Background: TREM2 is a microglial receptor, recently identified as a genetic risk factor for late onset AD.
Results: Sequential proteolytic processing of TREM2 involves ectodomain shedding and intramembranous cleavage by γ-secretase and affects signaling via its adaptor protein DAP12.
Conclusion: γ-Secretase-mediated intramembranous proteolysis modulates TREM2 signaling.
Significance: Inhibition of γ-secretase could impair TREM2 function in neuroinflammation.
Keywords: Alzheimer Disease, Intramembrane Proteolysis, Membrane, Presenilin, Protein Phosphorylation, Secretases, Signaling
Abstract
Triggering receptor expressed on myeloid cells-2 (TREM2) and its signaling adaptor protein TYROBP/DAP12 play important roles in signal transduction in dendritic cells, osteoclasts, tissue macrophages, and microglia. Recently, TREM2 variants have been shown to be linked to late onset Alzheimer disease. Here, we demonstrate that TREM2 undergoes sequential proteolytic processing by ectodomain shedding and intramembrane proteolysis. The C-terminal fragment (CTF) of TREM2 generated by ectodomain shedding is cleaved by γ-secretase. Importantly, pharmacologic and genetic γ-secretase inhibition resulted in accumulation of TREM2 CTF at the plasma membrane that also interacts with the signaling adaptor protein DAP12. Thus, the accumulated TREM2 CTF thereby might limit the interaction of DAP12 with the functional full-length receptor, resulting in decreased DAP12 phosphorylation and impaired metabolism of phosphatidylinositol 4,5-bisphosphate. Together, these data demonstrate γ-secretase-mediated intramembranous proteolysis of TREM2 and functionally link two Alzheimer disease-associated proteins in one signaling pathway.
Introduction
The triggering receptor expressed on myeloid cells-2 (TREM2)2 is a ∼40-kDa type I membrane glycoprotein with a single extracellular immunoglobulin-like domain, one transmembrane domain, and a short cytoplasmatic tail. Although the cytoplasmic domain of TREM2 does not contain an overt amino acid-based signaling motif, its transmembrane domain interacts with the adaptor protein TYROBP/DNAX-activating protein of 12 kDa (DAP12) via electrostatic interaction (1, 2). TREM2 was originally identified on monocyte-derived dendritic cells (3), but was later also detected on several myeloid cell types including osteoclasts, tissue macrophages, and microglia (1). TREM2 is localized predominantly in the Golgi complex, but also shuttles to and from the cell surface in endocytic and exocytic vesicles (4, 5).
DAP12 is a type I transmembrane protein, which acts as a signaling adaptor protein for TREM2 and a number of other cell surface receptors (6). The cytoplasmic domain of DAP12 contains an immunoreceptor tyrosine activation motif (ITAM) (1, 7). After activation of the interacting receptor, DAP12 undergoes phosphorylation at the two conserved ITAM tyrosine residues by Src kinases. Subsequent recruitment and activation of the Syk protein kinase trigger downstream signaling pathways, including the activation of mitogen-activated protein kinase (MAPK) and phospholipase Cγ (PLCγ) (7).
The physiological relevance of TREM2 and DAP12 is demonstrated by loss-of-function mutations of either gene that cause polycystic lipomembranous osteodysplasia with sclerosing leukoencephalopathy (PLOSL or Nasu-Hakola disease) associated with presenile dementia in the fourth decade of life in homozygous carriers (8, 9). Notably, rare variants of TREM2 in the heterozygous state have recently been identified by exome sequencing to increase the risk of late onset Alzheimer disease (LOAD) (10–14) and frontotemporal lobe dementia (15). Although the population frequency of the TREM2 R47H variant in the cohorts studied was much lower compared with the most common genetic risk factor, the apolipoprotein ϵ4 allele, the effects of both factors on the individual risk to develop LOAD were comparable (10, 11). TREM2 was also found to be up-regulated in microglia of amyloid precursor protein transgenic mouse models (10, 16, 17).
Here, we demonstrate that a C-terminal fragment of TREM2 generated by ectodomain shedding represents a novel substrate of the AD-associated γ-secretase. Interestingly, loss of γ-secretase activity leads to accumulation of TREM2 CTFs, impairing phosphorylation of the TREM2 adaptor protein DAP12. The combined data demonstrate a critical role of intramembranous proteolytic processing by γ-secretase in the signaling of TREM2 and thus, provide a functional link between both AD-associated factors.
EXPERIMENTAL PROCEDURES
Chemicals
Unless noted otherwise, all chemicals used for the described experiments were purchased in per analysi grade from Sigma-Aldrich, Roche Applied Science, Carl Roth (Karlsruhe, Germany), or Applichem (Darmstadt, Germany). Primers for cloning were obtained from Sigma-Aldrich, and restriction enzymes were from Fermentas (St. Leon-Rot, Germany). The radiochemicals were from Hartmann Analytic (Braunschweig, Germany). The following antibodies were used: anti-HA and anti-β-actin (Sigma-Aldrich), 9E10 against c-myc (Developmental Studies Hybridoma Bank, Iowa City, IA), anti-FLAG (Sigma-Aldrich), anti-GFP (Roche Applied Science), anti-mouse IgG and anti-rabbit IgG antibody conjugated with horseradish peroxidase (Sigma-Aldrich), anti-mouse IgG and anti-rabbit IgG antibody conjugated with Alexa Fluor 488 or Alexa Fluor 546 (Molecular Probes).
cDNA Constructs
For cloning of the different TREM2 and DAP12 constructs, specific primers containing HindIII, XhoI, or SfiI restriction sites were designed and used to amplify the corresponding DNA by PCR. For cloning of the phosphosite mutants of DAP12, internal primers were designed containing base exchanges generating base triplets for Phe at amino acid positions 92 and 103. The sequences of all primers are available upon request. Amplified DNA fragments were digested with HindIII, XhoI, or SfiI and ligated into pSecTag Hygro B vector using T4 ligase. Chemically competent Escherichia coli DH5α were transformed with plasmid DNAs and spread on LB agar supplemented with antibiotics. Single colonies were used to inoculate 2 ml of LB medium supplemented with antibiotics and grown overnight at 37 °C. After plasmid preparation, the sequence of the cloned DNA was checked by sequencing. The PIP2-sensor GFP-C1-PLCδ-PH was a gift from Tobias Meyer (Addgene plasmid 21179).
Cell Culture and Transfection
Both HEK293 and COS7 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) Glutamax containing 4.5 g/liter d-glucose (Invitrogen) supplemented with 10% fetal calf serum (FCS; PAN-Biotech, Aidenbach, Germany) and 1% penicillin/streptomycin solution (50 units/ml penicillin, 50 μg/ml streptomycin; Invitrogen) at 37 °C, 95% humidity, and 5% CO2. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. TREM2 was always co-transfected with DAP12 to allow efficient subcellular transport (2).
Immunocytochemistry
Cells, grown on poly-l-lysine-coated coverslips, were washed with PBS (140 mm NaCl, 10 mm Na2HPO4, 1.75 mm KH2PO4 in dH2O, pH 7.4) and fixed in 4% paraformaldehyde in PBS for 10 min. Cells were then washed three times in PBS and permeabilized with 0.25% Triton X-100 in PBS. Consecutively, cells were blocked with 10% BSA, 0.25% Triton X-100 in PBS and incubated with primary and secondary antibodies in 0.125% Triton X-100 in PBS supplemented with 5% BSA. Finally, cells were embedded on a microscope slide with ImmuMount (Thermo Scientific). To selectively detect cell surface proteins, cells were stained prior to fixation. For this purpose, cells were kept on ice during the whole staining procedure. For blocking, 10% BSA in DMEM was used. Primary and secondary antibodies were diluted in DMEM with 5% BSA. All washing steps were conducted with DMEM, and cells were finally fixed in 4% PFA in PBS. Cells were analyzed by fluorescence microscopy (AxioVert 200; Zeiss) equipped with a plan-Apochromat 63×/0.75 objective and an Axiocam MRm camera). Images were acquired and processed using AxioVision 4.8 software (Zeiss).
Protein Extraction
For total protein extraction, cells were rinsed in 0.8 ml of STEN lysis buffer (50 mm Tris, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 1% Triton X-100 in dH2O, pH 7.4) for 15 min on ice. The homogenates were cleared by centrifugation for 15 min at 16,000 rcf and 4 °C. For isolation of cellular membranes, cells were incubated in hypotonic buffer (10 mm Tris, 1 mm EDTA, 1 mm EGTA, dH2O, pH 7.6) for 15 min on ice, followed by passing the suspension through a needle 15 times. Cell debris and nuclei were pelleted by centrifugation (10 min at 300 rcf and 4 °C), and the supernatant was further centrifuged at 16,000 rcf for 60 min to obtain a membrane pellet. The pellet was then lysed in STEN lysis buffer for 15 min on ice.
In Vitro γ-Secretase Activity Assay
In vitro γ-secretase activity assays were performed as described previously (18). Briefly, isolated cellular membranes were resuspended in 150 mm citric buffer, pH 6.4. The reaction mixture was incubated for 2 h at 37 °C in the absence or presence of 10 μm DAPT and then centrifuged (16,000 rcf) for 1 h at 4 °C. Pellets and supernatants were separated by SDS-PAGE and proteins detected by Western immunoblotting.
Precipitation of Soluble Proteins with Trichloroacetic Acid (TCA)
Cell culture supernatants were collected and cleared from cellular debris by centrifugation for 10 min at 300 rcf. Sodium desoxycholic acid was added to a final concentration of 0.02% and incubated for 15 min. TCA was then added to a final concentration of 10%, and the mixtures were incubated for 1 h on ice. Precipitated proteins were collected by centrifugation for 15 min at 16,000 rcf and 4 °C and washed twice with ice-cold acetone. The washed pellets were air-dried, resuspended in 35 μl of Tris-SDS buffer (50 mm Tris, 1% SDS in dH2O), and incubated for 10 min at 50 °C. Finally, SDS-sample buffer was added, and samples were subjected to separation by SDS-PAGE.
Biotinylation of Cell Surface Proteins
Cells were cultured on poly-l-lysine-coated dishes. After two washes with ice-cold PBS, cells were incubated with 2 ml of biotin solution (0.5 mg/ml sulfo-NHS-biotin (Thermo Scientific) in PBS) for 30 min on ice under constant gentle shaking. Cells were then washed three times with 20 mm glycine in PBS; the last washing solution was kept on cells for 15 min. Cells were then lysed in 900 μl of STEN lysis buffer for 15 min on ice. After centrifugation for 10 min at 16,000 rcf and 4 °C, 50 μl of streptavidin-agarose (Invitrogen) was added to the lysates, and the mixture was incubated overnight at 4 °C on an overhead shaker. Finally, the streptavidin-agarose was washed four times for 10 min each with STEN buffer (50 mm Tris, 150 mm NaCl, 2 mm EDTA, 0.2% Nonidet P-40 in dH2O, pH 7.6), pelleted by centrifugation for 3 min at 600 rcf and 4 °C and resuspended in SDS-sample buffer and separated by SDS-PAGE.
Radiolabeling with 32Pi upon Activation of TREM2
Cells were grown on poly-l-lysine-coated dishes and starved in phosphate-free DMEM (Invitrogen) for 1 h. After incubation for 1 h in labeling medium (phosphate-free DMEM supplemented with 0.5 mCi/dish 32Pi) at 37 °C, 5% CO2, TREM2 was activated by 10 μg/ml anti-myc antibody. 200 μm orthovanadate was used to stabilize the DAP12 phosphorylation. The cells were then incubated for another hour, washed once with PBS, and washed in 1 ml of STEN lysis buffer. DAP12 was then precipitated from the cleared lysates with anti-HA antibody coupled to protein G-Sepharose beads (Invitrogen). The beads were pelleted by centrifugation for 2 min at 9300 rcf and 4 °C before they were washed once with STEN-NaCl buffer (50 mm Tris, 500 mm NaCl, 2 mm EDTA, 0.2% Nonidet P-40 in dH2O, pH 7.6) and twice with STEN buffer. Finally, beads were resuspended in 20 μl of SDS-sample buffer.
Analysis of PIP2
COS7 cells were plated on Ibidi μ-dishes and were transfected with the PIP2 sensor, TREM2, and DAP12. Cells were then incubated for 24 h in presence or absence of 10 μm DAPT. The fluorescence intensity of the PIP2 sensor was recorded every 10 min for 20 min in a time lapse setup. After adding c-myc antibody (10 μg/ml) to activate TREM2, fluorescence intensity was further recorded every 10 min for 100 min. For quantification, the average fluorescence intensity before antibody application was set as F0, and ΔF/F0 was calculated for every time point.
Densitometric Quantification and Statistical Analysis
Protein signals were quantified by densitometric analysis using Quantity One® software (Bio-Rad). Statistical analyses were carried out by a two-sided Student's t test (t test). For comparison of more than two individual groups, one-way analysis of variance with a Newman-Keuls ad hoc test was used. If not indicated otherwise, the diagrams show mean values ± corresponding S.E. of three independent experiments. Significance values are indicated by asterisks: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
Proteolytic Shedding of the TREM2 Ectodomain
TREM2 has been identified as a novel strong risk factor for LOAD, but it is unknown how TREM2 contributes to the disease (10, 11). To characterize the cellular metabolism of TREM2, we first analyzed whether this receptor undergoes proteolytic processing that might regulate its expression levels at the cell surface. Because the endogenous ligand of TREM2 is unknown and available antibodies do not reliably detect endogenously expressed TREM2, we first generated a TREM2 variant that carries a myc tag at the ectodomain, and an additional GFP-tag at the intracellular C terminus (myc-TREM2-GFP; Fig. 1A). To enable efficient transport of TREM2 to the cell surface, this TREM2 variant was co-expressed along with its adaptor molecule DAP12, which was tagged with a hemagglutinin epitope (DAP12-HA). As members of the matrix metalloprotease (MMP) family or the A Disintegrin And Metalloprotease (ADAM) family are major proteases involved in the cleavage of cell surface proteins, COS7 cells were incubated in absence or presence of batimastat, a well known inhibitor of proteases of both families (19, 20). In control cells, only low levels of cell surface-localized TREM2 were detected, although GFP detection indicated robust cellular expression (Fig. 1A). Interestingly, treatment with batimastat resulted in a ∼3-fold increase of cell surface-localized TREM2 (313.4 ± 18.8% versus 100 ± 10.7%; Fig. 1A).
FIGURE 1.
Shedding of the TREM2 ectodomain. A, COS7 cells were co-transfected with DAP12-HA and myc-TREM2-GFP (scheme). After 24 h, cells were treated with 10 μm batimastat or dimethyl sulfoxide as solvent control for 1 h. Surface localization of TREM2 was visualized by surface staining using anti-myc primary and Alexa Fluor 594-coupled secondary antibodies (see “Experimental Procedures”). Scale bars represent 20 μm. Bar chart shows quantification of TREM2 signals in randomly chosen areas of 75 × 75 pixels (Ctrl, n = 25; batimastat, n = 40 areas). B, HEK293 cells co-transfected with FLAG-DAP12-HA and myc-TREM2-GFP were incubated in the presence or absence of batimastat (10 μm) for 24 h in serum-free medium. Cellular membranes were isolated and conditioned medium precipitated with TCA. Proteins were separated by SDS-PAGE, and TREM2 was detected by Western immunoblotting with anti-GFP (membrane) or anti-myc antibodies (medium). The different bands detected for TREM2 in membrane fractions and for the secreted TREM2 ECD in medium might represent different glycosylation state variants. Bar charts show the quantification of TREM2 ECD/FL ratios (left panel) and TREM2 CTF/FL ratios (right panel) (n = 3). Statistical analysis was done by two-tailed t test. Error bars, S.E. **, p < 0.01; ***, p < 0.001.
Western immunoblotting further confirmed the inhibition of TREM2 ectodomain shedding by batimastat, as the treatment increased the levels of full-length TREM2 (TREM2 FL) in the membrane fraction and decreased the amount of the secreted TREM2 ectodomain variants (TREM2 ECD; 14.6 ± 2.4% versus 100 ± 14.6%; Fig. 1B). Because TREM2 contains putative glycosylation sites in its ectodomain, the different bands for the full-length receptor in the membrane and the extracellular fragment in the culture supernatant might represent different glycosylation variants. We also detected a lower molecular mass species of TREM2 of about 48 kDa, likely representing a C-terminal fragment of TREM2 tagged with GFP (TREM2 CTF-GFP; Fig. 1B). Consistent with decreased ectodomain shedding, levels of the TREM2 CTF were decreased upon incubation with batimastat (17.2 ± 0.5% versus 100 ± 10.6%; Fig. 1B). These combined results indicate that TREM2 undergoes ectodomain shedding by a protease of the ADAM or MMP family.
Intramembranous Cleavage of the TREM2 C-terminal Fragment by γ-Secretase
Having established the ectodomain shedding of TREM2, we next tested whether the resulting membrane-tethered TREM2 CTF represents a substrate for γ-secretase, a major protease complex involved in intramembranous cleavage of type I membrane proteins after precedent removal of globular ectodomains. To enable the specific detection of proteolytic fragments, we transfected HEK293 cells with a TREM2 FL variant carrying a FLAG- and myc/His epitope at its N and C terminus, respectively (Fig. 2A). Western immunoblotting with anti-c-myc antibodies revealed the expression of ∼35-kDa and ∼17-kDa TREM2 variants, consistent with the expected molecular masses of full-length TREM2 and its membrane-bound CTF, respectively (without a GFP tag, compare with Fig. 1). Interestingly, the pharmacological inhibition of γ-secretase with DAPT led to a significant increase of TREM2 CTFs (239.6 ± 10.8% versus 100 ± 18.2%; Fig. 2A). To further confirm these findings, the effect of genetic γ-secretase inhibition on TREM2 CTF accumulation was investigated. HEK293 cells stably overexpressing wild type (WT) or a dominant negative (DN) variant of presenilin 1 (PS1) were co-transfected with cDNAs encoding TREM2 and DAP12 (Fig. 2B). Consistent with the pharmacological γ-secretase inhibition, the expression of the PS1 DN mutant also led to significant TREM2 CTF accumulation compared with PS1 WT-expressing cells (184.9 ± 0.5% versus 100 ± 6%).
FIGURE 2.
Cleavage of TREM2 C-terminal fragments by γ-secretase. A–C, native HEK293 cells (A) or HEK293 cells stably overexpressing PS1 WT or PS1 DN variants (B and C) were co-transfected with FLAG-DAP12-HA and FLAG-TREM2-myc/His (A and B) or with FLAG-TREM2ΔECD-myc/His (C) and cultured for 24 h. Membrane proteins were separated by SDS-PAGE, and TREM2 variants were detected by Western immunoblotting with anti-myc antibody. Bar charts show the quantification of TREM2 CTF/FL ratios (A and B; n = 3) or relative levels of TREM2 CTFs (C; n = 2). Statistical determination was done by a two-tailed t test (A) (n = 3) or one-way analysis of variance ANOVA (B) ( n = 3). D, in vitro γ-secretase activity assay. Membranes of HEK293 cells expressing myc-TREM2-GFP were isolated and incubated for 2 h in the absence or presence of 10 μm DAPT. Reaction mixtures were separated by centrifugation into membrane (Membr.) and supernatant (Sup.) fractions. TREM2 CTF-GFP in the membrane fraction and soluble TREM2 ICD-GFP in the supernatant was detected by Western immunoblotting with anti-GFP antibodies. The soluble TREM2 ICD-GFP was strongly reduced after incubation with DAPT. Bar chart shows the quantification of TREM2 ICD/CTF ratios (n = 2). Error bars, S.E. **, p < 0.01; ***, p < 0.001.
It has been shown that γ-secretase selectively processes C-terminal stubs of proteins with short (10–15 amino acids) ectodomains (21, 22). Thus, we next generated a TREM2 variant lacking the TREM2 ectodomain (FLAG-TREM2ΔECD-myc/His). Expression of this construct revealed a marked accumulation in HEK293 PS1 DN compared with HEK293 PS1 WT cells (258.8 ± 43.5% versus 100 ± 16.7%; Fig. 2C). To further prove processing of TREM2 by γ-secretase, we performed in vitro γ-secretase cleavage assays using the myc-TREM2-GFP construct (Fig. 2D). By Western immunoblotting we detected the TREM2 CTF fused to GFP (∼48 kDa) in the pellet fraction containing cellular membranes. Notably, in the supernatant fraction that should contain soluble products released from the membrane upon γ-secretase cleavage, we detected a TREM2 band of ∼35 kDa, fitting well with the putative size of the TREM2 ICD fused to GFP. This band was strongly decreased in presence of DAPT (Fig. 2D). Together, these data demonstrate that the C-terminal fragment of TREM2 generated by ectodomain shedding represents a substrate for γ-secretase.
Accumulation of TREM2 CTFs at the Cell Surface upon Inhibition of γ-Secretase
γ-Secretase cleaves its substrates mainly at the cell surface or intracellular endocytic compartments (23–25). To specifically test whether inhibition of γ-secretase results in accumulation of TREM2 CTF at the cell surface, we first carried out cell surface biotinylation experiments upon cell incubation in absence or presence of the γ-secretase inhibitor DAPT (Fig. 3A). The pulldown of biotinylated proteins and subsequent detection of TREM2 FL or its CTF demonstrated a strong accumulation of the TREM2 CTF and a less pronounced increase of TREM2 FL at the cell surface upon γ-secretase inhibition (Fig. 3A). To further confirm the specific accumulation of TREM2 CTF at the cell surface, COS7 cells were co-transfected with FLAG-tagged TREM2ΔECD-myc/His and DAP12-HA and incubated in absence or presence of DAPT for 24 h. The specific immunocytochemical detection of the TREM2 CTF at the plasma membrane via its extracellular FLAG epitope revealed a punctuated pattern in the control cells, typical for cell surface-localized proteins (Fig. 3B). Cell treatment with DAPT increased the surface levels of TREM2 CTFs (319.2 ± 40.8% versus 100 ± 10.32%). Together with the biochemical experiments (Fig. 3A), these data demonstrate a selective accumulation of TREM2 CTF at the cell surface upon γ-secretase inhibition.
FIGURE 3.
TREM2 CTFs accumulate at the cell surface after γ-secretase inhibition. A, HEK293 cells were transfected with FLAG-TREM2-myc/His in combination with FLAG-DAP12-HA and incubated for 24 h in absence or presence of 10 μm DAPT. Surface proteins were labeled with sulfo-NHS-biotin for 30 min. Cells were lysed and biotin-labeled proteins precipitated with streptavidin-coated agarose beads. Aliquots of cell lysates and streptavidin-precipitates were separated by SDS-PAGE, and TREM2 was detected by Western immunoblotting with anti-myc antibodies. Treatment with DAPT increases the cell surface expression of TREM2. Bar graphs show the quantification of TREM2 CTF/FL ratios (n = 3). B, COS7 cells were co-transfected with DAP12-HA and FLAG-TREM2ΔECD-myc/His and incubated for 24 h in presence or absence of 10 μm DAPT. TREM2 CTF was visualized at the cell surface of living cells by anti-FLAG primary and Alexa Fluor 488-coupled secondary antibodies. Bar graph shows the quantification of TREM2 CTF-positive structures in five randomly chosen areas of 75 × 75 pixels in 10 cells each. Insets show enlarged images of boxed areas. Scale bars represent 20 μm. Statistical analyses were done by using a two-tailed t test. Error bars, S.E. *, p < 0.05; ***, p < 0.001.
Inhibition of γ-Secretase Impairs the Interaction of TREM2 with Its Signaling Adaptor Protein DAP12
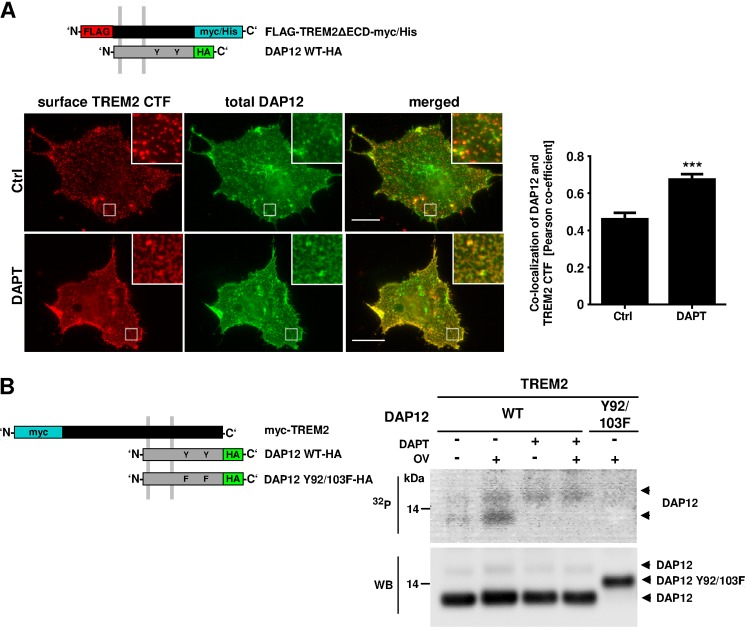
TREM2 interacts with its co-receptor DAP12 via charged amino acid residues in their transmembrane domains (2). This electrostatic interaction is critical for the signaling of activated TREM2 to its adaptor protein DAP12. Thus, we next tested whether γ-secretase inhibition affects the interaction of TREM2 with DAP12. To allow specific detection of cell surface-localized complexes of TREM2 CTF and DAP12, we co-expressed the TREM2ΔECD variant with a FLAG tag in the ectodomain and the DAP12-HA-tagged variant (Fig. 4A). After DAPT treatment, living cells were incubated with anti-FLAG antibodies on ice, fixed, and permeabilized. DAP12 localization was visualized with a specific antibody against the HA tag. TREM2 CTF at the cell surface was localized mainly in punctae in untreated control cells (Fig. 4A). Upon γ-secretase inhibition, the co-localization between TREM2 CTF and DAP12 increased and changed from punctuated pattern to larger, spiry structures that covered substantial areas of the plasma membrane (0.671 ± 0.0029 versus 0.458 ± 0.0034; Fig. 4A). Thus, accumulated TREM2 CTFs upon γ-secretase inhibition induced a redistribution of its interaction partner DAP12.
FIGURE 4.
Inhibition of γ-secretase alters the interaction of TREM2 with DAP12 and impairs DAP12 phosphorylation. A, COS7 cells expressing DAP12-HA and FLAG-TREM2ΔECD-myc/His (scheme) were incubated in the presence or absence of 10 μm DAPT for 24 h. Surface TREM2 CTF was visualized using anti-FLAG primary and Alexa Fluor 594-coupled secondary antibodies. After the specific cell surface staining, cells were fixed and permeabilized, and total DAP12 was stained using anti-HA primary and Alexa Fluor 488-coupled secondary antibodies. Co-localization of DAP12 and TREM2 CTF was analyzed by the Pearson co-efficient in four randomly chosen areas of four different cells. Insets show enlarged images of boxed areas. Scale bars represent 20 μm. Statistical analysis was done by two-tailed t test (p < 0.001). Error bars, S.E. ***, p < 0.001. B, HEK293 cells co-expressing myc-TREM2 and DAP12-HA were incubated in presence or absence of 10 μm DAPT for 24 h. The DAP12 Y92F/Y103F-HA variant lacking potential phosphorylation sites in the ITAM domain was co-transfected with TREM2 as a control (last lane). Cells were starved for 30 min in phosphate-free medium and then incubated with 32Pi for 1 h. TREM2 was activated by incubation with 10 μg/ml anti-myc antibody. After further incubation for 1 h in the presence or absence of 200 μm orthovanadate (OV), cells were lysed, and DAP12 was precipitated with anti-HA antibodies. Proteins were separated on a Tris-Tricine gel and transferred onto a nitrocellulose membrane. Radiolabeled proteins were visualized by autoradiography (upper panel, 32P). After exposure, DAP12-HA was detected on the same membrane by Western immunoblotting with anti-HA antibodies (lower panel, WB).
The ligand-induced activation of DAP12-interacting full-length receptors results in phosphorylation of DAP12 within its ITAM domain by Src kinase. Therefore, the phosphorylation of DAP12 upon activation of TREM2 was analyzed with or without γ-secretase inhibition by in vivo radiolabeling with 32Pi. After cell lysis, DAP12 was immunoprecipitated and phosphate incorporation analyzed by autoradiography. In control cells, two weakly phosphorylated variants of DAP12 were detected (Fig. 4B). 32P radiolabeling strongly increased after the inhibition of tyrosine phosphatases by orthovanadate, indicating efficient phosphorylation and rapid dephosphorylation of DAP12 upon activation of TREM2 (Fig. 4B). A variant of DAP12 with mutated phosphorylation sites (DAP12 Y92F/Y103F) did not undergo phosphorylation, demonstrating the specific labeling of these tyrosine residues. Interestingly, the inhibition of γ-secretase led to a complete suppression of phosphate incorporation into the lower migrating variant of DAP12, even in the presence of orthovanadate (Fig. 4B), demonstrating impaired signaling of TREM2 to its adaptor protein DAP12.
Accumulation of the TREM2 C-terminal Fragment Impairs Phosphatidylinositol Metabolism
DAP12 has been previously linked to the activation of PLCγ, which hydrolyzes PIP2 to inositol 1,4,5-trisphosphate and diacylglycerol (7, 26). Thus, impairment of DAP12 signaling by γ-secretase inhibition might decrease PLCγ activity thereby increasing PIP2 levels in the cell. To test this hypothesis, we analyzed changes in PIP2 levels in a time lapse experiment after activation of TREM2-DAP12 signaling by cross-linking TREM2 with anti-myc antibodies. To visualize PIP2 in this experimental setup, we took advantage of a previously described PIP2 sensor, consisting of the pleckstrin homology domain of PLCδ fused to GFP (27, 28). COS7 cells were co-transfected with this PIP2 sensor, myc-TREM2-mCherry and DAP12-HA and incubated for 24 h in the presence or absence of DAPT. In response to TREM2-DAP12 activation by antibody cross-linking, the fluorescence intensity remained constant in DAPT-treated cells, with only a small decrease due to photobleaching (Fig. 5A, blue line). In contrast, the fluorescence intensity in cells incubated without DAPT steadily decreased after application of antibody (Fig. 5A, red line), indicating a reduction of PIP2 levels. Cells expressing the PIP2 sensor alone did not respond to antibody treatment (Fig. 5A, gray line), indicating specific signal transduction via TREM2. Together, these results indicate that the inhibition of γ-secretase interferes with TREM2-dependent regulation of PIP2 levels at the plasma membrane.
FIGURE 5.
Impaired DAP12 signaling upon γ-secretase inhibition affects PIP2 metabolism. A, COS7 cells expressing the PIP2 sensor PH-PLCδ-GFP in combination with myc-TREM2-mCherry and DAP12-HA (scheme) were incubated for 24 h in presence or absence of 10 μm DAPT. Fluorescence of the PIP2 sensor was measured every 10 min for 120 min. c-myc antibodies (10 μg/ml) were applied after 20 min (dashed line). The graph shows the quantification of fluorescence intensities plotted as ΔF/F0 against time for cells incubated with (blue line) or without DAPT (red line). The fluorescence in cells expressing the PIP2 sensor alone served as control (gray line). Statistical determination was done by two-way analysis of variance (+DAPT, n = 4; −DAPT, n = 6). Error bars, S.E. **, p < 0.01; ***, p < 0.001. ns, not significant. B, schematic drawing shows the proposed role of TREM2 processing in membrane-proximal signaling with either active (upper panel) or inhibited γ-secretase (lower panel). In absence of ligand (upper panel, left), the ectodomain of membrane-associated TREM2 FL is cleaved by a sheddase. The resultant TREM2 CTF is then cleaved by γ-secretase. In the presence of ligand, TREM2 signals to DAP12 (upper panel, right). Upon inhibition of γ-secretase, TREM2 CTFs accumulate after ectodomain shedding (lower panel, left). Accumulated CTFs trap DAP12 co-receptors, thereby preventing the interaction with full-length TREM2 receptors (lower panel, right; see “Discussion” for details). L, ligand; γ, γ-secretase; P, phosphate.
DISCUSSION
Here, we show that TREM2 represents a novel substrate for γ-secretase and thereby unraveled a functional connection of two important factors involved in the pathogenesis of AD. The γ-secretase complex contains presenilin-1 or -2 as the catalytically active component (29–32). Importantly, mutations in the presenilins are the most common cause of familial early onset AD and have been linked to frontotemporal lobe dementia as well (33). γ-Secretase cleaves the amyloid precursor protein and thus, plays a direct role in amyloid-β generation (34, 35). Besides amyloid precursor protein, many additional protein substrates of γ-secretase have been identified, but their relation and potential functional connections to AD pathogenesis remain largely unknown.
The present data demonstrate that the inhibition of γ-secretase leads to a strong accumulation of the TREM2 CTF at the cell surface and trapping of its adaptor TYROBP/DAP12, which then impairs the signaling function of full-length TREM2. We also show that the full-length TREM2 undergoes ectodomain shedding before γ-secretase cleavage and thus represents a canonical substrate for regulated intramembrane proteolysis.
The initial cleavage of TREM2 by a shedding enzyme results in the liberation of a globular ectodomain which is a prerequisite for further intramembrane cleavage by γ-secretase. Although the responsible sheddase remains to be determined, the efficient inhibition of ectodomain shedding by batimastat suggests the involvement of metalloproteases of the MMP and/or ADAM family. Both classes of proteases are highly expressed in microglia cells (36) and can cleave several important receptors and cytokines (37).
Soluble variants of TREM2 and its homolog TREM1 have been detected in human cerebrospinal fluid (CSF) (38) or in blood of septic shock patients (39, 40). Although our data clearly support a release of soluble TREM2 by proteolytic shedding, additional mechanisms have also been proposed, including alternative splicing of the TREM2 mRNA that could lead to shortened variants without the transmembrane domains (41). However, as metalloprotease inhibitors also decreased the levels of soluble TREM1 variants (39, 42), proteolytic shedding appears to be a general mechanism for the release of both TREM receptors. After shedding of the ectodomain, the remaining TREM2 CTF is further degraded by γ-secretase (Fig. 5B). Interestingly, presenilin has been previously shown to be involved in microglial phagocytosis and migration, but the molecular mechanisms remain to be identified (43, 44). Thus, it will be important to investigate further whether these observed effects also involve misprocessing of TREM2.
Our data demonstrate that decreased processing of TREM2 CTFs by γ-secretase indeed interferes with TREM2-dependent signaling. The findings also indicate that the accumulation of TREM2 CTFs traps the adaptor DAP12 at the cell membrane, thereby decreasing its availability to interact with functional full-length TREM2 (see Fig. 5B). It is important to note that the TREM2 CTF generated by ectodomain shedding still contains the motif for the electrostatic interaction with DAP12. Interestingly, the phosphorylation of DAP12 upon activation of the full-length receptor was strongly decreased in cells with accumulated TREM2 CTF upon γ-secretase inhibition. Moreover, upon stimulation of TREM2, PIP2 metabolism was impaired by inhibition of γ-secretase. Alterations in phosphoinositide levels have been previously observed in cell lines expressing presenilin-1 FAD-associated mutations, but the molecular mechanisms were not identified (45, 46). Because DAP12 can negatively regulate phosphoinositol 3-kinase (PI3K) (47–49) and activate PLCγ (7, 26), our data suggest that impaired activation of DAP12 upon inhibition of γ-secretase could either result in decreased suppression of PI3K or decreased activity of the PLCγ and thus, elevation of PIP2 levels at the plasma membrane.
It is interesting to note that the recently identified rare variants of TREM2, which significantly increase the risk of AD, might result in a loss of function, either by introduction of a stop codon (p.Q33X variant) or by affecting ligand binding (p.R47H variant) (10, 11). Thus, these mutations might also impair signaling to DAP12.
Together, the data from our study demonstrate that TREM2, a novel risk factor for LOAD, and the presenilins, the most common factor for familial early onset AD, are both involved in the same signaling pathway. Thus, it will be interesting to further investigate the role of TREM2 and γ-secretase in microglia and neuroinflammatory processes associated with AD and frontotemporal lobe dementia.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB695 and KFO177 to J. W.; FOR1336, SFB704, and KFO177 to H. N.) and by the Hertie-Foundation (to H. N.).
- TREM2
- triggering receptor expressed on myeloid cells-2
- AD
- Alzheimer disease
- ADAM
- A Disintegrin And Metalloprotease
- CTF
- C-terminal fragment
- ICD
- intracellular domain
- DN
- dominant negative
- ECD
- extracellular domain
- FL
- full-length
- ITAM
- immunoreceptor tyrosine activation motif
- LOAD
- late onset Alzheimer disease
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PLC
- phospholipase C
- PS1
- presenilin 1
- rcf
- relative centrifugal force
- Tricine
- 2-[[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]amino]acetic acid.
REFERENCES
- 1. Colonna M. (2003) TREMs in the immune system and beyond. Nat. Rev. Immunol. 3, 445–453 [DOI] [PubMed] [Google Scholar]
- 2. Bouchon A., Dietrich J., Colonna M. (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J. Immunol. 164, 4991–4995 [DOI] [PubMed] [Google Scholar]
- 3. Bouchon A., Hernández-Munain C., Cella M., Colonna M. (2001) A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J. Exp. Med. 194, 1111–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sessa G., Podini P., Mariani M., Meroni A., Spreafico R., Sinigaglia F., Colonna M., Panina P., Meldolesi J. (2004) Distribution and signalling of TREM2/DAP12, the receptor system mutated in the human PLOSL dementia. Eur. J. Neurosci. 20, 2617–2628 [DOI] [PubMed] [Google Scholar]
- 5. Prada I., Ongania G. N., Buonsanti C., Panina-Bordignon P., Meldolesi J. (2006) Triggering receptor expressed in myeloid cells 2 (TREM2) trafficking in microglial cells: continuous shuttling to and from the plasma membrane regulated by cell stimulation. Neuroscience 140, 1139–1148 [DOI] [PubMed] [Google Scholar]
- 6. Lanier L. L., Corliss B. C., Wu J., Leong C., Phillips J. H. (1998) Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391, 703–707 [DOI] [PubMed] [Google Scholar]
- 7. Lowell C. A. (2011) Src family and Syk kinases in activating and inhibitory pathways in innate immune cells: signaling cross-talk. Cold Spring Harb. Perspect. Biol. 3, a002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paloneva J., Kestilä M., Wu J., Salminen A., Böhling T., Ruotsalainen V., Hakola P., Bakker A. B., Phillips J. H., Pekkarinen P., Lanier L. L., Timonen T., Peltonen L. (2000) Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat. Genet. 25, 357–361 [DOI] [PubMed] [Google Scholar]
- 9. Paloneva J., Manninen T., Christman G., Hovanes K., Mandelin J., Adolfsson R., Bianchin M., Bird T., Miranda R., Salmaggi A., Tranebjaerg L., Konttinen Y., Peltonen L. (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am. J. Hum. Genet. 71, 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guerreiro R., Wojtas A., Bras J., Carrasquillo M., Rogaeva E., Majounie E., Cruchaga C., Sassi C., Kauwe J. S., Younkin S., Hazrati L., Collinge J., Pocock J., Lashley T., Williams J., Lambert J. C., Amouyel P., Goate A., Rademakers R., Morgan K., Powell J., St George-Hyslop P., Singleton A., Hardy J., and Alzheimer Genetic Analysis Group (2013) TREM2 variants in Alzheimer's disease. N. Engl. J. Med. 368, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jonsson T., Stefansson H., Steinberg S., Jonsdottir I., Jonsson P. V., Snaedal J., Bjornsson S., Huttenlocher J., Levey A. I., Lah J. J., Rujescu D., Hampel H., Giegling I., Andreassen O. A., Engedal K., Ulstein I., Djurovic S., Ibrahim-Verbaas C., Hofman A., Ikram M. A., van Duijn C. M., Thorsteinsdottir U., Kong A., Stefansson K. (2013) Variant of TREM2 associated with the risk of Alzheimer's disease. N. Engl. J. Med. 368, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benitez B. A., Cooper B., Pastor P., Jin S. C., Lorenzo E., Cervantes S., Cruchaga C. (2013) TREM2 is associated with the risk of Alzheimer's disease in Spanish population. Neurobiol. Aging 34, 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang T., Yu J. T., Zhu X. C., Tan L. (2013) TREM2 in Alzheimer's disease. Mol. Neurobiol. 48, 180–185 [DOI] [PubMed] [Google Scholar]
- 14. Pottier C., Wallon D., Rousseau S., Rovelet-Lecrux A., Richard A. C., Rollin-Sillaire A., Frebourg T., Campion D., Hannequin D. (2013) TREM2 R47H variant as a risk factor for early-onset Alzheimer's disease. J. Alzheimers Dis. 35, 45–49 [DOI] [PubMed] [Google Scholar]
- 15. Guerreiro R. J., Lohmann E., Bras J. M., Gibbs J. R., Rohrer J. D., Gurunlian N., Dursun B., Bilgic B., Hanagasi H., Gurvit H., Emre M., Singleton A., Hardy J. (2013) Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia-like syndrome without bone involvement. JAMA Neurol. 70, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frank S., Burbach G. J., Bonin M., Walter M., Streit W., Bechmann I., Deller T. (2008) TREM2 is up-regulated in amyloid plaque-associated microglia in aged APP23 transgenic mice. Glia 56, 1438–1447 [DOI] [PubMed] [Google Scholar]
- 17. Melchior B., Garcia A. E., Hsiung B. K., Lo K. M., Doose J. M., Cameron Thrash J. C., Stalder A. K., Staufenbiel M., Neumann H., Carson M. J. (2010) Dual induction of TREM2 and tolerance-related transcript, Tmem176b, in amyloid transgenic mice: implications for vaccine-based therapies for Alzheimer's disease. ASN Neuro. 2, e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tamboli I. Y., Hampel H., Tien N. T., Tolksdorf K., Breiden B., Mathews P. M., Saftig P., Sandhoff K., Walter J. (2011) Sphingolipid storage affects autophagic metabolism of the amyloid precursor protein and promotes Aβ generation. J. Neurosci. 31, 1837–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hernandez-Pando R., Orozco H., Arriaga K., Pavön L., Rook G. (2000) Treatment with BB-94, a broad spectrum inhibitor of zinc-dependent metalloproteinases, causes deviation of the cytokine profile towards type-2 in experimental pulmonary tuberculosis in Balb/c mice. Int. J. Exp. Pathol. 81, 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sahin U., Weskamp G., Kelly K., Zhou H. M., Higashiyama S., Peschon J., Hartmann D., Saftig P., Blobel C. P. (2004) Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hemming M. L., Elias J. E., Gygi S. P., Selkoe D. J. (2008) Proteomic profiling of γ-secretase substrates and mapping of substrate requirements. PLoS Biol. 6, e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Struhl G., Adachi A. (2000) Requirements for presenilin-dependent cleavage of Notch and other transmembrane proteins. Mol. Cell 6, 625–636 [DOI] [PubMed] [Google Scholar]
- 23. Walter J., Kaether C., Steiner H., Haass C. (2001) The cell biology of Alzheimer's disease: uncovering the secrets of secretases. Curr. Opin. Neurobiol. 11, 585–590 [DOI] [PubMed] [Google Scholar]
- 24. Peraus G. C., Masters C. L., Beyreuther K. (1997) Late compartments of amyloid precursor protein transport in SY5Y cells are involved in β-amyloid secretion. J. Neurosci. 17, 7714–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaether C., Lammich S., Edbauer D., Ertl M., Rietdorf J., Capell A., Steiner H., Haass C. (2002) Presenilin-1 affects trafficking and processing of βAPP and is targeted in a complex with nicastrin to the plasma membrane. J. Cell Biol. 158, 551–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mao D., Epple H., Uthgenannt B., Novack D. V., Faccio R. (2006) PLCγ2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J. Clin. Invest. 116, 2869–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stauffer T. P., Ahn S., Meyer T. (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8, 343–346 [DOI] [PubMed] [Google Scholar]
- 28. Várnai P., Balla T. (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 143, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. De Strooper B., Annaert W. (2010) Novel research horizons for presenilins and γ-secretases in cell biology and disease. Annu. Rev. Cell Dev. Biol. 26, 235–260 [DOI] [PubMed] [Google Scholar]
- 30. Selkoe D. J., Wolfe M. S. (2007) Presenilin: running with scissors in the membrane. Cell 131, 215–221 [DOI] [PubMed] [Google Scholar]
- 31. Sisodia S. S., St George-Hyslop P. H. (2002) γ-Secretase, Notch, Aβ, and Alzheimer's disease: where do the presenilins fit in? Nat. Rev. Neurosci. 3, 281–290 [DOI] [PubMed] [Google Scholar]
- 32. Steiner H., Haass C. (2000) Intramembrane proteolysis by presenilins. Nat. Rev. Mol. Cell Biol. 1, 217–224 [DOI] [PubMed] [Google Scholar]
- 33. Mendez M. F., McMurtray A. (2006) Frontotemporal dementia-like phenotypes associated with presenilin-1 mutations. Am. J. Alzheimers Dis. Other Demen. 21, 281–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Strooper B., Saftig P., Craessaerts K., Vanderstichele H., Guhde G., Annaert W., Von Figura K., Van Leuven F. (1998) Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 391, 387–390 [DOI] [PubMed] [Google Scholar]
- 35. Wolfe M. S., Xia W., Ostaszewski B. L., Diehl T. S., Kimberly W. T., Selkoe D. J. (1999) Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- 36. Nuttall R. K., Silva C., Hader W., Bar-Or A., Patel K. D., Edwards D. R., Yong V. W. (2007) Metalloproteinases (MMPs and ADAMs) are enriched in microglia compared to leukocytes and they link microglia activation with cytokine levels. Glia 55, 516–526 [DOI] [PubMed] [Google Scholar]
- 37. Reiss K., Saftig P. (2009) The “A Disintegrin And Metalloprotease” (ADAM) family of sheddases: physiological and cellular functions. Semin. Cell Dev. Biol. 20, 126–137 [DOI] [PubMed] [Google Scholar]
- 38. Piccio L., Buonsanti C., Cella M., Tassi I., Schmidt R. E., Fenoglio C., Rinker J., 2nd, Naismith R. T., Panina-Bordignon P., Passini N., Galimberti D., Scarpini E., Colonna M., Cross A. H. (2008) Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain 131, 3081–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gibot S., Kolopp-Sarda M. N., Bené M. C., Bollaert P. E., Lozniewski A., Mory F., Levy B., Faure G. C. (2004) A soluble form of the triggering receptor expressed on myeloid cells-1 modulates the inflammatory response in murine sepsis. J. Exp. Med. 200, 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mahdy A. M., Lowes D. A., Galley H. F., Bruce J. E., Webster N. R. (2006) Production of soluble triggering receptor expressed on myeloid cells by lipopolysaccharide-stimulated human neutrophils involves de novo protein synthesis. Clin. Vaccine Immunol. 13, 492–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gingras M. C., Lapillonne H., Margolin J. F. (2002) TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol. Immunol. 38, 817–824 [DOI] [PubMed] [Google Scholar]
- 42. Gómez-Piña V., Soares-Schanoski A., Rodríguez-Rojas A., Del Fresno C., García F., Vallejo-Cremades M. T., Fernández-Ruiz I., Arnalich F., Fuentes-Prior P., López-Collazo E. (2007) Metalloproteinases shed TREM-1 ectodomain from lipopolysaccharide-stimulated human monocytes. J. Immunol. 179, 4065–4073 [DOI] [PubMed] [Google Scholar]
- 43. Farfara D., Trudler D., Segev-Amzaleg N., Galron R., Stein R., Frenkel D. (2011) γ-Secretase component presenilin is important for microglia β-amyloid clearance. Ann. Neurol. 69, 170–180 [DOI] [PubMed] [Google Scholar]
- 44. Lee J., Chan S. L., Mattson M. P. (2002) Adverse effect of a presenilin-1 mutation in microglia results in enhanced nitric oxide and inflammatory cytokine responses to immune challenge in the brain. Neuromolecular Med. 2, 29–45 [DOI] [PubMed] [Google Scholar]
- 45. Landman N., Jeong S. Y., Shin S. Y., Voronov S. V., Serban G., Kang M. S., Park M. K., Di Paolo G., Chung S., Kim T. W. (2006) Presenilin mutations linked to familial Alzheimer's disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc. Natl. Acad. Sci. U.S.A. 103, 19524–19529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Di Paolo G., Kim T. W. (2011) Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat. Rev. Neurosci. 12, 284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang K., Zhong B., Gilvary D. L., Corliss B. C., Vivier E., Hong-Geller E., Wei S., Djeu J. Y. (2002) Syk regulation of phosphoinositide 3-kinase-dependent NK cell function. J. Immunol. 168, 3155–3164 [DOI] [PubMed] [Google Scholar]
- 48. Peng Q., Malhotra S., Torchia J. A., Kerr W. G., Coggeshall K. M., Humphrey M. B. (2010) TREM2- and DAP12-dependent activation of PI3K requires DAP10 and is inhibited by SHIP1. Sci. Signal. 3, ra38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zompi S., Hamerman J. A., Ogasawara K., Schweighoffer E., Tybulewicz V. L., Di Santo J. P., Lanier L. L., Colucci F. (2003) NKG2D triggers cytotoxicity in mouse NK cells lacking DAP12 or Syk family kinases. Nat. Immunol. 4, 565–572 [DOI] [PubMed] [Google Scholar]