads to abrogation of the estrogen response and reduced expression of some ERα-target genes, further highlighting the functional importance of CARM1 in ERα-regulated gene expression.[13] The enzyme-defective CARM1 knock-in mice have defects similar to the CARM1 knockout counterparts, underlining the indispensability of enzymatic activity of CARM1 for its in vivo functions.[14] Moreover, our lab has shown CARM1 to be a unique ERα coactivator that can simultaneously inhibit cell proliferation and induce differentiation through global regulation of ERα-regulated genes in ERα-positive breast cancer cells.[15]
In addition to its significance in breast cancer and the estrogen signaling pathway, CARM1 also plays important roles in other biological processes. CARM1 is essential for cartilage development and endochondral ossification,[16] and is required for proper differentiation of adipocytes,[17] myocytes,[18] and pulmonary alveolar cells.[19] The expression and the associated methyltransferase activity of CARM1 were also reported to be necessary for regulating genes involved in glycogen metabolism in skeletal muscle cells and human glycogen storage diseases.[20] Furthermore, CARM1 was recently implicated in normal T cell cellularity and differentiation, functioning as a key epigenetic regulator of fetal hematopoiesis and thymocyte development.[21] Given the crucial roles of CARM1, small-molecule modulators able to enhance or inhibit enzymatic activity of CARM1 will be useful chemical tools for the mechanistic study of CARM1 in physiological and pathological processes.
Numerous strategies have been pursued to screen small-molecule inhibitors of CARM1 and other methyltransferases, including an in vitro methylation assay, microfluidic capillary electrophoresis, an enzyme-coupled continuous spectrophotometric assay or an AlphaScreen assay.[22] These assays, restricted by sensitivity, throughput and workflow, were not applicable for high-throughput screening (HTS) of potent small-molecule modulators of CARM1. To circumvent these problems, we developed an HTS compatible, homogenous LanthaScreen® cellular assay using time-resolved Förster resonance energy transfer (TR-FRET) technology, for monitoring CARM1 cellular activity. The time-resolved detection circumvents the issues that green fluorescence has light scatter and compound could have autofluorescence. The LanthaScreen® TR-FRET technology has been used for monitoring p53 acetylation[23] and histone H3 lysine site-specific modifications.[24] To our knowledge, it has not been used for monitoring arginine methylation, nor for HTS of a large compound library. In this report, we showed that cellular PABP1 methylation is a suitable reporter for CARM1 cellular activity. A TR-FRET assay was developed based on the methylation of GFPPABP1 and several key parameters have been optimized for HTS. Moreover, we validated that the TR-FRET signal appropriately responded to the addition of methyltransferase inhibitor or synthetic CARM1 activators, and performed well in a pilot screen using the National Institutes of Health (NIH) Clinical Collection Library. The results indicate that this TR-FRET platform is suitable for HTS to identify small-molecule activators of CARM1.
Results
A TR-FRET assay for monitoring CARM1 cellular activity
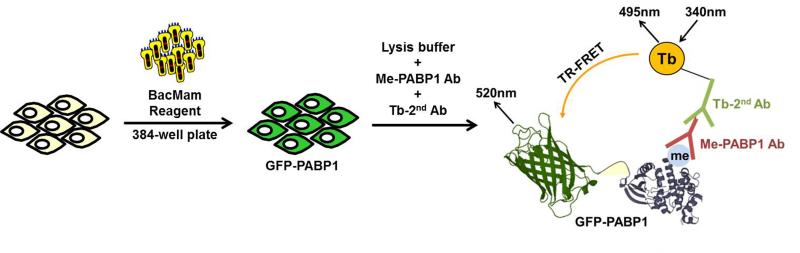
Although several assays have been reported for the discovery of small-molecule inhibitors of CARM1, most of them relied on biochemical assays using purified CARM1 protein and its protein or peptide substrates; these assays do not recapitulate CARM1 cellular activity. In order to screen for chemical activators of CARM1 in the biologically-relevant cellular milieu, we took advantage of LanthaScreen®, a high throughput compatible TR-FRET method developed by Life Technologies Incorporation.[23-24] LanthaScreen® utilizes a terbium (Tb)-labeled antibody to detect modifications of a GFP-fused substrate protein in cell lysates. We adopted LanthaScreen® to monitor CARM1 activity in cells using a Tb-labelled antibody to detect methylated GFP-PABP1 (Me-GFP-PABP1) (Figure 1). PABP1 is asymmetrically methylated by CARM1 at R455 and R460,[12] and our lab generated a rabbit polyclonal antibody to the methylated PABP1 (Me-PABP1 Ab). We used a Tb-labeled goat anti-rabbit secondary antibody (Tb-2nd Ab) as a donor fluorophore. GFP-PABP1 fusion proteins (acceptor) were generated as BacMam virus and transiently expressed in MCF7 cells. Although we are aware that stable cell lines are preferred for high throughput screening (HTS), the BacMam system (Life Technologies Inc., Madison) has been used for gene delivery of targets into cells for drug discovery by the pharmaceutical industry.[25] 24 hours after BacMam GFPPABP1 virus infection, cells were lysed directly in the assay plate with lysis buffer supplemented with both the Me-PABP1 Ab and Tb-labelled 2nd Ab. After 1 hour incubation, the formation of the ternary complex containing Me-GFP-PABP1, Me-PABP1 Ab, and Tb-2nd Ab was detected by fluorescence energy transfer between the Tb donor and the GFP acceptor. The TR-FRET ratio of the GFP-specific signal measured at 520 nm to Tb-specific signal measured at 495 nm reflected the level of Me-GFP-PABP1.
Figure 1. Schematic representation of Me-GFP-PABP1 TR-FRET assay for detecting CARM1 cellular activity.
MCF7 cells are infected with BacMam virus encoding a GFP-PABP1 fusion protein and plated in a 384-well assay plate. 24 hours after infection, cells are lysed and incubated for 1 hour in the presence of Me-PABP1 specific primary antibody (Me-PABP1 Ab) and goat anti-rabbit IgG chemically labeled with a terbium chelate (Tb-2nd Ab). After incubation, Tb is excited at 340 nm, and TR-FRET emission ratio of 520 nm over 495 nm is obtained. The association between the antibody and Me-GFP-PABP1 allows energy transfer to occur between the excited-state donor fluorophore Tb and acceptor fluorophore GFP, leading to an increase of the TR-FRET ratio. TR-FRET ratio obtained with Tb-2nd Ab alone is subtracted as background signal.
Cellular Me-PABP1 level is proportional to CARM1 expression level and the enzymatic activity
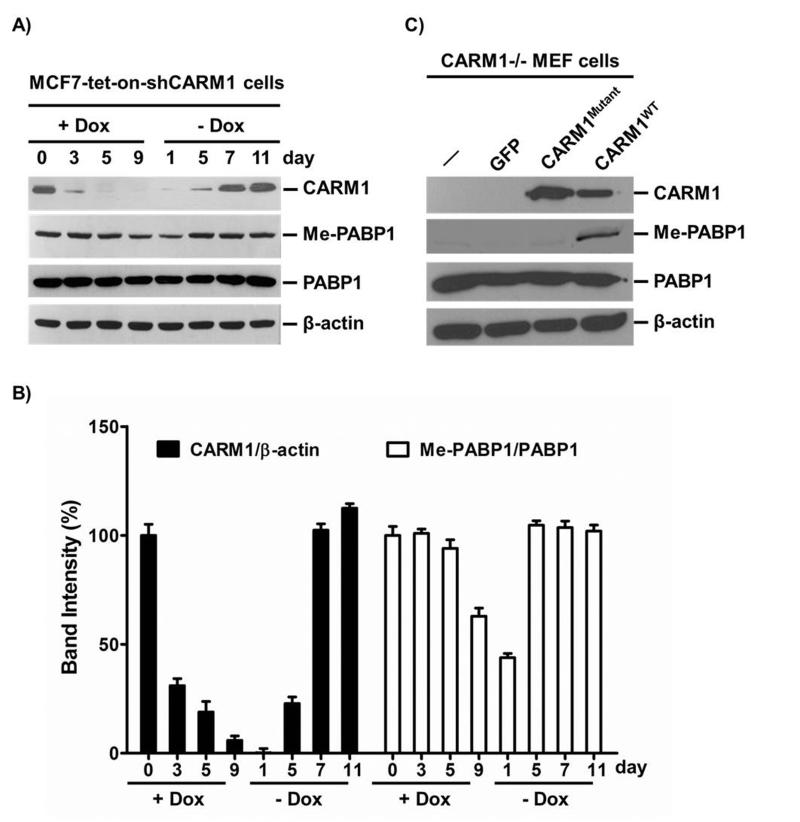
The specificity of the Me-GFP-PABP1 TR-FRET assay depends on the specificity and efficacy of association between substrate and the Me-PABP1 Ab. Although PABP1 is a major cellular substrate of CARM1, we needed to determine whether the level of Me-PABP1 detected by the antibody reflected differences in CARM1 expression or enzymatic activity. We had previously established a CARM1 shRNA inducible expressing MCF7 cell line (MCF7-tet-on-shCARM1), where CARM1 expression level can be dynamically regulated by the addition and withdrawal of doxycyclin (Dox).[15] As shown in Figure 2A and 2B, endogenous CARM1 was gradually depleted in the presence of Dox and was almost undetectable after 5 days of Dox treatment as measured by Western blot. Ten days post-treatment, Dox was removed by replacing with regular medium to allow CARM1 recovery, and the endogenous CARM1 level gradually recovered up to the normal level after 7 days. Correspondingly, the Me-PABP1 level was decreased at day 9 with CARM1 knockdown and recovered by day 5 when CARM1 was restored (Figures 2A & 2B). The slow and incomplete decrease of Me-PABP1 levels may be due to passive loss of methylated PABP1 accompanied with cell division, as no arginine demethylating enzyme has been discovered to date.[26] Consistent with this rationale, a more dramatic decrease of Me-PABP1 was observed after 15 days of Dox treatment (Figure S1). Total PABP1 levels were not affected by the loss or recovery of CARM1 (Figure 2A). To further determine if CARM1 enzymatic activity is required for restoration of Me-PABP1, wild-type CARM1 (CARM1WT) or enzyme-defective mutant CARM1 (CARM1Mutant)[27] were expressed at similar levels in CARM1 null mouse embryonic fibroblast (CARM1−/− MEF) cells which were shown to have undetectable Me-PABP1 levels (Figure 2C). Expression of CARM1WT but not CARM1Mutant restored the endogenous Me-PABP1 level, which was specifically recognized by our Me-PABP1 Ab. Collectively, these results suggest that cellular Me-PABP1 level is a bona fide reporter for CARM1 expression level and enzymatic activity, and that our Me-PABP1 Ab is specific and effective for recognizing Me-PABP1.
Figure 2. Cellular Me-PABP1 level is proportional to CARM1 expression level and the enzymatic activity.
(A) MCF7-tet-on-shCARM1 cells were treated with 500 ng/ml Dox for the indicated time. 10 days post-treatment, Dox was removed by replacing with regular medium to allow CARM1 recovery. CARM1, Me-PABP1 and total PABP1 were examined by Western blots. β-actin was used as a loading control. The Me-PABP1 level was reduced by day nine of Dox treatment and restored by day 5 after removal of Dox. (B) Quantification of CARM1 and Me-PABP1 Western blots in (A). The X-ray films were scanned and digitalized. The density of each band was measured using the Quantity One 1-D analysis software (Bio-Rad). The band intensity of CARM1 was normalized to that of β-actin, and the band intensity of Me-PABP1 was normalized to that of PABP1, in a time-point respective manner. The relative band intensity of day 0 was set as 100%. Data are mean ± SD of three independent measurements of the same X-ray film. (C) CARM1 null mouse embryonic fibroblast (MEF) cells were infected with GFP, wild-type CARM1 (CARM1WT) or enzyme-defective mutant (VLD189-191-AAA) CARM1 (CARM1Mutant) BacMam virus. Western blots showed that Me-PABP1 can be restored by expression of CARM1WT but not CARM1Mutant.
BacMam virus-mediated GFP-PABP1 expression and TR-FRET measurement in MCF7 cells
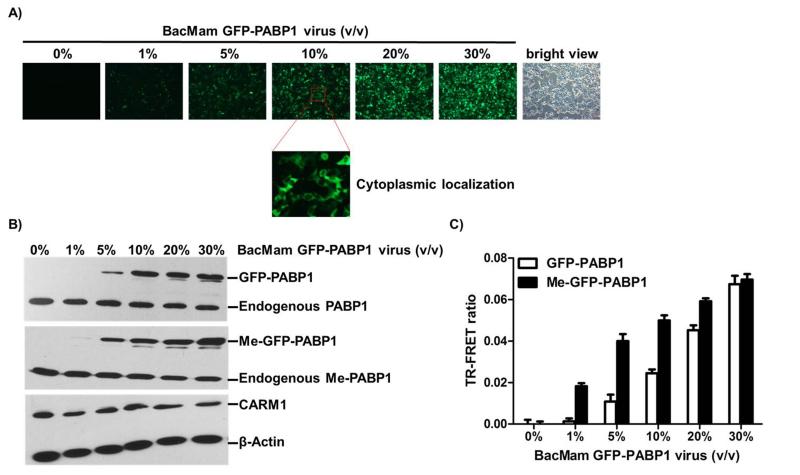
To determine whether BacMam virus-mediated GFP-PABP1, when expressed in MCF7 cells, retains the physiological properties of the endogenous PABP1 in terms of its cellular localization and methylation by CARM1, MCF7 cells were infected with a range of doses of BacMam GFP-PABP1 virus (depicted as % volume in medium) in a 6-well plate for 24 hours. GFP imaging (Figure 3A) and Western blots (Figure 3B, top panel) showed that BacMam virus-mediated GFP-PABP1 was expressed in a virus dose-dependent manner and localized to cytoplasm (Figure 3A) as reported by others.[28] Western blots with Me-PABP1 Ab further showed that BacMam virus-mediated GFPPABP1 was methylated as was the endogenous PABP1 (Figure 3B, middle panel). Moreover, 10% BacMam GFP-PABP1 virus infection achieved similar expression and methylation levels of GFP-PABP1 as the endogenous PABP1. BacMam virus infection did not perturb the endogenous CARM1 levels (Figure 3B, bottom panel). Taken together, these results suggest that freshly expressed GFP-PABP1 protein can be methylated by CARM1.
Figure 3. BacMam virus-mediated GFP-PABP1 protein can be methylated by CARM1 and TR-FRET signal can be measured in MCF7 cells.
(A) MCF7 cells were infected with the indicated amounts of BacMam GFP-PABP1 virus (4000 virus particles per microliter, indicated as volume/volume) in a 6-well plate for 24 hours. GFP imaging showed that BacMam virus-mediated GFP-PABP1 was expressed in a virus dose-dependent manner. (B) MCF7 cells were infected as in (A). Whole cell extracts were analyzed by western blots using antibodies against PABP1, Me-PABP1, CARM1 and β-actin. (C) MCF7 cells were infected as in (A) in a 384-well assay plate (7500 cells per well, 30 μl in total). TR-FRET measurement with Me-PABP1 or PABP1 primary antibody and Tb-2nd antibody indicated the increase of TR-FRET ratio as the dose of BacMam GFP-PABP1 virus increased. Background signal from Tb-2nd antibody alone was subtracted. The fold of Me-GFP-PABP1/ GFP-PABP1 activation is marked on the top of each condition. Data are mean ± SD of three independent experiments.
We next sought to measure the expression and methylation of GFP-PABP1 in MCF7 cells using the TR-FRET technology in a miniaturized 384-well format (Figure 3C). We first performed the TR-FRET assay with Tb-labeled 2nd Ab alone to detect the background signals resulting from the nonspecific proximity of Tb donor and GFP acceptor (Figure S2). In the following TR-FRET measurements, this background signal was subtracted. TR-FRET measurements with Me-PABP1 primary Ab in combination with Tb-labeled 2nd Ab showed increased TR-FRET ratios with increased doses of BacMam GFP-PABP1 virus (Figure 3C, black bars). Similar measurements were performed with the non-methyl specific PABP1 Ab to demonstrate the virus dose-dependent expression of GFP-PABP1 (Figure 3C, white bars). The ratio of Me-GFPPABP1 to GFP-PABP1 FRET signals was calculated and is shown in Figure 3C. As expected, at the lower dosages of BacMam GFP-PABP1 virus, the Me-GFP-PABP1/ GFP-PABP1 ratios were higher. The background signal from non-methylated GFPPABP1 increased when more viruses were used for infection and more GFP-PABP1 proteins were expressed (Figure 3C). Together, these results support that the TR-FRET assay enables the specific and quantitative detection of Me-GFP-PABP1. Based on the above data, we chose to use 10% BacMam GFP-PABP1 virus for the subsequent TR-FRET assay optimization in the 384-well format.
Optimization of key parameters of Me-GFP-PABP1 TR-FRET assay for HTS in the 384-well format
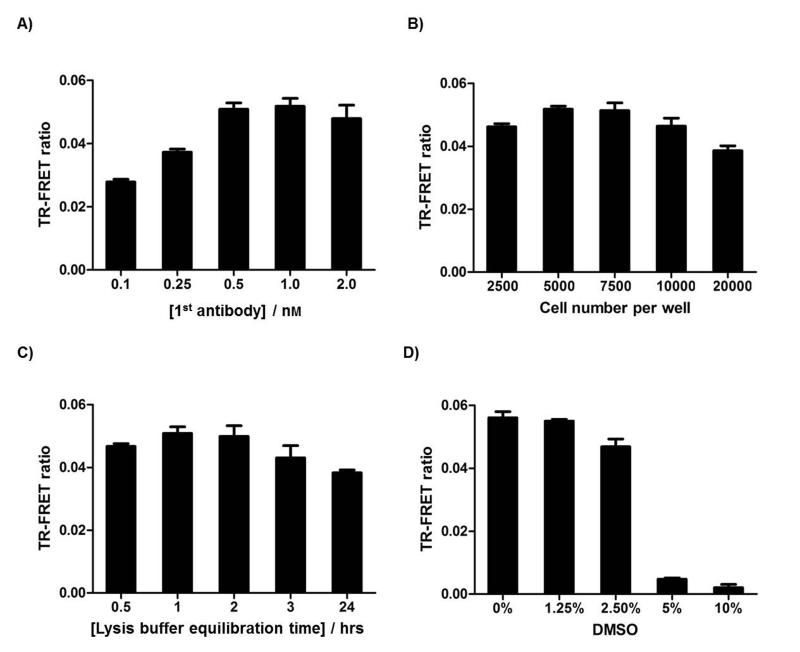
Because the efficiency and signal of cell-based TR-FRET assay can be influenced by various parameters, we proceeded to optimize several key parameters for HTS, including Me-PABP1 Ab concentration, cell number per well, and TR-FRET signal stability over time (Figure 4). Tb-2nd Ab concentration was set at 2 nM while other parameters are optimized. All optimization experiments were performed with 10% BacMam GFP-PABP1 virus, the dosage at which GFP-PABP1 was expressed at a similar level as endogenous PABP1 (Figure 3B). Optimizations with additional doses of BacMam GFP-PABP1 virus were also performed as shown in Figure S3. From the optimization experiments, the sensitivity of the TR-FRET assay increased following the increase of Me-PABP1 Ab concentration, and saturated at 0.5 nM Me-PABP1 Ab concentration (Figure 4A). Reducing or increasing cell number per well led to the decrease of the Me-GFP-PABP1 TR-FRET ratio, with 5000 to 7500 cells per well judged optimal for the Me-GFP-PABP1 TR-FRET assay (Figure 4B). The TR-FRET ratio remained stable for 0.5 to 2 hrs after cell lysis (Figure 4C), which is beneficial for scale-up of the assay for larger screens. Taken together, we used 7500 cells per well, 0.5 nM Me-PABP1 Ab, 2 nM Tb-2nd Ab, and 1 hr incubation time to achieve the optimal signals for the remaining studies.
Figure 4. Optimization of key parameters for the Me-GFP-PABP1 TR-FRET assay.
(A) Optimization of Me-PABP1 antibody concentration. 7500 MCF7 cells were seeded in a 384-well plate and infected with 10% BacMam GFP-PABP1 virus for 24 hours. TR-FRET signals were detected with indicated Me-PABP1 Ab concentration, 2 nM Tb-2nd Ab, and 1 hour incubation time. (B) Optimization of cell number per well with 0.5 nM Me-PABP1 Ab, 2 nM Tb-2nd Ab, and 1 hour incubation time. (C) Optimization of lysis buffer equilibration time with 7500 cells per well, 0.5 nM Me-PABP1 Ab, and 2 nM Tb-2nd Ab. (D) DMSO tolerance of Me-GFP-PABP1 TR-FRET assay with 7500 cells per well, 0.5 nM Me-PABP1 Ab, 2 nM Tb-2nd Ab, and 1 hour incubation time. Data are mean ± SD of three independent experiments.
DMSO is generally used to dissolve compound in small-molecule libraries, so the DMSO tolerance of Me-GFP-PABP1 TR-FRET assay was titrated to determine the appropriate final concentration of DMSO in HTS (Figure 4D). Me-GFP-PABP1 TR-FRET signal remained stable up to 1.25% (v/v) DMSO, and exhibited a partial loss in 2.5% DMSO but total loss in 5% or higher concentration of DMSO (Figure 4D). The total loss of Me-GFP-PABP1 TR-FRET signal in high DMSO conditions was due to the cell death, resulting in loss of GFP-PABP1 expression (Figure S4). For HTS, compounds were diluted 100-fold, leading to a final concentration of 1% DMSO which did not interfere with the cell viability or TR-FRET readout.
Me-GFP-PABP1 TR-FRET signal is reflective of CARM1 expression level and the enzymatic activity
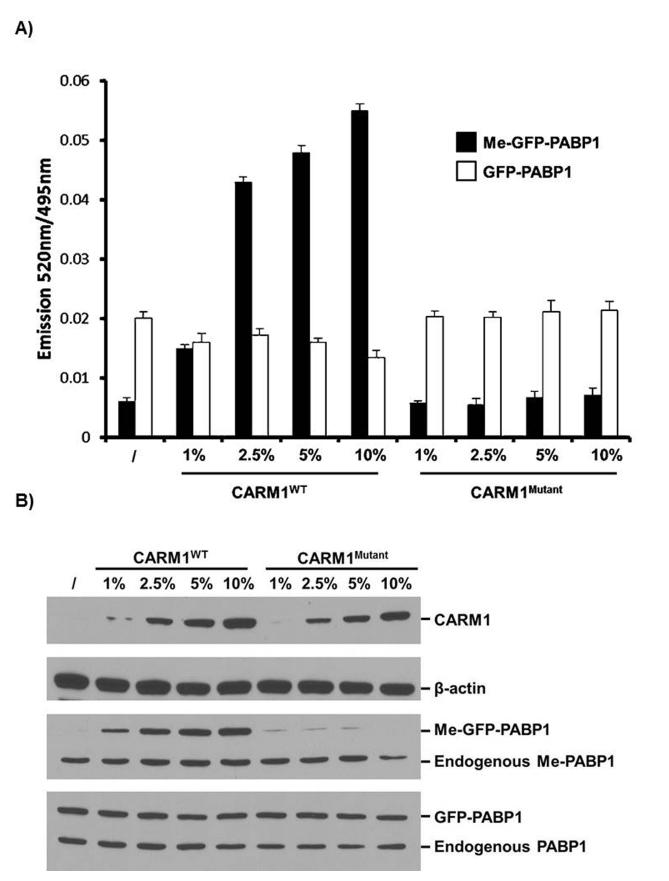
To validate that the Me-GFP-PABP1 TR-FRET assay can detect differences in cellular CARM1 expression level or enzymatic activity, the MCF7-tet-on-shCARM1 cell line was utilized. The cells were pre-treated with Dox to knockdown endogenous CARM1 (Figure 2A). Then BacMam viruses expressing GFP-PABP1 and either CARM1WT or CARM1Mutant were used to infect the cells. 24 hours later, Me-GFP-PABP1 specific and total GFP-PABP1 specific TR-FRET signals were detected (Figure 5A), and in parallel Me-GFP-PABP1 and total GFP-PABP1 were examined by Western blots (Figure 5B). The restoration of Me-GFP-PABP1 level was increased upon CARM1WT virus infection in a dose-dependent manner. In contrast, CARM1Mutant, although expressed at similar levels as CARM1WT, failed to induce methylation of GFP-PABP1 as measured by both TR-FRET assay and Western blots. These data suggest that the Me-GFP-PABP1 based TR-FRET assay provides a valid and sensitive tool for detecting the changes of both cellular CARM1 expression level and enzymatic activity. We found that the level of methylation of exogenous GFP-PABP1 achieved in 24 hrs responded to the level of CARM1 whereas the endogenous PABP1 was constitutively methylated (Figure 5B, middle panel). For this reason, we chose to introduce the CARM1 substrate transiently rather than making a stable cell line expressing GFP-PABP1.
Figure 5. Me-GFP-PABP1 TR-FRET signal is reflective of CARM1 expression level and enzymatic activity.
MCF7-tet-on-shCARM1 cells were pre-treated with Dox for 7 days to knockdown endogenous CARM1. (A) Pre-treated cells were seeded in a 384-well plate and infected with 10% BacMam GFP-PABP1 virus as well as the indicated doses of BacMam CARM1WT or CARM1Mutant virus for 24 hours. TR-FRET signals for both Me-GFPPABP1 (black bar) and total GFP-PABP1 (white bar) were obtained (n=3, SD). (B) Pre-treated cells were infected as in (A) in 6-well plate. Cells were lysed and western blots were performed with whole cell extract to show the expression of CARM1, GFP-PABP1, and level of Me-GFP-PABP1.
As demonstrated in Figure 5A, 1% BacMam CARM1WT virus infection achieved a Me-GFP-PABP1 TR-FRET ratio 2-3 fold above the background of no CARM1WT virus infection control, but still ~4 fold below the maximum ratio of ~0.06. This low level of methylation provided a sensitized genetic background upon which compounds could be screened for activators of CARM1 that lead to increased methylation of GFP-PABP1. Hence in screens for CARM1 activators, endogenous CARM1-depleted MCF7 cells will be batch-infected with 10% BacMam GFP-PABP1 virus along with 1% BacMam CARM1WT virus to generate large pools of cells that will be seeded into 384-well assay plates for compound screening.
Me-GFP-PABP1 TR-FRET responds to dose titration of a general methylation inhibitor and synthetic CARM1 activators
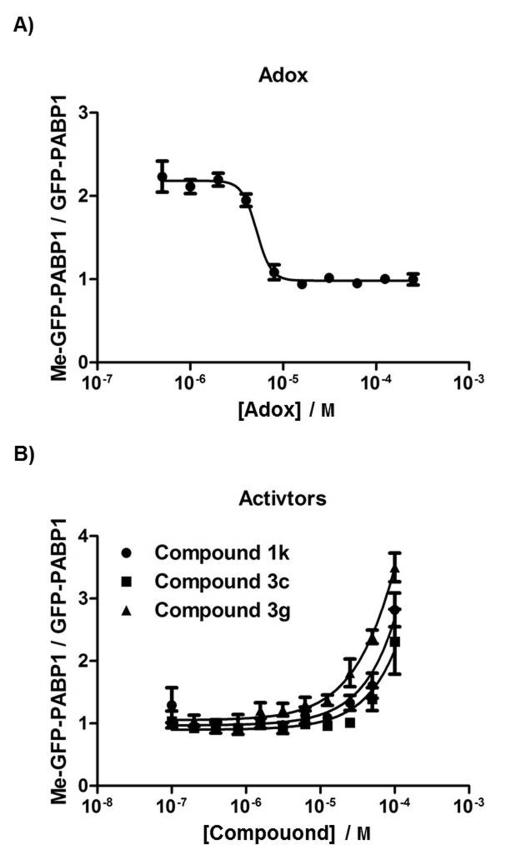
To demonstrate that the Me-GFP-PABP1 TR-FRET assay is specific and can quantitatively measure the inhibition or activation of methylation of GFP-PABP1 by known inhibitor or activators, we tested the TR-FRET assay in the presence of the indirect methyltransferase inhibitor adenosine dialdehyde (Adox) or synthetic CARM1 activators 1k, 3c, or 3g, which were recently reported by Dr. Gianluca Sbardella and colleagues (Figure 6).[29] Adox inhibits S-adenosylhomocysteine hydrolase, leading to the elevated levels of adenosylhomocysteine that block utilization of S-adenosylmethionine by methyltransferases.[30] As shown in Figure 6A, the Me-GFPPABP1 TR-FRET assay in MCF7 cells exhibited a dose-dependent response to the addition of Adox, with a representative IC50 value for Adox calculated as 5.2 μM. Uracandolates (aryl ureido acetamido indole carboxylates) 1k, 3c, and 3g were recently shown able to increase the methylation of PABP1 induced by CARM1 at 50 μM in in vitro methylation assays and in cells, although the activation mechanism is still unclear.[29] As shown in Figure 6B, the TR-FRET assays in the defined genetic background exhibited dose-dependent responses to all the three activators, although the EC50 values for these activators are determined to be 259 μM, 272 μM, and 306 μM, respectively. Taken together, these results validated that the TR-FRET assay responds appropriately to the addition of a chemical, ensuring a reliable readout for HTS.
Figure 6. Me-GFP-PABP1 TR-FRET responds to the addition of a known methyltransferase inhibitor and CARM1 activators.
(A) MCF7 cells were plated in a 384-well plate, infected with 10% BacMam GFP-PABP1 virus, and treated with serially-diluted protein methyltransferase inhibitor Adox for 24 hours. Me-GFP-PABP1 TR-FRET signal was normalized to total GFP-PABP1 TR-FRET signal to depict the inhibition curve. (B) MCF7-tet-on-shCARM1 cells were pre-treated with Dox for 7 days to knockdown endogenous CARM1. Pre-treated cells were seeded in a 384-well plate, infected with 10% BacMam GFP-PABP1 virus and 1% BacMam CARM1WT virus, and treated with serially-diluted known CARM1 activator (compound 1k, 3c, or 3g) for 24 hours. Me-GFP-PABP1 TR-FRET signal was normalized to total GFPPABP1 TR-FRET signal to depict the activation curves. Data are mean ± SD of three independent experiments.
A pilot screen with the NIH Clinical Collection Library
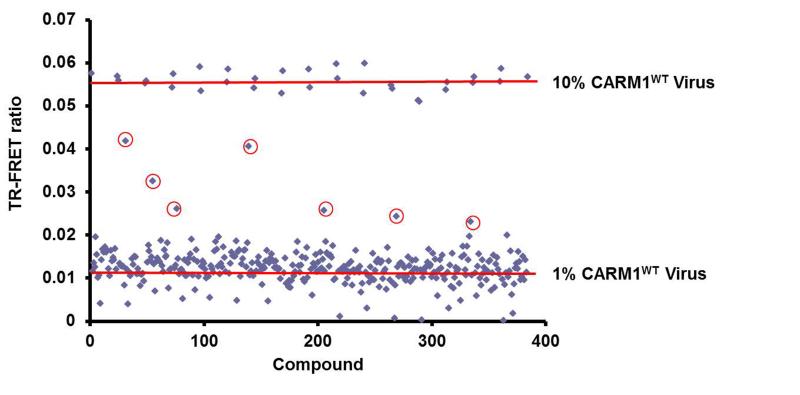
We next proceeded to conduct a pilot screen using 320 compounds in the NIH Clinical Collection Library to validate the overall performance of Me-GFP-PABP1 TR-FRET assay (Figure 7). Cells and viruses were prepared in the sensitized genetic background described above (Figure 5A). Compounds were screened at 10 μM in order to identify more potent CARM1 activators, and all wells in the 384-well plate received the same amount of DMSO as vehicle control or with compound. We used multiple wells with 1% and 10% BacMam CARM1WT virus infections to render a range of Me-GFP-PABP1 TR-FRET ratios for calculating Z’ and S/B ratio. Figure 7 shows that such a setting yielded an average S/B ratio of 7.2 and a Z’-factor of 0.76 which is suitable for a single point HTS.[31] Compounds 1k, 3c, and 3g were not used as positive controls in the pilot screen because they were inactive in the TR-FRET assay at 10 μM concentration (Figure 6B). Compounds providing a 520/495 nm TR-FRET ratio for Me-GFP-PABP1 greater than threefold the standard deviation above the mean of the vehicle controls were defined as the primary hits. Based on this criterion, seven compounds were selected as primary hits from the pilot screen (Figure 7, denoted with circles). We anticipated that the most likely false positive mechanism could be a non-specific increase in the fluorescence or induced increase of BacMam virus-mediated GFPPABP1 expression. Therefore, these seven selected compounds were counter-screened using the TR-FRET assay in which the TR-FRET signal was generated using the antibody to total PABP1 but not to the methyl-specific PABP1 (Figure 3C and Figure 5A). The compounds which increased the Me-GFP-PABP1 TR-FRET signal also increased the GFP-PABP1 TR-FRET signal by the same fold, indicating that none of the seven selected hits was CARM1 activator (data not shown). Nonetheless, the pilot screen results (Figure 7) imply that the Me-GFP-PABP1 TR-FRET assay provides a suitable and reliable platform for large-scale compound screening in pursuit of CARM1 activators.
Figure 7. A Pilot screen was performed using the NIH Clinical Collection Library.
MCF7-tet-on-shCARM1 cells were pre-treated with Dox for 7 days to knockdown endogenous CARM1. Pre-treated cells were batch-infected with 10% BacMam GFPPABP1 virus and 1% BacMam CARM1WT virus, and then aliquoted 30 μl into 384-well plate. 0.3 μl 1 mM compounds were added with a robot to a final 10 μM concentration. 24 hours post-treatment, Me-GFP-PABP1 TR-FRET ratios were obtained and plotted as scatter plots. 10% BacMam CARM1WT virus infection was used to provide the maximum activation range for Me-GFP-PABP1 TR-FRET ratio, and the Z’ factor was 0.76 for the pilot screen. Seven primary hits were denoted with circles.
Discussion
In this report, we successfully designed and implemented a LanthaScreen® TR-FRET cellular assay to monitor CARM1 activity in MCF7 breast cancer cells, which provides a robust assay format for HTS of small-molecule activators or inhibitors of CARM1 (Figure 1). Although the LanthaScreen® TR-FRET has been applied to monitor other post-translational modifications including acetylation, phosphorylation and lysine methylation,[23-24] this is the first application monitoring arginine methylation. Our initial attempt to use H3R17me2 as a reporter in TR-FRET assay did not work, probably because CARM1 is not the only PRMT to modify H3 at R17 (personal communication with Mark Bedford). PABP1 was chosen as the reporter of CARM1 activity because it has been identified as the major cytoplasmic substrate of CARM1 and the methylation sites have been mapped to R455 and R460.[12] We first confirmed that the level of PABP1 methylation can reflect the cellular CARM1 level (Figure 2 and Figure S1), and confirmed the specificity of methyl-PABP1 specific antibody which is the central determinant for the TR-FRET assay (Figure 2 and Figure 3). Although stable cell lines are widely accepted for HTS, we showed that endogenous PABP1 was constitutively methylated at a relatively high level even if only a trace amount of CARM1 existed in cells (Figure 5B), which restricts the assay window for the activation of PABP1 methylation by a CARM1 activator. Therefore, GFP-PABP1 was transiently expressed in MCF7 cells via the BacMam gene delivery system which has been adapted in a drug discovery setting in the pharmaceutical industry.[25] The BacMam system also enables the application of the TR-FRET assay in other cell types to monitor CARM1 activity in different cellular settings. In order to maintain GFP-PABP1 methylation at a low starting level which allows for the activation by a potential CARM1 activator, a small amount of CARM1 was introduced into endogenous CARM1-depleted MCF7 cells (Figure 5). Furthermore, this system proved suitable and robust enough for screening CARM1 activators as shown in the dose-dependent response to the addition of synthetic CARM1 activators 1k, 3c and 3g, and a pilot screen of NIH Clinical Collection Library (Figure 6B and Figure 7).
In the past years, several biochemical assays were established for discovery of small-molecule modulators of histone H3 modifying enzymes such as HDACs and methyltransferases.[3a] These in vitro assays used recombinant enzymes and the protein or peptide substrates, which could not preserve the cellular complexity and pathway specificity of the enzymes in the intracellular conditions.[22c] Moreover, they are not readily scalable to HTS of large quantity of compounds in terms of their detection methods. On the other hand, the TR-FRET assay is fully homogenous with no needs for washing, lysate transfer, or separation procedures, providing a reliable tool to screen for selective, potent, and cell-permeable activators of CARM1 in a large pool of compound libraries. Given that it is more difficult to get positive hits in the small-molecule activator screens in comparison to traditional inhibitor screens,[4] the false-positives may be less than the inhibitor screen. The main caveat of the Me-GFP-PABP1 TR-FRET assay results from the indirect detection method involving Me-PABP1 Ab and Tb-2nd Ab because the primary Me-PABP1 Ab was too diluted to be directly labeled with Tb. Nonetheless, our TR-FRET ratio in the range of 0.01-0.06 using this indirect method is reproducible and comparable to other published LanthaScreen® TR-FRET assays employing different proteins and substrates.[23-24]
CARM1 has been implicated in a variety of physiological and pathological processes, and the enzymatic activity of CARM1 has been reported to be crucial for the endochondral ossification and cartilage development,[16] glycogen metabolism,[20] and proper differentiation of adipocytes,[17] myocytes,[18] and pulmonary epithelial cells.[19] Moreover, in contrast to previous studies in which CARM1 was reported to be protumorigenic,[6b, 32] our lab found that overexpression of CARM1 triggers differentiation process in ERα-positive breast cancer cells through global regulation of ERα-target genes.[15] Accordingly, activators of CARM1 may provide a means to further test our novel hypothesis that activating enzymatic activity of CARM1 with a small molecule activator could mimic the effects of overexpression, phenotypically as well as regulating genes at a global level. Small-molecule activators of CARM1 identified from future HTS will also help establish the roles of CARM1 in reprogramming the epigenome in breast cancer cells, and also be beneficial in other disease studies, for example in skeletal muscle glycogen metabolism. Although compounds 1k, 3c and 3g have been shown to increase CARM1-mediated methylation of PABP1, both in the in vitro methylation assay and in the TR-FRET assay here, these compounds do not bind to CARM1 in the absence of substrates or cofactor and do not increase histone H3 methylation catalyzed by CARM1.[29] Moreover, the activation effects of these compounds were achieved at high concentrations (Figure 6B), which are likely to have off-target effects. Therefore it is advantageous to screen for more potent, selective activators of CARM1 which bind to allosteric site(s) on CARM1 in order to study the epigenetic regulation of CARM1 in breast cancer and other diseases.
In conclusion, the TR-FRET strategy applied in this study provides a promising tool to screen potent, selective, and cell-permeable small-molecule activators of CARM1. CARM1 is a coactivator of estrogen receptor α (ERα), the main target in human breast cancer. Small molecule activators identified from our screening will help elucidate the roles of CARM1 in vivo in cancer and epigenetic regulation. This methodology has potential to be extended to other PRMTs by screening either inhibitors or activators, through the development of appropriate substrate GFP-fusion proteins and methylation-site-specific antibodies. PRMT-mediated arginine methylation is involved in a variety of functional processes including transcriptional regulation, RNA metabolism and shuttling, and DNA damage repair.[5, 6c, 26] Small chemical modulators of PRMTs will facilitate studies of the structure-function relationships of PRMT substrate recognition and tuning their activities to control the appropriate biological processes.
Experimental Section
Cells and tissue culture
Human breast cancer cell line MCF7 was purchased from American Type Culture Collection (ATCC) and used within 6 months. MCF7-tet-on-shCARM1 inducible knockdown cell line was generated in our lab as previously described.[15] CARM1 null mouse embryonic fibroblast (CARM1−/− MEF) cells were provided by Dr. Mark Bedford. All cell lines were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies, Carlsbad, CA) supplemented with fetal bovine serum (FBS, Life Technologies, 10%), and incubated at 37 °C in a humidified atmosphere containing CO2 (5%).
Reagents and antibodies
Doxycycline (Dox) was purchased from Clontech (Mountain View, CA) and used at a final concentration (500 ng/ml) for MCF7-tet-on-shCARM1 cell line. Dimethyl sulfoxide (DMSO) and adenosine dialdehyde (Adox) were purchased from Sigma (St. Louis, MO). Synthetic aryl ureido acetamido indole carboxylates compounds 1k, 3c, and 3g were provided by Dr. Gianluca Sbardella and dissolved in DMSO. Compounds in the NIH Clinical Collection Library were provided by the Small Molecule Screening and Synthesis Facility (SMSSF) at UW-Madison. LanthaScreen® 6x Cellular Assay Lysis Buffer (catalog # A14298) and LanthaScreen® Terbium-labeled Anti-rabbit Antibody (catalog # PV3773, Tb-2nd Ab) were provided by Life Technologies. Peptide antibodies against CARM1, PABP1, and Me-PABP1 were custom-generated by Genemed Synthesis Inc., TX. The peptide used as antigen for generating me-PABP1 Ab is CPGAIR(CH3)2 PAAPR(CH3)2PPFST. β-actin antibody was purchased from Sigma (St. Louis, MO).
Plasmid construction and production of BacMam virus
To generate a GFP-PABP1 BacMam expression vector, the cDNA for full-length human PABP1 was subcloned into a pDEST8-CMV-n-GFP vector by Gateway cloning technology (Life Technologies). To generate the CARM1WT or CARM1Mutant BacMam expression vector, the cDNA for full-length human CARM1 encoding wild-type or the enzyme-defective mutant (VLD189-191-AAA)[27] protein was subcloned into an analogous vector without the GFP open reading frame. The resulting expression constructs were then transformed into DH10BAC bacterial strain for the preparation of bacmid DNA. PCR-qualified bacmid DNA was then transfected into SF9 insect cells for BacMam virus production according to Bac-to-Bac® Baculovirus Expression Systems manual (Life Technologies). Each lot of BacMam virus was produced as a stock (500 ml), and virus titer was determined to be ~4000 virus particles per microliter of stock solution.
TR-FRET cellular assay and data analysis
MCF7 cells or Dox-pre-treated MCF7-tet-on-shCARM1 cells were plated in 384-well assay plates (Corning, MA, # 3570; 30 μl /well) and infected with the indicated amounts of BacMam virus. When pilot screening was performed, compound (0.3 μl) dissolved in DMSO (100%) was added directly into the cell-virus suspension in each well using a Beckman Biomek FX robot. 24 hours post-infection, LanthaScreen® 6x Cellular Assay Lysis Buffer was supplemented with Me-PABP1 Ab or PABP1 Ab, Tb-2nd Ab (12 nM), and protease inhibitor cocktails (Sigma). Subsequently, the complete 6x lysis buffer was added to each well (6 μl /well) and the plates were incubated at room temperature in the dark for 1-3 hours. The plate was then read on an EnVision® multilabel plate reader (PerkinElmer, Waltham, MA) with excitation at 340 nm, emission at 520 nm and 495 nm, 100 μs lag time, and 200 μs integration time. The GFP-specific emission was measured at 520 nm, the Tb-specific emission was measured at 495 nm and the TR-FRET ratio was calculated as emission at 520 nm over emission at 495 nm. Background signal from the TR-FRET measurement with Tb-2nd Ab alone was subtracted from TR-FRET ratio using specific antibody. All the data were plotted by the GraphPad Prism 5 software, and the IC50 for Adox and EC50 for compounds 1k, 3c, or 3g were calculated by fitting the data to a sigmoidal dose-response equation with varying slope. The Z’-factor value was calculated as [Eq. (1)]:
Western blots
Cells were cultured and treated as described above. To perform Western blots, cells were collected by trypsinization, washed with Dulbecco’s phosphate buffer saline (DPBS) (Life Technologies), and lysed by suspension in lysis buffer [Tris (50 mM) pH 8.0, NaCl (400 mM), glycerol (10%), triton X-100 (0.5%), protease inhibitors, and benzonase]. After centrifugation, total protein was quantified using the BioRad Protein Assay (BioRad), and protein (20 μg) was resolved using SDS–PAGE and polyacrylamide gels (8%). Proteins were transferred to a nitrocellulose membrane for 1.5 hrs at 350 mA. Membranes were blocked with nonfat milk (5%) at room temperature for 1 hour and incubated overnight with anti-CARM1 antibody (1: 5000), Me-PABP1 Ab (1: 2000), PABP1 Ab (1: 2000), or anti-β-actin antibody (1:10000) at 4 °C. Membranes were then washed with DPBS and incubated with HRP-conjugated goat-anti-rabbit IgG secondary antibody for 1 hour at room temperature, and detected by ECL reaction and X-ray film exposure.
Quantification of CARM1 and Me-PABP1 Western blots
The X-ray films were scanned and digitalized. The density of each band was measured using the Quantity One 1-D analysis software (Bio-Rad). The band intensity of CARM1 was normalized to that of β-actin, and the band intensity of Me-PABP1 was normalized to that of PABP1, in a time-point respective manner. The relative band intensity of day 0 was set as 100% for normalization of band intensities at other time points.
Supplementary Material
Acknowledgements
We thank Jeanne Dudek and Leisha Kopp (Life Technologies Corporation) for technical support on BacMam virus production and TR-FRET assay development. We would like to thank Song Guo at SMSSF of UW-Madison for help on the pilot screen. This project is supported by National Institutes of Health RO1CA125387, Shaw Scientist Award, Villas Associate Award, and DOD ERA of HOPE Scholar Award to W.X.
References
- [1].Rodriguez-Paredes M, Esteller M. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- [2].Rangwala S, Zhang C, Duvic M. Future Med Chem. 2012;4:471–486. doi: 10.4155/fmc.12.6. [DOI] [PubMed] [Google Scholar]
- [3]a).Janzen TJWWP, Jin J, Frye SV. Drug Discov Today Technol. 2010;7:e59–e65. doi: 10.1016/j.ddtec.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heightman TD. Expert Opin Ther Targets. 2011;15:729–740. doi: 10.1517/14728222.2011.561786. [DOI] [PubMed] [Google Scholar]
- [4].Julie A Zorn JAW. Nature Chemical Biology. 2010;6:179–188. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
- [5].Bedford MT, Richard S. Molecular cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- [6]a).Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, Imhof R, Bedford MT, Natoli G, Hottiger MO. EMBO J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Frietze S, Lupien M, Silver PA, Brown M. Cancer research. 2008;68:301–306. doi: 10.1158/0008-5472.CAN-07-1983. [DOI] [PubMed] [Google Scholar]; c) Teyssier C, Le Romancer M, Sentis S, Jalaguier S, Corbo L, Cavailles V. Trends in endocrinology and metabolism: TEM. 2010;21:181–189. doi: 10.1016/j.tem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- [7].Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- [8].Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Sims RJ, 3rd, Rojas LA, Beck D, Bonasio R, Schuller R, Drury WJ, 3rd, Eick D, Reinberg D. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chevillard-Briet M, Trouche D, Vandel L. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]a).Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. The Journal of biological chemistry. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]; b) Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M. Molecular and cellular biology. 2006;26:2273–2285. doi: 10.1128/MCB.26.6.2273-2285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cheng D, Cote J, Shaaban S, Bedford MT. Molecular cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- [12].Lee J, Bedford MT. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Proc Natl Acad Sci U S A. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, Bedford MT. J Biol Chem. 2010;285:1147–1152. doi: 10.1074/jbc.M109.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Al-Dhaheri M, Wu J, Skliris GP, Li J, Higashimato K, Wang Y, White KP, Lambert P, Zhu Y, Murphy L, Xu W. Cancer research. 2011;71:2118–2128. doi: 10.1158/0008-5472.CAN-10-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ito T, Yadav N, Lee J, Furumatsu T, Yamashita S, Yoshida K, Taniguchi N, Hashimoto M, Tsuchiya M, Ozaki T, Lotz M, Bedford MT, Asahara H. BMC Dev Biol. 2009;9:47. doi: 10.1186/1471-213X-9-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yadav N, Cheng D, Richard S, Morel M, Iyer VR, Aldaz CM, Bedford MT. EMBO reports. 2008;9:193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen SL, Loffler KA, Chen D, Stallcup MR, Muscat GE. J Biol Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- [19].O’Brien KB, Alberich-Jorda M, Yadav N, Kocher O, Diruscio A, Ebralidze A, Levantini E, Sng NJ, Bhasin M, Caron T, Kim D, Steidl U, Huang G, Halmos B, Rodig SJ, Bedford MT, Tenen DG, Kobayashi S. Development. 2010;137:2147–2156. doi: 10.1242/dev.037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang SC, Dowhan DH, Eriksson NA, Muscat GE. Biochem J. 2012;444:323–331. doi: 10.1042/BJ20112033. [DOI] [PubMed] [Google Scholar]
- [21].Li J, Zhao Z, Carter C, Ehrlich LI, Bedford MT, Richie ER. J Immunol. 2012 doi: 10.4049/jimmunol.1102513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]a).Wigle TJ, Provencher LM, Norris JL, Jin J, Brown PJ, Frye SV, Janzen WP. Chem Biol. 2010;17:695–704. doi: 10.1016/j.chembiol.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Quinn AM, Allali-Hassani A, Vedadi M, Simeonov A. Mol Biosyst. 2010;6:782–788. doi: 10.1039/b921912a. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Luo M. ACS Chem Biol. 2012;7:443–463. doi: 10.1021/cb200519y. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Dorgan KM, Wooderchak WL, Wynn DP, Karschner EL, Alfaro JF, Cui Y, Zhou ZS, Hevel JM. Anal Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- [23].Robers MB, Loh C, Carlson CB, Yang H, Frey EA, Hermanson SB, Bi K. Molecular bioSystems. 2011;7:59–66. doi: 10.1039/c0mb00026d. [DOI] [PubMed] [Google Scholar]
- [24].Machleidt T, Robers MB, Hermanson SB, Dudek JM, Bi K. Journal of biomolecular screening. 2011;16:1236–1246. doi: 10.1177/1087057111422943. [DOI] [PubMed] [Google Scholar]
- [25].Kost TA, Condreay JP, Ames RS, Rees S, Romanos MA. Drug discovery today. 2007;12:396–403. doi: 10.1016/j.drudis.2007.02.017. [DOI] [PubMed] [Google Scholar]
- [26].Bedford MT, Clarke SG. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen D, Ma H, Hong H, Koh SS, huang S-M, Schurter BT, Aswad DW, Stallcup MR. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- [28]a).Afonina E, Stauber R, Pavlakis GN. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]; b) Salaun C, MacDonald AI, Larralde O, Howard L, Lochtie K, Burgess HM, Brook M, Malik P, Gray NK, Graham SV. Journal of virology. 2010;84:8539–8548. doi: 10.1128/JVI.00668-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Castellano S, Spannhoff A, Milite C, Dal Piaz F, Cheng D, Tosco A, Viviano M, Yamani A, Cianciulli A, Sala M, Cura V, Cavarelli J, Novellino E, Mai A, Bedford MT, Sbardella G. J Med Chem. 2012;55:9875–9890. doi: 10.1021/jm301097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].O’Dea RF, Mirkin BL, Hogenkamp HP, Barten DM. Cancer Res. 1987;47:3656–3661. [PubMed] [Google Scholar]
- [31].Zhang TDCJH, Oldenburg KR. Journal of biomolecular screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- [32]a).Ou CY, LaBonte MJ, Manegold PC, So AY, Ianculescu I, Gerke DS, Yamamoto KR, Ladner RD, Kahn M, Kim JH, Stallcup MR. Mol Cancer Res. 2011;9:660–670. doi: 10.1158/1541-7786.MCR-10-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) El Messaoudi S, Fabbrizio E, Rodriguez C, Chuchana P, Fauquier L, Cheng D, Theillet C, Vandel L, Bedford MT, Sardet C. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13351–13356. doi: 10.1073/pnas.0605692103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.