Abstract
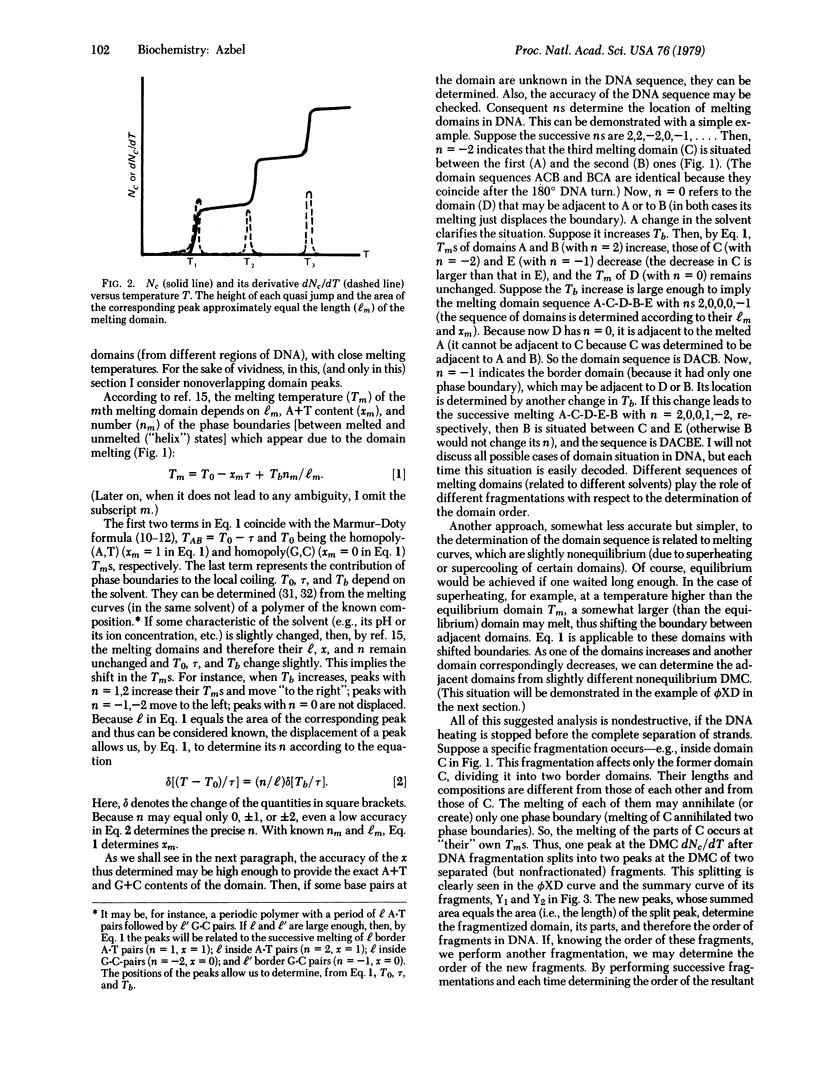
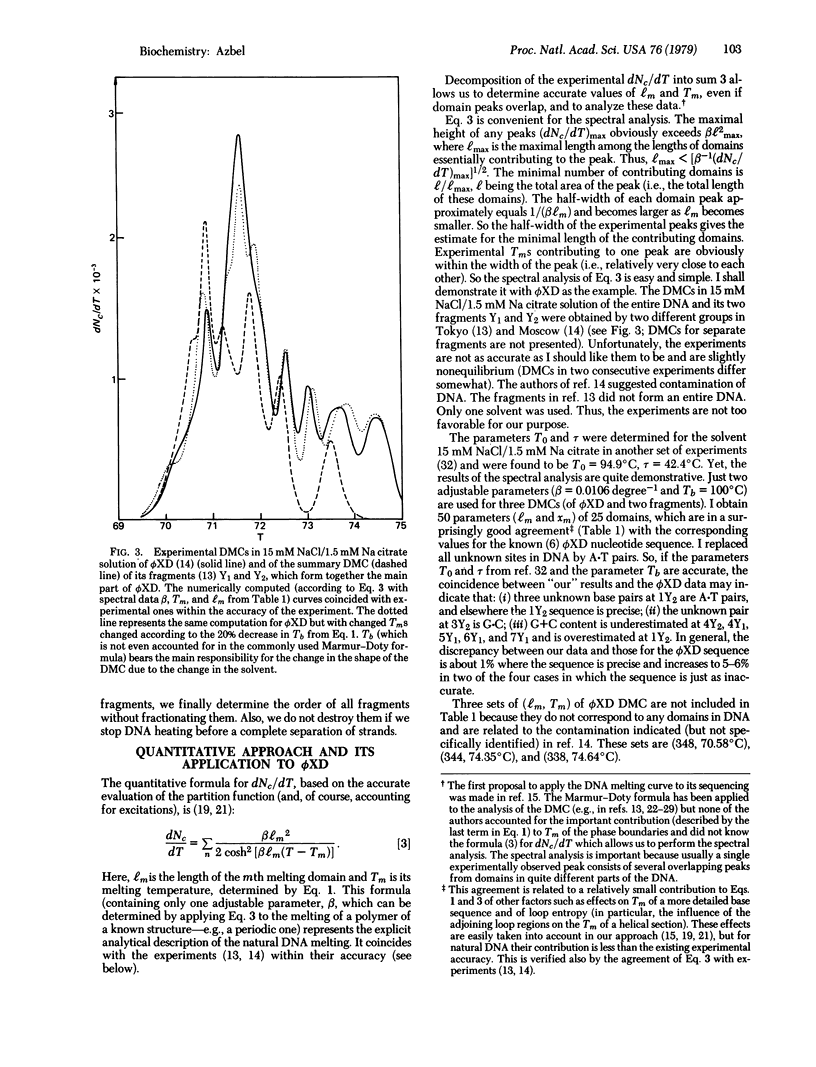
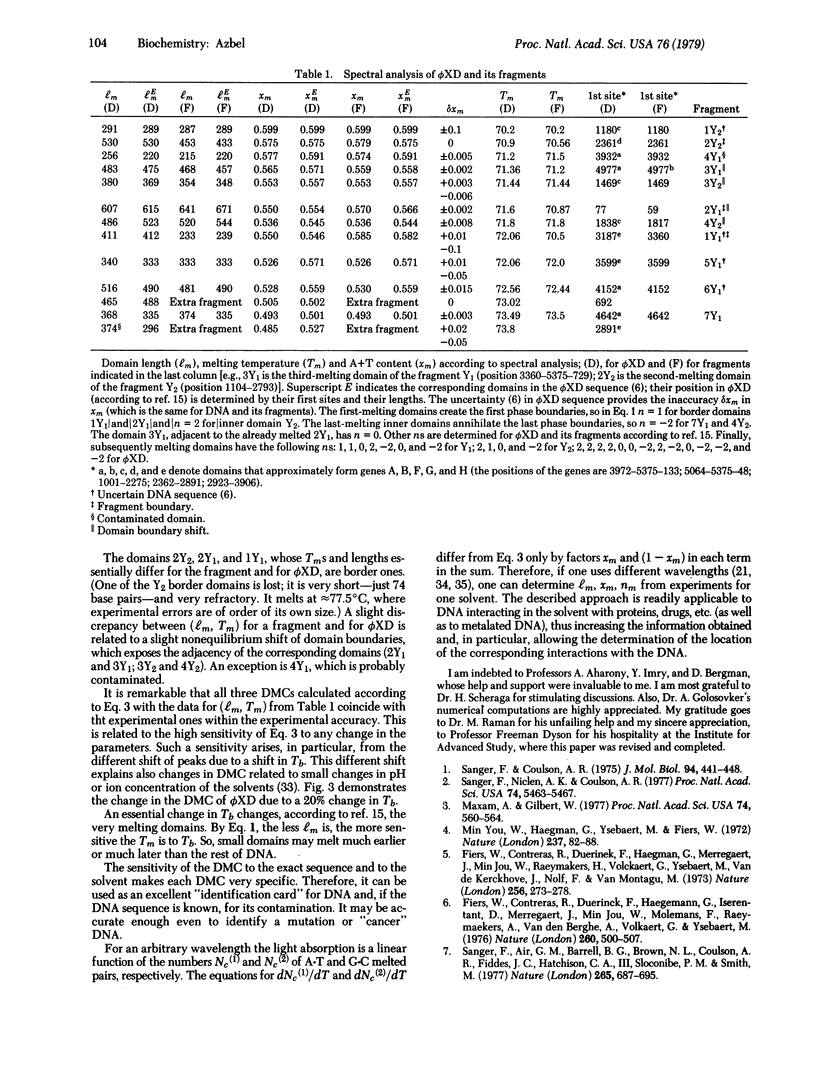
The dependence of DNA absorbance (for light at about 260 nm) on temperature is related to a specific DNA sequence structure in the vicinity of DNA thermal denaturation (the so-called DNA melting or coiling). A straightforward analysis of the experimental DNA melting curve allows us to determine the lengths, the A+T content, and the location in DNA of certain domains. In the case of a specific DNA fragmentation, the order of fragments in DNA can be learned from this analysis, nondestructively and quickly, without fractionating the fragments and other methods of fragmentation. If the DNA nucleotide sequence is known except for some sites and uncertain portions, the analysis determines these sites and the accuracy of the sequence at the portions. This information may complement exact methods of DNA sequencing. The proposed analysis is applied to bacteriophage phiX174, whose melting curve is known. The results are compared to and found to be in an excellent agreement with the known phiX174 nucleotide sequence.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama C., Gotoh O., Wada A. Spectral analysis on the melting fine structure of is lambda DNA and T2 DNA. Biopolymers. 1977 Feb;16(2):427–435. doi: 10.1002/bip.1977.360160215. [DOI] [PubMed] [Google Scholar]
- Ansevin A. T., Vizard D. L., Brown B. W., McConathy J. High-resolution thermal denaturation of DNA. I. Theoretical and practical considerations for the resolution of thermal subtransitions. Biopolymers. 1976 Jan;15(1):153–174. doi: 10.1002/bip.1976.360150111. [DOI] [PubMed] [Google Scholar]
- Azbel M. Y. The inverse problem for DNA. Biopolymers. 1973;12(7):1591–1609. doi: 10.1002/bip.1973.360120712. [DOI] [PubMed] [Google Scholar]
- Blake R. D., Lefoley S. G. Spectral analysis of high resolution direct-derivative melting curves of DNA for instantaneous and total base composition. Biochim Biophys Acta. 1978 Apr 27;518(2):233–246. doi: 10.1016/0005-2787(78)90180-6. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegeman G., Iserentant D., Merregaert J., Min Jou W., Molemans F., Raeymaekers A., Van den Berghe A. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976 Apr 8;260(5551):500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Duerinck F., Haegmean G., Merregaert J., Jou W. M., Raeymakers A., Volckaert G., Ysebaert M., Van de Kerckhove J. A-protein gene of bacteriophage MS2. Nature. 1975 Jul 24;256(5515):273–278. doi: 10.1038/256273a0. [DOI] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Gotoh O., Husimi Y., Yabuki S., Wada A. Hyperfine structure in melting profile of bacteriophage lambda DNA. Biopolymers. 1976 Apr;15(4):655–670. doi: 10.1002/bip.1976.360150406. [DOI] [PubMed] [Google Scholar]
- Hirschman S. Z., Felsenfeld G. Determination of DNA composition and concentration by spectral analysis. J Mol Biol. 1966 Apr;16(2):347–358. doi: 10.1016/s0022-2836(66)80178-x. [DOI] [PubMed] [Google Scholar]
- Hirschman S. Z., Gellert M., Falkow S., Felsenfeld G. Spectral analysis of the intramolecular heterogeneity of lambda-DNA. J Mol Biol. 1967 Sep 28;28(3):469–477. doi: 10.1016/s0022-2836(67)80097-4. [DOI] [PubMed] [Google Scholar]
- Lyubchenko Y. L., Vologodskii A. V., Frank-Kamenetskii M. D. Direct comparison of theoretical and experimental melting profiles for RF II phiX174 DNA. Nature. 1978 Jan 5;271(5640):28–31. doi: 10.1038/271028a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Jou W., Haegeman G., Ysebaert M., Fiers W. Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein. Nature. 1972 May 12;237(5350):82–88. doi: 10.1038/237082a0. [DOI] [PubMed] [Google Scholar]
- Poland D. Recursion relation generation of probability profiles for specific-sequence macromolecules with long-range correlations. Biopolymers. 1974;13(9):1859–1871. doi: 10.1002/bip.1974.360130916. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M., Van Assel S. Base compisition heterogeneity in kinetoplast DNA FROM FOUR SPECIES OF HEMOFLAGELLATES. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1249–1255. doi: 10.1016/s0006-291x(74)80418-3. [DOI] [PubMed] [Google Scholar]
- Tachibana H., Wada A., Gotoh O., Takanami M. Location of the cooperative melting regions in bacteriophage fd DNA. Biochim Biophys Acta. 1978 Feb 16;517(2):319–328. doi: 10.1016/0005-2787(78)90198-3. [DOI] [PubMed] [Google Scholar]
- Vizard D. L., Ansevin A. T. High resolution thermal denaturation of DNA: thermalites of bacteriophage DNA. Biochemistry. 1976 Feb 24;15(4):741–750. doi: 10.1021/bi00649a004. [DOI] [PubMed] [Google Scholar]
- Wada A., Tachibana H., Gotoh O., Takanami M. Long range homogeneity of physical stability in double-stranded DNA. Nature. 1976 Sep 30;263(5576):439–440. doi: 10.1038/263439a0. [DOI] [PubMed] [Google Scholar]
- Wada A., Tachibana H., Ueno S., Husimi Y., Machida Y. Melting fine structure of DNA fragments of known base sequence from theta X174. Nature. 1977 Sep 22;269(5626):352–353. doi: 10.1038/269352a0. [DOI] [PubMed] [Google Scholar]
- Yabuki S., Gotoh O., Wada A. Fine structures in denaturation curves of bacteriophage lambda DNA. Their relation to the intramolecular heterogeneity in base compositon. Biochim Biophys Acta. 1975 Jul 7;395(3):258–273. doi: 10.1016/0005-2787(75)90196-3. [DOI] [PubMed] [Google Scholar]



