Background: l-to-d conversion of an amino acid in a neuropeptide can be required for bioactivity.
Results: A new d-amino acid-containing peptide (DAACP), GdFFD, shows stereospecific bioactivity in the feeding circuit.
Conclusion: Our findings broaden the importance of this unusual post-translational modification, providing new methods to accelerate DAACP discovery.
Significance: GdFFD is the first DAACP showing bioactivity in a well defined circuit.
Keywords: Analytical Chemistry, Invertebrates, Mass Spectrometry (MS), Peptides, Post-translational Modification, Aplysia, D-Amino Acid-containing peptides, Epimer, MALDI-TOF/TOF MS, Stereoselective
Abstract
During eukaryotic translation, peptides/proteins are created using l-amino acids. However, a d-amino acid-containing peptide (DAACP) can be produced through post-translational modification via an isomerase enzyme. General approaches to identify novel DAACPs and investigate their function, particularly in specific neural circuits, are lacking. This is primarily due to the difficulty in characterizing this modification and due to the limited information on neural circuits in most species. We describe a multipronged approach to overcome these limitations using the sea slug Aplysia californica. Based on bioinformatics and homology to known DAACPs in the land snail Achatina fulica, we targeted two predicted peptides in Aplysia, GFFD, similar to achatin-I (GdFAD versus GFAD, where dF stands for d-phenylalanine), and YAEFLa, identical to fulyal (YdAEFLa versus YAEFLa), using stereoselective analytical methods, i.e. MALDI MS fragmentation analysis and LC-MS/MS. Although YAEFLa in Aplysia was detected only in an all l-form, we found that both GFFD and GdFFD were present in the Aplysia CNS. In situ hybridization and immunolabeling of GFFD/GdFFD-positive neurons and fibers suggested that GFFD/GdFFD might act as an extrinsic modulator of the feeding circuit. Consistent with this hypothesis, we found that GdFFD induced robust activity in the feeding circuit and elicited egestive motor patterns. In contrast, the peptide consisting of all l-amino acids, GFFD, was not bioactive. Our data indicate that the modification of an l-amino acid-containing neuropeptide to a DAACP is essential for peptide bioactivity in a motor circuit, and thus it provides a functional significance to this modification.
Introduction
Many peptides, particularly neuropeptides, undergo post-translational modification (PTM).4 An unusual PTM that is gaining increasing attention is the generation of d-amino acid-containing peptides (DAACPs) in animals (1). Since the isolation of the first animal peptide having a d-isomerization of an amino acid, the highly analgesic dermorphin from the skin of the South American frog Phyllomedusa (2, 3), more than 30 bioactive DAACPs (including hormones and antimicrobial peptides) have been discovered in frogs (2, 4–8), mollusks (9–24), crustaceans (25, 26), spiders (27, 28), and even mammals (29, 30).
DAACPs in animals, usually having one residue in the d-amino acid form, appear to be derived through a PTM because the d-residue is encoded by a normal codon for the corresponding l-amino acid. Importantly, peptide isomerases that convert an l-amino acid to a d-amino acid have been characterized in spider, frog, and platypus (31–33). DAACPs are particularly interesting because most studies to date have shown that they are functionally important; compared with the all-l-amino acid epimer, the d-amino acid-containing epimer confers distinct and, at times, dramatically enhanced receptor binding affinity (2, 6).
Despite the progress in DAACP analysis, multiple unresolved issues remain. First, although it is hypothesized that more DAACPs might exist in animals, they are rarely investigated, perhaps because of a lack of approaches to characterize this PTM in an untargeted, high throughput manner. Moreover, methods that are compatible with small quantities of tissue samples are desirable as neuropeptide studies oftentimes are sample limited. Second, although DAACPs have been shown to be more bioactive in the CNS in several animals (1, 34), it is not known how d-amino acid-containing neuropeptides might ultimately act on a specific neural circuit to affect network output and behavior. In fact, to the best of our knowledge, no studies have examined the effects of DAACPs on a defined neural circuit, likely due to the limited information on well defined neural circuits in most species. Here, we describe approaches to overcome these two limitations using a well known model animal, the sea slug Aplysia californica (35–37).
Like other PTMs, identification of DAACPs requires the use of biochemical and/or analytical chemical methods (1). However, this PTM is challenging to detect as it does not change the elemental composition, molecular formula, or even the mass of the peptide. Diastereomer separation techniques such as chromatography and capillary electrophoresis have been developed using synthetic peptides and used reliably to confirm the presence of DAACPs in various samples (1). Recently, we showed that the fragmentation pattern generated during MS/MS sequencing can be useful for distinguishing neuropeptide epimers in identified neurons known to express candidate DAACP prohormones (38). Here, we combined MS/MS with off-line multidimensional chromatographic separations to create a screening assay for potential endogenous DAACPs in small volume tissue extracts. To identify candidate DAACPs, we applied bioinformatics as a data mining technique to identify peptides from Aplysia that are similar to known DAACPs; Aplysia is well suited as the model because of the large amount of genomic information available for this animal from the RNAseq transcriptome assembly, the Broad Institute Genomic Database, and others (35, 39, 40).
Based on the results of our data mining, we targeted two putative neuropeptides in Aplysia (35), GFFD and YAEFL-amide (YAEFLa), which are structurally similar to two known DAACPs in the land snail Achatina fulica: achatin-I (GdFAD versus GFAD) (9) and fulyal (YdAEFLa versus YAEFLa), respectively (24). In fact, achatin-I is the first putative neuropeptide to be identified as a DAACP (9). Here, we demonstrate that GFFD in Aplysia has a DAACP epimer, GdFFD, similar to achatin-I in Achatina. In contrast, and to our surprise, we found the YAEFLa in the Aplysia CNS exists predominantly in the all l-form conformation, as the DAACP epimer was not detected in any ganglion in Aplysia. The lack of YdAEFLa and the presence of GdFFD may have implications for the evolution of peptide functions and demonstrates that sequence similarity is not enough to conclude that the PTM present in the peptide from one species is also present in another. We then examined the bioactivity of GFFD/GdFFD in the well described Aplysia feeding circuit (41–48). We found that only GdFFD is bioactive and, moreover, that it functions as an extrinsic modulator of the feeding circuit.
EXPERIMENTAL PROCEDURES
Animals
A. californica (120–170 g) were purchased from the University of Miami/National Institutes of Health National Resource for Aplysia or Marinus Scientific and kept in an aquarium containing aerated and filtered artificial seawater (Instant Ocean, Aquarium Systems Inc.) at ∼14 °C until used. Prior to dissection, animals were anesthetized by injection of isotonic MgCl2 (30–50% of body weight) into the body cavity.
Materials
Water was purified from a Milli-Q system (Millipore); the antibiotics, 2,5-dihydroxybenzoic acid, and acetonitrile (ACN) containing 0.01% TFA, were obtained from Sigma. The peptide standards, YAEFLa, GFFD, and the corresponding epimers with d-amino acids in the second position were synthesized by the Protein Sciences Facility of the Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign.
In Situ Hybridization
In situ hybridization was performed as described previously (42, 49, 50). Ganglia were digested with 1% protease type IX (Sigma) in 10 ml of artificial sea water (ASW: 460 mm NaCl, 10 mm KCl, 55 mm MgCl2, 11 mm CaCl2, and 10 mm HEPES, pH 7.6) for 3 h at room temperature (with rocking) to facilitate removal of the sheath. After digestion, the ganglia were washed with ASW and fixed overnight at 4 °C with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS. The ganglia were then washed, desheathed, and dehydrated in an ascending ethanol series and then rehydrated. Following rehydration, the ganglia were prehybridized and then hybridized overnight at 50 °C in hybridization buffer (50% formamide, 5 mm EDTA, 5 × SSC, 1 × Denhardt's solution, 0.1% Tween 20, and 0.5 mg/ml yeast tRNA) containing 2 μg/ml digoxigenin-labeled cRNA probes made from the corresponding cDNA templates. Following wash-out of the probes, the ganglia were then incubated overnight at 4 °C with a 1:200 dilution of alkaline phosphatase-conjugated anti-digoxigenin antibody (Roche Applied Science) in PBS containing 0.1% Tween, 0.2% BSA, and 1% normal goal serum. After washes with poly(butylene terephthalate) to remove unbound antibody, the ganglia were washed with detection buffer (100 mm NaCl, 50 mm MgCl2, 0.1% Tween 20, 1 mm levamisole, and 100 mm Tris-HCl, pH 9.5) and developed with 4.5 μl of nitro blue tetrazolium and 3.5 μl of 5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science) in 1 ml of detection buffer. The staining reaction was monitored visually and stopped by washing with poly(butylene terephthalate) when the level of staining was adequate. The stained ganglia were photographed using a Nikon microscope (Morrell Instruments) with epi-illumination against a white background. Photographs were taken with a Nikon CoolPix 990 digital camera, imported into Photoshop (Adobe Systems Inc.), and compiled into figures.
Cell Sampling for MALDI-TOF MS
In situ images were used as guidance for the isolation of neurons expressing the GFFD prohormone. The abdominal ganglia were dissected and incubated in 1% (w/v) protease (type IX, Bacterial; Sigma) in artificial sea water (ASW: 460 mm NaCl, 10 mm KCl, 10 mm CaCl2, 22 mm MgCl2, 6 mm MgSO4, and 10 mm HEPES, pH 7.8) supplemented with antibiotics (penicillin G, gentamycin, and streptomycin) and maintained at 34 °C for 45 min to loosen the connective tissue sheath. Following a 1–3-h rinse in ASW with antibiotics to remove the bulk of the protease, the ganglia were stretched onto a silicone elastomer (Sylgard, Dow Corning) layer in a Petri dish containing 3–4 ml of the ASW/antibiotic medium using 0.15 mm diameter tungsten needles (WPI); the connective tissue was surgically removed to expose neurons.
Clusters of GFFD-expressing neurons were identified according to their relative position to prominent morphological landmarks in the ganglion and excised with tungsten needles. The clusters were quickly transferred via a plastic micropipette filled with Milli-Q water to remove excess salts and deposited onto the MALDI target. Excess liquid was aspirated from the target, and spot-to-spot cell transfers were performed for sampling of individual neurons as described previously (38, 51). Matrix (0.3–0.5 μl of 2,5-dihydroxybenzoic acid, 10 mg/ml in 50% ACN, 0.01% TFA) was added to each spot. An equal volume of matrix and 1 mg/ml synthetic peptides dissolved in water were mixed and deposited on the target.
MALDI TOF/TOF MS Analysis of G(l/d)FFD Standards and Individual Aplysia Neurons
This analysis adopted protocols reported previously (38). Briefly, MS measurements were performed using an UltrafleXtremeTM MALDI TOF/TOF mass spectrometer (Bruker Daltonics). MS/MS measurements were performed in “LIFT” mode with the collision-induced dissociation (CID) function turned on. With 27-keV collisional energy, under a source pressure of 6 × 10−6 mbars, a potential rise in the LIFT chamber assists with the pseudo-CID process.
For these measurements, three spectra were generated from each spot on a MALDI plate, with each spectrum being an accumulation of at least 5000 laser pulses surveying all around the spot in a random pattern. Peak intensity and area were used for analysis. Laser intensity was held constant across all synthetic peptide spots (six for each epimer), including between the pair of all l- and d-amino acid-containing peptides. For cell samples, laser intensity was adjusted to observe peaks with a signal-to-noise ratio higher than 100 but not to saturate the detector, and once optimized, this parameter was kept consistent for the cell samples. Three individual neurons from two animals were evaluated for GdFFD and GFFD content using the MALDI MS/MS method. All samples were intentionally analyzed in a random order between the two isomeric forms by MS.
The observed fragment ion peaks were mass matched to predicted fragment ions using Protein Prospector. The MS/MS spectra were analyzed using the FlexAnalysis 3.4 and Biotools 3.2 software packages (Bruker Daltonics). Seven major fragment peaks with a signal-to-noise ratio ≥ = 10 were identified from the peak table for further analysis. The peak area of each fragment ion was normalized to the sum of all major fragment ions produced from the particular sample spot. Mass assignment accuracy was expressed as parts per million (ppm).
Principal Component Analysis (PCA) of Fragmentation Spectra
We used the sum of the peak areas of all major product ions from the MALDI MS/MS analysis because it includes all of the detected fragment ions instead of only two chirality- indicating fragments and may provide a greater discrimination of different fragmentation profiles between peptide epimers. JMP Pro 10 software (SAS Institute Inc.) was used for the multivariate analysis. Peak area data from synthetic peptides and cells were first extracted from FlexAnalysis 3.4 into an Excel spreadsheet (Microsoft Corp.). To improve data consistency, all peak area values were pretreated by normalizing to the sum of all major product ions (b2, b3, y2, y3, a2, Phe-immonium, and MH+-H2O ions) generated from the respective replicate. All normalized peak area values from these product ions were used for the analysis. The default estimation method, restricted maximum likelihood, was used. Summary plots, including an Eigenvector plot and an Eigenvalues table, were generated from the JMP Pro software.
Preparation of Neuropeptide Extracts
Peptide extracts for LC-MS/MS analysis have been prepared from either isolated neurons or from a whole ganglion, as specified under “Results.” In the case of isolated neurons, 20 μl of acidified acetone (40:6:1 acetone/H2O/HCl, v/v/v) was added to the isolated neurons in a centrifuge tube. After brief sonication, the extract was then dried in a vacuum evaporator (SpeedVacTM, Savant Instruments) and reconstituted in 20 μl of 5% ACN in water (95% H2O, 5% CH3CN, 0.01% TFA (v/v/v)). The sample was stored at 4 °C before analysis. When extracting from the whole cerebral and buccal ganglia, 500 μl of acidified acetone was used. The sample was then sonicated, centrifuged (Centrifuge 5804R, Eppendorf), and the supernatant collected. The tissue pellet was resuspended in an additional 500 μl of acidified acetone, vortexed, and stored at 4 °C overnight and centrifuged; the secondary supernatant was collected and combined with the primary supernatant. The combined extract was then dried in the SpeedVac and reconstituted in 20 μl of 5% ACN in water.
First Stage Microbore HPLC Fractionation of Peptide Extracts
Fractionations of abdominal, cerebral, and pedal ganglia extracts were performed with a microbore HPLC system (Magic 2002, Michrom Bioresources) with a reversed-phase C18 column (150 × 1.0 mm inner diameter, 5-μm particle size, 300-Å pores, Dionex) at a flow rate of 20 μl/min prior to LC-MS analysis. Fractionation of the buccal ganglia extracts was performed with an analytical HPLC system (BreezeTM 2 HPLC, Waters) with a reversed-phase Symmetry C18 column (75 × 4.6 mm ID, 3.5-μm particle size, 100-Å pores, Waters).
A UV-visible detector set at 220 nm was used for detection. A binary solvent system was used with solvents A (95% H2O, 5% ACN, 0.1% formic acid (FA), 0.01% TFA (v/v/v/v)) and B (95% ACN, 5% H2O, 0.1% FA, 0.01% TFA (v/v/v/v)). The solvent gradient started by increasing B from 1 to 5% over 5 min, up to 50% over the next 45 min, and followed by a 20-min gradient from 50 to 80% B. Fractions were manually collected using a fraction collector (FC 203B, Gilson).
LC-MS Analysis of Synthetic G(d/l)FFD and Y(d/l)AEFLa Peptides and Tissue Extracts
Synthetic peptide epimers, GFFD and GdFFD or YAEFLa and YdAEFLa, were prepared in three solutions, 1 μg/ml l-epimer, 1 μg/ml d-epimer, and a mixture of both. Two instrumental combinations of an HPLC system and electrospray ionization (ESI)-ion trap (IT) mass spectrometer were used for the separations, with both validated using appropriate standards.
For the initial experiments on all samples except the buccal ganglia extracts, the HPLC fraction of interest was analyzed using a capillary HPLC (capLC)-ESI-IT MS setup. The capillary HPLC system (CapLC, Waters) was equipped with a Dionex C18 PepMap column (150 × 0.3 mm inner diameter, 3-μm particle size, 100-Å pore size) running at a flow rate of 2.5 μl/min. Gradient elution was adopted (A: 0.05% TFA (v/v); B: 80% ACN, 20% H2O, and 0.04% TFA). The gradient started with 5% B, rising to 50% B in 20 min and then to 95% B in another 5 min. The gradient was dropped down from 95% B at 30 min to 5% at 31 min toward the end of a 50-min run. The eluent was interfaced on line with an ESI-IT mass spectrometer (HCTultra PTM Discovery SystemTM, Bruker Daltonics). We acquired data in the positive-ion mode with detection ranges of m/z 300–1500 and 50–2000 for MS and MS/MS, respectively. The two most intense precursor ions (preferred charge of +2) were automatically picked every minute for CID fragmentation.
To optimize the separation of YAEFLa and YdAEFLa, a hypercarb column (100 × 0.32 mm inner diameter, 5-μm particle size, Thermo Scientific), using the same gradient and solvent system as described above for the capLC on a C18 column, was interfaced to the HCTultra mass spectrometer.
The later confirmation of the GFFD/GdFFD peptides in the buccal ganglia used a newer instrument combination. For buccal ganglia extracts, each analytical HPLC fraction of interest was analyzed using the same capillary HPLC system and column as described above, although with some modifications. A flow rate of 4.0 μl/min was employed with gradient elution (A: 95% H2O, 5% ACN, 0.05% FA; B: 80% ACN, 20% H2O, 0.04% FA). The gradient started with 1% B, rising to 20% B in 20 min, then 45% B in 7 more min, and then to 90% B in another 17 min. The gradient was dropped from 90% B to 1% B toward the end of the 60-min run. This eluent was interfaced on line with a different ESI-IT mass spectrometer (amaZon speedTM, Bruker Daltonics). In addition, a viscosity calibration was performed prior to this separation; overall, these changes led to a different retention time for the peptides. However, to enable a comparison of results between the earlier and later separations, careful identification and calibrations using appropriate peptide standards were performed in all cases.
Antibodies and Immunostaining
GdFAD was used to generate the antibodies in rats as described previously (42, 52). Briefly, the antigen was prepared by coupling synthetic peptide (SynPep) to BSA (catalog no. A0281; Sigma) using 1-ethyl-3-(dimethylaminopropyl)carbodiimide (catalog no. E7750; Sigma). The coupled antigen was purified and used to inoculate the rats. Immunocytochemistry was performed as described previously (42, 52). Tissues were fixed in freshly prepared fixative (4% paraformaldehyde, 0.2% picric acid, 25% sucrose, and 0.1 m NaH2PO4, pH 7.6). Tissue was permeabilized and blocked by overnight incubation in blocking buffer (10% normal donkey serum, 2% Triton X-100, 1% BSA, 154 mm NaCl, 50 mm EDTA, 0.01% thimerosal, and 10 mm Na2HPO4, pH 7.4). The primary antibody was diluted 1:250 in the blocking buffer and incubated with the tissue for 4–7 days. The tissue was then washed twice per day for 2–3 days with washing buffer (2% Triton X-100, 1% BSA, 154 mm NaCl, 50 mm EDTA, 0.01% thimerosal, and 10 mm Na2HPO4, pH 7.4). After the washes, the tissue was incubated with a 1:500 dilution of secondary antibody (lissamine rhodamine donkey anti-rat; Jackson ImmunoResearch) for 2–3 days. The tissue was then washed twice with the washing buffer for 1 day and four times with storage buffer (1% BSA, 154 mm NaCl, 50 mm EDTA, 0.01% thimerosal, and 10 mm Na2HPO4, pH 7.4) for 1 day.
Electrophysiology
Intracellular and extracellular recordings of the physiological activity from CNS preparations (including the cerebral and buccal ganglia) were performed as described previously (53). The ganglia were desheathed, transferred to a recording chamber containing ∼1.5 ml of ASW, continuously perfused at 0.3 ml/min, and maintained at 14–17 °C. Peptides were dissolved in ASW immediately before each physiological test, and the peptide/ASW solution was perfused into the recording chamber. Intracellular recordings were obtained using 5–10 megohms sharp microelectrodes filled with 0.6 m K2SO4 plus 60 mm KCl.
Electrophysiological recordings were digitized on line using Axoscope (Molecular Devices, Inc.) and plotted with CorelDraw (Corel Inc.). Bar graphs were plotted using Axum (Adept Scientific). Data are expressed as mean ± S.E. Statistical tests (repeated measures one-way analysis of variance) were performed using Prism (GraphPad Software). When the data showed significant effects in analysis of variance, further individual comparisons were performed with Bonferroni's correction.
RESULTS
Identifying Putative DAACP-encoding Genes in A. californica
Orthologous genes oftentimes encode neuropeptide prohormones with similar functions in different species (54). Commonly, only amino acid sequences and associated PTMs required for function are conserved, while the rest of the prohormone differs among the species. For this study, we looked at existing research on A. fulica because three DAACPs, achatin-I (GdFAD) (9), fulicin (FdNEFVa) (10), and fulyal (YdAEFLa) (24), and their precursors, have been identified in Achatina. Achatin-I belongs to one precursor, named achatin-I precursor (55), whereas both fulicin and fulyal belong to another precursor, named fulcin gene-related peptide precursor (FGRP) (56). Because A. fulica and A. californica are both gastropod mollusks, there is a distinct possibility that DAACPs in Achatina may have homologs in Aplysia.
Indeed, previous large scale sequencing projects in Aplysia (35) have already identified two precursors that are similar to the above two precursors, named Aplysia achatin-like neuropeptide precursor (abbreviated as apALNP in this report, accession NP_001191589) and Aplysia fulicin-like neuropeptide precursor (abbreviated as apFLNP, NP_001191590) (Fig. 1). Despite this information, until this work no prior study predicted possible peptides from these two precursors, let alone characterized the actual peptides produced in Aplysia.
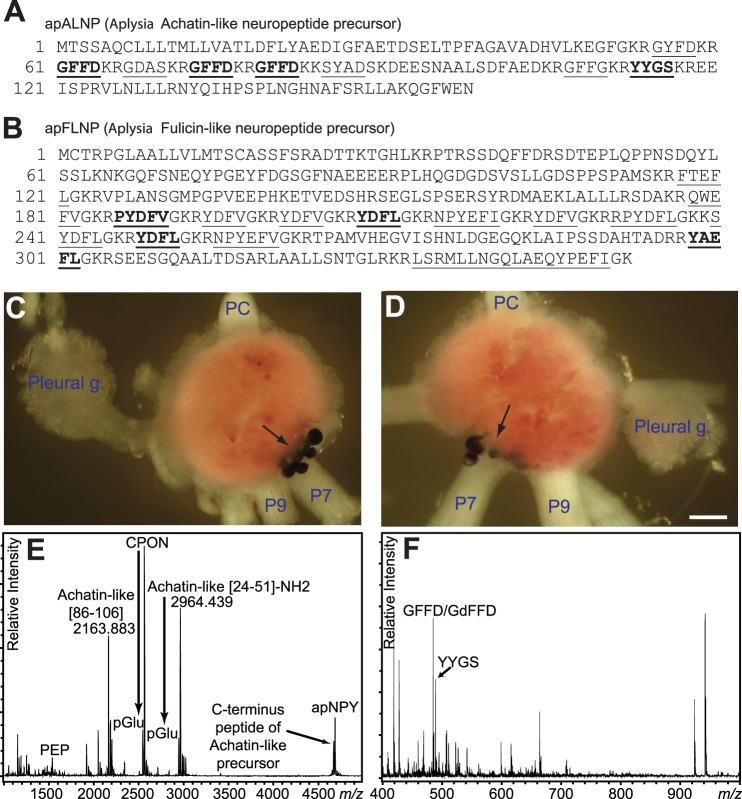
FIGURE 1.
Sequence and localization of expression of apALNP and apFLNP, and peptide profiles from apALNP-positive cells. A and B, precursor protein sequences of apALNP and apFLNP (35). Predicted peptides are underlined, and peptides that have been verified chemically in this report are shown in boldface. A, apALNP includes the predicted peptide GFFD; B, apFLNP includes the predicted peptide YAEFLa. All peptides from apFLNP are amidated. C and D, in situ hybridization showed that apALNP (GFFD) mRNA was expressed in a cluster of neurons near the root of pedal nerve 7 (P7) and pedal nerve 9 (P9) on the dorsal surface of the pedal ganglion. C, left pleural-pedal ganglia; D, right pleural-pedal ganglia. In both panels, the pedal commissure (PC) is facing upward, and the pleural ganglion (Pleural g.) is facing to either the left or right. Calibration, 200 μm. E and F, peptide profile detected by MALDI-TOF MS from the cells stained positive for cDNA corresponding to apALNP (GFFD/GdFFD) in in situ hybridization. E, several of the larger molecular weight peptide products from apALNP are detected from these cells by accurate mass match. Achatin-like (86–106)- and achatin-like (24–51)-NH2 were detected at 44 and 12 ppm accuracy, respectively. F, detection of peptide products from apALNP using MALDI-TOF MS settings optimized for detection of smaller molecular weight species. Several products, including GFFD/GdFFD and YYGS, are detected from the cells by accurate mass matching. GFFD/GdFFD and YYGS were detected at 206 and 818 ppm accuracy, respectively.
We began our analysis by first comparing apALNP against the achatin-I precursor as well as several related, nonredundant protein sequences from GenBankTM, PDB, SwissProt, PIR, and PRF using blastp. We found that apALNP has overall similarity to achatin-I precursor from A. fulica (50%), neuropeptide GFAD precursor from Helix lucorum (44%), and achatin from Lehmannia valentiana (42%). Next, we created a library of putative encoded peptides using the bioinformatics tool NeuroPred (57, 58), which predicts cleavage sites based on models trained on Aplysia prohormone cleavages and the Known Motif model. Sequence alignment showed seven repeats of GFAD in achatin-I precursor, and at those loci there were several similar sequences to achatin-like neuropeptide, including GYFD (predicted m/z 501.2), GFFD (m/z 485.2, with three copies), GDAS (m/z 349.1), SYAD (m/z 455.2), and GFFG (m/z 427.2) (Fig. 1A). In addition, YYGS was also predicted. Among these predicted peptides from apALNP, GFFD was the most abundant, and more importantly, it was most similar to GFAD. Thus, we hypothesized that GFFD in Aplysia might have a DAACP epimer, GdFFD, similar to achatin-I (GdFAD) in Achatina.
We then performed a similar analysis on apFLNP with FGRP. A sequence alignment using blastp indicated that apFLNP has 37% similarity to FGRP. Predicted neuropeptides included LSRMLLNGQLAEQYPEFIa (m/z 2121.1), RPYDFLa (m/z 809.4), NPYEFIa (m/z 781.4), NPYEFVa (m/z 767.4), QWEFVa (m/z 707.3), FTEFLa (m/z 655.3), SYDFLa (m/z 643.3), YAEFLa (m/z 641.3), PYDFVa (m/z 639.3), YDFLa (m/z 556.3, 2 copies), and YDFVa (m/z 542.2, 3 copies) (Fig. 1B). In addition, there were two other peptides, RPYDFLa and NPYEFVa. Among these predicted peptides from apFLNP, FTEFLa is the analog of the all l-form of FNEFVa. In our previous MALDI MS peptide mass profiling experiments of this prohormone, we have not detected the peptide FTEFLa (data not shown). Thus, we decided to focus on YAEFLa, and hypothesized that Aplysia YAEFLa might have a DAACP epimer, YdAEFLa, identical to the Achatina fulyal. We tested these two hypotheses concerning DAACPs in Aplysia, as described below.
Expression of Putative DAACP-encoding Prohormones in the Aplysia CNS
To determine whether apALNP (GFFD) and apFLNP (YAEFLa) are expressed in the Aplysia CNS, we performed in situ hybridization experiments using the antisense apALNP (n = 3) or apFLNP (n = 3) cRNA (generated from the whole sequences shown in Fig. 1, A and B) made with T7 or SP6 RNA polymerase. Control experiments included only vectors, and the antisense apALNP or apFLNP cRNA sequences were omitted. These controls resulted in no staining (n = 3).
For apALNP, we found that the most obvious staining of neuronal somata was in the pedal ganglion, i.e. a cluster of large neurons in the vicinity of the peripheral pedal nerve 7. These neurons were located primarily on the dorsal surface but were also partially visible on the ventral surface (Fig. 1, C and D) (n = 3). No somata were stained in the cerebral and buccal ganglia, which are involved in the generation of feeding behavior.
For apFLNP, we observed somata staining on the ventral surfaces of the cerebral ganglion, i.e. a single neuron near the root of the cerebral-buccal connective and a cluster of small neurons between the roots of the cerebral-pedal connective and cerebral-pleural connective (Fig. 2A) (n = 3). We also observed some staining in the abdominal ganglion (data not shown), but we found no staining of neuronal somata in the buccal or the pleural-pedal ganglia. In addition to showing that both apALNP and apFLNP are expressed in the Aplysia CNS, the above data also provided us with potential targets for isolating specific neurons for chemical identification of putative DAACPs.
FIGURE 2.
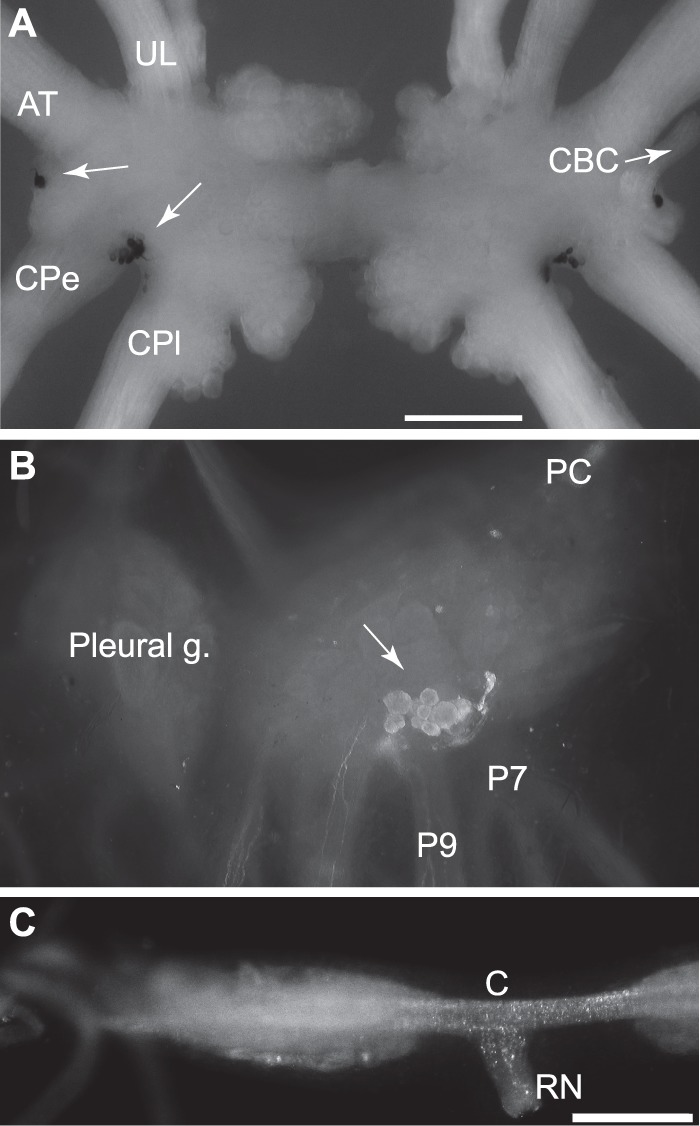
Localization of expression of apALNP and apFLNP. A, in situ hybridization shows that apFLNP (YAEFLa) mRNA is expressed on the ventral surfaces of the cerebral ganglion. A single neuron is located in the E cluster near the root of the cerebral-buccal connective (CBC and white arrow, only present at the right. The left CBC was accidentally cut.), and a cluster of small neurons is located between the roots of the cerebral-pedal connective (CPe) and cerebral-pleural connective (CPl). White arrows point to the labeled cells at the left side. AT, anterior tentacular nerve; UL, upper labial nerve. Calibration, 500 μm. B and C, immunocytochemistry with GdFAD antibody. B, dorsal surface of the pedal ganglion. The cluster of positive neurons matched in situ hybridization with apALNP, suggesting that GdFAD antibody can label apALNP-positive neurons. C, buccal ganglion. Consistent with in situ hybridization, there are no GdFAD-positive somata in the buccal ganglion, but there are GdFAD-positive fibers and varicosities, e.g. in the neuropile, commissure (C), and the radula nerve (RN). Presumably, the staining suggests that apALNP-positive fibers and varicosities are in the buccal ganglion. Calibration, 500 μm.
Chemical Characterization of Putative DAACPs (GdFFD and YdAEFLa)
In this series of experiments, we first sought to characterize GFFD and putative GdFFD from the Aplysia CNS using several small volume neuropeptide characterization approaches involving MS (59, 60). Based on the distribution of apALNP-positive neurons shown by in situ hybridization, we isolated these neurons from the pedal ganglion (Fig. 1, C and D). Using MALDI-TOF MS peptide mass profiling, we detected predicted peptides and their truncated forms from apALNP by mass match (Fig. 1E), including m/z 484.9, matching that of GFFD, as well as YYGS (Fig. 1F); GFFD was detected with 206 ppm accuracy under conditions optimized for detection of lower molecular weight peptides. Based on previous reports identifying the peptide epimerization process as a PTM (31–33), GdFFD would be generated by isomerization of GFFD after synthesis and cleavage from the prohormone. Therefore, both GFFD and GdFFD could be present in the extracts, but only as a single peak in the MALDI-TOF MS spectrum.
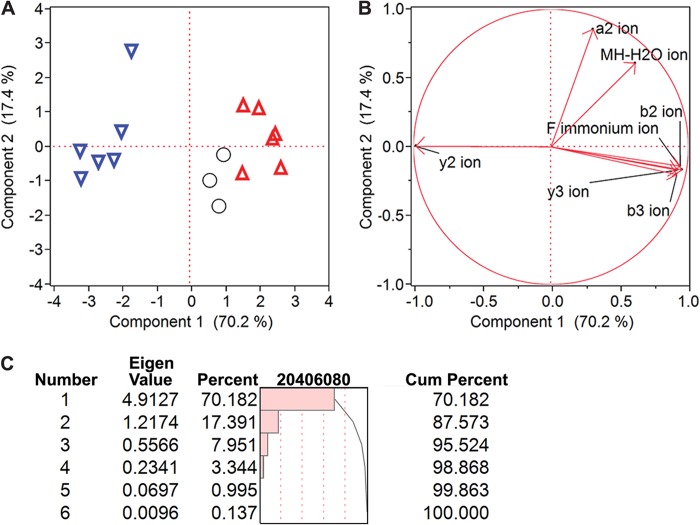
We and others have demonstrated previously that fragmentation of d- and l-epimers by CID on a MALDI-TOF/TOF instrument results in different ratios of specific fragment ions, referred to as reporters, which aid epimer differentiation during MS/MS analysis (38, 61, 62). Thus, we used this method to screen whether GdFFD was possibly present in the samples. To apply this approach for analysis of GFFD peptide chirality, we synthesized GFFD and GdFFD, investigated their characteristic fragmentation patterns, and found that y2 and b3 fragment ions served as reporters for discriminating between these two epimers (n = 6, Fig. 3). The ratios of y2/b3 peak intensities, as well as peak areas, were lower for GdFFD (Fig. 3A). Likewise, the Phe-immonium (Fi) ion was observed at a higher intensity in the case of GdFFD, and the y2/Fi ratio was lower for GdFFD. In addition, MS/MS analysis of isolated neurons (n = 3) revealed a fragmentation pattern that did not resemble that of the GFFD standard, suggesting that the endogenous peptide could be GdFFD (Fig. 3, A and B). To demonstrate the difference between epimers using the entire fragmentation profile, we applied multivariate analysis using all major product ions from the fragmentation spectra. PCA revealed a clustering of the spectra from isolated cells to the spectra from the GdFFD standard, distinct from the spectra from the GFFD standard, based primarily on principal component (PC) 1 (Fig. 4A). Seven fragment ions were used in the PCA analysis (Fig. 4B), including the ions b3, Fi, and y2. Component 1 accounts for over 70% of the variance, and two groups are formed by this component. The isolated neurons are grouped more closely with the synthetic GdFFD peptide, meaning the fragmentation profile more closely fits GdFFD than GFFD (Fig. 4C). The Eigenvector plot in Fig. 4B shows the ions that contributed to the discrimination. The y2 ion with an arrow pointing toward the left in Fig. 4B indicates the more abundant y2 ion is observed in the synthetic GFFD peptide, and this has contributed to the assignment of these samples to the group on the left of the PCA plot (Fig. 4A). The more abundant Fi, y3, b3, and b2 ions contribute to the assignment of a sample to the right-hand side of the plot. The largest contrast of fragment ion ratios came from Fi, y3, b3, and b2 ions to the y2 ion, and this is consistent with our initial choice of y2/b3 and y2/Fi being our chirality reporting ratios. PCA revealed a distinct grouping of the GFFD standard from the GdFFD standard and isolated neurons, suggesting a resemblance of features in the fragmentation spectra of isolated neurons with the GdFFD standard. Given these results, we decided to confirm the presence of GdFFD in these neurons.
FIGURE 3.
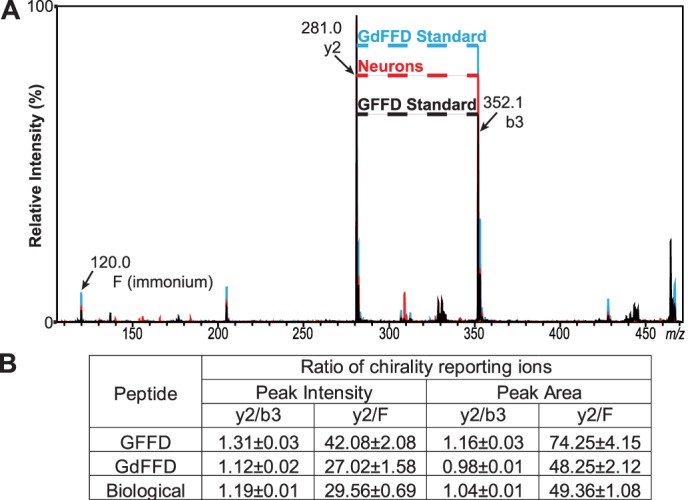
GFFD and GdFFD can be distinguished by their different fragmentation patterns using MALDI-TOF/TOF MS. A, overlaid representative MS/MS spectra from GFFD standard (black), GdFFD standard (blue), and isolated pedal neurons (red), normalized to the y2 ion as % intensity. B, each epimer, i.e. GFFD or GdFFD, produces a unique ratio using two of the most discriminative fragment ions, y2 and b3. The ratio measured from the isolated neurons appears to be closer to the value for GdFFD, suggesting that the neurons contain both GFFD and GdFFD.
FIGURE 4.
Statistical analysis demonstrates that peptides from isolated neurons are more similar to GdFFD, such that GdFFD is likely the major contributor to m/z 484. 9 in MALDI-TOF MS. A, PCA demonstrates the clustering of GdFFD standard (inverted triangle), GFFD standard (upright triangle), and biological samples (circles) based substantially on two principal components (PCs), with the biological samples clustering more closely with the GdFFD standard based primarily on PC1. Each symbol corresponds to the fragmentation profile of one sample. The average peak area from three acquisitions of each sample, normalized to the sum of the peak areas of all ions in the respective MS/MS spectrum, is used for PCA analysis. B, seven fragment ions are the variables used in the PCA analysis. Their impact regarding to the two principal components are shown in the figure. There are two distinctive groups regarding PC1. Fragment ions such as b3 and the Fi ion are in one group showing similar impact in group forming, and fragment ion y2 is another. The differences in these two groups of variables have contributed to assigning the two clusters. C, table of Eigenvalues shows that only PC1 and PC2 have an Eigenvalue higher than 1, and these two are the components that explain the majority of the variations. Cum, cumulative.
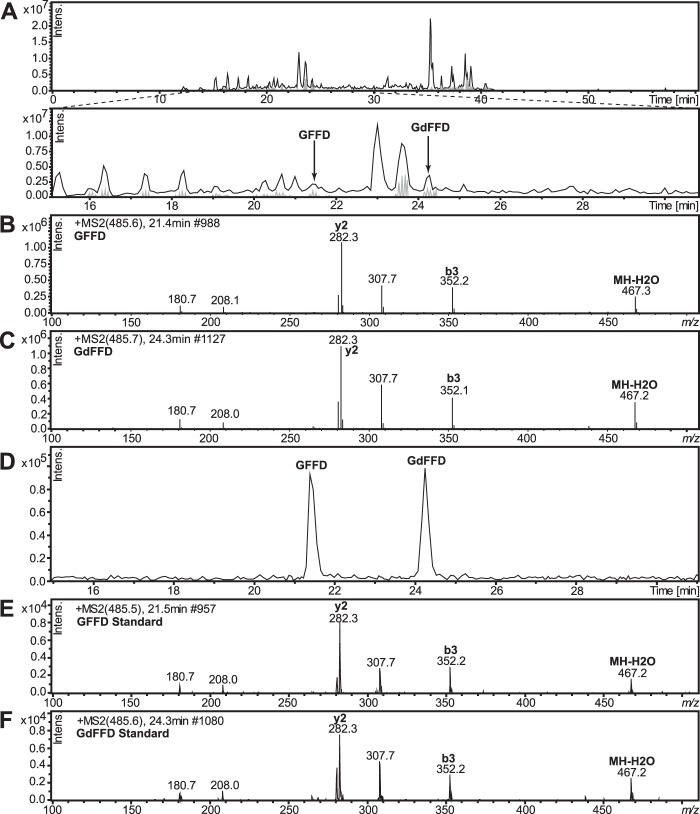
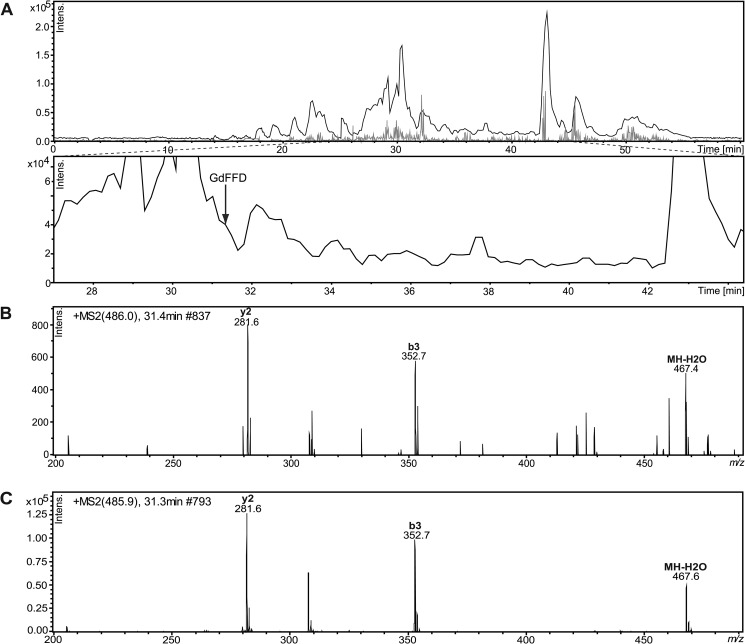
Because peptide epimers can be separated on C18 reversed-phase columns without a chiral selector (1), we also used a second method, LC-MS/MS, to confirm whether GFFD and GdFFD were both present in the samples. We analyzed extracts from whole pedal ganglia, instead of individual neurons, using ESI-IT MS preceded by chromatographic separation, to confirm the chirality of GFFD peptide. We used two separate instrument/separation conditions (see “Experimental Procedures”). One LC-MS/MS method achieved base-line separation of the synthetic GFFD and GdFFD mixture, with retention times of 21.4 and 24.3 min, respectively (Fig. 5, D–F), and the other had retention times of 30.3 and 31.4 min, respectively (Fig. 6). The LC-MS chromatogram in Fig. 5A identifies both GFFD and GdFFD in the neurons isolated from the pedal ganglia extracts. The retention times and fragmentation ions obtained from the samples match those from synthetic epimers. The mechanisms of molecular dissociation are distinct in our two MS systems and so the MS/MS profiles are different (compare the CID and the TOF/TOF fragmentation patterns shown in Fig. 5 with Fig. 3). Regardless, the intensities of the fragment ions m/z 282.3 (y2), m/z 307.7 (x2), and m/z 352.2 (b3) show characteristic patterns for each epimer. In addition, GdFFD was detected in cerebral ganglia extracts (data not shown) and whole buccal ganglia extracts, seen in the LC-MS chromatogram in Fig. 6A. Overall, the results from two independent methods, i.e. MALDI MS fragmentation analysis and LC-MS/MS using three instrumental platforms, verify that both GFFD and GdFFD are present in the Aplysia CNS.
FIGURE 5.
GFFD and GdFFD are detected from neurons isolated from the pedal ganglia from A. californica using LC-MS. A, LC separation of GFFD and GdFFD. B, MS/MS fragmentation spectrum of endogenous GFFD (21.4 min). C, MS/MS fragmentation spectrum of endogenous GdFFD (24.3 min). D, retention times on the LC and fragmentation patterns (E and F) of both peaks match well with those of synthetic peptides, which confirm the identification. E, MS/MS fragmentation spectrum of synthetic GFFD (21.4 min). F, MS/MS fragmentation spectrum of synthetic GdFFD (24.3 min).
FIGURE 6.
GdFFD is detected in whole buccal ganglia extracts from A. californica using LC-MS. A, LC separation of buccal ganglia peptides. Arrow points to a peak at the retention time matching that of the GdFFD standard. B, MS/MS of GdFFD (31.4 min). The fragmentation patterns and retention time of GdFFD matches the synthetic peptide in C, which confirms the identification.
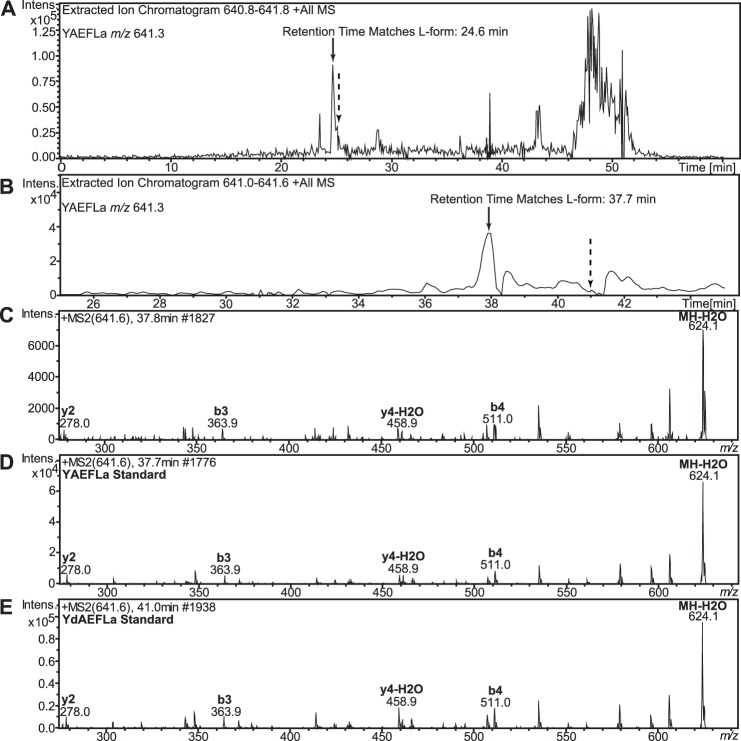
Next, we sought to characterize YAEFLa and putative YdAEFLa from the Aplysia CNS, because apFLNP in Aplysia predicts a peptide identical to the Achatina DAACP, fulyal (YdAEFLa), among others. The sparse expression pattern of apFLNP in the Aplysia CNS, as shown by in situ hybridization, rendered sampling for MALDI difficult. Therefore, we did not perform MALDI MS fragmentation analysis. Instead we decided to investigate the presence and chirality of the YAEFLa peptide using our LC-MS-based platform without prior MALDI-TOF/TOF MS structure verification. Synthetic YAEFLa and YdAEFLa were used to optimize the separation and MS conditions. First we used a Dionex C18 PepMap column, but the epimers had only a slight difference (0.8 min) in retention times (Fig. 7A), and so we tried a stationary phase with a different selectivity, the Thermo Scientific hypercarb column. The difference in retention time increased to 3.4 min, which improved our confidence in the chirality assignment via retention time. Two-stage HPLC separation was performed on the cerebral ganglia extract to identify Y(l/d)AEFLa (Fig. 7B). To comprehensively assay the fractions in an efficient manner, 20 fractions from the first stage were scrambled and combined for second-stage analysis (e.g. fractions 1, 7, and 13 were combined; fractions 2, 8, and 14 were combined). Only the first stage scrambled fraction contained a mass matching that of YAEFLa, based on the retention times of the YAEFLa and YdAEFLa standards. An MS/MS fragmentation profile (Fig. 7C) that closely resembled the synthetic YAEFLa provided additional support for this identification (Fig. 7, D and E).
FIGURE 7.
YAEFLa, but not YdAEFLa, is detected using LC-MS to analyze A. californica cerebral ganglia peptide extracts. A, solid arrow points to the retention time for YAEFLa, and dotted arrow points to the retention time for YdAEFLa. Although the separation in A used a C18 column, the retention times were too close for confident assignment, and a hypercarb column was used as the stationary phase for the separations in B, where the solid arrow points to a peak matching the retention time of the YAEFLa standard (37.7 min) and the dotted arrow points to a retention time expected for YdAEFLa (41.0 min), although no m/z 641.3 is observed at the mass spectrum at this retention time. C, MS/MS fragmentation spectrum of YAEFLa. D, MS/MS fragmentation spectrum of synthetic YAEFLa (37.7 min). E, MS/MS fragmentation spectrum of synthetic YdAEFLa (41.0 min). The MS/MS fragmentation pattern of the endogenous YAEFLa matches the expected fragmentation pattern of Y(l/d)AEFLa at a retention time matching the YAEFLa standard, confirming that this peptide is predominantly in the all l-form.
In addition, two other peptides from the same apFLNP prohormone, PYDFVa and YDFLa, were detected and sequenced, further lending support to the correct identification of m/z 641.3 as YAEFLa (data not shown). No signal satisfying our criteria for peak selection could be identified as YdAEFLa at the corresponding retention time window. Thus, we conclude that the l-form is the predominant isomeric form of YAEFLa in the cerebral ganglia of Aplysia. We also scanned HPLC fractions from the pedal and abdominal ganglia using the same LC-MS methods for YdAEFLa, but YdAEFLa was not detected. To summarize at this point, our data indicate that although YAEFLa is predominantly or completely in the all l-form in the Aplysia CNS, both GFFD and GdFFD are present in significant amounts, providing a basis for examining the actions of DAACPs in the Aplysia feeding circuit.
GdFFD, an Extrinsic Neuromodulator of the Feeding Circuit
In these experiments, we sought to determine whether GFFD, GdFFD, or both, are bioactive in the Aplysia feeding circuit. In situ hybridization with apALNP suggested that GFFD/GdFFD-containing neurons were primarily distributed in the pedal ganglion (Fig. 1, C and D). We performed immunocytochemistry on the pedal ganglion and the buccal ganglion using an antibody raised against GdFAD (Fig. 2, B and C). Immunostaining of pedal ganglia (Fig. 2B) showed that the antibody labeled a group of neurons that is similar to the cluster of neurons labeled with in situ hybridization by apALNP (Fig. 1, C and D), suggesting that the antibody is immunoreactive to peptides from apALNP. The antibody provided information about neurons expressing GdFAD, but it may also have labeled cells with GFFD (as the chiral selectivity of the antibody was not tested). The labeled cells matched the same cells in the apALNP in situ hybridization experiments. Immunostaining of the buccal ganglion (Fig. 2C) did not label any neuronal somata, consistent with our in situ hybridization experiments that revealed no positive staining of somata in the buccal ganglion. However, the immunostaining did show that there were immuno-positive fibers and varicosities in the buccal commissure and the neuropile of the buccal ganglion in which the central pattern generator for feeding is located. Indeed, we were able to detect the presence of GdFFD in peptide extracts from the whole buccal ganglion (Fig. 6). These data suggest that peptides (including GFFD/GdFFD) that originate from apALNP-positive somata of other ganglia, e.g. the pedal ganglion, innervate the buccal ganglion and therefore may act as extrinsic modulators of the feeding circuit.
To test this hypothesis, we examined the potential actions of GFFD and GdFFD on activity in the feeding network. First, we investigated the possibility that these peptides can directly activate the feeding circuit. After a 5–10-min resting period, when there was typically no spontaneous expression of motor programs (Fig. 8A), we perfused ASW that contained either GFFD or GdFFD to the bath for 8–15 min. We found that GFFD did not produce observable effects at 10−5 m. However, GdFFD was bioactive (Fig. 8B). In two experiments in which the rostral surface of the buccal ganglion was desheathed, we found inconsistent results with GdFFD application. Therefore, in the remaining preparations (n = 4), we only desheathed the caudal surface of the buccal ganglion.
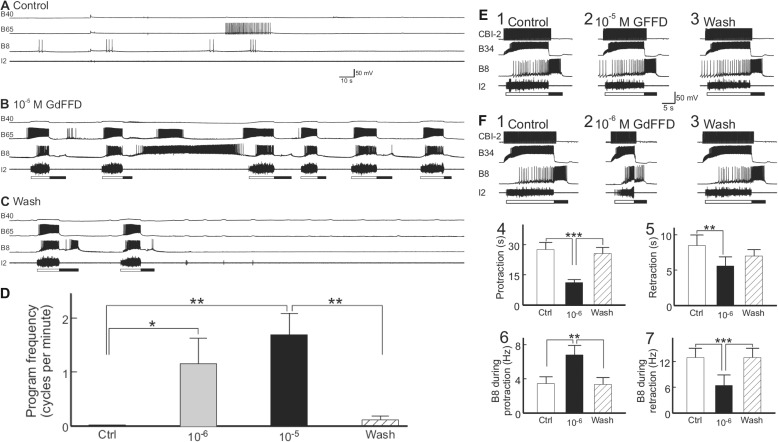
FIGURE 8.
GdFFD induces cyclic activity in the buccal central pattern generator. Each cycle of programs consists of a protraction (open bar, marked by I2 nerve activity) − retraction (closed bar, marked by hyperpolarization of B65 interneuron) sequence. A, control: at rest, there was little activity in the buccal CPG protraction interneurons B40 and B65, radula closure motoneuron B8, or the I2 nerve. B, during the perfusion of 10−5 m GdFFD, the feeding network became cyclically active. The motor programs induced were mostly egestive because B8 activity occurred during the protraction phase (represented by B65 bursting). Consistently, B40, a neuron important for ingestive programs, was not active, although it received cyclic synaptic inputs. C, after washout of the peptide, the preparation gradually returned to the inactive state, beginning with the trace, 4.2 min after the start of the washout. D, summary data (n = 4) showing the effects of 10−6 and 10−5 m GdFFD on the activity of the buccal CPG. The frequency of the motor programs is the number of cycles per min measured over a period of 7 min or more in each experimental condition. Both 10−6 and 10−5 m GdFFD increased the frequency of spontaneously occurring motor programs. Bonferroni post hoc test (with selected comparisons between peptide applications versus the control (Ctrl) or the wash) is as follows: *, p < 0.05; **, p < 0.01. Error bars represent S.E. E and F, effects of GFFD and GdFFD on motor programs elicited by the command-like interneuron CBI-2. B34 is a buccal protraction interneuron active during protraction (open bar, marked by I2 nerve activity); CBI-2 is an interneuron that triggers programs by activating protraction interneurons in the buccal ganglion, which, in turn, activate the retraction neurons. In addition, CBI-2 receives inhibitory feedback during retraction (65), and it is not or only weakly active during retraction. Thus, single cycle motor programs were elicited by stimulation of CBI-2 at 9 Hz throughout the protraction phase with inter-trial intervals of 1 min. E1–3, representative examples. E1, control: CBI-2 stimulation induced an ingestive motor program, in which B8 was predominantly active during retraction (filled bar, marked by hyperpolarization of B34 interneuron). E2, perfusion of 10−5 m GFFD had no obvious effect on the CBI-2-elicited program. E3, washout of the peptide. F1–3, representative examples. F1 and F3 are the same as E1 and E3, except with the perfusion of (F2) 10−6 m GdFFD, which increased B8 firing during protraction and reduced B8 firing during retraction. It also decreased the duration of protraction. F4–7, group data (n = 4) showing the effects of GdFFD on (F4) protraction duration, (F5) retraction duration, (F6) B8 activity during protraction, and (F7) B8 activity during retraction. Bonferroni post hoc test is as follows: **, p < 0.01; ***, p < 0.001.
In these four preparations, 10−6 and 10−5 m GdFFD caused statistically significant increases in the expression of motor programs (Fig. 8D, n = 4, F3,9 = 10.17; p < 0.01). The feeding circuit generates at least two types of motor programs, ingestive and egestive (43, 44, 63, 64). Each cycle of motor programs consists of a protraction-retraction sequence. Motor programs were classified based on the activity phase of radula closure motor neuron B8 relative to protraction-retraction. Motor programs are considered as ingestive when B8 is predominantly active during retraction, and egestive when B8 is predominantly active during protraction (63, 64). The motor programs evoked by GdFFD were mostly egestive, as B8 was predominantly active during protraction (Fig. 8B). Upon washout of GdFFD, the buccal ganglia gradually returned to an inactive state (Fig. 8C).
In the next series of experiments, we tested the effects of the peptides on motor programs elicited by the command-like interneuron CBI-2. CBI-2 can be activated by food stimuli and, when active, drives feeding in semi-intact preparations or feeding motor programs in the isolated CNS (65). Similar to the above experiments, we observed no obvious effect with GFFD, even with a concentration of 10−5 m (Fig. 8E). In contrast, 10−6 m GdFFD was effective in changing the parametric features of CBI-2-elicited programs. We did not test the effects of 10−5 m GdFFD because there was too much activity evoked by GdFFD. Fig. 8F shows a representative example. In control conditions, stimulation of CBI-2 throughout protraction evoked a motor program in which B8 was active predominantly during retraction; thus the program was ingestive (Fig. 8F1). However, when we perfused 10−6 m GdFFD, the parametric features of the motor program changed, e.g. protraction duration was shorter, and B8 was more active during protraction and less active during retraction (Fig. 8F2). Upon washout of the GdFFD, the motor program elicited by CBI-2 was similar to the control (Fig. 8F3). Group data (n = 4, Fig. 8F, 4 bar graphs) showed that 10−6 m GdFFD exerted statistically significant effects on protraction duration (Fig. 8F4, F2,6 = 41.53; p < 0.001), retraction duration (Fig. 8F5, F2,6 = 11.43; p < 0.01), B8 activity during protraction (Fig. 8F6, F2,6 = 25.32; p < 0.01), and B8 activity during retraction (Fig. 8F7, F2,6 = 41.77; p < 0.001). Essentially, these parametric changes indicate that motor programs elicited by CBI-2 became more egestive with GdFFD. This is consistent with the observation that motor programs elicited by the perfusion of GdFFD are mostly egestive (Fig. 8B).
Overall, our data indicated that 10−6 m GdFFD was the effective concentration at which GdFFD acted to induce egestive programs in quiescent preparations and to modify parametric features of CBI-2-elicited programs. In contrast, GFFD was without any observable effects in the Aplysia feeding circuit.
DISCUSSION
Approaches for Identifying DAACPs in Biological Tissue Samples
The overall goals of this study were to develop a multidisciplinary approach using bioinformatics, biochemistry, analytical chemistry, and electrophysiological tools to identify the presence of a d-amino acid in predicted DAACPs and to examine their potential physiological actions in neural circuits that control behaviors. DAACPs in animals are derived through an isomerase-mediated PTM by converting an l-amino acid to a d-amino acid. Similar to PTMs such as amidation, this PTM may serve a variety of functions in animals as follows. First, this modification allows the generation of a larger number of products from a single gene. Second, a variety of physiological functions can be achieved through differential binding to the peptide's cognate receptor. Third, the altered structure can result in a distinct (usually greater) resistance to protease degradation and clearance, thus conferring a different effective time frame after release.
However, in contrast to peptides with chemical PTMs, relatively few peptides are currently known to undergo endogenous isomerization. As a case in point, in one of the most studied invertebrate model systems, the mollusk Aplysia, several hundred neuropeptides, many with chemical PTMs, have been characterized to date (35, 41, 42, 48–50, 52, 66–70). Despite this, prior to this study, the cardioexcitatory neuropeptide NdWFa was the only known DAACP in Aplysia (11). The lack of a systematic approach for identifying putative DAACPs slows the exploration of their complements and roles in signaling in the animal kingdom.
Our initial goal, therefore, was to adopt an MS-based chemical identification technique for DAACP discovery. Previously, we reported a nontargeted screening approach using aminopeptidase to screen for potential DAACPs based on their resistance to proteolytic degradation in crude cell lysates (71). d-Amino acids, especially those in the second position from the N terminus, and unusual N-terminal PTMs, such as polyglutamylation, cause peptides to be particularly resistant to degradation by this enzyme. This method, however, works best when combined with additional techniques for peptide structural characterization. Gas phase basicities of the diastereomers can be differentiated when they break apart into fragments in an MS/MS experiment, and this difference can be manifested as different branching ratios among product ions. It has been shown that several different MS techniques can be used to probe this thermochemical difference between diastereomers (34, 61, 62, 72–74). We have also shown that simultaneous identification and characterization of potential DAACPs by MALDI-TOF/TOF MS can be applied to distinguish d-isomer peptides in biological samples as small as individual neurons (38). In this context, it is notable that historically, achatin-I was isolated from 30,000 snails using structural characterization via NMR (9). By comparison, our MS-based approach allowed low mass/small volume sampling and structural analysis of GdFFD.
A second goal was to empower MS-based proteomics techniques for the characterization of putative DAACPs from complex tissue extracts. Proteomics relies on LC-MS/MS techniques but benefits greatly from bioinformatics, i.e. the availability of genome, transcriptome, and proteome information for a growing number of species that allow in silico discovery of new signaling peptides. Homology searches that exploited structural and functional similarities between prohormone sequences in other species (75) allowed us to generate a hypothesis for the existence of two Aplysia putative DAACPs encoding the prohormones GdFFD and YdAEFLa. Prior to MS characterization, we confirmed the expression and distribution of these prohormones in the CNS by molecular techniques and investigated their final gene products in isolated neurons using proven MALDI-TOF/TOF MS. We took advantage of the distinct fragmentation patterns that can arise between peptide epimers using both MALDI MS/MS- and LC-ESI MS/MS-based approaches (34, 38, 61, 73, 74) to distinguish DAACPs and their all l-peptide counterparts. Our identification of GdFFD in Aplysia, similar to achatin-I (GdFAD) in Achatina, validates our homology-based MS approach as being effective for identifying novel DAACPs in related animals. As more genetic information in different animals becomes available, we expect that the utility of this systematic approach will increase. Similar to bioactive achatin-I, GdFFD contains a d-phenyl residue at the second position, indicating that this PTM is conserved; such conserved structures suggest a functional context, such as receptor binding. For example, the DAACP Ocp-1, an ortholog of achatin-I, is an excitatory peptide in the octopus cardiovascular system that acts as a neuropeptide and/or neurohormone (22).
In contrast, despite an identical sequence to fulyal (YdAEFLa) in Achatina, YAEFLa in Aplysia was only observed in the l-form, suggesting that homology is not enough to assign a peptide as a DAACP. At present, we cannot exclude the possibility that YdAEFLa in Aplysia may exist in small quantities, but given the signal-to-noise ratio of YAEFLa and assuming both epimers have a similar limit of detection, YAEFLa is clearly the predominant form in Aplysia. We also have not detected the predicted homolog of Achatina fulicin (FdNEFVa), FTEFLa, in Aplysia. Regardless, one implication of this result is that it is essential to perform chemical identification experiments for the suspected DAACPs, even for the peptides that are identical in structure in related animals. Although the mechanisms are presently unknown, we speculate that the absence of YdAEFLa in Aplysia may be due to the absence of an isomerase or the specificity of the isomerase in the neurons that contain apFLNP (including YAEFLa). Future identification of the isomerase in Aplysia and its distribution in the CNS may provide an answer to this question. In addition, the apparent different processing of YAEFLa between Aplysia and Achatina raises both functional and evolutionary questions. What is the functional significance of this difference? Is there an evolutionary reason(s) for the different processing of the similar peptide in different but related animals? Perhaps future discovery of FGRP orthologs and putative DAACPs in other species will help resolve these issues.
Functional Significance of DAACPs in the Aplysia Feeding Circuit
Previous work has shown that most currently known DAACPs have potent bioactivity in various pathways, including in the peripheral and central nervous systems, with l-form peptides exhibiting less or no bioactivity (1). However, until this work, it was not known whether (or how) DAACPs can modify the activity of a defined neural circuit. Here, by taking advantage of the well studied Aplysia feeding circuit, we describe one of first studies to provide evidence that a DAACP, i.e. GdFFD but not GFFD, is bioactive in the feeding circuit. This finding is therefore consistent with the previous work demonstrating that an isomerization can change the bioactivity of a peptide and demonstrates the importance of isomerization on circuit function. Notably, the only other DAACP uncovered in Aplysia, NdWFa, which contains amidation of the C terminus in addition to a d-amino acid residue, was shown to be much more effective in inducing heart beat in Aplysia than the amidated l-epimer NWFa (11).
We would like to emphasize that the choice of the feeding circuit for the study of circuit function of DAACPs is not random, because the expression of specific feeding behavior, which is a motivated behavior, is modulated by many intrinsic and extrinsic factors (36, 76) that are often mediated by actions of neuromodulators (36, 77–79). Consistent with this complexity, previous work has shown that the feeding circuit is extensively modulated by neuromodulators, including neuropeptides that have been discovered in Aplysia (41, 42, 48, 49, 66, 67, 69). This study constitutes the first example of a DAACP acting as a neuromodulator in the Aplysia feeding circuit. Notably, many of the peptides identified before, e.g. apNPY (80), SCP (81), FRF-amide/FMRF-amide (82), and apUII (48), are intrinsic modulators because they are localized in either sensory/motor neurons or higher order interneurons of the feeding circuit, which involves the cerebral and buccal ganglia. In contrast, GFFD/GdFFD-positive neurons are localized in the pedal ganglion, which is not directly involved in feeding. Thus, the actions of GdFFD on the feeding circuit classify it as an extrinsic modulator of the feeding circuit. Along this line, GdFFD is similar to myomodulin gene 2-derived peptides (MMG2-DPs) (66), which are also located in the pedal ganglion and are extrinsic modulators of the feeding circuit. However, GdFFD appeared to exert more robust actions in the feeding circuit, as it was capable of evoking network activity in quiescent preparations, whereas MMG2-DPs were not. Indeed, although it may not be surprising that intrinsic modulators of the feeding circuit, e.g. FCAP (69) and SCP (81), can directly evoke network activity, GdFFD appears to be the first known extrinsic peptide neuromodulator capable of such an action in Aplysia.
The Aplysia feeding network is multifunctional in that it can generate at least two types of feeding motor programs, i.e. ingestive and egestive. From this perspective, GdFFD also differs from MMG2-DPs in that GdFFD promotes egestive programs, whereas MMG2-DPs make the programs less egestive. Thus, GdFFD is similar to several intrinsic neuromodulators, e.g. apNPY, SCP, and FRF-amide/FMRF-amide, that promote egestive programs and may serve to interrupt ingestion of food when animals become satiated. Although we do not yet know under what circumstances GdFFD-positive neurons would be activated, it is attractive to speculate that GdFFD may function to interrupt ingestion when locomotion is active because the pedal ganglion is involved in the control of locomotion (83). In this sense, in contrast to the actions of intrinsic modulators apNPY and SCP, the extrinsic modulator GdFFD might provide a new context in which a modulator may bias the feeding programs toward egestion in the Aplysia feeding system (41, 47). To the best of our knowledge, GdFFD represents the first example of a DAACP acting on a defined neural circuit that ultimately influences behavior.
In summary, we describe a multidisciplinary approach that allowed us to identify a novel DAACP, GdFFD, in a well known model system, providing important evidence that post-translational conversion of the Phe residue to d-Phe in this peptide is required for circuit actions. We expect that our multifaceted technique can be applied to other organisms, including vertebrates, and should accelerate studies of PTMs, which are critical for the expression of functional peptides in animals. Indeed, our study indicates that it may be fruitful in the future to examine other peptides found within neural networks to determine whether peptides with no known bioactivity when testing with synthetic all-l-form peptides may in fact have a d-amino acid that is required for their activity.
This work was supported, in whole or in part, by National Institutes of Health Grants NS031609, NS066587, and NS070583 from NINDS, 5T32-GM070421 from NIGMS, and P30 DA018310 from NIDA. This work was also supported by Grant 31371104 from National Science Foundation of China and Grant CHE-11-11705 from the National Science Foundation, Division of Chemistry (with co-funding from the Division of Biological Infrastructure).
- PTM
- post-translational modification
- DAACP
- d-amino acid-containing peptide
- PCA
- principal component analysis
- ACN
- acetonitrile
- CID
- collision-induced dissociation
- FA
- formic acid
- MMG2-DP
- myomodulin gene 2-derived peptide
- ESI
- electrospray ionization
- IT
- ion trap
- Fi
- Phe-immonium.
REFERENCES
- 1. Bai L., Sheeley S., Sweedler J. (2009) Analysis of endogenous d-amino acid-containing peptides in metazoa. Bioanal. Rev. 1, 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Broccardo M., Erspamer V., Falconieri Erspamer G., Improta G., Linari G., Melchiorri P., Montecucchi P. C. (1981) Pharmacological data on dermorphins, a new class of potent opioid peptides from amphibian skin. Br. J. Pharmacol. 73, 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Montecucchi P. C., de Castiglione R., Piani S., Gozzini L., Erspamer V. (1981) Amino acid composition and sequence of dermorphin, a novel opiate-like peptide from the skin of Phyllomedusa sauvagei. Int. J. Pept. Protein Res. 17, 275–283 [DOI] [PubMed] [Google Scholar]
- 4. Barra D., Mignogna G., Simmaco M., Pucci P., Severini C., Falconieri-Erspamer G., Negri L., Erspamer V. (1994) [d-Leu2]Deltorphin, a 17-amino acid opioid peptide from the skin of the Brazilian hylid frog, Phyllomedusa burmeisteri. Peptides 15, 199–202 [DOI] [PubMed] [Google Scholar]
- 5. Erspamer V., Melchiorri P., Falconieri-Erspamer G., Negri L., Corsi R., Severini C., Barra D., Simmaco M., Kreil G. (1989) Deltorphins: a family of naturally occurring peptides with high affinity and selectivity for δ opioid binding sites. Proc. Natl. Acad. Sci. U.S.A. 86, 5188–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kreil G., Barra D., Simmaco M., Erspamer V., Erspamer G. F., Negri L., Severini C., Corsi R., Melchiorri P. (1989) Deltorphin, a novel amphibian skin peptide with high selectivity and affinity for δ opioid receptors. Eur. J. Pharmacol. 162, 123–128 [DOI] [PubMed] [Google Scholar]
- 7. Mor A., Delfour A., Sagan S., Amiche M., Pradelles P., Rossier J., Nicolas P. (1989) Isolation of dermenkephalin from amphibian skin, a high-affinity δ-selective opioid heptapeptide containing a d-amino acid residue. FEBS Lett. 255, 269–274 [DOI] [PubMed] [Google Scholar]
- 8. Mignogna G., Simmaco M., Kreil G., Barra D. (1993) Antibacterial and haemolytic peptides containing d-alloisoleucine from the skin of Bombina variegata. EMBO J. 12, 4829–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kamatani Y., Minakata H., Kenny P. T., Iwashita T., Watanabe K., Funase K., Sun X. P., Yongsiri A., Kim K. H., Novales-Li P. (1989) Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Férussac containing a d-amino acid residue. Biochem. Biophys. Res. Commun. 160, 1015–1020 [DOI] [PubMed] [Google Scholar]
- 10. Ohta N., Kubota I., Takao T., Shimonishi Y., Yasuda-Kamatani Y., Minakata H., Nomoto K., Muneoka Y., Kobayashi M. (1991) Fulicin, a novel neuropeptide containing a d-amino acid residue isolated from the ganglia of Achatina fulica. Biochem. Biophys. Res. Commun. 178, 486–493 [DOI] [PubMed] [Google Scholar]
- 11. Morishita F., Nakanishi Y., Kaku S., Furukawa Y., Ohta S., Hirata T., Ohtani M., Fujisawa Y., Muneoka Y., Matsushima O. (1997) A novel d-amino-acid-containing peptide isolated from Aplysia heart. Biochem. Biophys. Res. Commun. 240, 354–358 [DOI] [PubMed] [Google Scholar]
- 12. Jacobsen R., Jimenez E. C., Grilley M., Watkins M., Hillyard D., Cruz L. J., Olivera B. M. (1998) The contryphans, a d-tryptophan-containing family of Conus peptides: interconversion between conformers. J. Pept. Res. 51, 173–179 [DOI] [PubMed] [Google Scholar]
- 13. Jacobsen R. B., Jimenez E. C., De la Cruz R. G., Gray W. R., Cruz L. J., Olivera B. M. (1999) A novel d-leucine-containing Conus peptide: diverse conformational dynamics in the contryphan family. J. Pept. Res. 54, 93–99 [DOI] [PubMed] [Google Scholar]
- 14. Jimenez E. C., Craig A. G., Watkins M., Hillyard D. R., Gray W. R., Gulyas J., Rivier J. E., Cruz L. J., Olivera B. M. (1997) Bromocontryphan: post-translational bromination of tryptophan. Biochemistry 36, 989–994 [DOI] [PubMed] [Google Scholar]
- 15. Jimenéz E. C., Olivera B. M., Gray W. R., Cruz L. J. (1996) Contryphan is a d-tryptophan-containing Conus peptide. J. Biol. Chem. 271, 28002–28005 [DOI] [PubMed] [Google Scholar]
- 16. Jimenez E. C., Watkins M., Juszczak L. J., Cruz L. J., Olivera B. M. (2001) Contryphans from Conus textile venom ducts. Toxicon 39, 803–808 [DOI] [PubMed] [Google Scholar]
- 17. Buczek O., Jimenez E. C., Yoshikami D., Imperial J. S., Watkins M., Morrison A., Olivera B. M. (2008) I(1)-superfamily conotoxins and prediction of single d-amino acid occurrence. Toxicon 51, 218–229 [DOI] [PubMed] [Google Scholar]
- 18. Buczek O., Yoshikami D., Bulaj G., Jimenez E. C., Olivera B. M. (2005) Post-translational amino acid isomerization: a functionally important d-amino acid in an excitatory peptide. J. Biol. Chem. 280, 4247–4253 [DOI] [PubMed] [Google Scholar]
- 19. Dutertre S., Lumsden N. G., Alewood P. F., Lewis R. J. (2006) Isolation and characterisation of conomap-Vt, a d-amino acid containing excitatory peptide from the venom of a vermivorous cone snail. FEBS Lett. 580, 3860–3866 [DOI] [PubMed] [Google Scholar]
- 20. Han Y., Huang F., Jiang H., Liu L., Wang Q., Wang Y., Shao X., Chi C., Du W., Wang C. (2008) Purification and structural characterization of a d-amino acid-containing conopeptide, conomarphin, from Conus marmoreus. FEBS J. 275, 1976–1987 [DOI] [PubMed] [Google Scholar]
- 21. Pisarewicz K., Mora D., Pflueger F. C., Fields G. B., Marí F. (2005) Polypeptide chains containing d-γ-hydroxyvaline. J. Am. Chem. Soc. 127, 6207–6215 [DOI] [PubMed] [Google Scholar]
- 22. Iwakoshi E., Hisada M., Minakata H. (2000) Cardioactive peptides isolated from the brain of a Japanese octopus, Octopus minor. Peptides 21, 623–630 [DOI] [PubMed] [Google Scholar]
- 23. Fujisawa Y., Ikeda T., Nomoto K., Yasuda-Kamatani Y., Minakata H., Kenny P. T., Kubota I., Muneoka Y. (1992) The FMRFamide-related decapeptide of Mytilus contains a d-amino acid residue. Comp. Biochem. Physiol. C. 102, 91–95 [DOI] [PubMed] [Google Scholar]
- 24. Yasuda-Kamatani Y., Kobayashi M., Yasuda A., Fujita T., Minakata H., Nomoto K., Nakamura M., Sakiyama F. (1997) A novel d-amino acid-containing peptide, fulyal, coexists with fulicin gene-related peptides in Achatina atria. Peptides 18, 347–354 [DOI] [PubMed] [Google Scholar]
- 25. Yasuda A., Yasuda Y., Fujita T., Naya Y. (1994) Characterization of crustacean hyperglycemic hormone from the crayfish (Procambarus clarkii): multiplicity of molecular forms by stereoinversion and diverse functions. Gen. Comp. Endocrinol. 95, 387–398 [DOI] [PubMed] [Google Scholar]
- 26. Soyez D., Van Herp F., Rossier J., Le Caer J. P., Tensen C. P., Lafont R. (1994) Evidence for a conformational polymorphism of invertebrate neurohormones. d- amino acid residue in crustacean hyperglycemic peptides. J. Biol. Chem. 269, 18295–18298 [PubMed] [Google Scholar]
- 27. Heck S. D., Kelbaugh P. R., Kelly M. E., Thadeio P. F., Saccomano N. A., Stroh J. G., Volkmann R. A. (1994) Disulfide bond of ω-agatoxins IVB and IVC: Discovery of a d-serine residue in ω-agatoxin IVB. J. Am. Chem. Soc. 116, 10426–10436 [Google Scholar]
- 28. Kuwada M., Teramoto T., Kumagaye K. Y., Nakajima K., Watanabe T., Kawai T., Kawakami Y., Niidome T., Sawada K., Nishizawa Y. (1994) ω-Agatoxin-TK containing d-serine at position 46, but not synthetic ω-[l-Ser46]agatoxin-TK, exerts blockade of P-type calcium channels in cerebellar Purkinje neurons. Mol. Pharmacol. 46, 587–593 [PubMed] [Google Scholar]
- 29. Torres A. M., Menz I., Alewood P. F., Bansal P., Lahnstein J., Gallagher C. H., Kuchel P. W. (2002) d-Amino acid residue in the C-type natriuretic peptide from the venom of the mammal, Ornithorhynchus anatinus, the Australian platypus. FEBS Lett. 524, 172–176 [DOI] [PubMed] [Google Scholar]
- 30. Torres A. M., Tsampazi C., Geraghty D. P., Bansal P. S., Alewood P. F., Kuchel P. W. (2005) d-Amino acid residue in a defensin-like peptide from platypus venom: effect on structure and chromatographic properties. Biochem. J. 391, 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heck S. D., Faraci W. S., Kelbaugh P. R., Saccomano N. A., Thadeio P. F., Volkmann R. A. (1996) Posttranslational amino acid epimerization: enzyme-catalyzed isomerization of amino acid residues in peptide chains. Proc. Natl. Acad. Sci. U.S.A. 93, 4036–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shikata Y., Watanabe T., Teramoto T., Inoue A., Kawakami Y., Nishizawa Y., Katayama K., Kuwada M. (1995) Isolation and characterization of a peptide isomerase from funnel web spider venom. J. Biol. Chem. 270, 16719–16723 [DOI] [PubMed] [Google Scholar]
- 33. Torres A. M., Tsampazi M., Tsampazi C., Kennett E. C., Belov K., Geraghty D. P., Bansal P. S., Alewood P. F., Kuchel P. W. (2006) Mammalian l-to-d-amino-acid-residue isomerase from platypus venom. FEBS Lett. 580, 1587–1591 [DOI] [PubMed] [Google Scholar]
- 34. Soyez D., Toullec J.-Y., Montagné N., Ollivaux C. (2011) Experimental strategies for the analysis of d-amino acid containing peptides in crustaceans: a review. J. Chromatogr. B analyt. Technol. Biomed. Life Sci. 879, 3102–3107 [DOI] [PubMed] [Google Scholar]
- 35. Moroz L. L., Edwards J. R., Puthanveettil S. V., Kohn A. B., Ha T., Heyland A., Knudsen B., Sahni A., Yu F., Liu L., Jezzini S., Lovell P., Iannucculli W., Chen M., Nguyen T., Sheng H., Shaw R., Kalachikov S., Panchin Y. V., Farmerie W., Russo J. J., Ju J., Kandel E. R. (2006) Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 127, 1453–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jing J., Gillette R., Weiss K. R. (2009) Evolving concepts of arousal: insights from simple model systems. Rev. Neurosci. 20, 405–427 [DOI] [PubMed] [Google Scholar]
- 37. Cropper E. C., Evans C. G., Hurwitz I., Jing J., Proekt A., Romero A., Rosen S. C. (2004) Feeding neural networks in the mollusc Aplysia. Neurosignals 13, 70–86 [DOI] [PubMed] [Google Scholar]
- 38. Bai L., Romanova E. V., Sweedler J. V. (2011) Distinguishing endogenous d-amino acid-containing neuropeptides in individual neurons using tandem mass spectrometry. Anal. Chem. 83, 2794–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heyland A., Vue Z., Voolstra C. R., Medina M., Moroz L. L. (2011) Developmental transcriptome of Aplysia californica. J. Exp. Zool. B Mol. Dev. Evol. 316B, 113–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fiedler T. J., Hudder A., McKay S. J., Shivkumar S., Capo T. R., Schmale M. C., Walsh P. J. (2010) The transcriptome of the early life history stages of the California Sea Hare Aplysia californica. Comp. Biochem. Physiol. Part D Genomics Proteomics 5, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jing J., Vilim F. S., Horn C. C., Alexeeva V., Hatcher N. G., Sasaki K., Yashina I., Zhurov Y., Kupfermann I., Sweedler J. V., Weiss K. R. (2007) From hunger to satiety: reconfiguration of a feeding network by Aplysia neuropeptide Y. J. Neurosci. 27, 3490–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jing J., Sweedler J. V., Cropper E. C., Alexeeva V., Park J. H., Romanova E. V., Xie F., Dembrow N. C., Ludwar B. C., Weiss K. R., Vilim F. S. (2010) Feedforward compensation mediated by the central and peripheral actions of a single neuropeptide discovered using representational difference analysis. J. Neurosci. 30, 16545–16558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jing J., Weiss K. R. (2001) Neural mechanisms of motor program switching in Aplysia. J. Neurosci. 21, 7349–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jing J., Weiss K. R. (2002) Interneuronal basis of the generation of related but distinct motor programs in Aplysia: implications for current neuronal models of vertebrate intralimb coordination. J. Neurosci. 22, 6228–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Proekt A., Jing J., Weiss K. R. (2007) Multiple contributions of an input-representing neuron to the dynamics of the Aplysia feeding network. J. Neurophysiol. 97, 3046–3056 [DOI] [PubMed] [Google Scholar]
- 46. Hurwitz I., Kupfermann I., Susswein A. J. (1997) Different roles of neurons B63 and B34 that are active during the protraction phase of buccal motor programs in Aplysia californica. J. Neurophysiol. 78, 1305–1319 [DOI] [PubMed] [Google Scholar]
- 47. Wu J. S., Vilim F. S., Hatcher N. G., Due M. R., Sweedler J. V., Weiss K. R., Jing J. (2010) Composite modulatory feedforward loop contributes to the establishment of a network state. J. Neurophysiol. 103, 2174–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Romanova E. V., Sasaki K., Alexeeva V., Vilim F. S., Jing J., Richmond T. A., Weiss K. R., Sweedler J. V. (2012) Urotensin II in invertebrates: from structure to function in Aplysia californica. PLoS ONE 7, e48764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vilim F. S., Sasaki K., Rybak J., Alexeeva V., Cropper E. C., Jing J., Orekhova I. V., Brezina V., Price D., Romanova E. V., Rubakhin S. S., Hatcher N., Sweedler J. V., Weiss K. R. (2010) Distinct mechanisms produce functionally complementary actions of neuropeptides that are structurally related but derived from different precursors. J. Neurosci. 30, 131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vilim F. S., Alexeeva V., Moroz L. L., Li L., Moroz T. P., Sweedler J. V., Weiss K. R. (2001) Cloning, expression and processing of the CP2 neuropeptide precursor of Aplysia. Peptides 22, 2027–2038 [DOI] [PubMed] [Google Scholar]
- 51. Li L., Garden R. W., Sweedler J. V. (2000) Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 18, 151–160 [DOI] [PubMed] [Google Scholar]
- 52. Furukawa Y., Nakamaru K., Wakayama H., Fujisawa Y., Minakata H., Ohta S., Morishita F., Matsushima O., Li L., Romanova E., Sweedler J. V., Park J. H., Romero A., Cropper E. C., Dembrow N. C., Jing J., Weiss K. R., Vilim F. S. (2001) The enterins: a novel family of neuropeptides isolated from the enteric nervous system and CNS of Aplysia. J. Neurosci. 21, 8247–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jing J., Sasaki K., Perkins M. H., Siniscalchi M. J., Ludwar B. C., Cropper E. C., Weiss K. R. (2011) Coordination of distinct motor structures through remote axonal coupling of projection interneurons. J. Neurosci. 31, 15438–15449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Predel R., Neupert S. (2007) Social behavior and the evolution of neuropeptide genes: lessons from the honeybee genome. BioEssays 29, 416–521 [DOI] [PubMed] [Google Scholar]
- 55. Satake H., Yasuda-Kamatani Y., Takuwa K., Nomoto K., Minakata H., Nagahama T., Nakabayashi K., Matsushima O. (1999) Characterization of a cDNA encoding a precursor polypeptide of a d-amino acid-containing peptide, achatin-I and localized expression of the achatin-I and fulicin genes. Eur. J. Biochem. 261, 130–136 [DOI] [PubMed] [Google Scholar]
- 56. Yasuda-Kamatani Y., Nakamura M., Minakata H., Nomoto K., Sakiyama F. (1995) A novel cDNA sequence encoding the precursor of the d-amino acid-containing neuropeptide fulicin and multiple alpha-amidated neuropeptides from Achatina fulica. J. Neurochem. 64, 2248–2255 [DOI] [PubMed] [Google Scholar]
- 57. Southey B. R., Amare A., Zimmerman T. A., Rodriguez-Zas S. L., Sweedler J. V. (2006) NeuroPred: a tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 34, W267–W272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hummon A. B., Hummon N. P., Corbin R. W., Li L., Vilim F. S., Weiss K. R., Sweedler J. V. (2003) From precursor to final peptides: a statistical sequence-based approach to predicting prohormone processing. J. Proteome Res. 2, 650–656 [DOI] [PubMed] [Google Scholar]
- 59. Hummon A. B., Amare A., Sweedler J. V. (2006) Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom. Rev. 25, 77–98 [DOI] [PubMed] [Google Scholar]
- 60. Li L., Sweedler J. V. (2008) Peptides in the brain: Mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem. 1, 451–483 [DOI] [PubMed] [Google Scholar]
- 61. Adams C. M., Kjeldsen F., Zubarev R. A., Budnik B. A., Haselmann K. F. (2004) Electron capture dissociation distinguishes a single d-amino acid in a protein and probes the tertiary structure. J. Am. Soc. Mass Spectrom. 15, 1087–1098 [DOI] [PubMed] [Google Scholar]
- 62. Sachon E., Clodic G., Galanth C., Amiche M., Ollivaux C., Soyez D., Bolbach G. (2009) d-Amino acid detection in peptides by MALDI-TOF-TOF. Anal. Chem. 81, 4389–4396 [DOI] [PubMed] [Google Scholar]
- 63. Morton D. W., Chiel H. J. (1993) In vivo buccal nerve activity that distinguishes ingestion from rejection can be used to predict behavioral transitions in Aplysia. J. Comp. Physiol. A 172, 17–32 [DOI] [PubMed] [Google Scholar]
- 64. Morgan P. T., Jing J., Vilim F. S., Weiss K. R. (2002) Interneuronal and peptidergic control of motor pattern switching in Aplysia. J. Neurophysiol. 87, 49–61 [DOI] [PubMed] [Google Scholar]
- 65. Jing J., Weiss K. R. (2005) Generation of variants of a motor act in a modular and hierarchical motor network. Curr. Biol. 15, 1712–1721 [DOI] [PubMed] [Google Scholar]
- 66. Proekt A., Vilim F. S., Alexeeva V., Brezina V., Friedman A., Jing J., Li L., Zhurov Y., Sweedler J. V., Weiss K. R. (2005) Identification of a new neuropeptide precursor reveals a novel source of extrinsic modulation in the feeding system of Aplysia. J. Neurosci. 25, 9637–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lloyd P. E., Mahon A. C., Kupfermann I., Cohen J. L., Scheller R. H., Weiss K. R. (1985) Biochemical and immunocytological localization of molluscan small cardioactive peptides in the nervous system of Aplysia californica. J. Neurosci. 5, 1851–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Phares G. A., Lloyd P. E. (1996) Immunocytological and biochemical localization and biological activity of the newly sequenced cerebral peptide 2 in Aplysia. J. Neurosci. 16, 7841–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sweedler J. V., Li L., Rubakhin S. S., Alexeeva V., Dembrow N. C., Dowling O., Jing J., Weiss K. R., Vilim F. S. (2002) Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of Aplysia. J. Neurosci. 22, 7797–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fujisawa Y., Furukawa Y., Ohta S., Ellis T. A., Dembrow N. C., Li L., Floyd P. D., Sweedler J. V., Minakata H., Nakamaru K., Morishita F., Matsushima O., Weiss K. R., Vilim F. S. (1999) The Aplysia mytilus inhibitory peptide-related peptides: identification, cloning, processing, distribution, and action. J. Neurosci. 19, 9618–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ewing M. A., Wang J., Sheeley S. A., Sweedler J. V. (2008) Detecting d-amino acid-containing neuropeptides using selective enzymatic digestion. Anal. Chem. 80, 2874–2880 [DOI] [PubMed] [Google Scholar]
- 72. Cooks R. G., Wong P. S. H. (1998) Kinetic method of making thermochemical determinations: Advances and applications. Acc. Chem. Res. 31, 379–386 [Google Scholar]
- 73. Serafin S. V., Maranan R., Zhang K., Morton T. H. (2005) Mass spectrometric differentiation of linear peptides composed of l-amino acids from isomers containing one d-amino acid residue. Anal. Chem. 77, 5480–5487 [DOI] [PubMed] [Google Scholar]
- 74. Redeker V., Toullec J.-Y., Vinh J., Rossier J., Soyez D. (1998) Combination of peptide profiling by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and immunodetection on single glands or cells. Anal. Chem. 70, 1805–1811 [DOI] [PubMed] [Google Scholar]
- 75. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 76. Kupfermann I. (1974) Dissociation of the appetitive and consummatory phases of feeding behavior in Aplysia: a lesion study. Behav. Biol. 10, 89–97 [DOI] [PubMed] [Google Scholar]
- 77. Blitz D. M., Nusbaum M. P. (2012) Modulation of circuit feedback specifies motor circuit output. J. Neurosci. 32, 9182–9193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Taghert P. H., Nitabach M. N. (2012) Peptide neuromodulation in invertebrate model systems. Neuron 76, 82–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Strand F. L. (1999) in Neuropeptides: Regulators of Physiological Processes (Stevens C. F., ed) pp. 121–140, MIT Press, Cambridge, MA [Google Scholar]
- 80. Rajpara S. M., Garcia P. D., Roberts R., Eliassen J. C., Owens D. F., Maltby D., Myers R. M., Mayeri E. (1992) Identification and molecular cloning of a neuropeptide Y homolog that produces prolonged inhibition in Aplysia neurons. Neuron 9, 505–513 [DOI] [PubMed] [Google Scholar]
- 81. Sossin W. S., Kirk M. D., Scheller R. H. (1987) Peptidergic modulation of neuronal circuitry controlling feeding in Aplysia. J. Neurosci. 7, 671–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schaefer M., Picciotto M. R., Kreiner T., Kaldany R.-R., Taussig R., Scheller R. H. (1985) Aplysia neurons express a gene encoding multiple FMRFamide neuropeptides. Cell 41, 457–467 [DOI] [PubMed] [Google Scholar]
- 83. Jing J., Vilim F. S., Cropper E. C., Weiss K. R. (2008) Neural analog of arousal: persistent conditional activation of a feeding modulator by serotonergic initiators of locomotion. J. Neurosci. 28, 12349–12361 [DOI] [PMC free article] [PubMed] [Google Scholar]