Summary
Comprehensive analysis of cis-regulatory elements is key to understanding the dynamic gene regulatory networks that control embryonic development. While transgenic animals represent the gold standard assay, their generation is costly, entails significant animal usage, and in utero development complicates time-course studies. As an alternative, embryonic stem (ES) cells can readily be differentiated in a process that correlates well with developing embryos. Here, we describe a highly effective platform for enhancer assays using an Hsp68/Venus reporter cassette that targets to the Hprt locus in mouse ES cells. This platform combines the flexibility of Gateway® cloning, live cell trackability of a fluorescent reporter, low background and the advantages of single copy insertion into a defined genomic locus. We demonstrate the successful recapitulation of tissue-specific enhancer activity for two cardiac and two haematopoietic enhancers. In addition, we used this assay to dissect the functionality of the highly conserved Ets/Ets/Gata motif in the Scl+19 enhancer, which revealed that the Gata motif is not required for initiation of enhancer activity. We further confirmed that Gata2 is not required for endothelial activity of the Scl+19 enhancer using Gata2−/− Scl+19 transgenic embryos. We have therefore established a valuable toolbox to study gene regulatory networks with broad applicability.
Keywords: ES cells, Enhancer, Haematopoiesis, Transcription
Introduction
The intricate process of embryonic development involves dynamic interactions of transcription factors with gene regulatory elements within gene regulatory networks (GRNs) (Davidson, 2010; Pimanda and Göttgens, 2010). Deciphering the underlying mechanisms and identifying the participants of GRNs is therefore paramount for elucidating normal developmental processes. Key interactions involve the combinatorial binding of transcription factors to cis-regulatory elements, and fundamental insights can be acquired from their in silico and molecular dissection. Comprehensive experimental interrogation requires rigorous functional analyses for which the use of transgenic animals has historically been considered the gold standard assay.
For the mouse model this can be expensive in animal usage since variable copy-numbers of the transgene and integration positional effects require multiple transgenic lines in order to establish reproducible expression patterns. This variability can be circumvented by exploiting embryonic stem (ES) cell lines with a defective hypoxanthine guanine phosphoribosyl transferase 1 (Hprt) gene, since strategies that restore Hprt function enable selection for single copy integration. As a ubiquitously expressed gene, Hprt resides in a favourable chromatin environment and it has been demonstrated that inclusion of tissue-specific promoter elements into Hprt targeting constructs results in transgene expression entirely under the control of exogenous regulatory elements. For example, Flt-1, vWF and Tie-2 regulatory elements inserted as single-copy reporter transgenes into the Hprt gene locus all displayed appropriate expression patterns in transgenic mice (Evans et al., 2000; Minami et al., 2002). However, unlike the two-week time frame of analysing F0 transgenic embryos generated by microinjection, the generation of Hprt transgenic reporter mice takes a minimum of four months. Moreover, regardless of the procedure employed for acquiring transgenic mouse embryos their intra-uterine development complicates time-course studies.
Alternative in vitro methods are therefore highly desirable, not only to accelerate scientific progress, but also in light of significant animal welfare issues associated with large-scale generation of transgenic mouse lines. Mouse ES cells offer a distinct advantage due to the ease with which they can be manipulated, their ability to differentiate into cell types from all three germ layers, and for the way in which quantitative information of the activity of regulatory elements can be generated during in vitro differentiation time-courses. Using a LacZ reporter gene we have previously shown that the temporal activity of the well-characterised Scl/Tal1 stem cell enhancer (Scl+19), inserted into the Hprt locus correlates well with endogenous gene expression (Smith, A. M. et al., 2008). However, while the traditional choice of LacZ reporter genes offers the advantage of performing histological studies with relative ease, the complex protocols required for flow cytometric analyses using this reporter limits the more advanced cellular experiments that are possible with alternative reporters such as GFP.
Here, we introduce a highly effective and adaptable toolkit that can be used to explore both wild type and perturbed enhancer activity at high-throughput, using mouse ES cell differentiation. We have replaced the LacZ reporter for a fluorescent reporter gene (Venus-YFP), enabling continuous time-course experiments, as well as substituting standard cloning strategies with Gateway® cloning (Hartley et al., 2000). This offers a rapid pipeline for more streamlined cloning together with a flexible and trackable reporter system. Here, we demonstrate the reproducibility of this approach for successful recapitulation of tissue-specific enhancer activity in both cardiac and haematopoietic lineages. Further advantages of using this technology become evident when we undertake focused dissection of Gata and Ets motifs that are known to influence the Scl+19 enhancer activity (Göttgens et al., 2002). We have gained new insights into the influence of individual transcription factors on tissue-specific activity.
Results
Reporter construction and minimal promoter selection
To streamline flow cytometric analyses of reporter gene activity, we replaced the LacZ reporter gene used previously (Smith, A. M. et al., 2008) with a yellow fluorescent reporter gene, Venus (Nagai et al., 2002) to generate an Hprt targeting cassette containing the SV40 minimal promoter followed by the Venus reporter. However, a high proportion (∼80%) of differentiated ES cells targeted with the resulting SV40/Venus constructs showed YFP expression even without an enhancer (data not shown), indicating that the SV40 minimal promoter is leaky in this context and therefore inappropriate for our assay. Recent large-scale transgenic studies of enhancers have made use of the Hsp68 minimal promoter (Pennacchio et al., 2006; May et al., 2012; Visel et al., 2013), which has long been recognised as having a low background in enhancer assays (Kothary et al., 1989). We therefore replaced the SV40 minimal promoter to produce an Hsp68/Venus Hprt targeting construct and then generated multiple independent Hsp68/Venus ES cell clones. In contrast to the SV40/Venus clones, Hsp68/Venus clones showed much lower background YFP (data not shown), thus suggesting that a suitable platform for enhancer analyses with fluorescent reporters had been established.
Cardiac enhancers display activity in ES cell-derived beating cardiomyocytes
To test whether Hsp68/Venus constructs are effective for assessing tissue-specific enhancer activity in live cells, we selected previously characterised enhancers that drive reporter gene expression in the heart. We chose two enhancers using the VISTA Enhancer Browser (http://enhancer.lbl.gov; Visel et al., 2007); mm75 and mm77. Enhancer mm75 is located on chromosome 2 and is flanked by the bone morphogenetic protein BMP7 and Tfap2d, a transcription factor that is expressed in the developing heart (Zhao et al., 2003). Enhancer mm77 is located on chromosome 12 between the transcription factor Cux2 and the cardiac-specific gene Myl2 (Chien et al., 1993). Both enhancers show strong and very specific activity in the developing heart (Visel et al., 2007).
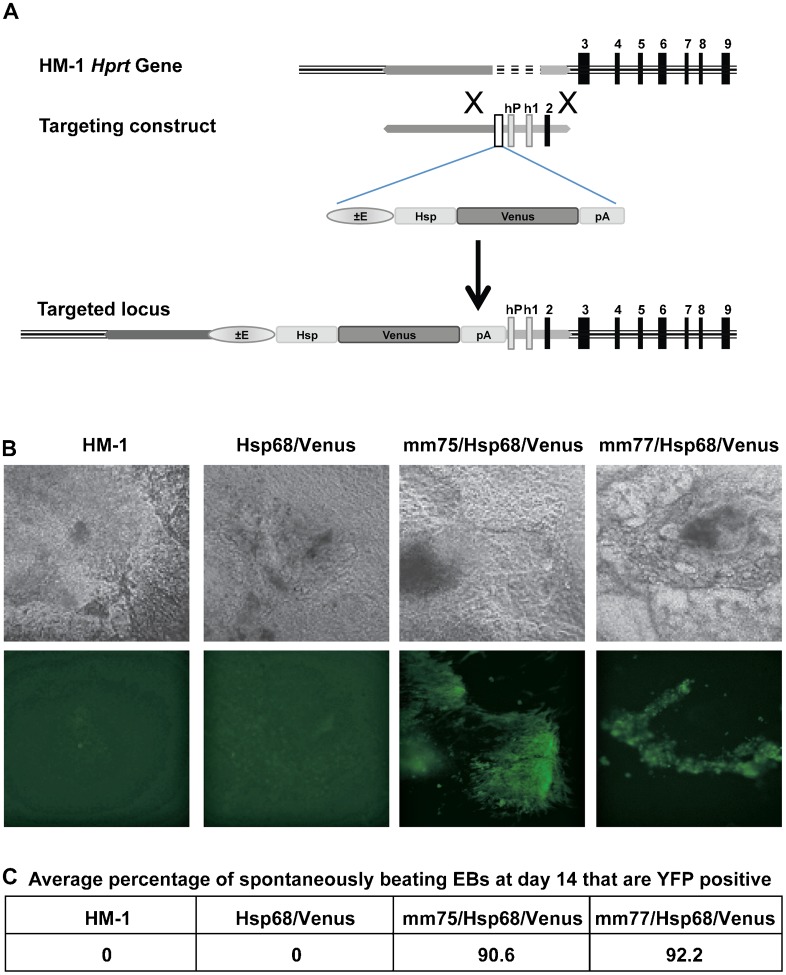
Using a cloning strategy that we have described previously (Smith, A. M. et al., 2008) we produced mm75/Hsp68/Venus and mm77/Hsp68/Venus reporter constructs and generated targeted HM-1 ES cell lines (Fig. 1A). After differentiating ES cells for eight days into embryoid bodies (EBs), cardiomyocytes are formed and spontaneously start beating. We therefore differentiated the ES cells for up to 14 days by which time cardiomyocytes are prevalent. We monitored YFP expression and found that, in agreement with expression patterns identified in the transgenic mice (Visel et al., 2007), both enhancers show highly specific activity in cardiomyocytes (Fig. 1B; supplementary material Movies 1–4). In order to quantitate this, we counted the number of spontaneously contracting EBs and then determined the percentage of these that expressed YFP. Over 90% of beating cardiomyocytes were YFP positive for each of the enhancer constructs (Fig. 1C). Importantly, no YFP fluorescence was observed in spontaneously contracting EBs derived from ES cell clones containing the enhancer-less Hsp68/Venus transgene. Robust cardiomyocyte expression of both heart enhancers therefore validated our in vitro enhancer assay as a potential alternative to conventional transgenic analyses.
Fig. 1. Hprt targeting strategy and cardiac enhancer activity in beating cardiomyocytes.
(A) The Hprt locus in mouse HM-1 ES cells lacks the Hprt promoter, exon 1 and exon 2. The Hprt targeting vector contains the enhancer of interest, the Hsp68 minimal promoter and Venus fluorescent reporter gene upstream of the human HPRT promoter, human exon 1 and mouse exon 2 and these are all flanked by Hprt locus homology arms. Homologous recombination of this targeting construct in HM-1 ES cells results in reconstitution of the Hprt locus and insertion of the enhancer/Hsp68/Venus reporter upstream of the promoter. Use of Hprt substrate analogues to select for HM-1 with deficient or reconstituted Hprt provides a stringent selection method for selection of correctly targeted clones. (B) Bright field (above) and fluorescent (below) images of representative beating embryoid bodies (EBs) at day 14, from HM-1, Hsp68/Venus control, mm75/Hsp68/Venus and mm77/Hsp68/Venus clones. (C) Average percentage of spontaneously beating EBs that are YFP positive at day 14 for the above clones, average of two independent differentiation experiments.
Increased efficiency with Gateway® cloning
Having established that the Hsp68/Venus Hprt targeting reporter gene constructs offer a powerful alternative to study tissue-specific enhancer activity, we next sought to improve the efficiency of construct generation. Gateway® Cloning Technology provides a highly efficient method of shuttling DNA sequences between multiple vector systems (Hartley et al., 2000) as it exploits recombination through specific attachment (att) sites identified from lambda bacteriophage interactions (Landy, 1989) and does not have the limitations of classic cloning strategies involving restriction sites. We therefore inserted Hsp68/Venus downstream of a Gateway® cassette and transferred this cassette into the Hprt targeting vector. Simple PCR amplification of enhancer sequences with primers that incorporate att sites now allows the generation of pDONR vectors from where the enhancers can be efficiently transferred into the new Gateway® adapted Hsp68/Venus Hprt targeting vector.
A Gfi1 enhancer shows activity within haematopoietic-fated mesoderm lineage
The transcription factor Gfi1 is expressed in multiple tissues, including haematopoietic progenitors and stem cells, lymphoid and myeloid cells (Karsunky et al., 2002; Hock et al., 2004; Yücel et al., 2004; Rosenbauer and Tenen, 2007; Wilson et al., 2010a; Lancrin et al., 2012). We have previously identified an enhancer that is located around 35 kb upstream of Gfi1 (referred to as the Gfi1-35 enhancer) within an intron of the neighbouring gene Evi5 (Wilson et al., 2010a). In transgenic mice this enhancer shows specific activity in the dorsal aorta and foetal liver, in a pattern that reflects endogenous gene expression (Wilson et al., 2010a; Lancrin et al., 2012). As the Gfi1-35 enhancer is regulated by six haematopoietic transcription factors that are critical for haematopoiesis (Wilson et al., 2010b; Wilson et al., 2010a), this element is of particular interest in terms of gene regulatory networks that control the emergence of early blood progenitors. We therefore inserted the Gfi1-35 element into the Hsp/Venus vector to further analyse its function using the ES cell in vitro differentiation model of haematopoiesis.
ES cells can readily differentiate towards the haematopoietic lineage in a process that closely resembles in vivo development (Keller et al., 1993), and which can be monitored using well-characterised cell surface markers (Mitjavila-Garcia et al., 2002; Mikkola et al., 2003). For example, the transition of Flk1 (Kdr) positive cells of the haemangioblast towards CD41 (Itg2a) positive haematopoietic progenitors from mouse embryonic stage E6.5–10.5, is reflected by the distribution of these cell surface markers during day 3–7 of ES cell differentiation (supplementary material Fig. S1). We initially followed YFP expression by fluorescent imaging of differentiating EBs from ES cells targeted with Gfi1-35/Hsp/Venus (and Hsp/Venus and HM-1 controls) from day 3 to 6 (Fig. 2A). YFP expression appeared brightest at day 4, but continued to day 6.
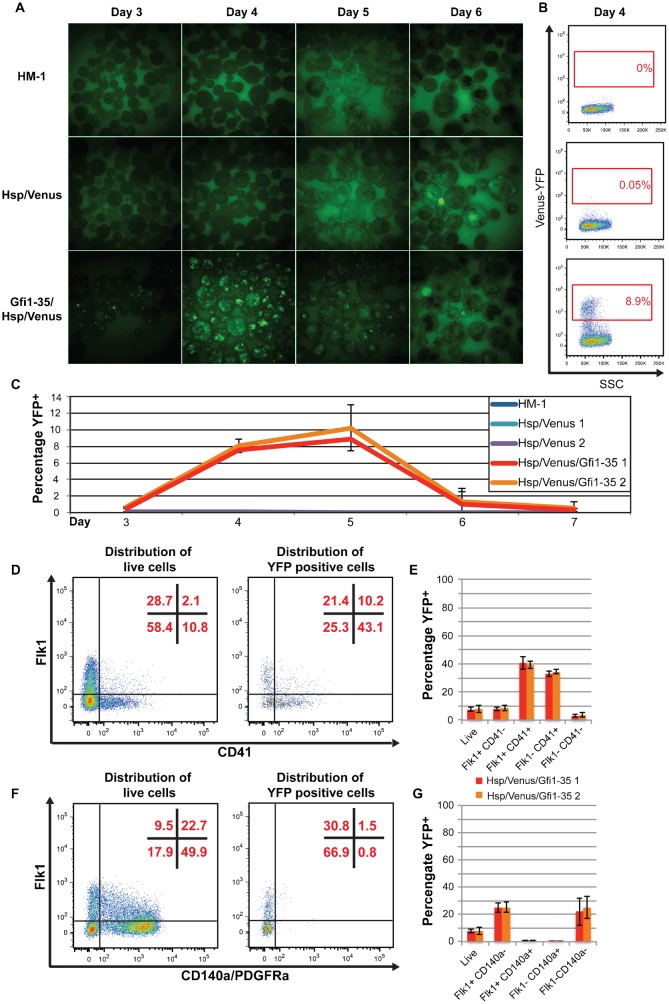
Fig. 2. Gfi1-35 activity marks haematopoietic-fated cells during embryoid body differentiation.
(A) Fluorescent images of day 3–6 EBs from the representative clones of HM-1 (top), Hsp/Venus control (middle) and Gfi1-35/Hsp/Venus (bottom) lines. (B) Representative flow cytometry plots of Venus-YFP vs side scatter (SSC) for HM-1 (top), Hsp/Venus control (middle) and Gfi1-35/Hsp/Venus (bottom) day 4 EB cells with the YFP positive gate, and its percentage of the live cell population, shown in red. (C) Percentage of the YFP positive population (gating shown in B) from day 3–7 for HM-1 (blue), two representative Hsp/Venus control clones (aqua and purple) and two representative Gfi1-35/Hsp/Venus (red and orange) clones, showing average of three independent differentiation experiments ± standard deviation. (D) Representative flow cytometry plots showing distribution of Gfi1-35/Hsp/Venus day 4 EB cells in Flk1/CD41 quadrants for all live cells (left plot) and YFP positive cells only (right plot), with the percentage of cells in each quadrant shown in red. (E) Percentage of YFP positive cells in each of the Flk1/CD41 quadrants of (D) for Gfi1-35/Hsp/Venus (clone 1 in red, clone 2 in orange). Average of three independent differentiation experiments ± standard deviation. (F) As in (D), but for Flk1/CD140a quadrants. (G) As for (E), but for Flk1/CD140a quadrants in (F).
To quantify this YFP expression, we disaggregated EBs and analysed YFP expression by flow cytometry, initially at day 4. A distinct YFP positive population was apparent above background levels in the controls (Fig. 2B). We determined the size of the YFP+ population by flow cytometry from day 3 to 7, and found that it peaks at day 5 at 8–10%. However, as seen by fluorescent microscopy, YFP expression level was highest at day 4 (Fig. 2C). At day 4, these YFP+ EB cells are found within Flk1+CD41−, Flk+CD41+ and Flk1−CD41+ populations (specifically enriched in the later two populations), but not the CD140a+ cardiac lineage population (Fig. 2D), consistent with the specific activity of the Gfi1-35 enhancer in haematopoietic specification, and the previously reported embryonic staining pattern in mouse embryos (Wilson et al., 2010a; Lancrin et al., 2012). However, previous analyses of the Gfi1-35 enhancer were limited to a single time point (E11.5 of mouse embryo development), and analyses of endogenous Gfi1 expression using knockin mice is hampered by the limited numbers of early blood progenitors that can be obtained from early mouse embryos. By contrast, as well as readily permitting the generation of early blood progenitor populations, the ES cell-based Gfi1-35 reporter system enables us to follow the entire developmental program from embryonic mesoderm to multi-potential blood progenitors. As a consequence, our new Gfi1-35 enhancer reporter system provides an unprecedented level of detail about enhancer activity during haematopoietic specification, and suggests that Gfi1-35 is active in Flk1+ mesoderm fated to haematopoietic rather than cardiac specification, prior to CD41 expression, remaining active during the specification of committed early haematopoietic cells (Flk1−CD41+).
The Scl+19 enhancer marks mesoderm with haematopoietic and cardiac potential
To further assess the ability of the Hsp68/Venus Hprt targeting approach to recapitulate in vivo enhancer data and dissect the functionality of regulatory elements, we chose to concentrate on the Scl+19 enhancer, due to its simple and highly conserved Ets/Ets/Gata motif, well-characterised activity in transgenic embryos and luciferase assays (Göttgens et al., 2002; Göttgens et al., 2004; Silberstein et al., 2005; Spensberger et al., 2012). Scl/Tal1 is a critical transcription factor for developmental patterning in the mouse conceptus, necessary for primitive and definitive haematopoiesis (Shivdasani et al., 1995; Porcher et al., 1996; Robb et al., 1996; Barton et al., 2001; Kassouf et al., 2008) and antagonising cardiac fate of embryonic endothelium (Ismailoglu et al., 2008; Van Handel et al., 2012). Its correct spatiotemporal expression is therefore critical for normal development. In transgenic mouse embryos the Scl+19 enhancer is active in embryonic endothelium, haemangioblasts, and committed haematopoietic progenitors (Sánchez et al., 1999; Göttgens et al., 2002; Silberstein et al., 2005).
Using the Gateway® strategy outlined above we targeted the Scl+19 enhancer into the Hprt gene locus in HM-1 ES cells. Analysis of differentiating EBs using fluorescent imaging suggested the Scl+19 enhancer was active from day 3 to 6 with a large percentage of EB cells expressing YFP during this period (Fig. 3A). To further investigate the Scl+19 activity, we disaggregated the EBs at day 4 and analysed YFP expression by flow cytometry. As with SV/LacZ reporters of the Scl+19 in transgenic animals, baseline fluorescence of enhancer-targeted cells was above the HM-1 control (Fig. 2B, Fig. 3B). However, YFP+ cells were readily identifiable as a distinct population (Fig. 3B). We further analysed this YFP+ population during the EB differentiation by flow cytometry. Similar to the Gfi1-35, the YFP+ population was seen from day 3 to 5, peaking at day 4 when 10–14% of the differentiating cells were YFP+ (Fig. 3C). By day 6, only background level YFP expression was seen.
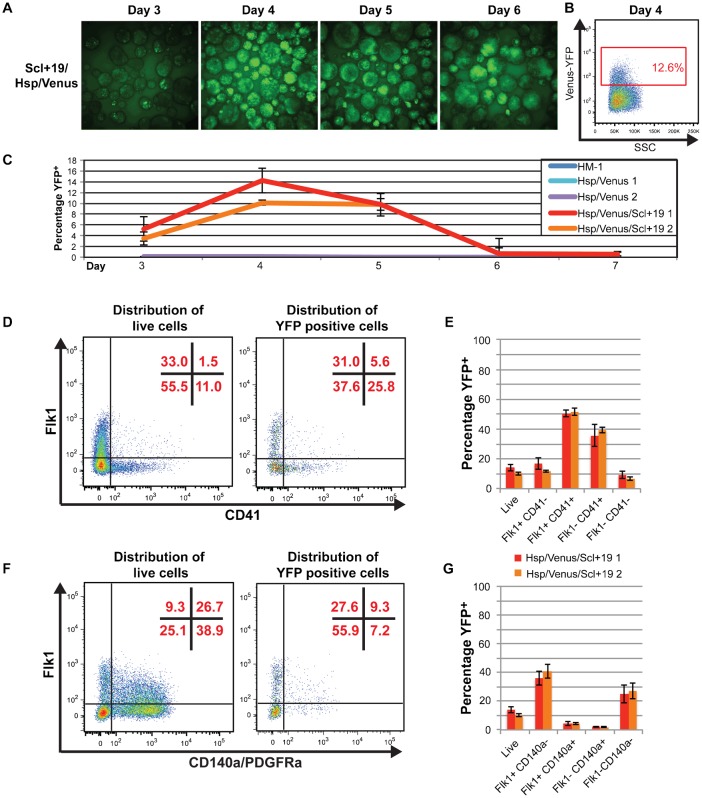
Fig. 3. Scl+19 activity marks haematopoietic cells and cardiac mesoderm in differentiating embryoid bodies.
(A) Fluorescent images of day 3–6 EBs from a representative Scl+19/Hsp/Venus clone. (B) Representative flow cytometry plots of Venus-YFP vs side scatter (SSC) of day 4 EBs for Scl+19/Hsp/Venus line with the YFP positive gate, and its percentage of the live cell population, shown in red. (C) Percentage of the YFP+ population (gating shown in B) from day 3–7 for HM-1 (blue), two representative Hsp/Venus control clones (aqua and purple) and two representative Scl+19/Hsp/Venus clones (red and orange), showing the average of three independent differentiation experiments ± standard deviation. (D) Representative flow cytometry plots showing distribution of Scl+19/Hsp/Venus day 4 EB cells in Flk1/CD41 quadrants for all live cells (left plot) and YFP positive cells only (right plot), with the percentage of cells in each quadrant shown in red. (E) Percentage of YFP positive cells in each of the Flk1/CD41 quadrants of (D) for Scl+19/Hsp/Venus (clone 1 in red, clone 2 in orange). Average of three independent differentiation experiments ± standard deviation. (F) As in (D), but for Flk1/CD140a quadrants. (G) As for (E), but for Flk1/CD140a quadrants in (F).
We next looked at the distribution of YFP+ cells at day 4 EB, using the surface markers Flk1, CD140a and CD41 to chart developmental progression towards a haematopoietic fate. YFP+ cells were enriched in the Flk1+CD41−, Flk1+CD41+ and Flk1−CD41+ populations during haematopoietic specification. YFP+ cells were relatively depleted amongst CD140a+ cells (65.6% of live cells were CD140a+ compared with 16.5% of YFP+ cells (Fig. 3E)). While this result suggests some enrichment for activity in the non-cardiogenic mesoderm this was much less pronounced than for the Gfi1-35 enhancer, where only 2.3% of YFP+ cells expressed CD140a (Fig. 2E). Importantly, consistent differences between the Scl+19 and Gfi1-35 enhancers were observed using multiple independent ES cell clones, with the Scl+19 enhancer showing relatively high activity in developmentally more immature cells.
The cellular resolution afforded by the Hsp68/Venus reporter system therefore provides detailed developmental stage-specific information on enhancer activity that would be very difficult to obtain using transgenic mice.
Quantitative dissection of the Scl+19 enhancer by motif mutations
Having demonstrated the ability to identify distinct activity patterns for two different haematopoietic enhancers, we next explored the utility of the Hsp68/Venus Hprt targeting approach to compare activities of wild type and mutant enhancers. Transcription factor binding events at regulatory elements are predicted by the presence of conserved binding motifs and the mutation of such motifs allows the specific activity of a class of transcription factors to be determined. However, current methods of mutational analysis are limited to population level luciferase assays in transformed cell lines or qualitative analysis in transgenic embryos. Previous analysis of the Scl+19 by these methods suggested that mutation of any of the two highly conserved Ets motifs or the single Gata motif results in loss of activity (Göttgens et al., 2002). Here we exploit the aforementioned advantages of the Hprt targeting approach and use this for quantitative analyses to compare the effects of motif mutations on enhancer activity. The YFP reporter facilitates analyses at the single cell level and the close resemblance of differentiating cell types in vitro and in vivo enable us to model the effects in the developing embryo.
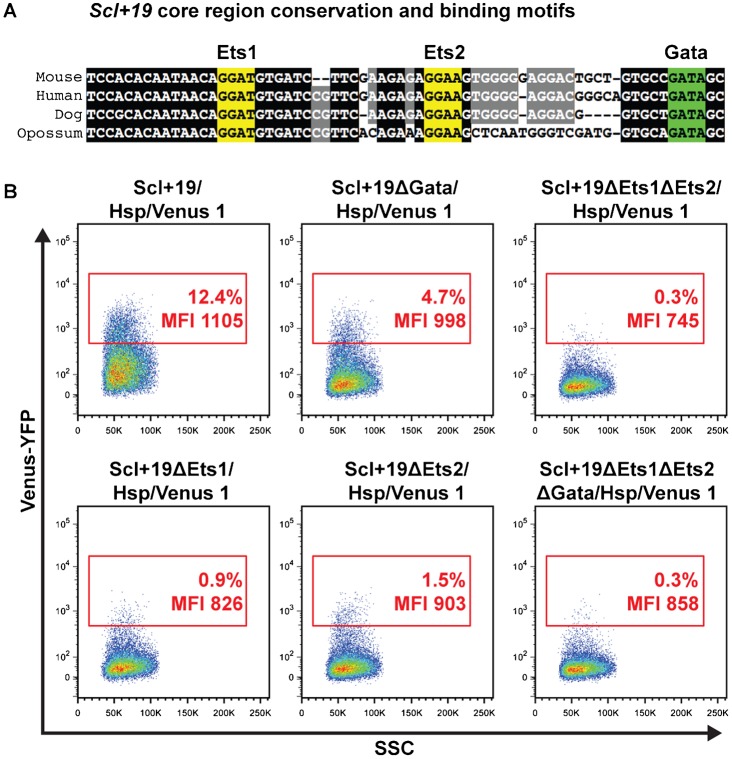
To assess the contribution of the Ets and Gata motifs found within the Scl+19 (Fig. 4A), we introduced Scl+19 motif mutations created previously (Göttgens et al., 2002) into Hsp/Venus Hprt targeting constructs using Gateway® cloning, and targeted these mutated enhancers to the Hprt locus in HM-1 ES cells. As wild type Scl+19 activity peaks at day 4 of the differentiation, we assessed the effects of the motif mutations on YFP expression at this time point by flow cytometry (Fig. 4B). Mutation of the Gata motif (Scl+19ΔGata/Hsp/Venus) reduced the percentage of YFP+ cells from ∼12% to ∼5%. Mutation of the Ets motifs (Scl+19ΔEts1/Hsp/Venus and Scl+19ΔEts2/Hsp/Venus) individually caused a more severe reduction in the YFP+ population to 0.9 and 1.5%, respectively. Mutation of all three motifs (Scl+19ΔEts1ΔEts2ΔGata/Hsp/Venus) or only both Ets motifs simultaneously (Scl+19ΔEts1ΔEts2/Hsp/Venus) caused complete loss of the YFP signal to background levels. In accordance with the reduction in the size of the YFP+ populations of the Scl+19 mutants, the mean fluorescent intensity (MFI) is also least affected in the Scl+19ΔGata/Hsp/Venus (998 vs 1105 in the wild type Scl+19/Hsp/Venus), while mutation of the Ets motifs results in an MFI that is more similar to the Scl+19ΔEts1ΔEts2ΔGata/Hsp/Venus (745–903 vs 858). To determine whether loss of the Gata motif was cell-type specific, we analysed YFP expression within CD41/Flk1 quadrants at day 4 EB (supplementary material Fig. S2). Relative to wild-type Scl+19 activity, YFP expression was lost in approximately half of Flk1+CD41− and Flk1+CD41+ populations, and approximately three-quarters of Flk1−CD41+ cells. This trend suggests the Gata motif may be more important, although still not necessary, for Scl+19 activity in committed CD41+ haematopoietic cells.
Fig. 4. Dissection of the Scl+19 enhancer by motif mutation.
(A) Sequence conservation of the Scl+19 core region in mouse, human, dog and opossum, with Ets motifs highlighted in yellow and the Gata motif highlighted in green. (B) Representative flow cytometry plots of Venus-YFP vs side scatter (SSC) of day 4 EBs for Scl+19/Hsp/Venus and Scl+19 mutant clones with the YFP positive gate, its percentage of the live cell population and mean fluorescence intensity (MFI), shown in red.
The use of single copy Venus-YFP reporter integration by this assay therefore affords a quantitative dissection of motif contribution to enhancer activity. Combined, these data suggest that all three motifs within the Scl+19 are necessary for its full activity. However, the motifs are dispensable for partial Scl+19 activity, as YFP+ cells are seen in all single motif mutation lines, but at a lower percentage. This observation suggests that increased transcription factor occupancy at the Scl+19 enhancer increases the likelihood that the enhancer will become active in a given cell. Moreover, mutation of the Gata motif caused a relatively mild reduction compared to mutation of the Ets motifs, suggesting that Gata factors are not critically required to activate the Scl+19.
Gata2 is not required for endothelial activity of the Scl+19 enhancer in vivo
The observation that initiation of Scl+19 activity during ES cell differentiation did not require an intact Gata motif was surprising given that we had shown previously that Gata2 binding is important for Scl+19 enhancer activity in haematopoietic cells (Göttgens et al., 2002; Pimanda et al., 2007). Gata2 was originally identified as an endothelial transcription factor (Wilson et al., 1990), and subsequently shown to be expressed in haemangioblasts and blood stem/progenitor cells (Suzuki et al., 2006; Lugus et al., 2007). To investigate whether Gata2 is dispensable for induction of the Scl+19 enhancer in vivo, we crossed transgenic mice carrying an Scl+19 enhancer LacZ reporter (Scl-3′enh/SV/lacZ) (Sánchez et al., 1999; Sánchez et al., 2001) with Gata2+/− mice, and then generated Gata2+/+, Gata2+/− or Gata2−/− E9.5 transgenic mouse embryos carrying the Scl+19 reporter (Fig. 5A).
Fig. 5. Gata2 is not required for endothelial activity of the Scl+19 enhancer in E9.5 transgenic mouse embryos.
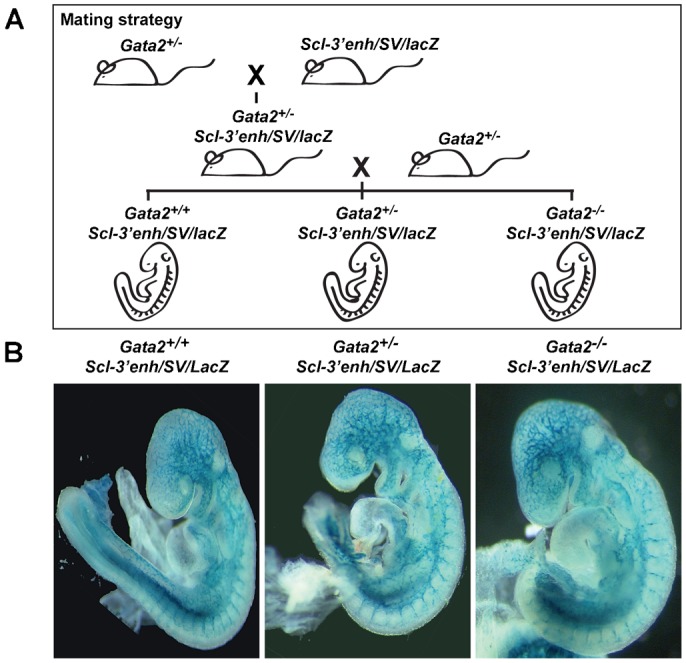
(A) Gata2+/− mice were crossed with mice carrying an Scl+19 reporter (Scl-3′enh/SV/lacZ) to produce Gata2+/− Scl-3′enh/SV/lacZ mice. These were re-crossed with Gata2+/− mice, from which E9.5 embryos were collected and genotyped for Gata2 and Scl-3′enh/SV/lacZ phenotypes. (B) Representative whole mount images of E9.5 mouse embryos, genotypes shown above, stained for lacZ expression using Xgal.
LacZ staining showed Scl+19 activity within the endothelium was similar in Gata2+/− and Gata2−/− E9.5 embryos compared to wild-type embryo staining, thus demonstrating Gata2 is not required for Scl+19 activity at this early stage of development. Due to the embryonic lethality of Gata2 deletion, the role of Gata2 in the activity of the Scl+19 at later stages of development, such as during definitive haematopoiesis, could not be determined. Nevertheless, in vivo analysis at E9.5 is consistent with our analyses of Hsp68/Venus reporter ES cell lines, which had shown Ets activity to be more important than Gata factors to activate the Scl+19 enhancer.
Discussion
Here, we have validated a novel ES cell-based enhancer assay that combines an efficient Gateway® cloning and Hprt gene targeting strategy with the flexibility and speed of analysis afforded by in vitro ES cell differentiation models and live cell analysis capability via YFP reporter tracking. This robust system additionally offers quantitative analysis of enhancer motif mutations to dissect regulatory inputs. Beyond these technological advantages, this assay also provides significant financial and animal welfare benefits over current costly transgenic mouse methodologies.
While we present here only the use of our new Hprt targeting constructs for analysis of enhancers in cardiac and haematopoietic lineages, it is not limited to these. Numerous other cell types can be generated in EBs by altering the culture conditions, including endothelium, adipocytes, skeletal and smooth muscle, neurons, and hepatocytes, in differentiation pathways that are thought to mimic normal development (reviewed elsewhere (Höpfl et al., 2004) and summarised in supplementary material Table S1). We therefore believe this system has broad applicability to analyses and dissection of enhancer activity in many developmental pathways and provides an attractive alternative to current transgenic mouse based methods.
While surface marker expression during embryonic development of the haematopoietic system has been well defined, this is not the case for other cell types. However, numerous tissue-specific cis-regulatory elements have been identified during embryogenesis by the VISTA Enhancer Browser project (Visel et al., 2007). Combination of this knowledge with our Hsp68/Venus reporter ES cell targeting strategy allows YFP expression to be used as a surrogate marker for lineage commitment, providing a robust read out for optimisation of ES cell differentiation protocols or screening surface marker expression during organogenesis. The approach described here could also be applied to human development where it could find wide application in regenerative medical research, by adaption of the targeting approach. The HPRT (Zwaka and Thomson, 2003), ROSA26 (Irion et al., 2007) and OCT4 (Hockemeyer et al., 2009) loci have all been successfully targeted in human ES cells. More recently, the AAV1 site within protein phosphatase 1 regulatory subunit 12C (PPP1R12C) (Samulski et al., 1991) has been shown to be the preferred site for gene targeting in human ES and iPS cells (Smith, J. R. et al., 2008; Hockemeyer et al., 2009). PPP1R12C is ubiquitously expressed in most cell types and the gene locus is in an open and active chromatin state permitting high levels of transgene expression. Moreover, chromatin insulators at the AAVS1 site ensure that transgene insertion does not perturb neighbouring gene expression and that the targeted transgene expression is tightly controlled (Lombardo et al., 2011).
Using our novel enhancer assay, we present evidence that both the Scl+19 and Gfi1-35 are active from the Flk1+ mesoderm stage through to committed CD41+ haematopoietic cell stages of haematopoietic specification. Gfi1-35 and Scl+19 enhancer activities show similar kinetics and specificities, although the Scl+19 is active earlier in the differentiation, with YFP+ cells seen from day 3 of the EB differentiation while Gfi1-35 activity is only seen from day 4 of the EB differentiation. This later activity of the Gfi1-35 is not unsurprising considering Scl is thought to be an important upstream regulator of this element (Wilson et al., 2010b). While Scl plays a critical early role in haematopoiesis, where it is necessary for haemangioblast commitment from Flk1+ mesoderm, Gfi1 is thought to play a later role in haematopoiesis, during Flk1+CD41+ to committed CD41+ haematopoietic cells (Lancrin et al., 2012). However, our data suggest the Gfi1-35 is active as early as the Flk1+CD41− stage, implying the haematopoietic transcription factor network that includes Gfi1-35 is active prior to CD41 expression during haematopoietic specification, and may indeed mark cells fated to this lineage.
Unlike the broad haematopoietic and endothelial staining patterning of the Scl+19 seen in Fig. 5B and (Göttgens et al., 2002), Gfi1-35 activity marks only haematopoietic clusters in the dorsal aorta and cells in the foetal liver of developing embryos (Lancrin et al., 2012). Consistent with this, our data show greater specificity of Gfi1-35 activity compared to the Scl+19 in day 4 EBs for cells undergoing commitment to the haematopoietic lineage. Furthermore, Scl+19 activity is seen in 16% of CD140a+ cells at this time point, when CD140a expression is thought to mark prospective cardiac mesoderm and progenitors. Interestingly, Scl is thought to play a role in patterning of cardiac mesoderm (Van Handel et al., 2012), with Scl−/− endocardium spontaneously beating while Scl overexpression inhibits cardiac differentiation in EBs (Ismailoglu et al., 2008). Our data suggest Scl+19 is active in a subset of cardiac mesoderm, where Scl expression likely plays a role in cardiac vs endothelial fate.
In summary, we have developed an efficient and robust enhancer assay as an alternative to current transgenic mouse methods and have validated its ability to recapitulate tissue-specific enhancer activity in both cardiac and haematopoietic lineages.
Materials and Methods
Targeting vector construction
To generate a reporter gene construct containing Venus downstream of an Hsp68 promoter, the SV40 promoter was cut out of SV40/Venus/pA KS using a HindIII restriction digest, end filling then digesting with NotI. We then cloned in the Hsp68 promoter, which was isolated from Hsp68LacZ KS, using a BamHI digest, followed by end filling and digesting with NotI. Subsequently this Hsp68/Venus/PA construct was digested with SphI, end filled and digested with NotI in preparation for cloning the cardiac enhancers mm75 and mm77, both of which were available as HspLacZ KS reporter gene constructs (Lawrence Berkeley National Laboratory (Visel et al., 2007)). These constructs were digested with BamHI (mm75) or SmaI (mm77), end filled and then digested with NotI before cloning into the linearised Hsp68/Venus/PA vector. The resulting enhancer Hsp68/Venus reporter cassette was then cloned as a NotI-Asp718 (blunt ended) fragment into the Hprt targeting construct pMP8NEBdeltalacZ (Misra et al., 2001) digested with MluI (end filled) and NotI.
To adapt the cloning to Gateway technology, the reporter cassette was isolated from the Hsp68/Venus/PA construct as a SphI blunt ended fragment and cloned immediately downstream of the Gateway® Cassette A (Invitrogen Life Technologies). The Hsp68/Venus/Gateway® Cassette was then cut out as a XhoI fragment, filled and blunt end cloned into the Hprt targeting construct pSKB1 (Bronson et al., 1996), linearised with MluI and filled in. The resulting targeting (destination) vector, pSKB1-GW-Hsp68-Venus (Hsp/Venus Gateway®) contained the Gateway® reporter gene cassette flanked by longer Hprt homology arms than the above (pMP8NEBdeltalacZ) targeting construct thus enabling greater recombination efficiency.
The Gfi-35 element was PCR amplified using primers with attB sequences (underlined) upstream of enhancer specific sequence (gfi1_35attb1F GGGGACAAGTTTGTACAAAAAAGCAGGCTGAGGTTTTTAAGGCAGTGAATCAT, gfi1_35attb1R GGGGACCACTTTGTACAAGAAAGCTGGGTCACTAGAACCGAGTGCTGGA). This was then gel purified (Qiagen 78704) and used for BP recombination into the pDONRTM221 vector (Invitrogen Life Technologies). Successfully generated Gfi-35 pDONRTM221 vectors were recombined with the Hsp/Venus Gateway® destination vector using LR clonase (Invitrogen Life Technologies).
The Scl+19 wild type and mutated sequences were cloned into the Hsp/Venus Gateway® targeting construct from SV/Luc constructs from (Göttgens et al., 2002) primers with attB sequences (underlined), GGGGACAAGTTTGTACAAAAAAGCAGGCTATATTAATCCCTCACTCAACAGCA and GGGGACCACTTTGTACAAGAAAGCTGGGTTGAGGTAGGGCTTAGGGGG via the pDONRTM221 vector as described above. Plasmids were verified by sequencing.
The negative control vector containing the minimal promoter only (Hsp68/Venus) was generated by annealing complementary oligos containing adjacent attB sequences (attbF1 GGGGACAAGTTTGTACAAAAAAGCAGGCTACCCAGCTTTCTTGTACAAAGTGGTCCCC, attbR1 GGGGACCACTTTGTACAAGAAAGCTGGGTAGCCTGCTTTTTTGTACAAACTTGTCCCC). These were made up to 800 ng/ul each in 1× TE and 50 mM NaCl and heated to 95°C before gradually cooling to room temperature. The double stranded attB sequence was then used in the recombination reactions described above in order to generate a destination vector containing only the attB sites upstream of the Hsp68/Venus reporter. All plasmid vectors generated as part of this study are available on request.
ES cell culture
HM-1 cells (Magin et al., 1992) were cultured feeder-free on gelatinised tissue culture plates at 37°C in 5% CO2 in ES cell medium (Knockout Dulbecco's Modified Eagle Medium (KO-DMEM; Invitrogen) supplemented with 15% foetal bovine serum (Gibco), recombinant murine LIF (ORF Genetics), L-glutamine, Pen/Strep and β-mercaptoethanol).
Gene targeting
Prior to gene targeting, HM-1 cells were cultured in ES cell medium supplemented with 6-thiguanine (Sigma) for 7 days to select for Hprt-deficient cells. HM-1 cells and linearised targeting vector were electroporated using a Gene Pulser electroporator (BioRad) at 800 V, 3 µF, and plated at clonal density. After 24 h, medium was replaced with ES cell medium supplemented with HAT (100 µM hypoxanthine, 0.4 µM aminopterin, 16 µM thymidine; Sigma) to select for Hrpt expressing clones. After 10–12 days, individual HAT-resistant ES cell colonies were picked and expanded in ES cell medium supplemented with HT (100 µM hypoxanthine, 16 µM thymidine; Sigma). ES cells were expanded in HT ES cell medium for seven days and then normal ES cell medium. Correct gene targeting was confirmed by Southern blotting or RT-PCR for expression of the reactivated Hprt using human_HPRT_exon1_forward CAGGCGAACCTCTCGGCTTT and mouse_Hprt_exon3_reverse GTGATGGCCTCCCATCTCCTT primers.
Cardiac differentiation
Cardiac differentiation of ES cells was performed as described (Maltsev et al., 1993). Briefly ES cells were dissociated, washed twice and resuspended at a concentration of 400 cells/20 µl in Dulbecco's Modified Eagle Medium (Sigma) supplemented with 20% FCS (Hyclone), 5×10−5 M β-mercaptoethanol (Invitrogen) and 1× non-essential amino acids (Invitrogen). Cardiac differentiation by hanging drop method was initiated by pipetting 20 µl drops containing ES cells onto the lid of a 10 cm petri dish, which was then inverted and placed back on the dish containing PBS. Embryoid bodies were allowed to form in hanging drops for 2 days after which they were resuspended in 10 ml of culture medium in 10 cm petri dishes for a further 5 days. At day 7 of culture, single EBs were plated into individual wells of a 24 well plate and cultured for an additional 7 days. At day 14 of culture, individual EBs were scored for the presence of spontaneously contracting cells and expression of YFP.
Haematopoietic differentiation
ES cells were differentiated into EBs according to (Sroczynska et al., 2009). Briefly, ES cells were passaged 24 h prior to differentiation and cultured in Iscove's Modified Dulbecco's Medium (IMDM; HyClone) supplemented with FBS, LIF and monothioglycerol (MTG). For the differentiation, ES cells were dissociated, washed twice and resuspended in differentiation medium (IMDM supplemented with 15% FBS (HyClone), 10% protein-free hybridoma medium II, 2 mM L-glutamine, 0.3 mg/ml human transferrin, 0.3 mM acorbic acid and 0.3 mM MTG). ES cells were seeded in Ultra Low Attachment 6-well dishes (Corning) at 10,000 cells/ml in 3 ml of differentiation medium. Differentiation medium was replaced after 5 days. EBs were harvested at required time points, disaggregated using Trp-LE (Invitrogen), spun down and resuspended in PBS supplemented with 5% FBS.
Fluorescence imaging
Bright field and fluorescence images were taken using a LEICA DMI 3000B microscope with a Hamamatsu digital camera (Orca-Flash 4.0) and Optomorph (for still images) or HCLimageLive (for timelapse imaging) software.
Flow cytometric analysis
Disaggregated EB cells were blocked using purified CD16/CD32 antibody (BD) and stained with PE-Cy7-CD41 antibody (Biolegend), APC-Flk1 antibody (BD), PE-CD140a (BD) or PE-CD41 (BD). Dapi (Sigma) was used as a viability stain. Cells were analysed using a 5 laser LSRFortessa cell analyser (BD) and data analysed using FlowJo software.
Transgenic mouse breeding and embryo analysis
All mice were housed and maintained according to UK Home Office regulations. Gata2+/− mice (Tsai et al., 1994) and Scl-3′enh/SV/lacZ mice (Sánchez et al., 1999; Sánchez et al., 2001) were crossed and Gata2+/− Scl-3′enh/SV/lacZ mice confirmed by genotyping. Gata2+/− and Gata2+/− Scl+19/SV/lacZ mice were then crossed and E9.5 embryos collected, genotyped and stained as described previously (Tsai et al., 1994; Sánchez et al., 1999). The Scl-3′enh is a 5.5 kb genomic fragment that containing the 641 bp Scl+19 enhancer element (Sánchez et al., 1999; Göttgens et al., 2002). The Scl+19 element has previously been shown to be responsible for the activity of the Scl-3′enh region (Göttgens et al., 2002).
Supplementary Material
Acknowledgments
We gratefully acknowledge S. Orkin for providing Gata2+/− mice, S. Duncan for the pMP8NEBdeltaLacZ plasmid, and M. de Bruijn for the Hsp68/LacZ plasmid.
Footnotes
Competing interests: The authors have no competing interests to declare.
Funding
Research in the authors' laboratory is supported by the National Centre for the Replacement, Refinement and Reduction of Animals in Research, Leukemia and Lymphoma Research, The Leukaemia and Lymphoma Society, Cancer Research UK, the Biotechnology and Biological Sciences Research Council, the Medical Research Council and core support grants from the Wellcome Trust to the Cambridge Institute for Medical Research and Wellcome Trust–MRC Cambridge Stem Cell Institute. D.E.D. was supported by the National Heart Lung and Blood Institute [grant number 5T32HL098057], and L.A.P. by the National Institute of Neurological Disorders and Stroke [grant number R01NS062859A] and by the National Human Genome Research Institute [grant numbers R01HG003988 and U54HG006997].
References
- Barton L. M., Gottgens B., Gering M., Gilbert J. G., Grafham D., Rogers J., Bentley D., Patient R., Green A. R. (2001). Regulation of the stem cell leukemia (SCL) gene: a tale of two fishes. Proc. Natl. Acad. Sci. USA 98, 6747–6752 10.1073/pnas.101532998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronson S. K., Plaehn E. G., Kluckman K. D., Hagaman J. R., Maeda N., Smithies O. (1996). Single-copy transgenic mice with chosen-site integration. Proc. Natl. Acad. Sci. USA 93, 9067–9072 10.1073/pnas.93.17.9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien K. R., Zhu H., Knowlton K. U., Miller-Hance W., van-Bilsen M., O'Brien T. X., Evans S. M. (1993). Transcriptional regulation during cardiac growth and development. Annu. Rev. Physiol. 55, 77–95 10.1146/annurev.ph.55.030193.000453 [DOI] [PubMed] [Google Scholar]
- Davidson E. H. (2010). Emerging properties of animal gene regulatory networks. Nature 468, 911–920 10.1038/nature09645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans V., Hatzopoulos A., Aird W. C., Rayburn H. B., Rosenberg R. D., Kuivenhoven J. A. (2000). Targeting the Hprt locus in mice reveals differential regulation of Tie2 gene expression in the endothelium. Physiol. Genomics 2, 67–75. [DOI] [PubMed] [Google Scholar]
- Göttgens B., Nastos A., Kinston S., Piltz S., Delabesse E. C., Stanley M., Sanchez M. J., Ciau-Uitz A., Patient R., Green A. R. (2002). Establishing the transcriptional programme for blood: the SCL stem cell enhancer is regulated by a multiprotein complex containing Ets and GATA factors. EMBO J. 21, 3039–3050 10.1093/emboj/cdf286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttgens B., Broccardo C., Sanchez M. J., Deveaux S., Murphy G., Göthert J. R., Kotsopoulou E., Kinston S., Delaney L., Piltz S. et al. (2004). The scl +18/19 stem cell enhancer is not required for hematopoiesis: identification of a 5′ bifunctional hematopoietic-endothelial enhancer bound by Fli-1 and Elf-1. Mol. Cell. Biol. 24, 1870–1883 10.1128/MCB.24.5.1870-1883.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Temple G. F., Brasch M. A. (2000). DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 10.1101/gr.143000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H., Hamblen M. J., Rooke H. M., Schindler J. W., Saleque S., Fujiwara Y., Orkin S. H. (2004). Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature 431, 1002–1007 10.1038/nature02994 [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R. C., Katibah G. E., Amora R., Boydston E. A., Zeitler B. et al. (2009). Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 27, 851–857 10.1038/nbt.1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höpfl G., Gassmann M., Desbaillets I. (2004). Differentiating embryonic stem cells into embryoid bodies. Methods Mol. Biol. 254, 79–98 10.1385/1-59259-741-6:079 [DOI] [PubMed] [Google Scholar]
- Irion S., Luche H., Gadue P., Fehling H. J., Kennedy M., Keller G. (2007). Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 25, 1477–1482 10.1038/nbt1362 [DOI] [PubMed] [Google Scholar]
- Ismailoglu I., Yeamans G., Daley G. Q., Perlingeiro R. C. R., Kyba M. (2008). Mesodermal patterning activity of SCL. Exp. Hematol. 36, 1593–1603 10.1016/j.exphem.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Karsunky H., Zeng H., Schmidt T., Zevnik B., Kluge R., Schmid K. W., Dührsen U., Möröy T. (2002). Inflammatory reactions and severe neutropenia in mice lacking the transcriptional repressor Gfi1. Nat. Genet. 30, 295–300 10.1038/ng831 [DOI] [PubMed] [Google Scholar]
- Kassouf M. T., Chagraoui H., Vyas P., Porcher C. (2008). Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood 112, 1056–1067 10.1182/blood-2007-12-128900 [DOI] [PubMed] [Google Scholar]
- Keller G., Kennedy M., Papayannopoulou T., Wiles M. V. (1993). Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol. Cell. Biol. 13, 473–486 10.1128/MCB.13.1.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothary R., Clapoff S., Darling S., Perry M. D., Moran L. A., Rossant J. (1989). Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105, 707–714. [DOI] [PubMed] [Google Scholar]
- Lancrin C., Mazan M., Stefanska M., Patel R., Lichtinger M., Costa G., Vargel O., Wilson N. K., Möröy T., Bonifer C. et al. (2012). GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 120, 314–322 10.1182/blood-2011-10-386094 [DOI] [PubMed] [Google Scholar]
- Landy A. (1989). Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 58, 913–941 10.1146/annurev.bi.58.070189.004405 [DOI] [PubMed] [Google Scholar]
- Lombardo A., Cesana D., Genovese P., Di Stefano B., Provasi E., Colombo D. F., Neri M., Magnani Z., Cantore A., Lo Riso P. et al. (2011). Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods 8, 861–869 10.1038/nmeth.1674 [DOI] [PubMed] [Google Scholar]
- Lugus J. J., Chung Y. S., Mills J. C., Kim S. I., Grass J. A., Kyba M., Doherty J. M., Bresnick E. H., Choi K. (2007). GATA2 functions at multiple steps in hemangioblast development and differentiation. Development 134, 393–405 10.1242/dev.02731 [DOI] [PubMed] [Google Scholar]
- Magin T. M., McWhir J., Melton D. W. (1992). A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 20, 3795–3796 10.1093/nar/20.14.3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev V. A., Rohwedel J., Hescheler J., Wobus A. M. (1993). Embryonic stem cells differentiate in vitro into cardiomyocytes representing sinusnodal, atrial and ventricular cell types. Mech. Dev. 44, 41–50 10.1016/0925-4773(93)90015-P [DOI] [PubMed] [Google Scholar]
- May D., Blow M. J., Kaplan T., McCulley D. J., Jensen B. C., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C. et al. (2012). Large-scale discovery of enhancers from human heart tissue. Nat. Genet. 44, 89–93 10.1038/ng.1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H. K., Fujiwara Y., Schlaeger T. M., Traver D., Orkin S. H. (2003). Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood 101, 508–516 10.1182/blood-2002-06-1699 [DOI] [PubMed] [Google Scholar]
- Minami T., Donovan D. J., Tsai J. C., Rosenberg R. D., Aird W. C. (2002). Differential regulation of the von Willebrand factor and Flt-1 promoters in the endothelium of hypoxanthine phosphoribosyltransferase-targeted mice. Blood 100, 4019–4025 10.1182/blood-2002-03-0955 [DOI] [PubMed] [Google Scholar]
- Misra R. P., Bronson S. K., Xiao Q., Garrison W., Li J., Zhao R., Duncan S. A. (2001). Generation of single-copy transgenic mouse embryos directly from ES cells by tetraploid embryo complementation. BMC Biotechnol. 1, 12 10.1186/1472-6750-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitjavila-Garcia M. T., Cailleret M., Godin I., Nogueira M. M., Cohen-Solal K., Schiavon V., Lecluse Y., Le Pesteur F., Lagrue A. H., Vainchenker W. (2002). Expression of CD41 on hematopoietic progenitors derived from embryonic hematopoietic cells. Development 129, 2003–2013. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Pennacchio L. A., Ahituv N., Moses A. M., Prabhakar S., Nobrega M. A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K. D. et al. (2006). In vivo enhancer analysis of human conserved non-coding sequences. Nature 444, 499–502 10.1038/nature05295 [DOI] [PubMed] [Google Scholar]
- Pimanda J. E., Göttgens B. (2010). Gene regulatory networks governing haematopoietic stem cell development and identity. Int. J. Dev. Biol. 54, 1201–1211 10.1387/ijdb.093038jp [DOI] [PubMed] [Google Scholar]
- Pimanda J. E., Ottersbach K., Knezevic K., Kinston S., Chan W. Y. I., Wilson N. K., Landry J.-R., Wood A. D., Kolb-Kokocinski A., Green A. R. et al. (2007). Gata2, Fli1, and Scl form a recursively wired gene-regulatory circuit during early hematopoietic development. Proc. Natl. Acad. Sci. USA 104, 17692–17697 10.1073/pnas.0707045104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C., Swat W., Rockwell K., Fujiwara Y., Alt F. W., Orkin S. H. (1996). The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86, 47–57 10.1016/S0092-8674(00)80076-8 [DOI] [PubMed] [Google Scholar]
- Robb L., Elwood N. J., Elefanty A. G., Köntgen F., Li R. L., Barnett L. D., Begley C. G. (1996). The scl gene product is required for the generation of all hematopoietic lineages in the adult mouse. EMBO J. 15, 4123–4129. [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F., Tenen D. G. (2007). Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7, 105–117 10.1038/nri2024 [DOI] [PubMed] [Google Scholar]
- Samulski R. J., Zhu X., Xiao X., Brook J. D., Housman D. E., Epstein N., Hunter L. A. (1991). Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 10, 3941–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M., Göttgens B., Sinclair A. M., Stanley M., Begley C. G., Hunter S., Green A. R. (1999). An SCL 3′ enhancer targets developing endothelium together with embryonic and adult haematopoietic progenitors. Development 126, 3891–3904. [DOI] [PubMed] [Google Scholar]
- Sánchez M. J., Bockamp E. O., Miller J., Gambardella L., Green A. R. (2001). Selective rescue of early haematopoietic progenitors in Scl(-/-) mice by expressing Scl under the control of a stem cell enhancer. Development 128, 4815–4827. [DOI] [PubMed] [Google Scholar]
- Shivdasani R. A., Mayer E. L., Orkin S. H. (1995). Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 373, 432–434 10.1038/373432a0 [DOI] [PubMed] [Google Scholar]
- Silberstein L., Sánchez M. J., Socolovsky M., Liu Y., Hoffman G., Kinston S., Piltz S., Bowen M., Gambardella L., Green A. R. et al. (2005). Transgenic analysis of the stem cell leukemia +19 stem cell enhancer in adult and embryonic hematopoietic and endothelial cells. Stem Cells 23, 1378–1388 10.1634/stemcells.2005-0090 [DOI] [PubMed] [Google Scholar]
- Smith A. M., Sanchez M. J., Follows G. A., Kinston S., Donaldson I. J., Green A. R., Göttgens B. (2008). A novel mode of enhancer evolution: the Tal1 stem cell enhancer recruited a MIR element to specifically boost its activity. Genome Res. 18, 1422–1432 10.1101/gr.077008.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. R., Maguire S., Davis L. A., Alexander M., Yang F., Chandran S., ffrench-Constant C., Pedersen R. A. (2008). Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells 26, 496–504 10.1634/stemcells.2007-0039 [DOI] [PubMed] [Google Scholar]
- Spensberger D., Kotsopoulou E., Ferreira R., Broccardo C., Scott L. M., Fourouclas N., Ottersbach K., Green A. R., Göttgens B. (2012). Deletion of the Scl +19 enhancer increases the blood stem cell compartment without affecting the formation of mature blood lineages. Exp. Hematol. 40, 588–598, e1 10.1016/j.exphem.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sroczynska P., Lancrin C., Pearson S., Kouskoff V., Lacaud G. (2009). In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol. Biol. 538, 317–334 10.1007/978-1-59745-418-6_16 [DOI] [PubMed] [Google Scholar]
- Suzuki N., Ohneda O., Minegishi N., Nishikawa M., Ohta T., Takahashi S., Engel J. D., Yamamoto M. (2006). Combinatorial Gata2 and Sca1 expression defines hematopoietic stem cells in the bone marrow niche. Proc. Natl. Acad. Sci. USA 103, 2202–2207 10.1073/pnas.0508928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai F. Y., Keller G., Kuo F. C., Weiss M., Chen J., Rosenblatt M., Alt F. W., Orkin S. H. (1994). An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371, 221–226 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Van Handel B., Montel-Hagen A., Sasidharan R., Nakano H., Ferrari R., Boogerd C. J., Schredelseker J., Wang Y. L., Hunter S., Org T. et al. (2012). Scl represses cardiomyogenesis in prospective hemogenic endothelium and endocardium. Cell 150, 590–605 10.1016/j.cell.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Minovitsky S., Dubchak I., Pennacchio L. A. (2007). VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., Taher L., Girgis H., May D., Golonzhka O., Hoch R. V., McKinsey G. L., Pattabiraman K., Silberberg S. N., Blow M. J. et al. (2013). A high-resolution enhancer atlas of the developing telencephalon. Cell 152, 895–908 10.1016/j.cell.2012.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. B., Dorfman D. M., Orkin S. H. (1990). A nonerythroid GATA-binding protein is required for function of the human preproendothelin-1 promoter in endothelial cells. Mol. Cell. Biol. 10, 4854–4862 10.1128/MCB.10.9.4854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N. K., Timms R. T., Kinston S. J., Cheng Y. H., Oram S. H., Landry J. R., Mullender J., Ottersbach K., Gottgens B. (2010a). Gfi1 expression is controlled by five distinct regulatory regions spread over 100 kilobases, with Scl/Tal1, Gata2, PU.1, Erg, Meis1, and Runx1 acting as upstream regulators in early hematopoietic cells. Mol. Cell. Biol. 30, 3853–3863 10.1128/MCB.00032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N. K., Foster S. D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P. M., Kinston S., Ouwehand W. H., Dzierzak E. et al. (2010b). Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7, 532–544 10.1016/j.stem.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Yücel R., Kosan C., Heyd F., Möröy T. (2004). Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J. Biol. Chem. 279, 40906–40917 10.1074/jbc.M400808200 [DOI] [PubMed] [Google Scholar]
- Zhao F., Lufkin T., Gelb B. D. (2003). Expression of Tfap2d, the gene encoding the transcription factor Ap-2 delta, during mouse embryogenesis. Gene Expr. Patterns 3, 213–217 10.1016/S1567-133X(02)00067-4 [DOI] [PubMed] [Google Scholar]
- Zwaka T. P., Thomson J. A. (2003). Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 21, 319–321 10.1038/nbt788 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.