Abstract
Gastrointestinal stromal tumours (GIST) are thought to derive from the interstitial cells of Cajal (ICC) or an ICC precursor. Oncogenic mutations of the receptor tyrosine kinase KIT are present in most GIST. KIT K642E was originally identified in sporadic GIST and later found in the germ line of a familial GIST cohort. A mouse model harbouring a germline Kit K641E mutant was created to model familial GIST. The expression profile was investigated in the gastric antrum of the KitK641E murine GIST model by microarray, quantitative PCR and immunofluorescence. Gja1/Cx43, Gpc6, Gpr133, Pacrg, Pde3a, Prkar2b, Prkcq/Pkce, Rasd2, Spry4 and Tpbg/5T4 were found to be up-regulated. The proteins encoded by Gja1/Cx43, Pde3a, Prkcq/Pkce were localized in Kit-ir ICC in wild-type and KitK641E animals while Spry4 and Tpbg/5T4 were detected in Kit-ir cells only in KitK641E, but not in KitWT/WT animals. Most up-regulated genes in this mouse model belong to the gene expression profile of human GIST but also to the profile of normal Kit+ ICC in the mouse small intestine. Spry4 and Tpbg/5T4 may represent candidates for targeted therapeutic approaches in GIST with oncogenic KIT mutations.
Keywords: KIT, receptor tyrosine kinase, gastrointestinal stromal tumour, in vivo model, interstitial cells of Cajal
Introduction
KIT (a.k.a. c-kit) is a receptor tyrosine kinase (RTK). Its ligand stem cell factor (SCF) binding is required for the development of haematopoietic stem cells, melanocytes, mast cells, germ cells and interstitial cells of Cajal (ICC) of the digestive tract. The SCF/KIT system is involved in multiple biological processes, e.g. survival, proliferation and differentiation of KIT-expressing (Kit+) cell lineages.
KIT is proto-oncogene homologue to the v-kit oncogene of the Hardy-Zuckerman (HZ)-4 feline sarcoma retrovirus (for review: [1]). Oncogenic KIT mutations, i.e. leading to constitutive KIT activation, have been identified in various human neoplasms, including myeloproliferative syndromes, mastocytosis, germ cell tumours and gastrointestinal stromal tumours (GIST) (for review: [1]). GIST is the most common sarcoma of the gastrointestinal tract and exhibit ICC differentiation. ICC are tiny populations of mesenchymal cells located within the muscularis propria of the gastrointestinal tract, where they coordinate peristalsis through inherent pacemaker function and network formation [2]. Approximately 85% of GIST harbour oncogenic KIT mutations and 7% contain oncogenic platelet-derived growth factor receptor-α (PDGFRA) mutations.
Rare families have germline oncogenic KIT or PDGFRA mutations of exactly the same type as are seen in sporadic GIST. Individuals who harbour germline oncogenic KIT or PDGFRA mutations develop GIST with 100% penetrance. KIT K642E, a mutation resulting in a K-to-E substitution at codon 642 in the catalytic domain I of KIT, was originally identified in sporadic GIST [3] and has also been encountered as a germ-line mutation associated with hyperplasia of the ICC layer and GIST formation [4].
Transgenic mice harbouring Kit K641E, the murine homolog of human KIT K642E, have been generated by a knock-in gene targeting strategy, providing an in vivo GIST model with massive hyperplasia of Kit+ cells, especially in antrum and cecum [5].
We have used this model to assess the influence of Kit K641E on gene expression by comparing the transcriptome in the antrum of homozygous KitK641E:Neo/K641E:Neo mice and wild-type (WT) KitWT/WT littermates. To validate the gene expression profiles, differences in gene expression were confirmed by real-time quantitative PCR (qPCR) and proteins were localized by immunofluorescence (IF) in the muscularis propria of the antrum, with special attention to the Kit immunoreactive (Kit-ir) cells.
Despite the fact that human GIST overwhelmingly harbour different Kit mutations, most up-regulated genes identified in this study belong to the expression profile of human GIST and/or to the general gene expression profile of Kit+ ICC in the mouse small intestine, validating the relevancy of this mouse model and also further reinforcing the hypothesis that GIST originate from the ICC lineage.
Two up-regulated genes, detected only in KitK641E, but not in WT animals may represent candidates for targeted therapeutic approaches in GIST with oncogenic KIT mutations.
Materials and methods
Animals
Transgenic KitK641E mice [5] were maintained and experiments performed in accordance with the ethics committee for animal well-being of the Faculty of Medicine, Université Libre de Bruxelles, Brussels, Belgium.
In our colony, whereas KitWT/K641E:Neo heterozygous mice were viable and asymptomatic up to advanced age, most KitK641E:Neo/K641E:Neo homozygous mice died before weaning i.e.3 weeks after birth. Therefore we used 2-week-old (P14) KitK641E:Neo/K641E:Neo, KitWT/K641E:Neo and KitWT/WT littermates as well as adult (3–6-month old) KitWT/K641E:Neo and KitWT/WT animals for experiments as indicated below. Genotyping was performed as previously described [5].
Mice were killed by cervical dislocation. The stomach was removed and transferred to a Petri dish filled with 0.1 M phosphate buffered saline (PBS), pH 7.4 at room temperature (RT). Surrounding tissues (pancreas, mesentery, fat, etc.) were carefully removed without damaging the serosa. The antrum was delineated from the gastric corpus based on visual landmarks on the serosa.
Gene expression analysis
Microarray analysis was performed on antral tissue from three pairs of KitK641E:Neo/K641E:Neo and KitWT/WT littermates. Total RNA was extracted using the Mirvana kit (Ambion, Inc., Austin, TX, USA) according to manufacturer's instructions. cDNA was synthesized from 1 μg of RNA, followed by production of antisense RNA containing the modified nucleotide 5-(3-aminoallyl)-uridine triphosphate (UTP) using the Amino Allyl MessageAmp™ II aRNA Amplification kit (Ambion, Inc.). After labelling with Cy3 or Cy5 (GE Healthcare Bio-Sciences, Princeton, NJ, USA), sample pairs were hybridized onto mouse exonic evidence based oligonucleotide MEEBO slides (Stanford Functional Genomics Facility, Stanford, CA, USA). The oligonucleotide set consists of 38,784 70-mer probes that were designed using a transcriptome-based annotation of exonic structure for genomic loci. Hybridizations were replicated with dye swap.
Before use, cDNA samples were screened by qPCR for expression of pancreatitis-associated protein (Pap) in order to eliminate any sample contaminated by pancreatic tissue and for expression of the neomycin resistance cassette (NeoR1), which was confirmed to be only present in KitK641E transgenic animals and not in WT littermates.
Microarray data analysis
Slides were scanned using a Molecular Devices 4000B (Molecular Devices, Sunnyvale, CA, USA) laser scanner and expression levels were quantified using GenePix Pro 6.1 image analysis software (Axon Instruments, Inc., Union City, CA, USA). Image acquisitions were performed with automatic photomultiplier gains adjustment. Artefact-associated spots were eliminated by both visual and software-guided flags, as well as spots with a signal/background fluorescence ratio less than 2. The fluorescence data were imported into Acuity 4.0 software package (Molecular Devices). A non-linear locally weighted scatter plot normalization method applied to each individual block (print-tip option) was carried out using Acuity 4.0 software package (Molecular Devices). Data obtained from mean of the normalized log2-ratio calculated for each duplicate were used for the subsequent steps. A cut-off of absolute normalized fold change > 2 in at least one pair of the hybridized arrays was used to define a set of candidate genes for further investigation. A t-test based on the Bonferroni method was used to determine the P-value. Genes with P < 0.01 were selected. GENESIS software [6] was used for graphical representation (Fig. 1).
Fig. 1.
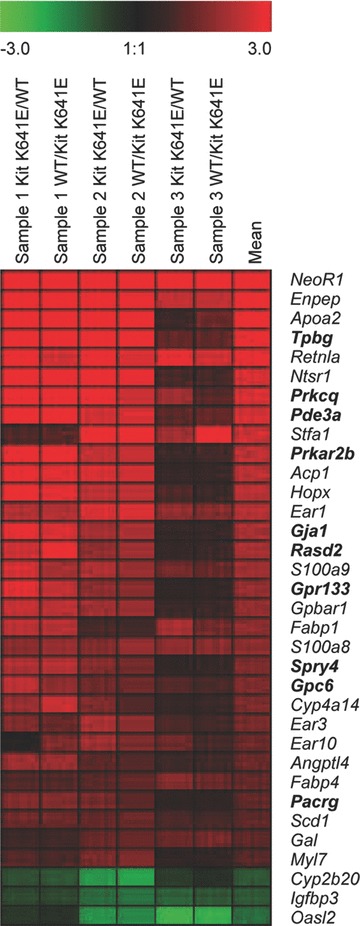
Expression pattern of 35 genes differentially expressed in the antrum of P14 KitK641E:Neo/K641E:Neo homozygous mice versus KitWT/WT littermates. Each row represents the relative level of expression for a single gene. Each column shows the expression level for a single sample replicated with dye swap centred at the mean of expression level across the three samples. The red and green colours indicate higher or lower expression levels, respectively. Significant differential expression has been confirmed by qPCR for the genes indicated in bold.
Candidate genes were also analysed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/), which provides a set of functional annotation tools for gene ontology.
Comparison of the gene expression profile in KitK641E:Neo/K641E:Neo antrum with expression profile in human GIST and in mouse ICC
We used the gene expression omnibus (GEO) database (http://www.ncbi.nlm;gov/entrez/query.fcgi?db=geo) to probe published microarray data from human GIST for genes found to be significantly up-regulated in KitK641E:Neo/K641E:Neo antrum. We also analysed the gene expression profiles of purified Kit-ir ICC from P9 mouse small intestine reported by Chen and colleagues [7].
Real-time quantitative PCR
The performance of the microarrays and statistical analyses used was assessed by qPCR on selected genes. A minimum of three different RNA samples of P14 KitK641E:Neo/K641E:Neo mice antrum and their respect KitWT/WT littermates was used for experimentation. Total RNA was extracted from mouse antrum using RNeasy Mini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Genomic DNA was removed with the DNA-free™ kit (Ambion, Inc.). RNA was reverse transcribed with 200 units of M-MLV Reverse Transcriptase (Invitrogen, Eugene, OR, USA) in a reaction containing 1 μg of random primers (Amersham Bioscience, Piscataway, NJ, USA), 10 mM each dNTP, 1× first-strand buffer, and 100 mM dithiothreitol followed by heat activation. The cDNA reverse transcription product was amplified with specific primers (Table 1) by qPCR using SYBR Green chemistry on a 7500 Realtime PCR system (Applied Biosystems, Foster City, CA, USA). Identical thermal profile conditions, namely: 95°C for 10 min.; then 40 cycles of 95°C for 15 sec. and 60°C for 1 min. were used for all primer sets. Emitted fluorescence was measured during the annealing/extension phase and amplification plots were generated using the sequence detection system. Transcriptional quantification relative to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) and β-actin housekeeping genes was performed with Qbase software [8].
Table 1.
Primers used for qPCR
| Acp1 F | TGTATGGATGAAAGCAATCTGAGAG | Pacrg R | CCTCGCTCATAACATTTTCGG |
| Acp1 R | TTTCTGTGGATCATAGCTCCCA | Pde3a F | CGGATACAGGGACATTCCTTATCA |
| Dog1 F | GGCTCTCCTATCCCTTCTCCC | Pde3a R | CACTTGGGAGGCCAGGAATC |
| Dog1 R | TTCATAGACTATTGTGCTCCGGG | Prkar 2b F | GTTCAACGCTCCAGTTATAAACCG |
| Gja1/Cx43 F | CCGAACTCTCCTTTTCCTTTGA | Prkar 2b R | TTATCCTGGACTCTGCATCGTCT |
| Gja1/Cx43 R | CCATGTCTGGGCACCTCTCT | Prkcq/Pkcθ F | TGGGCGGACAGAAATATGGT |
| Gapdh F | TGTGTCCGTCGTGGATCTGA | Prkcq /Pkcθ R | TCCTTCATTCTCAAACTCACTCATGT |
| Gapdh R | CCTGCTTCACCACCTTCTTGA | Prkcn/Prkd3 F | TGGAGATATGTTGGAAATGATTCTG |
| Gpc6 F | CGGACACAGCAAAGCCAGAT | Prkcn/Prkd3 R | AATTCCTCAAGGCAACAAGTATCTG |
| Gpc6 R | TGTCTGGCCGCGTGATG | Rab6 F | CAAGCTGGTGTTCCTGGGAG |
| Gpr 133 F | CCCAGGAACATCCAGGCTTT | Rab6 R | TGCCAATTGTTGCCTGATAGGT |
| Gpr 133 R | CGCTCCAGTTGTGTCCTGAAG | Rasd2 F | CATCCTCACAGGAGATGTCTTCAT |
| Hopx F | GGTGGAGATCCTGGAGTACAACTT | Rasd2 R | TTTTTATTCTTCAGGCAGGACTTGA |
| Hopx R | CGCTGCTTAAACCATTTCTGC | Spry2 F | AAAGCCGCGATCACGGA |
| Kit F | TGGGAGCTCTTCTCCTTAGGAA | Spry2 R | GGCTGCGACCCGTTGC |
| Kit R | TGCTCCGGGCTGACCAT | Spry 4 F | TGACTCTGCAGCTCCTCAAAGA |
| NeoR1 F | GGCCGCTTTTCTGGATTCAT | Spry 4 R | TCACAGGGACGCTGCTCTG |
| NeoR1 R | CCGCCAAGCTCTTCAGCA | Stfa1 F | CCAGATGATTGCTAACAAGGTCAG |
| Nrp1 F | TGTGCAAAACCAACAGACCTAGAT | Stfa1 R | GTGAATGAAACAACCATGACCTACAT |
| Nrp1 R | TTCTTGTCACCTTCCCCTTCTC | Tpbg/5T4 F | TCTACTGCTGCTTTGCTCACG |
| Pap F | ATTACTTTAACTGGGAGAGGAACCC | Tpbg/5T4 R | AGGATCGGATGAGCGACCT |
| Pap R | TGTCATATCTCTCCATTTTAGAAATCCA | β actin F | AACCGTGAAAAGATGACCCAGAT |
| Pacrg F | GCCATGATGAAAAACTCAGTCG | β actin R | GCCTGGATGGCTACGTACATG |
Primers were designed using the Primer Express 2.0 software (Applied Biosystems) and their specificity for the gene considered was verified using the Basic Local Alignment Search Tool (BLAST, http://www.ncbi.nlm.nih.gov/BLAST/). The amplification efficiency was assessed for all pairs of primers and was found to be greater than 95%. Statistical analysis was performed with the unpaired Student's t-test provided by Microsoft Office Excel 2003 and a P-value smaller than 0.05 was regarded as statistically significant.
Immunofluorescence
Specimens for IF were processed as described [9]. For Double IF staining slides were brought to RT, rinsed in 10 mM Tris (Merck-Belgolabo, Overijse, Belgium) and 0.15 M sodium chloride, pH 7.4 tris buffered saline (TBS), containing 0.1% (v/v) Triton-X 100 (TBS-TX), and incubated for 1 hr in 10% normal horse serum (NHS) (Hormonologie Laboratoire, Marloie, Belgium) and TBS-TX to reduce background staining. The slides were left overnight at RT in a humid chamber with the primary antibodies diluted in TBS-TX containing 2% NHS, rinsed in TBS and then incubated in the dark for 1 hr at RT in TBS containing the secondary antibodies. Slides were then rinsed in TBS and incubated in the dark for 1 hr at RT with Streptavidin coupled to NorthernLight™ (NL) 557 (R&D Systems, Abingdon, Oxon, UK) in TBS.
Primary antibodies raised in different species and secondary antibodies coupled with different fluorochromes were combined to specifically label one marker in green (Alexa 488), the other in red (NL557).
Primary and secondary antibodies used for immunohistochemistry are summarized in the table of antibodies (Table 2).
Table 2.
Primary and secondary antibodies used for IF
| Primary antibodies | Catalogue number | Host | Supplier | Dilution |
|---|---|---|---|---|
| Anti-c-Kit (C-14) | sc-1493 | goat | Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA | 1:1000 |
| Anti-c-Kit/CD117 | A4502 | rabbit | DAKO, Carpinteria, CA, USA | 1:500 |
| Anti-Spry4 | pab0230 | rabbit | Covalab, Villeurbanne, France | 1:3000 |
| Anti-Spry2 | #07—524 | rabbit | Upstate, Lake Placid, N Y, USA | 1:1000 |
| Anti-Tpbg/5T4 | ab45520 | rabbit | Abcam, Cambridge, UK | 1:200 |
| Anti-Pkrcq/Pkcθ | #2059 | rabbit | Cell Signaling Technology, Inc., Danvers, MA, USA | 1:100 |
| Anti-Pde3a | [10] | sheep | DSTT Unit, University of Dundee, Scotland | 1:3000 |
| Anti-Gja1/Cx 43 | #3512 | rabbit | Cell Signaling Technology, Inc. | 1:500 |
| Secondary antibodies | Catalogue number | Host | Supplier | Dilution |
|---|---|---|---|---|
| Anti-goat Alexa 448 | A11055 | donkey | Invitrogen | 1.200 |
| Anti-rabbit Alexa 448 | A21206 | donkey | Invitrogen | 1:400 |
| Anti-rabbit biotinylated | 711–065-152 | donkey | Jackson Immunoresearch, Cambridge, UK | 1:200 |
| Anti-sheep biotinylated | 713–065-147 | donkey | Jackson Immunoresearch | 1:200 |
The optimal working dilution has been determined empirically for each antibody. Omission of one of the primary or of one of the secondary antibodies resulted in the absence of immunoreactivity. The protocol used for double IF staining did not modify the distribution or the intensity of each individual labelling observed in corresponding single procedures.
Nuclei were stained with Hoechst, a nucleic acid-binding molecule fluorescent in the blue spectrum, 5 μM in Tris-HCl (Merck-Belgolabo) 0.05 M (pH 7.4), containing 0.5 mg/ml Ribonuclease A (type 1-AS from bovine pancreas), for 5 min. in the dark at RT.
Confocal microscopy
After three rinses in TBS, cover slips were mounted with FluorSave™ Reagent anti-fade mounting medium (Calbiochem, Nottingham, UK) in 50% glycerol and secured with nail polish before viewing under a LSM510 NLO multiphoton confocal microscope fitted on an Axiovert M200 inverted microscope equipped with C-Apochromat 40×/1.2 N.A. and 63×/1.2 N.A. water immersion objectives (Zeiss, Iena, Germany).
The 488-nm excitation wavelength of the Argon/2 laser, a main dichroic HFT 488 and a bandpass emission filter (BP500–550 nm) were used for selective detection of the green fluorochrome.
The 543 nm excitation wavelength of the HeNe1 laser, a main dichroic HFT 488/543/633 and a long-pass emission filter (LP560 nm) were used for selective detection of the red fluorochrome.
The nuclear stain Hoechst was excited in multiphotonic mode at 760 nm with a Mai Tai tunable broad-band laser (Spectra-Physics, Darmstad, Germany) and detected using a main dichroic HFT KP650 and a bandpass emission filter (BP435–485 nm).
Optical sections, 2-μm thick, were collected sequentially for each fluorochrome. The images generated (1522 × 1522 pixels, pixel size: 0.1 μm) were merged and displayed with the Zeiss LSM510 software and exported as TIFF image files.
All images show single optical sections across the regions of interest and are representative for a minimum of two preparations from three different animals for each genotype.
Results
Gene expression profiling of KitK641E:Neo/K641E:Neo antrum
Expression profiles based on calculated intensity ratio were obtained from RNA samples extracted from the antrum of three P14 KitK641E:Neo/K641E:Neo homozygous mice and their respective KitWT/WT littermates.
We found 47 genes significantly up-regulated (i.e. at least a twofold difference and P-value <0.01) and 4 genes significantly down-regulated in KitK641E:Neo/K641E:Neo antrum versus wild-type (Fig. 1; Table 3). The largest difference (ratio: 21.47) was observed for NeoR1, the neomycin resistance cassette present in KitK641E:Neo transgenic animals. 13 RIKEN sequences (12 up-regulated & 1 down regulated in KitK641E:Neo/K641E:Neo) were not further considered.
Table 3.
Microarray analysis of genes differentially expressed in KitK641E:Neo/K641E:Ne° antrum versus KitWT/WT littermates
| Gene symbol | Description | Fold change | t-Test | Unigene Id | ICC profile | GIST profile |
|---|---|---|---|---|---|---|
| NeoR | Neomycine R1 | 21.47 | 4.0E-06 | |||
| Enpep | Glutamyl aminopeptidase | 10.13 | 2.0E-04 | Mm.1193 | X | |
| Apoa2 | Mus musculus apolipoprotein A-ll (Apoa2), mRNA. | 8.98 | 5.1E-03 | Mm.389209 | ||
| Tpbg | Trophoblast glycoprotein | 7.52 | 2.3E-03 | Mm.432513 | X | |
| Retnla | Resistin like α | 7.12 | 3.0E-06 | Mm.441868 | ||
| Ntsrl | Neurotensin receptor 1 | 6.14 | 4.6E-03 | Mm.301712 | ||
| Prkcq | Protein kinase C, θ | 6.05 | 7.9E-04 | Mm.329993 | X | X |
| Pde3a | Phosphodiesterase 3A, cGMP inhibited | 6.05 | 2.1E-03 | Mm.103728 | X | X |
| Stfal | Stefin A1 | 5.56 | 3.7E-03 | Mm.390870 | ||
| Prkar2b | Protein kinase, cAMP dependent regulatory, type II β | 4.89 | 5.3E-03 | Mm.25594 | X | X |
| Acp1 | Acid phosphatase 1, soluble | 4.86 | 6.9E-03 | Mm.359831 | ||
| Hopx | Homeobox only domain | 4.82 | 4.5E-03 | Mm.181852 | ||
| Earl | Eosinophil-associated, ribonuclease A family, member 1 | 4.67 | 6.7E-05 | Mm.86948 | ||
| Gja1 | Gap junction membrane channel protein α1 (connexin 43, Cx43) | 4.08 | 7.7E-03 | Mm.378921 | X | |
| Rasd2 | RASD family, member 2 | 4.00 | 7.9E-03 | Mm.179267 | X | |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | 3.86 | 2.4E-04 | Mm.2128 | ||
| Gpr133 | G protein-coupled receptor 133 | 3.54 | 1.0E-02 | Mm.44050 | X | |
| Gpbarl | G protein-coupled bile acid receptor 1 | 3.48 | 4.2E-03 | Mm.246587 | ||
| Fabpl | Fatty acid binding protein 1, liver | 3.30 | 2.5E-03 | Mm.22126 | ||
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | 3.29 | 0.0E+00 | Mm.21567 | ||
| Spry4 | Sprouty homolog 4 (Drosophila) | 2.91 | 7.4E-03 | Mm.42038 | X | |
| Gpc6 | Glypican 6 | 2.89 | 1.1E-03 | Mm.440025 | X | |
| Cyp4a14 | Cytochrome P450, family 4, subfamily a, polypeptide 14 | 2.84 | 7.1E-03 | Mm.250901 | ||
| Ear3 | Eosinophil-associated, ribonuclease A family, member 3 | 2.71 | 8.5E-04 | Mm.327386 | ||
| Ear10 | Eosinophil-associated, ribonuclease A family, member 10 | 2.49 | 5.1E-03 | Mm.429087 | ||
| Angptl4 | Angiopoietin-like 4 | 2.48 | 3.3E-03 | Mm.196189 | ||
| Fabp4 | Fatty acid binding protein 4, adipocyte | 2.32 | 7.9E-05 | Mm.582 | ||
| Pacrg | Park2 co-regulated | 2.24 | 5.2E-03 | Mm.18889 | X | X |
| Scd1 | Stearoyl-Coenzyme A desaturase 1 | 2.23 | 5.0E-05 | Mm.267377 | ||
| Gal | Galanin | 2.21 | 3.2E-04 | Mm.4655 | ||
| Myl7 | Myosin, light polypeptide 7, regulatory | 2.03 | 4.6E-03 | Mm.46514 | ||
| Cyp2b20 | Cytochrome P450, family 2, subfamily b, polypeptide 20 | 0.50 | 8.8E-03 | Mm.218749 | ||
| Igfbp3 | Insulin-like growth factor binding protein 3 | 0.49 | 9.9E-05 | Mm.29254 | ||
| Oasl2 | 2′-5′ oligoadenylate synthetase-like 2 | 0.41 | 7.8E-03 | Mm.228363 |
Twofold or higher difference with a P-value <0.01 was regarded as significant. A ratio >2 indicates up-regulation, while values <0.5 indicate
down-regulation, in KitK641E:Neo/K641E:Neo antrum.
Significant differential expression has been confirmed by qPCR for the genes indicated in bold.
Genes belonging to a Kit-ir ICC profile [7] and genes up-regulated in human GIST (http://www.ncbi.nlm;gov/entrez/query.fcgi?db=geo) are indicated by ‘X’ in the two last columns at right.
A gene ontology description for these genes is provided in Table S1.
The list of all differentially expressed probe sets is provided as Supplementary Data 1 and the complete microarray data set has been deposited in GEO Database (http://www.ncbi.nlm;gov/entrez/query. fcgi?db = geo; accession number: GSE12931).
Confirmation of differential gene expression by qPCR
We used qPCR to further examine the expression of a subset of 13 genes selected for their putative roles in ICC development and maintenance, in Kit signalling pathways or in cancer biology. Means of relative level of expression in the antrum of P14 KitK641E:Neo/K641E:Neo mice and their respective KitWT/WT littermates and standard errors of log2-fold changes are shown in Fig. 2.
Fig. 2.
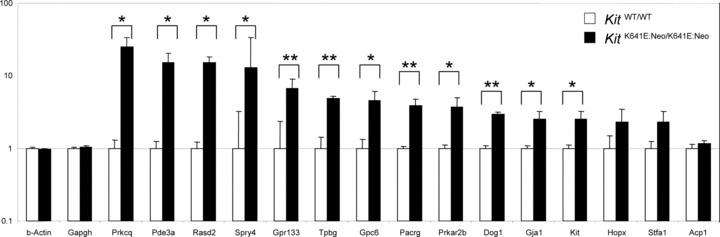
Difference in gene expression confirmed by qPCR. Ten genes identified in microarray analysis are significantly up-regulated in KitK641E:Neo/K641E:Neo antrum versus WT littermate: Pkrcq/Pkcθ; Pde3a; Rasd2; Spry4; Gpr133; Tpbg/5T4; Gpc6; Pacrg; Prkar2b and Gja1/C×43, were confirmed by qPCR (P< 0.05, indicated by *; P < 0.01, indicated by ** respectively). Hopx and Stfa1 did not reach the significance threshold (P= 0.07 and P= 0.2, respectively), while Acp1, despite an estimated 4.8-fold up-regulation suggested by microarray analysis, showed no difference by qPCR. Kit and Dog1 were also significantly up-regulated in KitK641E:Neo/K641E:Neo antrum.
qPCR confirmed a significant (Student's t-test P< 0.05) up-regulation for 10 out of 13 genes (indicated in ‘bold’ in Table 3) in KitK641E:Neo/K641E:Neo antrum: Protein kinase Cθ (Pkrcq/Pkcθ); Phosphodiesterase 3A, cGMP inhibited, (Pde3a); RASD family, member 2, (Rasd2); Sprouty homolog 4, (Spry4); G protein-coupled receptor 133, (Gpr133); Trophoblast glycoprotein, (Tpbg/5T4); Glypican 6, (Gpc6); Park2 co-regulated, (Pacrg); Protein kinase, cAMP dependent regulatory, type IIβ}, (Prkar2b); and Gap junction membrane channel protein α1, (Gja1/C×43).
Homeobox only domain (Hopx) and Stefin A1 (Stfa1) also appeared up-regulated but did not reach the significance threshold (P= 0.07 and P= 0.2, respectively) while Acid phosphatase 1, soluble (Acp1), despite an estimated 4.8-fold up-regulation suggested by microarray analysis, showed no difference (log2-fold change of homozygous animals = 1.17; P = 0.38) by qPCR.
While Spry4 and Pkrcq/Pkcθ were significantly up-regulated in KitK641E:Neo/K641E:Neo antrum, Sprouty homolog 2 (Spry2) (Fig. 4b), Protein kinase C, gama, (Pkrcc/Pkc gama) and Protein kinase C nu/Protein kinase D3 (Prkcn/Prkd3) showed no change (log2-fold change in homozygous animals = 1.3; P= 0.18; = 1.56; P= 0.13; = 1.19; P= 0.21, respectively).
Fig. 4.
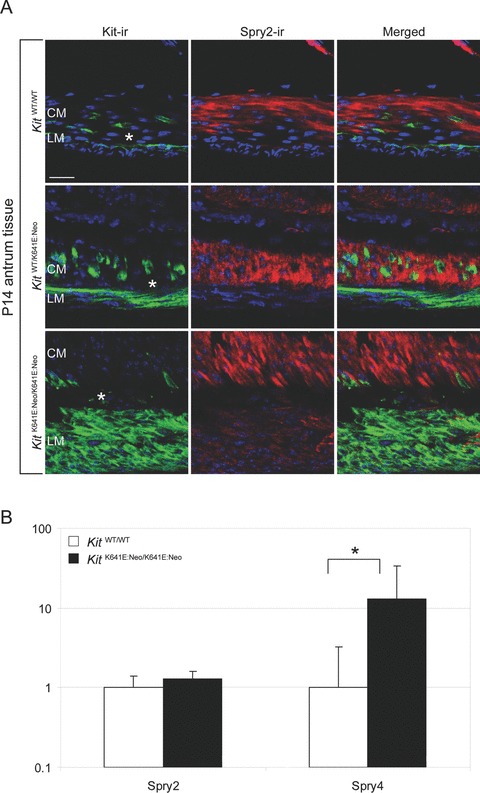
Spry2-ir is detected in KitK641E and KitWT/WT in smooth muscle cells. Spry2-ir (in red) was observed in smooth muscle cells – but not in Kit-ir cells (in green) – in all genotypes (A). By qPCR, Spry4 – but not Spry2 – was up- regulated in KitK641E:Neo/K641E:Neo (B). Figures are oriented with the circular muscle layer (CM) up and the longitudinal muscle layer (LM) down. Scale bar: 20 microns.
We also assessed by qPCR expression of Kit, which is the best established marker for ICC (for review see [11]) and the protooncogene most often activated in human GIST [12] and of Discovered on GIST 1 (Dog1), a gene strongly up-regulated in human GIST and also expressed in normal ICC [13]. Both Kit and Dog1 were significantly up-regulated in KitK641E:Neo/K641E:Neo antrum: log2-fold change was 2.5 and 2.95, respectively (Fig. 2).
To address the issue of stringency of the 2-fold cut-off selected for our microarray analysis, we tested by qPCR member of RAS oncogene family 6 (Rab6) which showed an estimated 1.7-fold change by microarray analysis. The difference by qPCR was not significant (log2-fold change of homozygous animals = 0.84; P= 0.13).
Genes up-regulated in KitK641E:Neo/K641E:Neo antrum belong to the gene profiles of murine ICC and human GIST
Nine genes significantly up-regulated in our study (Enpep; Pkrcq/Pkcθ; Pde3a; Prkar2b; Gja1/C×43; Rasd2; Gpr133; Gpc6; Pacrg) belong to the expression profile of ICC and six (Tpbg/5T4; Pkrcq/Pkcθ; Pde3a; Prkar2b; Spry4; Pacrg) belong to the expression profile of human GIST (Table 3).
Protein expression in Kit-ir ICC
The gut wall contains a variety of diverse cell types, the Kit-ir ICC representing only a very small percentage of the cells forming the muscularis propria. To further validate our findings, we used IF to investigate the localization of Pkrcq/Pkce, Pde3a, Gja1/C×43, Spry4, Tpgb/5T4 proteins in the muscularis propria of the mouse antrum, focusing on the Kit-ir ICC.
Prkcq/Pkcθ-ir, Pde3a-ir and Gja1/C×43-ir are present in Kit-ir ICC
We observed Prkcq/Pkcθ-ir in Kit-ir ICC at all ages and genotypes analysed. Prkcq/Pkcθ-ir was also moderately present in the myenteric plexus (Fig. S1).
Similarly, Pde3a-ir was also observed in Kit-ir ICC while no Pde3a-ir was detected in the myenteric plexus (Fig. S2).
In all conditions tested, Gja1/Cx43-ir, a marker for gap-junctions, was detected as tiny dots mostly present on, but not restricted to, Kit positive ICC, as Gja1/C×43-ir was also occasionally encountered in the regions of the myenteric plexus and between Kit negative smooth muscle cells in the antrum (Fig. S3).
Spry4-ir and Tpbg/5T4-ir are detected in Kit-ir ICC only in KitK641E
Spry4 and Tpbg/5T4 are important for embryonic development and their deregulation leads to malignant cell growth [14, 15]. Sprouty proteins represent a major class of ligand-inducible inhibitors of RTK-dependent signalling pathways [16].
Spry4-ir was observed in the Kit-ir hyperplasic layer in P14 homozygous KitK641E:Neo/K641ENeo antrum and in the Kit+ cell clusters in heterozygous KitWT/K641E:Neo P14 and adult antrum (Fig. 3).
Fig. 3.
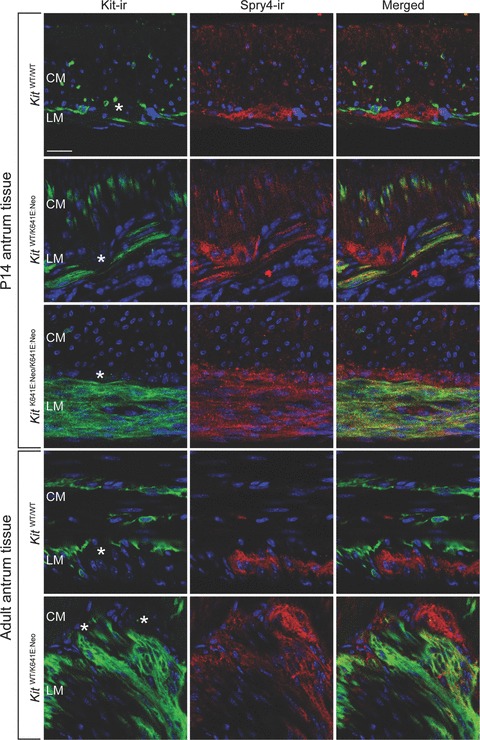
Spry4-ir in Kit-ir ICC is detected only in KitK641E. Spry4-ir (in red) was observed in Kit-ir ICC (in green) only in the presence of the Kit K641E:Neo allele but not in WT littermates. Spry4-ir was also consistently found in the myenteric plexus (indicated by *) in all genotypes. Figures are oriented with the circular muscle layer (CM) up and the longitudinal muscle layer (LM) down. Scale bar: 20 microns.
Conversely, strong Spry2-ir was present only in smooth muscle cells in all genotypes. Spry2-ir was not detected in Kit+ ICC in heterozygous KitWT/K641E:Neo and WT antrum while in homozygous KitK641E:Neo/K641E.Neo antrum, the Kit-ir layer exhibited a faint Spry2-ir (Fig. 4a).
The oncofetal protein, Tpbg/5T4, is a membrane tumour-associated protein expressed in various carcinomas [15]. Tpbg/5T4-ir was found in the Kit+ hyperplasic layer in P14 homozygous KitK641E:Neo/K641E.Neo antrum and in the Kit-ir cell clusters in heterozygous KitWT/K641E:Neo P14 and adult antrum (Fig. 5).
Fig. 5.
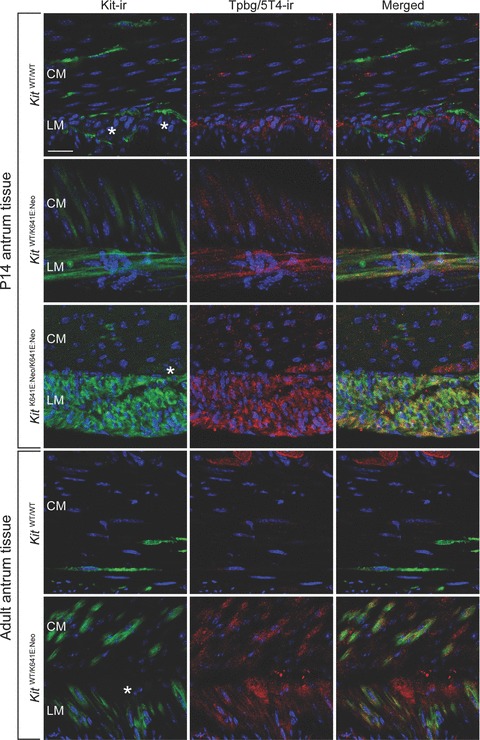
Tpbg/5T4-ir in Kit-ir ICC is detected only in KitK641E. Tpbg/5T4-ir (in red) was observed in Kit-ir ICC (in green) only in the presence of the Kit K641E:Neo allele but not in WT littermates. Tpbg/5T4-ir was also consistently found in the myenteric plexus (indicated by *) in all genotypes. Figures are oriented with the circular muscle layer (CM) up and the longitudinal muscle layer (LM) down. Scale bar: 20 microns.
In contrast, in KitWT/WT littermates, no Tpbg/5T4-ir was detected in Kit+ cells.
Myenteric plexus exhibited both Spry4-ir and Tpbg/5T4-ir in all conditions analysed. (Figs 3 and 5).
Discussion
Three knock-in mouse models have been generated to model familial forms of human GIST with germ-line oncogenic Kit mutation: Kit del-V558 [17], Kit K641E [5] and Kit D818Y [18]. In the Kit del-V558 model [17] tumour response to imatinib has been investigated using microarrays [19] but, to the best of our knowledge, this study in KitK641E is the first to address difference in the transcriptome between a GIST model and its normal counterpart.
The antrum was selected for its well characterized organization of muscle layers and Kit+ ICC and for the marked hyperplasia of Kit+ cells present in homozygous, KitK641E animals [5].
Kit-ir ICC represents less than 1% of the cell population of the highly heterogeneous gut wall. Modifications in gene expression occurring in ICC might thus vanish in the ambient ‘noise’. Nevertheless, comparisons between our microarray data and the expression profiles of mouse intestinal ICC [7] and human GIST (Table 3), as well as IF localization of selected proteins in Kit-ir cells (see below) support the relevancy of our approach.
Most so-called ‘GIST markers’ in the literature, like Pkrcq/PKCθ or Dog1, belong to the ICC profile. Their increased amount in the Kit antrum likely reflects the increased amount of Kit+ cells in the gut wall. The broad overlap between the gene expression profile in KitK641E antrum and the expression profiles of ICC purified from mouse small intestine [7] further supports the idea of a common origin for ICC and GIST (Table 3).
In this study we have identified three genes (Prkcq/Pkcθ, Pde3a, Gja1/C×43) overexpressed in KitK641E mouse antrum and associated with ICC gene expression profile [7] with restricted localization in Kit-ir cells. Prkcq/Pkce was very significantly up-regulated in KitK641ENeo/K641ENeo antrum, while two other Pkcs, Pkrcc/Pkc gama and Prkcn/Prkd3, were unchanged. In line with previous observations in guinea pig and human gut [20], Prkcq/Pkcθ-ir was observed in the Kit+ cells and in myenteric plexus in both KitWT/WTand KitK641ENeo/K641ENeoantrum (Fig. S1).
For the first time we have demonstrated here that Pde3a-ir is localized specifically in Kit-ir ICC in KitWT/WT and KitK641E:Neo/K641E:Neo antrum (Fig. S2) and could thus be regarded as a novel marker for ICC in the gut wall.
Kit-ir ICC forms networks with gap junctions between Kit-ir ICC and between Kit-ir ICC and smooth muscle cells. Gja1/C×43-ir we observed mostly – but not exclusively – in Kit-ir ICC, both in KitK641E:Neo and in KitWT/WT antrum (Fig. S3).
We also identified two genes, Tpbg/5T4 and Spry4, which do not belong to the normal ICC profile but are induced in Kit-ir cells by the KitK641E oncogenic mutant (Figs 3 and 5).
Tpbg/5T4 is a trophoblastic transmembrane glycoprotein initially defined by the monoclonal antibody 5T4 [21]. Tpbg/5T4 is not expressed in most adult normal tissues – including the small intestine – while Tpbg/5T4 is strongly expressed in trophoblast and in various (e.g. gastric and colorectal) carcinomas, where it is associated with metastatic behaviour and poor clinical prognosis [15]. Tpbg/5T4 affects cell motility through interactions with E-cadherin [22]. It is involved in cell migration during embryogenesis and in cell invasion associated with implantation. Due to its restricted expression profile, Tpbg/5T4 is regarded as a tumour-associated antigen [15] and is a promising target for anti-tumour vaccine development and for targeted therapy with exotoxin. The cancer vaccine TroVax, a modified vaccine Ankara encoding the tumour-associated antigen Tpbg/5T4, has been tested in phase I and II studies in colorectal cancer patients [23] and in an open-label phase 2 trial in hormone refractory prostate cancer patients [24]. Our finding of the induction of Tpbg/5T4 expression in Kit+ cells by KitK641E raises the possibility of a similar therapeutic approach in GIST with oncogenic Kit mutation but confirmation of Tpbg/5T4 expression in human GIST is a prerequisite.
The Sprouty (Spry) family is a highly conserved group of ligand-inducible inhibitors of RTK-induced signalling pathways originally described in drosophila. There are four mammalian orthologues (Spry1–4) whose modulation is growth factor- and cell context-dependent [16]. Spry proteins inhibit Ras–ERK MAPK cascades activated by RTK, leaving PI3K, Janus kinase (Jak) and other pathways unaffected [25]. Sprys become tyrosine phosphorylated after growth factor stimulation and tyrosine phosphorylation is required for Spry inhibitory activity [26]. Notably, tyrosine phosphorylation of Sprys induced by a growth factor vary among Sprys and cell types, suggesting that Spry are not functionally equivalent and might have adapted to particular signalling circuits.
Besides Spry2, minimal data is available about the other members of the Spry family. Our results showed a significant up-regulation of Spry4– but not Spry2– in KitK641E:Neo antrum, suggesting a differential effect also of oncogenic KitK641E mutation on the expression of the Spry family. Noteworthy, Spry4 is expressed in GIST, but not in other sarcomas [27] and Spry4 expression is down regulated by imatinib in GIST cells in parallel to the inhibition of KIT, AKT and Erk1/2 phosphorylation [28].
The Spry proteins appear as key negative regulators that limit the strength, duration and range of activation of RTKs, contributing to control of growth and differentiation and may thus be relevant in human carcinogenesis [14], as targeting the MAPK pathway has been proposed in the treatment of malignant melanoma [29].
In summary, we have shown expression of several established ICC related proteins in the antrum of KitK641E:Neo mice, supporting the conclusion that GIST arise from ICC or an ICC-precursor. Furthermore, we identify Tpbg/5T4 and Spry4, which do not belong to the normal ICC profile but are induced in Kit-ir cells by an oncogenic KitK641E. The expression patterns of Tpbg/5T4 and Spry4 should now be investigated in human material to determine their potential as novel specific markers and/or therapeutic targets for GIST with oncogenic KIT mutations.
Acknowledgments
We are indebted to Perrine Hagué and Huy Nguyen Tran for expert technical assistance and preparation of the figures, respectively.
Grant Support
National Fund for Scientific Research (Belgium), Télévie grant 7.4.558.07.F, jointly to J.M.V. and C.E.
National Fund for Scientific Research (Belgium), Fonds de la Recherche Scientifique Médicale grant 3.4.571.07.F to J.M.V.
National Fund for Scientific Research (Belgium), grant 7.4.529.08.F and Belgian State Policy PAIP (P6/28) to C.E. S.R. is bursary of the Belgian Kids Fund, 2006–2008.
J.M.V. F.L. are Research Director and Research Associate, respectively, of the National Fund for Scientific Research (Belgium).
B.P.R. is funded by a generous grant from The Life Raft Group.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary data 1
List of all differentially expressed genes in the antrum of P14 KitK641E:Neo/K641E:Neo homozygous mice and their WT littermates. Local ID, three independent experimental samples, their mean, t-test P-value, Genbank ID, Unigene ID, official symbol, Oligo ID Sequences, Product of the gene, Unigene description and Function description are indicated for all genes differentially expressed.
Table S1 Gene ontology description of transcripts differentially expressed in antrum P14 KitK641E:Neo/K641E:Neo homozygous mice and their WT littermates Genes differentially expressed were analysed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/).
Details of Cell component, Molecular function and Biological process are indicated for each differentially expressed gene.
Fig. S1 Prkcq/Pkcθ-ir in Kit-ir ICC in the mouse antrum. Prkcq/Pkcθ-ir, (in red) was present in Kit-ir ICC (in green) in the antrum of P14 and adult mice of all genotypes. In the myenteric plexus (indicated by *), Prkcq/Pkcθ-ir was also moderately present. Figures are oriented with the circular muscle layer (CM) up and the longitudinal muscle layer (LM) down. Scale bar: 20 microns
Fig. S2 Pde3a-ir in Kit-ir ICC in the mouse antrum. Pde3a-ir (in red) was present in Kit-ir ICC (in green) in the antrum of P14 and adult mice of all genotypes. Figures are oriented with the circular muscle layer (CM) up and the longitudinal muscle layer (LM) down. Scale bar: 20 microns.
Fig. S3 Gja1/C×43-ir in Kit-ir ICC in the mouse antrum. Gja1/C×43-ir (in red) was present in Kit-ir ICC (in green) in the antrum of P14 and adult mice of all genotypes. Gja1/C×43-ir was also occasionally present between myocytes in the circular muscle layer. Insert: close-up view of Gja1/C×43-ir gap junctions. Figures are oriented with the circular muscle layer (CM) up and the longitudinal muscle layer (LM) down. Scale bar: 20 microns
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1582-4934.2009.00768.x (This link will take you to the article abstract).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Kitamura Y. Gastrointestinal stromal tumors: past, present, and future. J Gastroenterol. 2008;43:499–508. doi: 10.1007/s00535-008-2200-y. [DOI] [PubMed] [Google Scholar]
- 2.Mazzone A, Farrugia G. Evolving concepts in the cellular control of gastrointestinal motility: neurogastroenterology and enteric sciences. Gastroenterol Clin North Am. 2007;36:499–513. doi: 10.1016/j.gtc.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Lux ML, Rubin BP, Biase TL, et al. KIT extracellular and kinase domain mutations in gastrointestinal stromal tumors. Am J Pathol. 2000;156:791–5. doi: 10.1016/S0002-9440(10)64946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isozaki K, Terris B, Belghiti J, et al. Germline-activating mutation in the kinase domain of KIT gene in familial gastrointestinal stromal tumors. Am J Pathol. 2000;157:1581–5. doi: 10.1016/S0002-9440(10)64795-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin BP, Antonescu CR, Scott-Browne JP, et al. A knock-in mouse model of gastrointestinal stromal tumor harboring kit K641E. Cancer Res. 2005;65:6631–9. doi: 10.1158/0008-5472.CAN-05-0891. [DOI] [PubMed] [Google Scholar]
- 6.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Ordog T, Chen J, et al. Differential gene expression in functional classes of interstitial cells of Cajal in murine small intestine. Physiol Genomics. 2007;31:492–509. doi: 10.1152/physiolgenomics.00113.2007. [DOI] [PubMed] [Google Scholar]
- 8.Hellemans J, Mortier G, De Paepe A, et al. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanderwinden JM, Rumessen JJ, De Laet MH, et al. CD34 immunoreactivity and interstitial cells of Cajal in the human and mouse gastrointestinal tract. Cell Tissue Res. 2000;302:145–53. doi: 10.1007/s004410000264. [DOI] [PubMed] [Google Scholar]
- 10.Pozuelo Rubio M, Campbell DG, Morrice NA, Mackintosh C. Phosphodiesterase 3A binds to 14–3-3 proteins in response to PMA-induced phosphorylation of Ser428. Biochem J. 2005;392:163–72. doi: 10.1042/BJ20051103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rumessen JJ, Vanderwinden JM. Interstitial cells in the musculature of the gastrointestinal tract: Cajal and beyond. Int Rev Cytol. 2003;229:115–208. doi: 10.1016/s0074-7696(03)29004-5. [DOI] [PubMed] [Google Scholar]
- 12.Isozaki K, Hirota S. Gain-of-function mutations of receptor tyrosine kinases in gastrointestinal stromal tumors. Curr Genomics. 2006;7:469–75. doi: 10.2174/138920206779315755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Espinosa I, Lee CH, Kim MK, et al. A novel monoclonal antibody against dog1 is a sensitive and specific marker for gastrointestinal stromal Tumors. Am J Surg Pathol. 2008;32:210–8. doi: 10.1097/PAS.0b013e3181238cec. [DOI] [PubMed] [Google Scholar]
- 14.Lo TL, Fong CW, Yusoff P, et al. Sprouty and cancer: the first terms report. Cancer Lett. 2006;242:141–50. doi: 10.1016/j.canlet.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Wang Y. 5T4 oncotrophoblast glycoprotein: janus molecule in life and a novel potential target against tumors. Cell Mol Immunol. 2007;4:99–104. [PubMed] [Google Scholar]
- 16.Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Sommer G, Agosti V, Ehlers I, et al. Gastrointestinal stromal tumors in a mouse model by targeted mutation of the Kit receptor tyrosine kinase. Proc Natl Acad Sci USA. 2003;100:6706–11. doi: 10.1073/pnas.1037763100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakai N, Ishikawa T, Nishitani A, et al. A mouse model of a human multiple GIST family with KIT-Asp820Tyr mutation generated by a knock-in strategy. J Pathol. 2007;214:302–11. doi: 10.1002/path.2296. [DOI] [PubMed] [Google Scholar]
- 19.Rossi F, Ehlers I, Agosti V, et al. Oncogenic Kit signaling and therapeutic intervention in a mouse model of gastrointestinal stromal tumor. Proc Natl Acad Sci USA. 2006;103:12843–8. doi: 10.1073/pnas.0511076103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Southwell BR. Localization of protein kinase C theta immunoreactivity to interstitial cells of Cajal in guinea-pig gastrointestinal tract. Neurogastroenterol Motil. 2003;15:139–47. doi: 10.1046/j.1365-2982.2003.00394.x. [DOI] [PubMed] [Google Scholar]
- 21.Hole N, Stern PL. A 72 kD trophoblast glycoprotein defined by a monoclonal antibody. Br J Cancer. 1988;57:239–46. doi: 10.1038/bjc.1988.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spencer HL, Eastham AM, Merry CL, et al. E-cadherin inhibits cell surface localization of the pro-migratory 5T4 oncofetal antigen in mouse embryonic stem cells. Mol Biol Cell. 2007;18:2838–51. doi: 10.1091/mbc.E06-09-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shingler WH, Chikoti P, Kingsman SM, Harrop R. Identification and functional validation of MHC class I epitopes in the tumor-associated antigen 5T4. Int Immunol. 2008;20:1057–66. doi: 10.1093/intimm/dxn063. [DOI] [PubMed] [Google Scholar]
- 24.Amato RJ, Drury N, Naylor S, et al. Vaccination of prostate cancer patients with modified vaccinia ankara delivering the tumor antigen 5T4 (TroVax): a Phase 2 Trial. J Immunother. 2008;31:577–85. doi: 10.1097/CJI.0b013e31817deafd. [DOI] [PubMed] [Google Scholar]
- 25.Gross I, Bassit B, Benezra M, Licht JD. Mammalian sprouty proteins inhibit cell growth and differentiation by preventing ras activation. J Biol Chem. 2001;276:46460–8. doi: 10.1074/jbc.M108234200. [DOI] [PubMed] [Google Scholar]
- 26.Mason JM, Morrison DJ, Bassit B, et al. Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop. Mol Biol Cell. 2004;15:2176–88. doi: 10.1091/mbc.E03-07-0503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Nielsen TO, West RB, Linn SC, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–7. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- 28.Frolov A, Chahwan S, Ochs M, et al. Response markers and the molecular mechanisms of action of Gleevec in gastrointestinal stromal tumors. Mol Cancer Ther. 2003;2:699–709. [PubMed] [Google Scholar]
- 29.Panka DJ, Atkins MB, Mier JW. Targeting the mitogen-activated protein kinase pathway in the treatment of malignant melanoma. Clin Cancer Res. 2006;12:2371s–5s. doi: 10.1158/1078-0432.CCR-05-2539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


