Background: The interaction between eukaryotic initiation factor 4G (eIF4G) and eIF3 promotes translation initiation in mammals.
Results: Human eIF3 subunits -c, -d, and -e interact with two subdomains in eIF4G.
Conclusion: Multiple contacts between eIF3 and eIF4G are required for mRNA recruitment to the human ribosome.
Significance: Characterizing the eIF3-eIF4G interface might reveal a new regulatory mechanism and provide novel therapeutic targets.
Keywords: mRNA, Protein Cross-linking, Protein-Protein Interactions, Ribosomes, Translation, Initiation, eIF3, eIF4G
Abstract
Recruitment of mRNA to the 40S ribosomal subunit requires the coordinated interaction of a large number of translation initiation factors. In mammals, the direct interaction between eukaryotic initiation factor 4G (eIF4G) and eIF3 is thought to act as the molecular bridge between the mRNA cap-binding complex and the 40S subunit. A discrete ∼90 amino acid domain in eIF4G is responsible for binding to eIF3, but the identity of the eIF3 subunit(s) involved is less clear. The eIF3e subunit has been shown to directly bind eIF4G, but the potential role of other eIF3 subunits in stabilizing this interaction has not been investigated. It is also not clear if the eIF4A helicase plays a role in stabilizing the interaction between eIF4G and eIF3. Here, we have used a fluorescence anisotropy assay to demonstrate that eIF4G binds to eIF3 independently of eIF4A binding to the middle region of eIF4G. By using a site-specific cross-linking approach, we unexpectedly show that the eIF4G-binding surface in eIF3 is comprised of the -c, -d and -e subunits. Screening multiple cross-linker positions reveals that eIF4G contains two distinct eIF3-binding subdomains within the previously identified eIF3-binding domain. Finally, by employing an eIF4G-dependent translation assay, we establish that both of these subdomains are required for efficient mRNA recruitment to the ribosome and stimulate translation. Our study reveals unexpected complexity to the eIF3-eIF4G interaction that provides new insight into the regulation of mRNA recruitment to the human ribosome.
Introduction
Recruitment of eukaryotic mRNAs to the ribosome requires coordinated interactions between initiation factors and the 40S ribosomal subunit. One possible recruitment model involves the initial selection of an mRNA by association of the eukaryotic initiation factor 4F (eIF4F)2 complex with the 5′-7-methyl guanosine cap structure (1–4). This complex consists of the cap-binding protein, eIF4E, the DEAD-box helicase, eIF4A, and the scaffold protein, eIF4G. The eIF4A component likely unwinds any cap-proximal secondary structure so that it can then bind to the 43S pre-initiation complex (43S-PIC). The ATP-dependent RNA unwinding activity of eIF4A is strongly stimulated by eIF4G and the accessory protein eIF4B (5–9). In addition to its cap binding function, eIF4E also serves to promote mRNA restructuring by stimulating the helicase activity of eIF4A (10). Once a single-stranded region of the mRNA is available it can stably bind into the decoding site of the 43S-PIC.
In mammals, the interaction between the eIF4F-mRNA complex and the 43S-PIC is likely stabilized by a direct interaction between eIF4G and eIF3 (11–14). The importance of this interaction for controlling translation is implied from the observation that the affinity between eIF4G and eIF3 is increased by insulin treatment in vivo (15). Specifically, the activation of mTOR complex 1 (mTORC1) is required to stabilize the interaction between these binding partners (15, 16). It has also been suggested that eIF4A and eIF3 may bind in a cooperative manner to eIF4G since a greater amount of eIF3 can be immunoprecipitated with eIF4G in the presence of eIF4A using purified components (11). However, this has not been confirmed by using a quantitative binding assay.
Human eIF4G has been shown to possess a distinct ∼90 amino acid domain that forms a high affinity interaction with the eIF3 complex in vitro (11). This binding domain can compete with full-length eIF4G for binding to eIF3, implying that this region is indeed sufficient for binding eIF3 (11). The importance of this interaction for translation has been investigated by in vitro and in vivo translation assays using an eIF4G construct containing a quadruple alanine mutation in the eIF3 binding domain (17). This study showed that the mutant is less active in stimulating translation in a protease treated in vitro reticulocyte lysate system (17). In contrast, there is little difference between the ability of this mutant eIF4G protein to stimulate translation in vivo compared with wild type eIF4G (17). It is difficult to interpret the full effect of this particular mutation on translation since no quantitative binding data has been generated to determine its effect on the affinity between eIF4G and eIF3. Nevertheless, these data may imply that a substantial amount of redundancy exists between initiation factor interactions. Therefore, it is reasonable to believe that no single interaction is essential for mRNA recruitment to the 40S subunit.
Because of its large molecular mass and complexity, it has been difficult to identify the exact binding site of eIF4G on the eIF3 complex. Human eIF3 possesses an anthropomorphic five-lobed structure that contains 13 non-identical subunits that range in size from ∼25 kDa to 170 kDa (18–22). It has been shown that eIF4G makes a direct interaction with the eIF3e subunit within the context of the eIF3 complex (23). To determine this, an affinity tagged minimum eIF3 binding region of eIF4G was used as bait to fish out eIF3 subunit fragments from a partially proteolyzed eIF3 complex. One limitation of this approach is that it does not rule out the possibility that other eIF3 subunits may be important in stabilizing the interaction with eIF4G when eIF3 is not proteolyzed.
Here, we have employed an anisotropy assay to quantitatively show that human eIF4G binds to eIF3 independently of eIF4A. To further characterize the eIF4G-binding surface in eIF3, we have employed two different cross-linking approaches that utilize a minimum binding domain in eIF4G and intact purified eIF3. Our data reveal that three subunits of eIF3 (eIF3c, -d, and -e) form a binding surface for eIF4G. We further show that the previously identified eIF3-binding domain in eIF4G is comprised of two distinct subdomains that interact with these eIF3 subunits. Finally, we use an eIF4G-dependent reticulocyte lysate system to show that both of these subdomains are required to stimulate translation initiation in vitro.
EXPERIMENTAL PROCEDURES
Recombinant Expression of eIF4G Constructs
The numbering of eIF4G constructs used in this work is based on UniProt entry Q04637, although our constructs contain a glutamine insertion between residues 696 and 697 that corresponds to an isoform no longer included in this database. Recombinant eIF4G constructs shown in Fig. 1 contain an N-terminal 6X-HIS-MBP tag (eIF4G711–1104) or 6×-HIS tag (eIF4G1011–1104) and C-terminal FLAG tag. Additionally, 6 native cysteine residues are mutated to alanine in eIF4G711–1104 to prevent interference during labeling. A single cysteine mutation is incorporated into both constructs at S1041 for site-specific modification. These mutations do not affect eIF3 or eIF4A binding in either construct (data not shown). The eIF4G constructs used in the boxB tethered assay are N-terminally tagged with 6×-HIS and the 22 amino acid sequence of the bacteriophage λ transcription anti-terminator protein N (λ) that binds the boxB RNA hairpin (14, 24). For expression, eIF4G constructs in pET28c are transformed into BL-21(DE3) Escherichia coli cells. Typically, 2–4 liters of BL-21(DE3) cells are grown to an OD of 0.4–0.6 in LB media, and induced with 0.5 μg/ml isopropyl β-d-1-thiogalactopyranoside (IPTG) for 3–5 h at 30 °C. Cells are centrifuged at 4 °C for 20 min at 4000 × g, suspended in buffer A (Buffer A: 20 mm Hepes pH 7.5, 400 mm KCl, 5 mm imidazole, 10% glycerol, 10 mm BME) and then lysed using a microfluidizer as described previously (5). Resulting lysates are clarified by centrifugation and purified by Ni-NTA chromatography (Qiagen), as previously described (5, 10). The eluted protein is incubated with TEV protease to remove MBP during overnight dialysis at 4 °C in buffer B (buffer B: 20 mm Hepes pH 7.5, 50 mm KCl, 10% glycerol, 1 mm DTT). Further purification is achieved using Mono S (10/10) (eIF4G1011–1104 and λ-eIF4G), or HiTrap Heparin HP (eIF4G711–1104 and λ-eIF4G) columns (GE Healthcare), as previously described (5, 10). For each of these constructs, we purified ∼1 mg protein per liter of culture.
FIGURE 1.
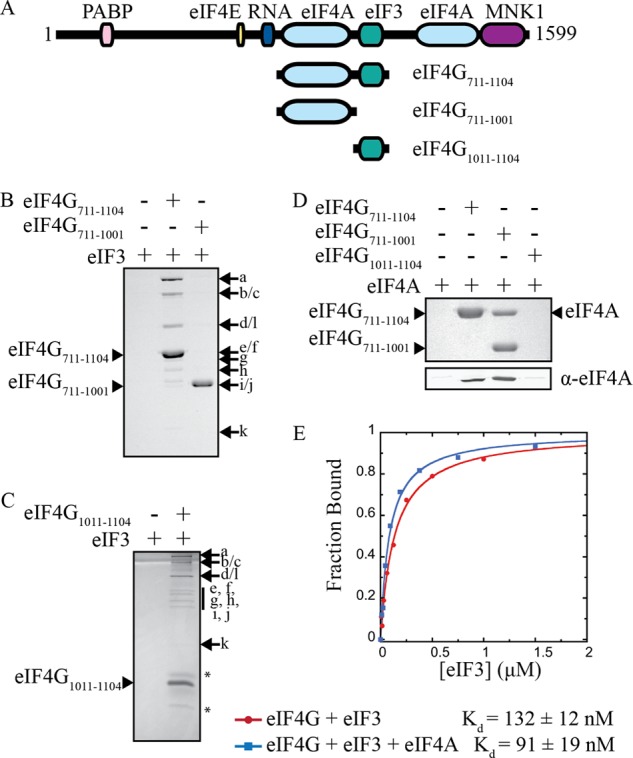
Characterization of eIF4G binding domains. A, domain map of full-length human eIF4G, and constructs used in this study. All constructs are C terminally FLAG-tagged, and eIF4G1011–1104 is also N terminally HIS-tagged. B and C, immunoprecipitation of FLAG-tagged eIF4G constructs and purified eIF3, as indicated. Eluted proteins are separated by SDS-PAGE and visualized with Coomassie Blue. Asterisks (*) denote degradation products. D, immunoprecipitation of FLAG-tagged eIF4G with purified eIF4A. Eluted proteins are separated by SDS-PAGE and visualized with Coomassie Blue (top panel) or by immunoblotting with eIF4A antiserum (bottom panel). E, representative plots of fluorescence polarization assays used to determine the equilibrium dissociation constant (Kd) of the eIF3-eIF4G711–1104 interaction in the absence or presence of a saturating amount of eIF4A. The Kd values shown are the average of at least 3 trials ± S.E.
Purification of eIF3 and eIF4A
HeLa cell cytosolic extracts were a kind gift from Dr. Robert Tjian (University of California, Berkeley). Human eIF3 is purified as previously described from HeLa cytosolic extracts (18, 21). Human eIF4A is expressed recombinantly in E. coli as previously described (5, 10).
In Vitro Modification of eIF4G with Sulfo-NHS-Diazirine or Fluorescein-5-malemide
Human eIF4G711–1104 is modified with fluorescein-5-malemide and eIF4G1011–1104 is modified with the photoactivatable cross-linker Sulfo-NHS-Diazirine (SDA) (Thermo Scientific) for fluorescence polarization and cross-linking assays, respectively. All eIF4G proteins are first dialyzed for 2 h at 4 °C into buffer C (buffer C: 20 Hepes, 300 mm KCl, 10% glycerol, 1 mm TCEP). eIF4G (∼35 μm) is then incubated with a 20-fold molar excess of fluorescein-5-malemide or SDA at room temperature for 30 min. Excess modification compounds are removed by dialysis or by purifying labeled proteins using HiTrap Heparin HP or SP-Sepharose columns (GE Healthcare).
Coimmunoprecipitation
EZ view Red Anti-FLAG M2 Affinity Gel (30 μl)(Sigma-Aldrich) is washed twice in 500 μl of buffer D (buffer D: 20 mm Hepes pH 7.5, 100 mm KCl, 10% glycerol, and 0.5 mm DTT). Resin is then incubated with 10 μg of recombinant eIF4G for at least 1 h in 50 μl of buffer D, washed twice in 100 μl buffer E (buffer E: 20 mm Hepes pH 7.5, 200 mm KCl, 10% glycerol, 0.5 mm DTT, and 0.5% TritonX-100) and once in 100 μl of buffer D. Human eIF4A (10 μg) or eIF3 (10 μg) is diluted in 30 μl of buffer D then added and incubated for 1 h at 4 °C. The resin is then washed four times in 75 μl of buffer E. Bound proteins are eluted by incubating the resin in 20 μl, 250 ng/μl FLAG peptide in buffer D for 1 h at 4 °C. Coimmuniprecipitated proteins are separated by SDS-PAGE then stained with Coomassie Blue or analyzed by immunoblotting with specific antiserum, as indicated in the figures.
Fluorescence Polarization Binding Assay
Fluorescence polarization is measured in a Victor X5 Multilabel Plate Reader (Perkin Elmer), similar to previously published studies (25–27). Each binding reaction is set up in buffer D and allowed to reach equilibrium at 30 °C for 5 min prior to fluorescence polarization measurements. Reactions contained 10 nm eIF4G-Fl, an eIF3 concentration in the range of 0–1.5 μm, and were conducted in the presence or absence of 1 μm eIF4A, which is a saturating amount based on previously published affinity data (28). Each Kd measurement is reported as the average of at least three experiments ± S.E. The figures show representative experiments.
BoxB Tethered Translation Assay
The transcription template for the BoxB tethered assay is a derivative of our previously described template, and was purchased from GenScript (10). The 5′-UTR, which is located between XhoI and HindIII restriction sites (underlined), has been replaced with the human beta globin 5′-UTR sequence as follows: CTCGAGACATTTGCTTCTGACACAACTGTGTTCACTAGCAACCTCAAACAGACACCAAGCTT. The template is transcribed in vitro and purified using phenol-chloroform extraction following standard protocols. Functional assays are carried out in 20-μl reactions containing 65% nuclease-treated rabbit reticulocyte lysate, 20 μm amino acid mixture minus leucine (Promega), 20 μm amino acid mixture minus methionine (Promega), 0.8 units/μl Recombinant RNasin Ribonuclease Inhibitor (Promega), 45 mm potassium chloride, 115 mm potassium acetate, 1.7 mm magnesium acetate, 0.5 μm uncapped RNA template, and 1 μm λ-eIF4G. Reactions are incubated at 30 °C for 30 min prior to luminescence measurement for 10 s on the Victor X5 Multilabel Plate Reader (Perkin Elmer) by addition of Renilla luciferase substrate (Promega), as described previously (10). We normalized luciferase activity to a reaction containing no λ-eIF4G, and presented the average of 4 reactions ± S.E.
Cotranslational Benzophenone Cross-linker Incorporation and Expression of eIF4G1011–1104 Using Amber Mutants
The benzophenone cross-linker, in the form of p-Benzoylphenylalanine (Bpa; BaChem), is incorporated into recombinant eIF4G1011–1104 using the pEVOL plasmid that was kindly provided by Dr. Peter Schultz (The Scripps Research Institute) (29). Single amber codon (TAG) mutations are introduced throughout the length of eIF4G1011–1104 through PCR site-directed mutagenesis at intervals of ∼10 amino acids. Amino acid substitutions are designated in the text by the single letter code for the endogenous residue, followed by the residue number and an “X” to indicate the amber mutant codon and location of Bpa incorporation. eIF4G amber mutant plasmids are cotransformed into BL-21(DE3) E. coli cells with pEVOL, which contains the orthogonal tRNA and tRNA synthetase pair required for Bpa incorporation at amber stop codons (29, 30). Cells are grown to A600 = 0.4–0.6, and protein expression is induced with 0.5 μg/ml IPTG, 0.02 mm arabinose, and 0.2 mm Bpa for 4 h at 30 °C or overnight at 16 °C. eIF4G-Bpa proteins are purified in the same way as unmodified eIF4G1011–1104. All amber mutants are expressed either as a mix of full-length eIF4G1011–1104 (with Bpa incorporation) and truncations of this construct (due to termination at the amber stop codon), or only the truncation product. These are successfully separated using ion exchange chromatography, as described for the unmodified protein. Only 6 amber mutant sites of the 10 cloned resulted in expressible full-length constructs. Successful sites expressed and purified include: M1011X, R1031X, S1041X, S1061X, K1071X, and W1093X. When Bpa is omitted from the expression media, only truncated forms of eIF4G1011–1104 are expressed. For each eIF4G-Bpa construct expressed, we purified ∼200–600 μg of protein per liter of culture.
Cross-linking
Cross-linking reactions are typically carried out in 20 μl incubations in clear, thin-wall PCR tubes. Each reaction contains 2.5 μm eIF4G1011–1104-SDA and 0.5 μm eIF3 or 2.5 μm eIF4G1011–1104-Bpa and 0.25 μm eIF3 in buffer D, unless otherwise stated. Following preincubation at 30 °C for 5 min, cross-linking is carried out using the 365 nm UV light from a AlphaImager HP gel imaging device (Cell Biosciences) for 5 min at room temperature, except in time course studies where the UV exposure time is as indicated. Cross-linked products are separated by SDS-PAGE and analyzed by immunoblotting with antiserum as indicated.
Western Blotting
Cross-linked products are transferred to a PVDF membrane (Millipore) using a Mini Trans-Blot Cell or the Trans-Blot Turbo Transfer System (Bio-Rad). Blots are incubated with primary antibodies diluted in TBST for 1.5 h at room temperature or overnight at 4 °C. Primary antibodies used in this study include: mouse monoclonal anti-FLAG M2 antibody (Sigma-Aldrich), rabbit anti-eIF3b (Santa Cruz Biotechnology, sc-28857), rabbit anti-eIF3c (produced by Christopher Fraser), mouse anti-eIF3c (Santa Cruz Biotechnology, sc-74507), mouse anti-eIF3d (Santa Cruz Biotechnology, sc-271515), rabbit anti-eIF3l (Abcam, PAB17471), mouse anti-eIF3e (Santa Cruz Biotechnology, sc-74505), and rabbit anti-eIF3f (Abcam ab74568). Blots are washed three times for 10 min each in TBST, followed by 1-hour incubation at room temperature with appropriate secondary antibodies. Secondary antibodies are HRP-conjugated (Santa Cruz Biotechnology) in cross-linking time course and competition experiments (Fig. 2) or fluorescently labeled with DyLight 800 or DyLight 680 (Thermo Scientific) in all other experiments. Blots are visualized using a Storm 860 Imager (Molecular Dynamics) for HRP-conjugated secondary antibodies or the Odyssey Infrared Imaging System (Li-Cor) for fluorescent secondary antibodies.
FIGURE 2.
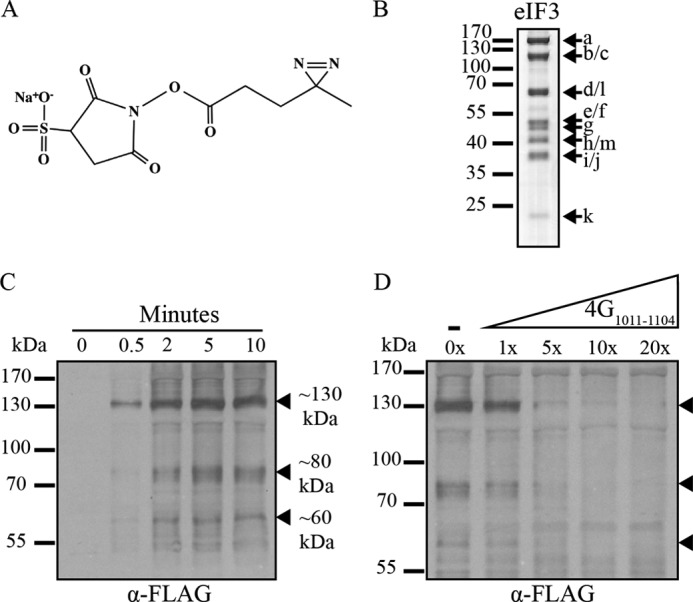
eIF4G1011–1104-SDA site-specifically cross-links to 3 distinct eIF3 subunits. A, chemical structure of the Sulfo-NHS-Diazirine (sulfo-SDA, Pierce) cross-linker used to site-specifically modify eIF4G1011–1104 at Ser-1041. B, human eIF3 subunits are separated by SDS-PAGE and visualized with Coomassie Blue staining. C, time course study of eIF4G1011–1104-SDA cross-linking with eIF3. Products are separated by SDS-PAGE and analyzed by immunoblotting for FLAG-tagged eIF4G1011–1104-SDA. Filled arrows designate cross-linked eIF3 subunits. D, unmodified eIF4G1011–1104 competes for eIF3 binding to reduce eIF4G1011–1104-SDA cross-linking to eIF3. Lanes are labeled with the fold excess of unmodified eIF4G1011–1104 competitor over eIF4G1011–1104-SDA.
RESULTS
Human eIF4G Binds to eIF3 Independently of eIF4A
To quantitatively determine if eIF4G binding to eIF3 is regulated by eIF4A in the absence of the 40S ribosomal subunit, we optimized an anisotropy-based equilibrium-binding assay that utilizes highly purified components. To this end, we generated a FLAG-tagged recombinant eIF4G truncation spanning amino acids 711 to 1104 (eIF4G711–1104), which includes the central eIF4A and eIF3 binding sites (Fig. 1A). To fluorescently modify this eIF4G truncation, we genetically encoded a single cysteine (S1041C) into the protein so that it can be site-specifically modified with fluorescein. To verify that this single cysteine eIF4G construct binds to eIF4A and eIF3 we employed a coimmunoprecipitation assay, as described under “Experimental Procedures.” Both eIF4A and eIF3 interact specifically with the FLAG-tagged eIF4G711–1104 truncation (Fig. 1, B and D). Removal of the eIF3 binding domain (eIF4G711–1001) does not prevent eIF4A binding, but completely abolishes eIF3 binding (Fig. 1, B and D). Consistent with this and previous data (11, 13), we find that a 93 amino acid region of eIF4G (eIF4G1011–1104) is sufficient for binding to eIF3 in our coimmunoprecipitation assay (Fig. 1C).
To quantitatively determine the affinity of the eIF4G-eIF3 interaction, we measured the anisotropy of fluorescein-labeled eIF4G711–1104 in the presence of an increasing concentration of purified eIF3. Anisotropy is converted into the fraction of eIF4G711–1104 bound at each eIF3 concentration (Fig. 1E). Binding studies were conducted in the absence or presence of a saturating amount of eIF4A to determine if eIF4A influences the affinity of eIF4G711–1104 to eIF3. Our data show that the 132 ± 12 nm equilibrium dissociation constant (Kd) for the eIF4G711–1104-eIF3 complex in the absence of eIF4A is only slightly reduced upon the addition of a saturating amount of eIF4A (91 ± 19 nm; Fig. 1E). This strongly implies that the eIF4A-binding domain directly upstream of the eIF3-binding domain does not contribute to eIF3 affinity, at least in the absence of the 40S subunit. This data does not eliminate the possibility that the C-terminal eIF4A binding site of eIF4G may influence eIF3 binding, although this is unlikely since this secondary binding site is known to have a much lower affinity for eIF4A (28).
Site-specific Cross-linking of eIF4G1011–1104 to Purified Human eIF3
Affinity purification of a human eIF4G1015–1118 truncation to partially proteolyzed eIF3 has identified the eIF3e subunit as a direct binding site for eIF4G (23). To circumvent the use of partially proteolyzed eIF3 to study the interaction between eIF4G and eIF3, we have generated a site-directed cross-linking strategy that successfully cross-links recombinant eIF4G1011–1104 to intact highly purified eIF3 containing all 13 subunits. To this end, we modified FLAG-tagged eIF4G1011–1104 with a photoactivatable cross-linker, sulfo-NHS-Diazirine (SDA; Fig. 2A), using a single cysteine engineered into the eIF3-binding domain of the protein (S1041C).
Photocross-linking is carried out between SDA-labeled eIF4G1011–1104 and purified eIF3 (Fig. 2B), as described under “Experimental Procedures.” The optimal UV exposure time was determined by a time course experiment using purified eIF3 (1 μm) and a 10-fold molar excess of eIF4G1011–1104-SDA to ensure maximum cross-linking efficiency. Covalently cross-linked species are identified by separation on SDS-PAGE followed by Western blotting with an anti-FLAG monoclonal antibody. Specific cross-linking between eIF4G and an eIF3 subunit is indicated by a molecular weight shift of an eIF3 subunit by the apparent molecular weight of FLAG-tagged eIF4G1011–1104-SDA (∼15 kDa). We observe a time-dependent appearance of 3 major cross-linked bands that possess apparent molecular masses of ∼130, 80, and 60 kDa (Fig. 2C, filled arrows). Importantly, each cross-linked product forms only in the presence of the SDA cross-linker and eIF3, confirming the formation of specific eIF3-eIF4G-bound complexes (data not shown). To further ensure that the eIF3-eIF4G cross-linked products are a result of a specific interaction not influenced by the presence of the SDA cross-linker, we performed a competition assay with unmodified eIF4G1011–1104. Increasing amounts of unmodified eIF4G1011–1104 effectively competes for the same eIF3 binding sites as eIF4G1011–1104-SDA, thereby reducing eIF4G1011–1104-SDA cross-linking (Fig. 2D). In addition, we note that any nonspecific binding remains largely unaffected, once again suggesting that both labeled and unlabeled eIF4G1011–1104 specifically interact with the same surface on eIF3. By subtracting the ∼15 kDa contributed by eIF4G1011–1104, our data imply that the largest cross-linked band corresponds to either eIF3b or eIF3c; the 80 kDa band corresponds to either eIF3d or eIF3l; and the 65 kDa band corresponds to eIF3e or eIF3f.
The eIF3 Binding Domain in eIF4G Interacts with eIF3c, -d, and -e Subunits
To unambiguously determine which eIF3 subunits specifically cross-link to eIF4G1011–1104-SDA we used a number of commercially available eIF3 subunit specific antibodies (Fig. 3). To establish the identity of the largest cross-linked band, we probed with eIF3b and eIF3c specific antisera. Although the majority of each eIF3 subunit migrates at their expected molecular weight (Fig. 3, A and B, lower band, empty arrow), we detect an appreciable amount of eIF3c migrating at an apparent molecular mass of ∼130 kDa when eIF4G1011–1104-SDA is present (Fig. 3A, lane 1, filled arrow). Since no corresponding change in eIF3b migration is detected, this indicates that eIF4G1011–1104 directly interacts with, or is positioned very close to, eIF3c upon eIF4G-eIF3 complex formation. Repeating this process with eIF3d and eIF3l specific antisera, we find that eIF3d is identified as the major ∼80 kDa cross-linked subunit (Fig. 3, C and D). We also find that a very small, but reproducible, amount of eIF3l is also detected in the ∼80 kDa cross-linked band. Although not as strong as the amount of eIF3d detected, this implies that one or both of eIF3d and eIF3l may be involved in binding eIF4G. Probing with eIF3e and eIF3f specific antiserum unambiguously identifies eIF3e as the ∼60 kDa cross-linked species (Fig. 3, E and F).
FIGURE 3.
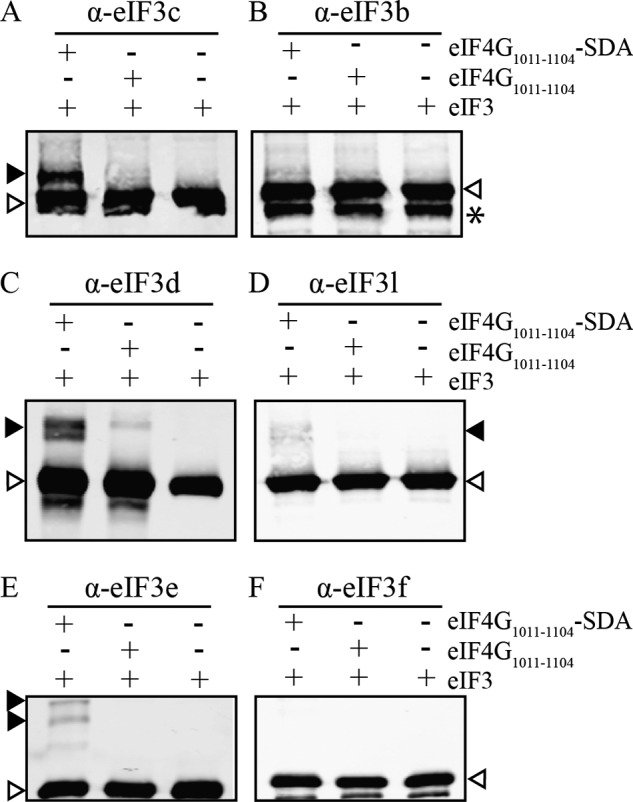
eIF4G1011–1104-SDA cross-links to eIF3 subunits -c, -d, and -e. A–F, eIF4G1011–1104 ± SDA was incubated with eIF3 and exposed to UV light as described under “Experimental Procedures.” Cross-linking reactions are separated by SDS-PAGE followed by immunoblot analysis for specific eIF3 subunits as indicated. Control lanes containing only eIF3 are shown in the third lane in each blot. Filled arrows in panels A, C, D, and E indicate eIF4G1011–1104-SDA cross-linking to subunits eIF3c, -d, -e, and to a lesser extent, -l. Empty arrows denote uncross-linked eIF3 subunits. The asterisk (*) in B denotes a degradation product of eIF3b, which is likely an artifact from the eIF3 purification process.
eIF4G Contains 2 Subdomains that Interact with eIF3c and -d or eIF3e
The diazirene active group of the SDA cross-linker is situated at the end of a 3.9 Å spacer arm. While this is a relatively short distance, it is possible that some of the subunit interactions we observe may be due to the length and flexibility of the cross-linker. To address this, we employed an alternate method to confirm which of the eIF3c, -d, -l, and -e subunits interact with eIF4G. The unnatural amino acid p-benzoylphenylalanine (Bpa) has been used recently to explore multi-factor interactions in vivo and in vitro in bacteria, yeast, and mammalian systems (31–36). Bpa is photoactivatable at 350–360 nm, preferentially reacts with C-H bonds versus water, and can be activated reversibly. If no suitable reaction partner is readily available following activation, Bpa can relax to its ground energy state (37, 38). Importantly, the Bpa cross-linker distance is ∼3 Å and is less flexible than SDA (39) (Fig. 4A). We have employed a recently developed method to site-specifically incorporate Bpa into a recombinant protein using a bacterial expression system (29). This approach employs an orthogonal tRNA/tRNA-synthetase pair for Bpa incorporation into a single amber stop codon mutant site that is engineered into the gene of interest.
FIGURE 4.
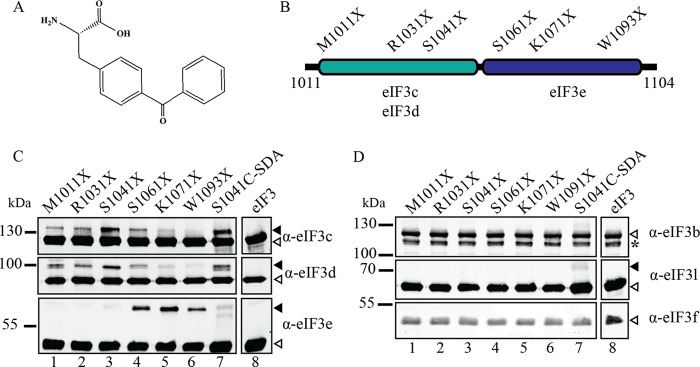
Identification of 2 independent eIF3-binding subdomains in eIF4G. A, chemical structure of the p-benzoyl-phenylalanine cross-linker (Bpa, Bachem) used for genetic incorporation. B, diagram of 4G1011–1104 with sites of amber mutations used for Bpa incorporation. The regions of maximal eIF3 subunit cross-linking from panels C and D are also depicted. C and D, results of eIF3 cross-linking with 4G1011–1104-Bpa labeled at single indicated sites (lanes 1–6), 4G1011–1104-SDA (lane 7), or eIF3 alone (lane 8) are shown. The eIF3 subunit specific antibody used for each immunoblot is indicated to the right of each panel. Filled arrows indicate cross-linked eIF3 subunits, while empty arrows indicate uncross-linked subunits. Asterisk (*) in D indicates an eIF3b degradation product.
We generated 10 unique amber mutant constructs of eIF4G1011–1104 mRNA for Bpa incorporation, as described under “Experimental Procedures.” Six of the ten mutants efficiently express in bacteria and appropriately span the length of the eIF3-binding domain in eIF4G (eIF4G1011–1104; Fig. 4B). Small gaps in the coverage of this span are due to constructs that we were unable to express with Bpa incorporation; these constructs are expressed only as a truncation ending at the site of the amber stop codon as would be expected if Bpa is not incorporated and peptide termination occurs. Cross-linking reactions using eIF4G1011–1104-Bpa are carried out as described for the cross-linking of eIF4G1011–1104-SDA to eIF3. Immunoblot analysis of the cross-linked products unambiguously identifies the close proximity of eIF3 subunits -c, -d, and -e to eIF4G (Fig. 4C, filled arrows). Immunoblots for eIF3b, -l, and -f confirm that these subunits do not directly interact with eIF4G (Fig. 4D). The weak eIF3l cross-linked band observed using eIF4G1011–1104-SDA (Fig. 3D, lane 1, Fig. 4D, lane 7) is not observed when eIF4G1011–1104-Bpa is used (Fig. 4D, middle panel). This strongly implies that the eIF3l subunit is near, but does not directly contribute to the eIF4G binding surface.
By testing several Bpa incorporation sites, we are able to reveal distinct local areas of preferential binding to the different eIF3 subunits. Subunits eIF3c and eIF3d bind to the N-terminal portion of the eIF3-binding region, with peak cross-linking intensity at S1041X (Fig. 4C, top panel, lane 3). Interestingly, S1041 is the same site used for attachment of the SDA cross-linker (Fig. 4C, lane 7) and for the fluorescence polarization assays (Fig. 1E). Subunit eIF3e shows strong cross-linking intensity toward to the C-terminal end of the eIF3 binding region (K1071X; Fig. 4C). It is worth noting that the doublets of cross-linked eIF3d and eIF3e observed with eIF4G1011–1104-SDA (Fig. 3, C and E) are not apparent with eIF4G1011–1104-Bpa. This indicates that the Bpa cross-linker is a more precise tool than the more flexible SDA cross-linker. These experiments confirm the formation of an eIF4G-binding surface consisting of eIF3c, -d, and -e subunits. Furthermore, our data establish distinct regions within eIF4G that preferentially bind each subunit at this interface, likely contributing to the overall stabilization of this interaction.
Both Subdomains in eIF4G Interact with eIF3 for Efficient Ribosome Recruitment
To test the importance of the eIF4G-eIF3 interaction on promoting mRNA recruitment to the ribosome, we have employed a tethered-mRNA assay previously used to identify the functional “core” region of eIF4G in vivo (14). This assay employs an engineered boxB hairpin in the 5′-UTR of an mRNA to specifically recruit a 22 amino acid sequence of the bacteriophage λ transcription anti-terminator protein N (λ N- amino acids 1–22). We have modified this assay for use in a reticulocyte lysate system so that we can precisely test the role of eIF4G truncations in promoting mRNA recruitment to the ribosome (Fig. 5, A and B). This approach avoids the prerequisite of manipulating endogenous eIF4G protein activity using a viral protease, as only λ-eIF4G can be recruited to the uncapped boxB reporter used in this assay (17). Nuclease-treated rabbit reticulocyte lysate is supplemented with 1 μm of a λ-eIF4G truncation together with 0.5 μm boxB reporter mRNA followed by measuring protein synthesis for 30 min at 30 °C. The amount of synthesized luciferase for each λ-eIF4G construct is measured and normalized to lysates supplemented with boxB reporter RNA in the absence of λ-eIF4G (Fig. 5C, Neg). Our data show that a truncation of eIF4G that begins at the 2A protease site and contains both the eIF4A and eIF3 binding domains stimulates translation roughly 35-fold (λ-eIF4G682–1104; Fig. 5C).
FIGURE 5.
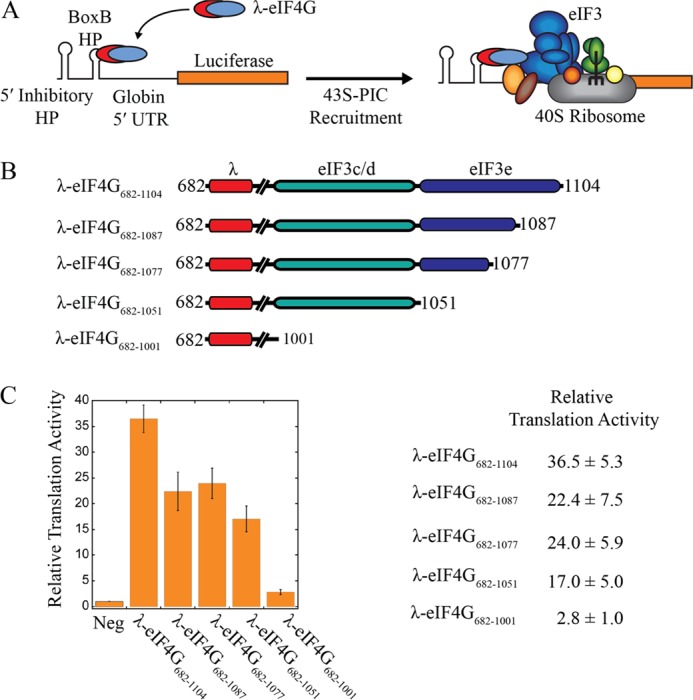
eIF3-binding subdomains in eIF4G both stimulate mRNA recruitment and translation. A, an eIF4G-dependent translation assay is depicted that mediates mRNA recruitment to the ribosome independent of cap binding. A boxB hairpin (BoxB HP) upstream of a beta globin 5′-UTR sequence specifically recruits λ-tagged eIF4G682–1104 to the Renilla luciferase reporter. A stable inhibitor hairpin at the 5′-end is positioned to prevent 5′-end dependent mRNA recruitment to the ribosome. B, diagrams of eIF4G constructs used, which are N-terminally HIS-tagged and fused with the RNA binding domain of bacteriophage λ transcription anti-terminator protein N. Amino acid numbering is shown to indicate each eIF4G truncation construct used. Line breaks in the diagrams represent the RNA and eIF4A binding domains of human eIF4G. C-terminal truncations of the eIF3-binding domain are used to define functional subdomains of this region. A summary of the relative luciferase translation over background (no λ-eIF4G added, Neg) that has been normalized to 1 is shown as the average of 4 trials ± S.E. C, plot of relative luciferase translation following incubation of the boxB Renilla luciferase reporter in nuclease treated reticulocyte lysate for 30 min at 30 °C. Each bar represents the relative luciferase activity in the absence (Neg) or presence of λ-eIF4G truncations, as indicated. Luciferase translation is shown as the average of 4 trials ± S.E.
To establish the relative contributions of the eIF3c, -d, and -e subunits in stimulating translation, we separated the eIF3-binding region into distinct eIF3c/d (eIF4G1011–1051) and eIF3e (eIF4G1052–1104) binding subdomains (Fig. 5B). To specifically determine the contribution of the eIF3e subunit-binding domain, we created three separate truncations that remove 18 amino acids (λ-eIF4G682–1087), 28 amino acids (λ-eIF4G682–1077), or the entire eIF3e-binding subdomain (λ-eIF4G682–1051). Partial or complete deletion of the eIF3e-binding domain retains roughly 50–65% translation activity compared with λ-eIF4G682–1104 (Fig. 5C). Further deletion of the eIF3c/d-binding domain (λ-eIF4G682–1001) completely abolishes luciferase translation, indicating that both eIF3c/d and eIF3e-binding domains are required for efficient mRNA recruitment and translation.
DISCUSSION
Human eIF4G is a scaffold protein that contains binding sites for other initiation factors, including PABP, eIF4E, eIF4A, and eIF3. The importance of individual initiation factor binding sites in eIF4G on mRNA translation has been investigated in vitro and in vivo (17). The relative contribution of the eIF4G-PABP interaction appears to be relatively modest in both yeast and mammalian mRNA translation (17, 40, 41). In contrast, the interactions of eIF4E and eIF4A with eIF4G play a critical role in mRNA recruitment, restructuring and translation (10, 42, 43). More recently, it has become evident that the interaction of eIF4G with RNA is also required for efficient mRNA recruitment and translation initiation (44, 45). The exact contribution of the eIF4G-eIF3 interaction in mRNA recruitment and scanning is less clear. No direct interaction between eIF4G and eIF3 has been identified in yeast, but a direct interaction has been identified in mammals and proposed to play an important role in mRNA recruitment and translation (12). Precise truncation analysis has revealed a ∼90 amino acid region of human eIF4G that constitutes the minimal eIF3-binding domain in the absence of the 40S ribosomal subunit (11, 13).
In this study, we have further characterized the direct interaction between human eIF4G and eIF3. Using a quantitative binding assay, we have established that the affinity between eIF4G and eIF3 is not altered by the presence of eIF4A, at least in the absence of the 40S ribosomal subunit (Fig. 1). This finding is in contrast to previous data using an immunoprecipitation assay that suggested mutually cooperative binding between eIF4A and eIF3 with eIF4G (11). Since it is not possible to determine the affinity between these initiation factors by using an immunoprecipitation assay, our data strongly suggest that eIF4A and eIF3 bind independently to the middle region of human eIF4G. It is worth noting that both of these studies did not include the C-terminal HEAT repeat that is known to provide a second binding site for eIF4A (46). It therefore remains to be determined if the C-terminal domain of eIF4G may in turn regulate eIF3 binding in an eIF4A-dependent manner.
Previous work employed a pull-down assay to determine that eIF4G binds directly to the eIF3e subunit of the eIF3 complex (23). However, this approach is hindered by the requirement to use partially proteolyzed eIF3 to identify eIF4G-interacting subunits. This raises a fundamental question about whether additional eIF3 subunits may help to stabilize the interaction with eIF4G. To address this, we generated two site-specific cross-linking assays to further explore which eIF3 subunits provide a binding surface for eIF4G. Importantly, these cross-linking assays use intact purified eIF3 so that a complete interaction map between these initiation factors can be obtained. Our data unambiguously show that eIF3c, eIF3d, and eIF3e together form a binding surface for eIF4G (Figs. 3 and 4). By using multiple cross-linker positions in the eIF3 binding domain of eIF4G, we are able to generate an interaction model that reveals two distinct binding subdomains in eIF4G that bind to different eIF3 subunits. One subdomain consists of a 40 amino acid region (1011–1051) that binds to eIF3c and eIF3d. An additional subdomain consists of a 52 amino acid region (1052–1104) that binds to eIF3e. Our data imply that the eIF3c and eIF3d-binding sites in eIF4G may overlap, although it should be noted that the resolution of our cross-linking approach is not able to unambiguously determine this. It is also possible that there is a mixture of either eIF3c or eIF3d binding to eIF4G in different cross-linked complexes. A subunit interaction map of human eIF3 previously identified eIF3c, eIF3d and eIF3e as interacting subunits (19). Using this model, our data provide new constraints to show the eIF4G binding interaction with eIF3c, eIF3d and eIF3e in the context of the complete eIF3 subunit interaction map (Fig. 6). It is interesting to note that eIF3l interacts with eIF3e (19, 47). This may account for the small amount of cross-linking observed between eIF4G and eIF3l when using the SDA cross-linker (Fig. 3). Interestingly, it was recently shown using a reconstituted eIF3 complex that the presence of eIF3d was necessary to provide a high affinity interaction with eIF4G (20). These data correspond well with our new interaction model that implicates eIF3d as an important component of the eIF4G binding site in the eIF3 complex.
FIGURE 6.
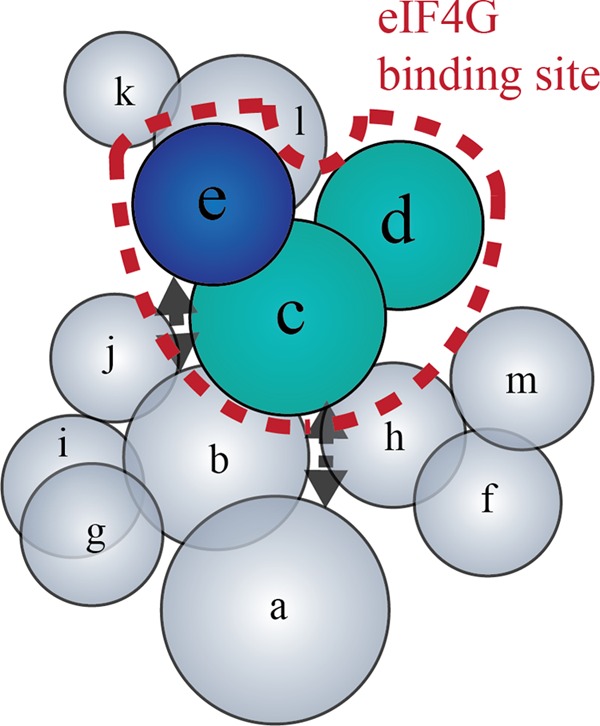
Model of eIF4G interaction sites on eIF3. The human eIF3 subunit interaction map is adapted from Zhou et al. (19). Placement of each subunit and the dotted black arrows reflect known intra-complex interactions between subunits. The dashed line encloses subunits involved in eIF4G binding. eIF3 subunits are colored according to the color schemes used in Figs. 4 and 5, which is based on the subdomain of eIF4G that each eIF3 subunit contacts.
We employed an eIF4G-dependent translation assay to establish the biological importance of our interaction model. Previous work suggested that the eIF3e subunit likely provides the highest affinity interaction with eIF4G (23). Surprisingly, deletion of the eIF3e-binding domain in eIF4G maintains 50–65% translation activity in our assay. Further removal of the eIF3c/d binding domain in eIF4G eliminates this remaining activity. This implies that both of the eIF3 interacting subdomains play important roles in providing the high affinity interaction with eIF3 that is necessary to promote translation initiation. It is important to note that our translation assay directly tethers eIF4G to the mRNA independently of the cap structure. It will therefore be important in the future to determine the exact roles of these two eIF3 interacting subdomains in eIF4G using a cap-dependent translation assay. This will likely require the generation of point mutations in eIF4G that can specifically disrupt individual eIF3 subunit interactions in the context of full-length eIF4G. We anticipate that our interaction model now provides a suitable framework to aid the design of such mutations. In addition, since the interaction is regulated in an mTOR-dependent manner, it is plausible that one or more phosphorylation sites on eIF4G and/or eIF3 may be responsible for altering the affinity between these factors. Our data now suggest that as well as eIF4G, the eIF3c, -d, and -e subunits of the eIF3 complex are possible candidates to be phosphorylated in an mTOR-dependent manner.
Acknowledgments
We thank Enoch Baldwin and the Fraser laboratory for many insightful comments during this study. We gratefully acknowledge David King and Sean Studer for helpful advice regarding cross-linking protocols. We are indebted to Peter Schultz for generously providing the pEVOL plasmid for benzophenone incorporation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM092927 and Training Grant T32 GM007377 from the NIGMS (to N. V.).
- eIF
- eukaryotic initiation factor
- PIC
- pre-initiation complex
- UTR
- untranslated region.
REFERENCES
- 1. Fraser C. S. (2009) The molecular basis of translational control. Prog. Mol. Biol. Transl. Sci. 90, 1–51 [DOI] [PubMed] [Google Scholar]
- 2. Gingras A. C., Raught B., Sonenberg N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913–963 [DOI] [PubMed] [Google Scholar]
- 3. Hinnebusch A. G., Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parsyan A., Svitkin Y., Shahbazian D., Gkogkas C., Lasko P., Merrick W. C., Sonenberg N. (2011) mRNA helicases: the tacticians of translational control. Nat. Rev. Mol. Cell Biol. 12, 235–245 [DOI] [PubMed] [Google Scholar]
- 5. Özes A. R., Feoktistova K., Avanzino B. C., Fraser C. S. (2011) Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 412, 674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. (1990) Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 10, 1134–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rogers G. W., Jr., Richter N. J., Lima W. F., Merrick W. C. (2001) Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276, 30914–30922 [DOI] [PubMed] [Google Scholar]
- 8. Abramson R. D., Dever T. E., Merrick W. C. (1988) Biochemical evidence supporting a mechanism for cap-independent and internal initiation of eukaryotic mRNA. J. Biol. Chem. 263, 6016–6019 [PubMed] [Google Scholar]
- 9. Bi X., Ren J., Goss D. J. (2000) Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry 39, 5758–5765 [DOI] [PubMed] [Google Scholar]
- 10. Feoktistova K., Tuvshintogs E., Do A., Fraser C. S. (2013) Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc. Natl. Acad. Sci. U.S.A. 110, 13339–13344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korneeva N. L., Lamphear B. J., Hennigan F. L., Rhoads R. E. (2000) Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 275, 41369–41376 [DOI] [PubMed] [Google Scholar]
- 12. Lamphear B. J., Kirchweger R., Skern T., Rhoads R. E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270, 21975–21983 [DOI] [PubMed] [Google Scholar]
- 13. Morino S., Imataka H., Svitkin Y. V., Pestova T. V., Sonenberg N. (2000) Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20, 468–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Gregorio E., Preiss T., Hentze M. W. (1999) Translation driven by an eIF4G core domain in vivo. EMBO J. 18, 4865–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris T. E., Chi A., Shabanowitz J., Hunt D. F., Rhoads R. E., Lawrence J. C., Jr. (2006) mTOR-dependent stimulation of the association of eIF4G and eIF3 by insulin. EMBO J. 25, 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thoreen C. C., Chantranupong L., Keys H. R., Wang T., Gray N. S., Sabatini D. M. (2012) A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hinton T. M., Coldwell M. J., Carpenter G. A., Morley S. J., Pain V. M. (2007) Functional analysis of individual binding activities of the scaffold protein eIF4G. J. Biol. Chem. 282, 1695–1708 [DOI] [PubMed] [Google Scholar]
- 18. Damoc E., Fraser C. S., Zhou M., Videler H., Mayeur G. L., Hershey J. W., Doudna J. A., Robinson C. V., Leary J. A. (2007) Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell Proteomics 6, 1135–1146 [DOI] [PubMed] [Google Scholar]
- 19. Zhou M., Sandercock A. M., Fraser C. S., Ridlova G., Stephens E., Schenauer M. R., Yokoi-Fong T., Barsky D., Leary J. A., Hershey J. W., Doudna J. A., Robinson C. V. (2008) Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. U.S.A. 105, 18139–18144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun C., Todorovic A., Querol-Audí J., Bai Y., Villa N., Snyder M., Ashchyan J., Lewis C. S., Hartland A., Gradia S., Fraser C. S., Doudna J. A., Nogales E., Cate J. H. (2011) Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl. Acad. Sci. U.S.A. 108, 20473–20478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Siridechadilok B., Fraser C. S., Hall R. J., Doudna J. A., Nogales E. (2005) Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515 [DOI] [PubMed] [Google Scholar]
- 22. Hashem Y., des Georges A., Dhote V., Langlois R., Liao H. Y., Grassucci R. A., Hellen C. U., Pestova T. V., Frank J. (2013) Structure of the Mammalian Ribosomal 43S Preinitiation Complex Bound to the Scanning Factor DHX29. Cell 153, 1108–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LeFebvre A. K., Korneeva N. L., Trutschl M., Cvek U., Duzan R. D., Bradley C. A., Hershey J. W., Rhoads R. E. (2006) Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 281, 22917–22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Gregorio E., Baron J., Preiss T., Hentze M. W. (2001) Tethered-function analysis reveals that elF4E can recruit ribosomes independent of its binding to the cap structure. RNA 7, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maag D., Lorsch J. R. (2003) Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 330, 917–924 [DOI] [PubMed] [Google Scholar]
- 26. Fraser C. S., Berry K. E., Hershey J. W., Doudna J. A. (2007) eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26, 811–819 [DOI] [PubMed] [Google Scholar]
- 27. Weeks K. M., Crothers D. M. (1992) RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry 31, 10281–10287 [DOI] [PubMed] [Google Scholar]
- 28. Korneeva N. L., Lamphear B. J., Hennigan F. L., Merrick W. C., Rhoads R. E. (2001) Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 276, 2872–2879 [DOI] [PubMed] [Google Scholar]
- 29. Young T. S., Ahmad I., Yin J. A., Schultz P. G. (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 395, 361–374 [DOI] [PubMed] [Google Scholar]
- 30. Chin J. W., Martin A. B., King D. S., Wang L., Schultz P. G. (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu D., Wowor A. J., Cole J. L., Kendall D. A. (2013) Defining the Escherichia coli SecA dimer interface residues through in vivo site-specific photo-cross-linking. J. Bacteriol. 195, 2817–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Abe R., Caaveiro J. M., Kozuka-Hata H., Oyama M., Tsumoto K. (2012) Mapping ultra-weak protein-protein interactions between heme transporters of Staphylococcus aureus. J. Biol. Chem. 287, 16477–16487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ozawa K., Horan N. P., Robinson A., Yagi H., Hill F. R., Jergic S., Xu Z. Q., Loscha K. V., Li N., Tehei M., Oakley A. J., Otting G., Huber T., Dixon N. E. (2013) Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, epsilon, theta, and beta reveals a highly flexible arrangement of the proofreading domain. Nucleic Acids Res. 41, 5354–5367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shiota T., Mabuchi H., Tanaka-Yamano S., Yamano K., Endo T. (2011) In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U.S.A. 108, 15179–15183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grunbeck A., Huber T., Sachdev P., Sakmar T. P. (2011) Mapping the ligand-binding site on a G protein-coupled receptor (GPCR) using genetically encoded photocrosslinkers. Biochemistry 50, 3411–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu C. C., Lin Y. C., Chen H. T. (2011) The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol. Cell. Biol. 31, 2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kauer J. C., Erickson-Viitanen S., Wolfe H. R., Jr., DeGrado W. F. (1986) p-Benzoyl-L-phenylalanine, a new photoreactive amino acid. Photolabeling of calmodulin with a synthetic calmodulin-binding peptide. J. Biol. Chem. 261, 10695–10700 [PubMed] [Google Scholar]
- 38. Dormán G., Prestwich G. D. (1994) Benzophenone photophores in biochemistry. Biochemistry 33, 5661–5673 [DOI] [PubMed] [Google Scholar]
- 39. Farrell I. S., Toroney R., Hazen J. L., Mehl R. A., Chin J. W. (2005) Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods 2, 377–384 [DOI] [PubMed] [Google Scholar]
- 40. Searfoss A., Dever T. E., Wickner R. (2001) Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21, 4900–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park E. H., Walker S. E., Lee J. M., Rothenburg S., Lorsch J. R., Hinnebusch A. G. (2011) Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. EMBO J. 30, 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pause A., Belsham G. J., Gingras A. C., Donzé O., Lin T. A., Lawrence J. C., Jr., Sonenberg N. (1994) Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371, 762–767 [DOI] [PubMed] [Google Scholar]
- 43. Pause A., Méthot N., Svitkin Y., Merrick W. C., Sonenberg N. (1994) Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 13, 1205–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Prévôt D., Decimo D., Herbreteau C. H., Roux F., Garin J., Darlix J. L., Ohlmann T. (2003) Characterization of a novel RNA-binding region of eIF4GI critical for ribosomal scanning. EMBO J. 22, 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yanagiya A., Svitkin Y. V., Shibata S., Mikami S., Imataka H., Sonenberg N. (2009) Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol. Cell. Biol. 29, 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Imataka H., Sonenberg N. (1997) Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 17, 6940–6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morris-Desbois C., Réty S., Ferro M., Garin J., Jalinot P. (2001) The human protein HSPC021 interacts with Int-6 and is associated with eukaryotic translation initiation factor 3. J. Biol. Chem. 276, 45988–45995 [DOI] [PubMed] [Google Scholar]


