Background: Genetic variants of NOD2 are linked to inflammatory bowel disease (IBD) etiology.
Results: DSS model of colitis in wild-type and inducible nitric-oxide synthase (iNOS) null mice revealed that NOD2-iNOS/NO-responsive microRNA-146a targets NUMB gene facilitating Sonic hedgehog (SHH) signaling.
Conclusion: miR-146a-mediated NOD2-SHH signaling regulates gut inflammation.
Significance: Identification of novel regulators of IBD provides new insights into pathophysiology and development of new therapy concepts.
Keywords: Inflammation, Inflammatory Bowel Disease, Innate Immunity, Macrophages, MicroRNA, Nitric Oxide, NOD-like Receptors (NLR), Signal Transduction, Signaling
Abstract
Inflammatory bowel disease (IBD) is a debilitating chronic inflammatory disorder of the intestine. The interactions between enteric bacteria and genetic susceptibilities are major contributors of IBD etiology. Although genetic variants with loss or gain of NOD2 functions have been linked to IBD susceptibility, the mechanisms coordinating NOD2 downstream signaling, especially in macrophages, during IBD pathogenesis are not precisely identified. Here, studies utilizing the murine dextran sodium sulfate model of colitis revealed the crucial roles for inducible nitric-oxide synthase (iNOS) in regulating pathophysiology of IBDs. Importantly, stimulation of NOD2 failed to activate Sonic hedgehog (SHH) signaling in iNOS null macrophages, implicating NO mediated cross-talk between NOD2 and SHH signaling. NOD2 signaling up-regulated the expression of a NO-responsive microRNA, miR-146a, that targeted NUMB gene and alleviated the suppression of SHH signaling. In vivo and ex vivo studies confirmed the important roles for miR-146a in amplifying inflammatory responses. Collectively, we have identified new roles for miR-146a that established novel cross-talk between NOD2-SHH signaling during gut inflammation. Potential implications of these observations in therapeutics could increase the possibility of defining and developing better regimes to treat IBD pathophysiology.
Introduction
The nucleotide-binding oligomerization domain (NOD)5 leucine-rich repeat containing receptor (NLR) protein family orchestrates intracellular surveillance to mediate innate immune responses and inflammation (1, 2). Among the several NOD leucine-rich repeat-containing receptor proteins, NOD2 acts as an important innate sensor that detects cytosolic pathogen-associated molecular patterns as well as damage-associated molecular patterns. Muramyl dipeptide (MDP), a component of bacterial cell wall, is recognized by NOD2 to effectuate immune responses including the production of important cytokines such as IL-1β, IL-6, TNF-α, and IL-10 via NF-κB (3). Interestingly, NOD2 plays a crucial role during several human disorders. Polymorphisms in NOD2 gene are shown to be associated with an assortment of inflammatory conditions such as IBDs (4, 5). Thus, the functional attributes of NOD variants gain immense importance as critical regulators of inflammation.
IBDs are a group of chronic and relapsing inflammatory conditions of the intestinal lining that are characterized by abdominal pain, diarrhea, and severe rectal bleeding. IBDs are broadly categorized into two subtypes among which Crohn disease (CD) represents the most severe form of the pathogenesis (6, 7). Notably, NOD2 is one of the first genes to be linked to IBD susceptibility, and more than 60 NOD2 genetic variants are reported to be associated with CD (8). NOD2 variants confer defective sensing of the agonist, MDP, and NF-κB activation by either not executing the role to restrain inflammation (loss of function) or directly activating expression of several proinflammatory genes (gain of function) (9). Consistent with these observations, monocytes obtained from CD patients (3020insC NOD2 frameshift mutant) show decreased ability to produce inflammatory cytokines such as IL-1β, IL-6, TNF-α, and IL-10, suggesting that 3020insC CD-associated mutation results in loss of function. However, other studies have reported that gain of function variant-associated CD macrophages or epithelial cells show elevated NF-κB activation and production of IL-1β, IL-6, and TNF-α (10). Further, patients with CD respond paradoxically to anti-TNF-α treatments, presenting an interesting conundrum about the involvement of NOD2 variants in the pathogenesis of CD (9). Despite the wealth of information available regarding the identities of NOD2 variants, the precise role and dynamics of NOD2 signaling and its contribution in initiating and controlling inflammation remain unclear.
Intriguingly, the pathology of IBDs is a result of the complex association between genetic and environmental factors that lead to exaggerated inflammatory immune responses and the destruction of intestinal mucosa (7). The knowledge of cellular programming in initiation, development, and manifestation of IBDs is insufficiently understood. However, several evidences implicate a vital role for deregulated host responses to gut microbiota during IBDs. This evidence includes excessive superoxides and NO produced by immune cells such as macrophages that culminate in defective intestinal mucosal barrier functions (11). Although NO is an important secondary messenger that modulates normal cellular functions and gut homeostasis, the sustained production of NO results in detrimental effects (12). Further, inducible nitric-oxide synthase (iNOS)-induced NO has been implicated to play a vital role in arbitrating the inflammatory response during colitis (13). Nevertheless, the studies on the molecular mechanism involved in NO-mediated immune homeostasis during IBD are scanty. In this regard, recent investigations have underscored the ability of NO to regulate key signaling networks including Sonic hedgehog (SHH) pathways (14). Remarkably, SHH signaling is associated with processes such as gut development and maintenance of gut homeostasis (15, 16). Additionally, inflammatory environments often trigger increased expression of SHH ligands (17, 18). In this perspective, the current study attempts to delineate the role for SHH signaling during the onset or establishment and the perpetuation of IBDs.
Here, we identified a novel cross-talk between NOD2 and SHH signaling pathways in macrophages. Notably, our data delineate a prominent role of miR-146a/SHH signaling in amplifying inflammatory responses and provide a mechanistic understanding of gain-of-function variants of NOD2 in the pathogenesis of human IBDs.
EXPERIMENTAL PROCEDURES
Cells and Mice
Primary macrophages were isolated from peritoneal exudates of C57BL/6 wild-type (WT) and iNOS−/− mice. Brewer thioglycollate (8%) was used to enrich macrophages in the peritoneal cavity of mice. The cells were cultured in DMEM (Life Technologies) containing 10% FBS (Sigma-Aldrich) for 6–8 h, and adherent cells were used as peritoneal macrophages. The purity of these macrophages was confirmed by F4/80 staining using FACS and was found to be >95%. All transfection studies were carried out with murine RAW 264.7 macrophage-like cells. All studies involving mice were carried out after the approval from the Indian Institute of Science, Institutional Ethics Committee for Animal Experimentation as well as from the Institutional Biosafety Committee.
Reagents and Antibodies
General laboratory chemicals were obtained from Sigma-Aldrich or Merck (Darmstadt, Germany). MDP, anti-iNOS, and anti-β-actin antibodies were purchased from Sigma-Aldrich. Anti-SHH, anti-GLI1, anti-PTCH1, anti-NUMB, anti-Ser-9 phospho-GSK-3β, and anti-p65 antibodies were purchased from Cell Signaling Technology (Danvers, MA). Anti-proliferating cell nuclear antigen antibody was purchased from Calbiochem. HRP-conjugated anti-rabbit IgG and HRP-conjugated anti-mouse IgG2a were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA).
Treatment with Pharmacological Reagents
The pharmacological reagents were purchased from Calbiochem. In all experiments, macrophages were treated with inhibitors for 1 h before experimental treatments at the following concentrations: SIN1 (20 μm), 1400W (100 μm), PP2 (10 μm), BAY 11-7082 (20 μm), betulinic acid (10 μm), and cyclopamine (10 μm). Dimethyl sulfoxide (DMSO) at 0.1% concentration was used as the vehicle control. In all experiments involving pharmacological reagents, a tested concentration of respective inhibitor was used after careful titration experiments assessing the viability of the macrophages using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.
RNA Isolation and Quantitative Real Time RT-PCR
Macrophages were treated as indicated, and total RNA was isolated using TRI reagent (Sigma-Aldrich). 2 μg of total RNA was converted into cDNA using first-strand cDNA synthesis kit (Bioline, London, UK). Quantitative real time RT-PCR was performed using SYBR Green PCR mixture (Kapa Biosystems Inc., Woburn, MA) for quantification of the target gene expression. All the experiments were repeated at least three times independently to ensure the reproducibility of the results. The primers used in the study are Gapdh forward 5′-gagccaaacgggtcatcatct-3′, Gapdh reverse 5′-gaggggccatccacagtctt-3′; Shh forward 5′-aaagctgacccctttagccta-3′, Shh reverse 5′-ttcggagtttcttgtgatcttcc-3′; Gli1 forward 5′-ccaagccaactttatgtcaggg-3′, Gli1 reverse 5′-agcccgcttctttgttaatttga-3′; Gli2 forward 5′-caacgcctactctcccagac-3′, Gli2 reverse 5′-gagccttgatgtactgtaccac-3′; Smo forward 5′-gagcgtagcttccgggacta-3′, Smo reverse 5′-ctgggccgattcttgatctca-3′; Ptch1 forward 5′-gccacagcccctaacaaaaat-3′, Ptch1 reverse 5′-acccacaatcaactcctcctg-3′; Il-12 forward 5′-gacttgaagatgtaccagacag-3′, Il-12 reverse 5′-gagatgagatgtgatgggag-3′; Tnf-α forward 5′-agcccacgtcgtagcaaaccaccaa-3′, Tnf-α reverse 5′-acacccattcccttcacagagcaat-3′; Ccl5 forward 5′-ttccctgtcatcgcttgctct-3′, Ccl5 reverse 5′-cggatggagatgccgatttt-3′; Cxcl9 forward 5′-tcttttcctcttgggcatcatctt-3′, Cxcl9 reverse 5′-tttccccctcttttgctttttctt-3′; and Il-6 forward 5′-cttcttgggactgatgctggtg-3′, Il-6 reverse 5′-caggatttcccagagaacatgtg-3′.
Quantification of miRNA Expression
For detection of miR-146a by quantitative real time RT-PCR, total RNA was isolated from treated or untreated macrophages. Quantitative real time RT-PCR for miR-146a was done using TaqMan miRNA assays (Applied Biosystems-Invitrogen) as per the manufacturer's instructions. U6 snRNA was used for normalization.
Immunoblotting
Macrophages were lysed in radioimmunoprecipitation assay buffer consisting of 50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1 mm PMSF, 1 μg/ml each of aprotinin, leupeptin, and pepstatin, 1 mm Na3VO4, and 1 mm NaF. An equal amount of protein from each cell lysate was resolved in a 12% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA) by the semidry transfer (Bio-Rad) method. The blots were blocked with 5% nonfat dry milk powder in TBST (20 mm Tris-HCl (pH 7.4), 137 mm NaCl, and 0.1% Tween 20) for 60 min to remove nonspecific binding. The blots were incubated overnight at 4 °C with primary antibody followed by incubation with goat anti-rabbit-HRP or anti-mouse-HRP secondary antibody in 5% BSA for 2 h. The immunoblots were developed with enhanced chemiluminescence detection system (PerkinElmer Life Sciences) as per the manufacturer's instructions. All immunoblots are representatives of at least three independent experiments.
Nuclear and Cytosolic Subcellular Fractionation
Macrophages were harvested and gently resuspended in Buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm DTT, and 0.5 mm PMSF). After incubation on ice for 15 min, cell membranes were disrupted with 10% Nonidet P-40. The cytosolic extract was separated by centrifugation at 13,000 rpm for 15 min at 4 °C. The pellet was lysed with Buffer C (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, and 1 mm PMSF), and the nuclear protein extract was collected. The nuclear and cytosolic fractions were resolved on denaturing polyacrylamide gel, and further processing was done as mentioned as described under “Immunoblotting.”
In Vivo Studies In Mice Using Murine DSS Model of Colitis
The murine colitis model of intestinal inflammation was established using low molecular weight dextran sodium sulfate (DSS) as described below. WT and iNOS−/− mice were divided into two groups containing six mice each. The test group was administered drinking water supplemented with low molecular weight DSS solution (2.5%), whereas the control group of mice was fed with autoclaved water for 9 days. Mice were carefully monitored every day for clinical symptoms such as weight loss, bloody stools, and diarrhea. After 7, 8, or 9 days of DSS treatment, the clinical symptoms of IBD were scored in WT and their iNOS−/− littermates. The clinical scores were given as follows: 0 = no symptoms; 1 = diarrhea; 2 = rectal bleeding; and 4 = death. At the end of DSS treatment, mice were euthanized, and colons and small intestines were dissected. The total length of colon in both groups of WT and iNOS−/− mice was measured, and colon was divided into three parts as ascending colon, transverse colon, and descending colon. Each of these samples was processed for total RNA isolation.
Transfection Studies
RAW 264.7 macrophage cells were transfected with 100 nm siRNA or miRNA mimic using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Transfection efficiency was found to be more than 50% in all the experiments as determined by counting the number of siGLO lamin A/C-positive cells in a microscopic field using a fluorescent microscope. 48 h after transfection, respective experiments were performed as indicated. Nod2, Rip2, Tak1, Shh, Gli1, and control siRNAs were obtained from Dharmacon (Waltham, MA) as siGENOMETM SMARTpool reagents, which contain a pool of four different double-stranded RNA oligonucleotides. miR-146a mimic, miR-146a inhibitor (Anti-miRTM), and control mimics were purchased from Ambion-Invitrogen. RAW 264.7 macrophages were transiently transfected with the following constructs using low molecular weight polyethylenimine (PEI) (Sigma-Aldrich): pcDNA3 vector, pcDNA3 SHH vector, pcDNA3 NUMB vector, pGVP2-NUMB 3′-UTR luciferase reporter construct, pGVP2-NUMB 3′-UTR reverse luciferase reporter construct, WT miR-146a promoter luciferase construct, and mutant miR-146a promoter luciferase for binding sites of transcription factors NF-κB, c-ETS, PU.1, c-Myc, HSF2, and Oct-1 constructs. 48 h after transfection, the cells were treated as described and processed as required.
Luciferase Assays
RAW 264.7 macrophages were transfected with NUMB 3′-UTR construct, NUMB 3′-UTR reverse construct, along with β-galactosidase vector and indicated miRNA mimics or treatments. After 48 h of transfection, cells were lysed in reporter lysis buffer (Promega, Madison, WI) and assayed for luciferase activity using luciferase assay reagent. The results were normalized for transfection efficiencies by assay of β-galactosidase activity.
Statistical Analysis
Levels of significance for comparison between samples were determined by the Student's t test distribution and one-way ANOVA. The data in the graphs are expressed as the mean ± S.E., and p values < 0.05 were defined as significant. GraphPad Prism 3.0 software (GraphPad software, San Diego, CA) was used for all the statistical analyses.
RESULTS
iNOS/NO Mediates NOD2-SHH Signaling Cross-talk
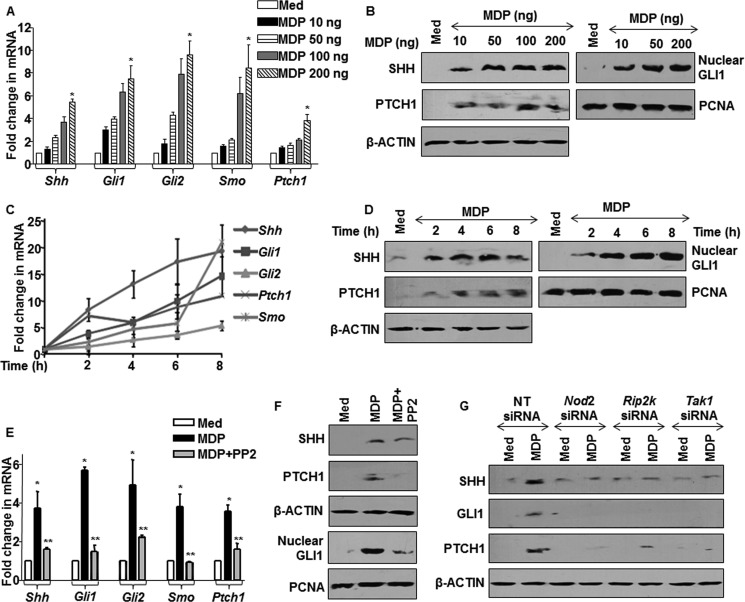
NOD2 signaling is a central cytosolic surveillance pathway as well as a crucial regulator of inflammation (2). However, NOD2-mediated molecular regulators of inflammatory responses have not been clearly identified and studied. In this regard, recent studies have indicated a strong role for SHH signaling during inflammation (19). We thus analyzed the potential role of SHH signaling as a NOD2-responsive pathway. The SHH signaling activation is marked by induced expression of SHH, glioma-associated oncogene family zinc finger (GLI)1, GLI2, smoothened (SMO), Patched 1 (PTCH1), and nuclear translocation of transcriptional activator GLI1 (20). Treatment of macrophages by MDP, a NOD2 agonist, showed robust induction as well as activation of SHH signaling with increase in NOD2 signaling activation (Fig. 1, A–D). To ascertain the involvement of NOD2-arbitrated RIP2-TAK1 signaling in controlling SHH pathway, the expression levels of SHH signaling molecules were assayed in the presence of the RIP2-specific inhibitor, PP2. The inactivation of RIP2 kinase activity by PP2 showed significant reduction in MDP-triggered SHH signaling (Fig. 1, E and F). The utilization of Nod2-, Rip2-, and Tak1-specific siRNAs validated the involvement of NOD2 signaling in regulating the activation of SHH signaling (Fig. 1G). Together, these observations strongly suggest that NOD2 signaling holds the capacity to control activation of SHH signaling.
FIGURE 1.
NOD2 pathway activates SHH signaling. A and B, mouse peritoneal macrophages were stimulated with MDP at the indicated concentrations. A and B show transcript and protein level changes in the SHH activation markers and nuclear translocation of GLI1. Med, medium; PCNA, proliferating cell nuclear antigen. C and D, time kinetics analysis of SHH signaling activation at both transcript (C) and protein levels (D) upon stimulation of NOD2 pathway is shown. E and F, macrophages were treated with PP2, a RIP2 kinase inhibitor, prior to MDP treatment and status of SHH signaling induction, and activation was assayed at transcript (E) or protein levels (F). G, RAW 264.7 cells were transfected with siRNAs specific to Nod2, Rip2, and Tak1, and activation of SHH signaling was assayed by immunoblotting. NT, nontargeted siRNA. *, p < 0.05 versus control; **, p < 0.05 versus MDP treatment (one-way ANOVA).
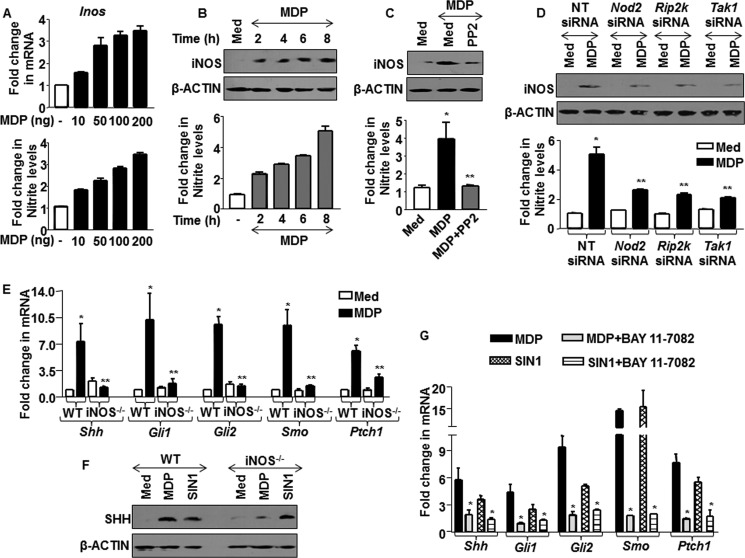
Recent studies have demonstrated that NOD2 signaling activation in macrophages increases the expression of TNF-α, IL-6, COX-2, and iNOS (21, 22). Further, NO initiates and mediates cross-talks with other signaling pathways including NOTCH and WNT signaling (21, 23). Thus, we hypothesized that NOD2-induced iNOS expression and sustained production of NO could be vital to arbitrate cross-talks with SHH signaling. From this perspective, kinetics analysis of iNOS expression in MDP-treated macrophages showed an induced expression of iNOS as well as NO production (Fig. 2, A and B). The loss of NOD2 signaling functions by perturbations mediated by pharmacological inhibitor of RIP2 or specific siRNAs to Nod2, Rip2, and Tak1 confirmed NOD2 signaling-dependent iNOS expression and NO production (Fig. 2, C and D and data not shown). This affirms NO as a mediator of NOD2 signaling. We further evaluated the role of NOD2-responsive NO to activate SHH signaling. Interestingly, iNOS null macrophages showed compromised ability to trigger NOD2-induced activation of SHH signaling as compared with WT macrophages (Fig. 2, E and F). The inadequacy of iNOS null macrophages to activate SHH signaling was not a result of generalized defect as the addition of an NO donor, SIN1, showed augmented activation of SHH signaling in iNOS null macrophages, which is comparable with that of WT macrophages (Fig. 2F and data not shown).
FIGURE 2.
NO mediates NOD2-driven activation of SHH signaling. A, macrophages were treated with various concentrations of MDP as indicated. mRNA levels of Inos were assayed by quantitative real time RT-PCR, and change in nitrate production was measured using Griess reagent. B, macrophages were treated with MDP, and kinetics of iNOS expression and NO production were assayed by immunoblotting and Griess reagent, respectively (mean ± S.E., n = 3). Med, medium. C and D, iNOS expression and NO production were analyzed by inhibition of NOD2 signaling by PP2 (C) or Nod2-, Rip2-, and Tak1-specific siRNAs (D). *, p < 0.05 versus control; **, p < 0.05 versus MDP treatment or MDP-treated nontargeted (NT) siRNA (one-way ANOVA). E and F, iNOS null and WT macrophages were treated with MDP or SIN1, and the activation status of SHH signaling was monitored at the transcript (E) and protein levels (F). WT, wild type; iNOS−/−, iNOS knock-out. G, BAY 11-7082, an IκB inhibitor, was used to monitor activation of SHH signaling upon activation of NOD2 pathway by MDP or with exogenous supply of NO by SIN1. *, p < 0.05 versus control; **, p < 0.05 versus WT MDP treatment (one-way ANOVA).
NOD2 receptors have been known to utilize NF-κB to regulate expression of downstream genes (24). To dissect the role of NF-κB during NOD2-iNOS/NO-mediated activation of SHH signaling, macrophages were treated with MDP or SIN1 in the presence or absence of a NF-κB inhibitor, BAY 11-7082 (which prevents IκB-α phosphorylation). As represented in Fig. 2G, inhibition of NF-κB activity showed a significant decrease in SHH signaling activation. As a proof of concept, we monitored NF-κB translocation from cytosol to nucleus with MDP or SIN1 treatment of macrophages (data not shown). These data conclusively suggest NO as a potential link for NOD2-SHH signaling cross-talk.
NOD2 Attenuates NUMB Gene Activity
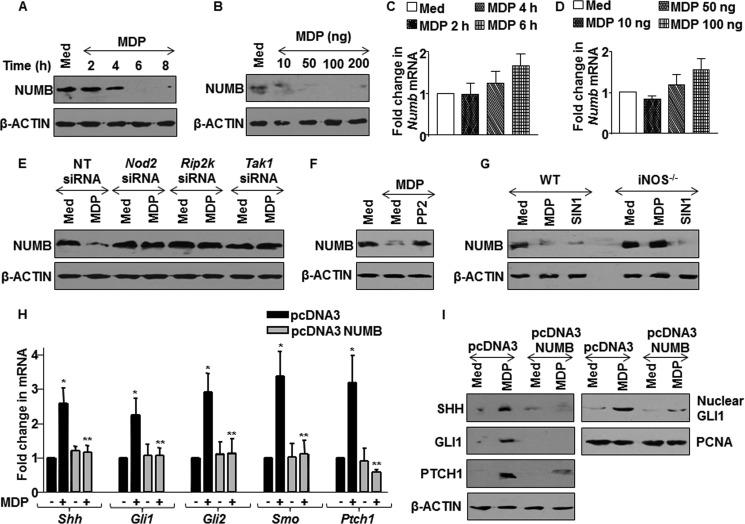
The binding of SHH ligand to PTCH1 receptors derepresses PTCH1-mediated suppression of SMO that culminates into destabilization of inhibitory complex and nuclear translocation of GLI1 to the promoters of SHH-responsive genes (20). However, checks and balances in terms of activation of SHH signaling are carefully regulated by the NUMB-mediated negative regulatory loop. Recent studies have implicated NUMB in GLI1 degradation through ITCH (Itchy E3 ubiquitin protein ligase)-dependent ubiquitination, sequestering active forms of GLI1 transcription factors and suppression of GLI transcriptional activity (25, 26). In this perspective, we analyzed the NOD2-mediated regulation of NUMB gene expression. The dosage dependence and time kinetics analysis demonstrated the progressive decrease in the NUMB protein levels with increase in NOD2-MDP engagement (Fig. 3, A and B). However, there was no significant change observed at transcript levels of NUMB (Fig. 3, C and D). Further, NOD2 signaling interference by Nod2-, Rip2-, or Tak1-specific siRNAs or PP2 led to the significant alleviation of MDP-induced decrease in NUMB levels (Fig. 3, E and F). In agreement with the previous results, the exogenous supply of NO by SIN1 showed a decrease in NUMB protein levels, whereas iNOS null macrophages failed to exhibit such effects (Fig. 3G). Gain-of-function studies were undertaken to further validate the role of NUMB in regulating NOD2-driven SHH signaling. The activation of SHH signaling in response to MDP was severely compromised in the NUMB protein overexpressing macrophages as compared with that of control cells (Fig. 3, H and I) as analyzed by expression of SHH, GLI1, GLI2, SMO, PATCH1, and nuclear translocation of transcriptional activator GLI1. Altogether, these results summarize and confirm that NOD2-iNOS/NO-triggered SHH signaling activation is mediated through down-regulation of NUMB protein.
FIGURE 3.
NOD2 pathway modulates NUMB expression to activate SHH signaling. A and B, expression of NUMB in macrophages with MDP treatment at the indicated (A) time points and (B) concentrations was assessed using immunoblotting. Med, medium. C and D, macrophages were treated with various concentrations of MDP (C) or at different time points (D) as indicated. The change in mRNA levels of Numb was assayed by quantitative real time RT-PCR. E and F, Nod2-, Rip2-, and Tak1-specific siRNA-transfected RAW 264.7 macrophages (E) or PP2-pretreated macrophages (F) were analyzed for NUMB expression. NT, nontargeted siRNA. G, WT or iNOS null macrophages were treated with MDP or SIN1, and expression of NUMB upon MDP treatment was determined. WT, wild type; iNOS−/−, iNOS knock-out. H and I, pcDNA3 NUMB-transfected macrophages were assessed for activation of SHH signaling at transcript levels (H) as well as at protein levels and by nuclear translocation of GLI1 (I). *, p < 0.05 versus control; **, p < 0.05 versus pcDNA3 MDP treatment (one-way ANOVA).
NOD2-induced miR-146a Potentiates SHH Signaling Activation by Targeting NUMB Gene
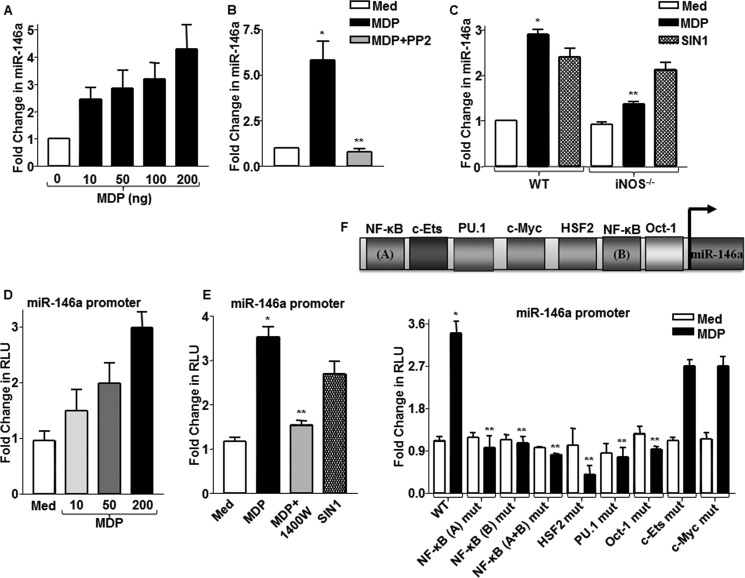
The down-regulation of NUMB upon NOD2 signaling at the protein levels but not at the transcript levels implicated the possible intervention of post-transcriptional regulations mediated by microRNAs. In this regard, NUMB has been identified as a potential target of miR-146a (27). Besides, miR-146a is one of the first identified inflammatory miRNAs whose expression was found to be elevated during various inflammatory conditions (28, 29). Further, extensive bioinformatic algorithms validated that miR-146a targets an 8-mer site located at residues spanning 2899–2906 in mouse NUMB 3′-UTR. In this perspective, we analyzed the contribution of miR-146a, if any, in modulating NOD2/NO-mediated SHH signaling. As illustrated in Fig. 4A, miR-146a levels increased gradually in macrophages with increasing doses of MDP. NOD2 signaling interference with PP2 showed significant reduction in MDP-induced expression of miR-146a (Fig. 4B). Further, iNOS null macrophages showed compromised ability to induce miR-146a expression. Concordantly, SIN1-induced expression of miR-146a was comparable in both iNOS null and WT macrophages (Fig. 4C). The miR-146a promoter luciferase reporter assays validated the crucial role of NOD2-iNOS signaling in the expression of miR-146a. (Fig. 4, D and E). Promoter luciferase analysis with WT and mutant miR-146a promoters revealed NF-κB along with HSF2, PU.1, Oct1, c-ETS, and c-Myc as vital transcription factors that orchestrate NOD2-mediated expression of miR-146a (Fig. 4F).
FIGURE 4.
NOD2/NO axis regulates expression of miR-146a. A, miR-146a expression was determined in macrophages stimulated with MDP using quantitative real time RT-PCR. B, macrophages pretreated with RIP2 kinase inhibitor, PP2, were assayed for miR-146a expression after MDP treatment. *, p < 0.05 versus control; **, p < 0.05 versus MDP treatment (one-way ANOVA). Med, medium. C, iNOS null or WT macrophages were treated with MDP or SIN1, and expression levels of miR-146a were analyzed as described. WT, wild type; iNOS−/−, iNOS knock-out. D and E, miR-146a promoter-transfected cells were treated with the indicated concentrations of MDP (D) or with 1400W, iNOS activity inhibitor, prior to MDP stimulation (E), and miR-146a promoter luciferase activity was measured. Alternatively, cells were stimulated with SIN1 in panel E, and miR-146a promoter activity was assayed. *, p < 0.05 versus WT control; **, p < 0.05 versus WT MDP (one-way ANOVA). RLU, relative luciferase unit. F, WT miR-146a promoter luciferase construct or mutant miR-146a promoter luciferase constructs for binding sites of indicated transcription factors were used to assess MDP-induced miR-146a promoter reporter activity by luciferase assay (mean ± S.E., n = 3). *, p < 0.05 versus WT control; **, p < 0.05 versus WT MDP (one-way ANOVA).
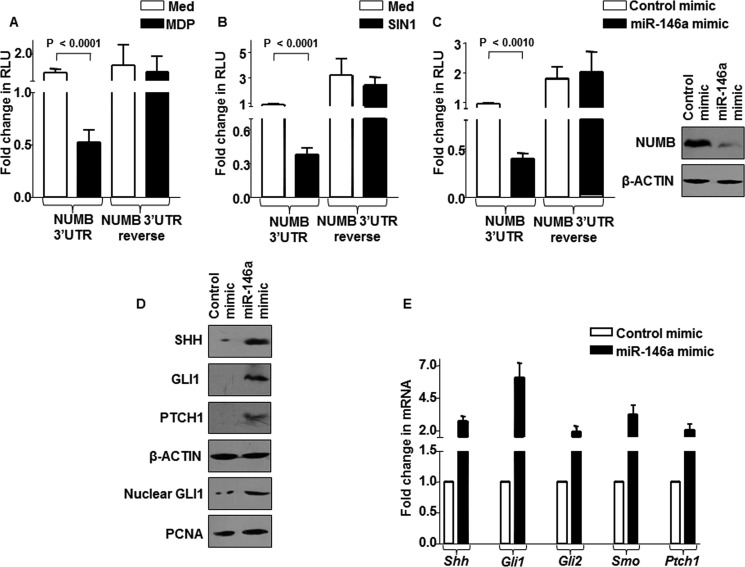
To ascertain the miR-146a-NUMB interactions, we utilized WT NUMB 3′-UTR and NUMB 3′-UTR reverse (luciferase construct that harbors NUMB 3′-UTR in reverse direction). As represented in Fig. 5, A and B, MDP or SIN1 treatment considerably reduced WT NUMB 3′-UTR luciferase activity, but not NUMB 3′-UTR reverse luciferase activity. Importantly, enforced miR-146a expression through miR-146a mimics markedly reduced NUMB 3′-UTR luciferase activity as well as NUMB protein levels, validating NUMB as a direct target of miR-146a (Fig. 5C). The positive regulation of SHH signaling by miR-146a was confirmed by enforced expression of miR-146a in macrophages, which triggered activation of SHH signaling (Fig. 5, D and E).
FIGURE 5.
miR-146a modulates SHH signaling by targeting NUMB. A–C, pGVP2-NUMB 3′-UTR luciferase reporter construct or pGVP2-NUMB 3′-UTR reverse luciferase reporter construct (3′-UTR is in reverse orientation)-transfected macrophages were treated with MDP (A) or SIN1 (B) or co-transfected with miR-146a mimics (C) to analyze NUMB 3′-UTR luciferase activity (mean ± S.E., n = 3) (one-way ANOVA) or NUMB protein levels. Med, medium; RLU, relative luciferase unit. D and E, miR-146a mimic-transfected macrophages were assessed for activation of SHH signaling at protein (D) and transcript levels (E).
NOD2-iNOS/NO-miR-146a-mediated SHH Signaling Regulates Inflammatory Gene Expression
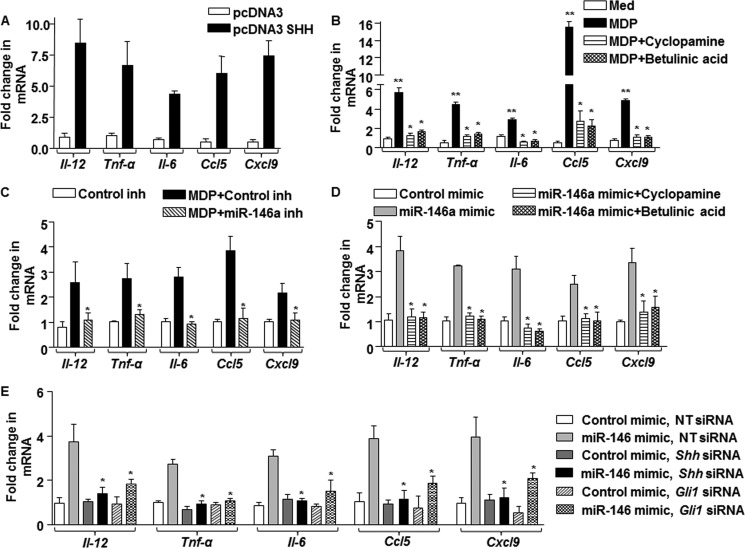
As mentioned earlier, several independent studies have suggested that NOD2 signaling and SHH signaling are involved in modulating the inflammatory responses (2, 13, 19). Hence, we analyzed the role for the miR-146a-SHH signaling axis in regulating the cytokine milieu or inflammatory gene expression during NOD2 signaling. As shown in Fig. 6A, overexpression of SHH in macrophages resulted in the induced expression of Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9. Corroborating these data, inhibition of SHH signaling by pharmacological inhibitors such as cyclopamine (SMO inhibitor) or betulinic acid (GLI inhibitor) significantly reduced the ability of MDP-NOD2 signaling to induce the expression of these cytokines (Fig. 6B). Further, to establish the role of miR-146a during NOD2-induced cytokine expression, we have utilized miR-146a inhibitors (Anti-MiRs) and mimics. Accordingly, miR-146a inhibitor-transfected macrophages were severely compromised in their ability to induce the expression of Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9 on MDP treatment (Fig. 6C), whereas overexpression of miR-146a in macrophages was sufficient to induce these genes (Fig. 6D). Substantiating this observation, utilization of SHH signaling-specific pharmacological inhibitors (Fig. 6D) or knockdown of SHH signaling by Shh- or Gli1-specific siRNAs (Fig. 6E) significantly repressed the miR-146a mimic-induced expression of Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9.
FIGURE 6.
miR-146a-mediated SHH signaling regulates inflammatory responses. A, pcDNA3 SHH-transfected macrophages were assessed for expression of proinflammatory genes, Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9. B, macrophages pretreated with pharmacological inhibitors of SHH signaling, cyclopamine (inhibits SMO), and betulinic acid (inhibits GLI), were analyzed for the indicated genes after MDP stimulation. **, p < 0.05 versus control; *, p < 0.05 versus MDP (one-way ANOVA). Med, medium. C, expression of MDP-induced proinflammatory genes was analyzed in macrophages transfected with miR-146a inhibitor and Control inhibitor. *, p < 0.05 versus MDP + Control inhibitor (MDP+Control inh) (one-way ANOVA). D and E, miR-146a mimic-transfected macrophages were either treated with SHH signaling-specific pharmacological inhibitor such as cyclopamine or betulinic acid (D) or co-transfected with Shh- or Gli1-specific siRNA (E). Quantitative real time RT-PCR for analysis of inflammatory cytokines Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9 was performed. *, p < 0.05 versus miR-146a mimic (one-way ANOVA). NT, nontargeted siRNA.
iNOS/NO-mediated SHH Signaling Is Necessary for IBD Immunopathology in Vivo
To establish the in vivo significance of NOD2-iNOS/NO-miR-146a-SHH signaling-induced inflammatory responses, we utilized the well established DSS-induced colitis murine model of mucosal inflammation. The DSS model mimics human IBD with respect to its etiology, pathogenesis, and therapeutic response (30, 31).
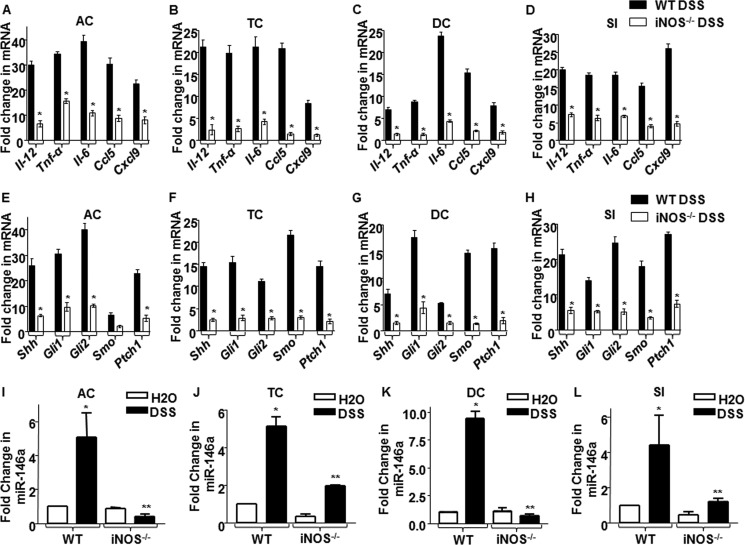
iNOS and its product NO are key regulators of acute and chronic phases of inflammatory responses during IBDs (13). The use of iNOS/NO inhibitors has been shown to be effective in reducing inflammation during IBD (32). In agreement with these previous studies (13, 32), we observed that DSS-administered WT mice showed significant decrease in body weight and survival as well as colon length with DSS treatment as compared with results in iNOS null mice (data not shown). Because IBDs are characterized by elevated proinflammatory cytokines that play key roles in orchestrating remittance and exacerbation of inflammatory responses, we analyzed the cytokine milieu or the inflammatory signatures of DSS-mediated inflamed colons. As shown in Fig. 7, A–D, WT mice exhibited elevated levels of Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9. On the contrary, iNOS null mice failed to effectuate inflammatory responses on DSS treatment, suggesting the essential requirement of iNOS/NO to mediate inflammatory responses in IBDs.
FIGURE 7.
Inflammatory responses during DSS-induced IBD immunopathology are controlled by iNOS/NO-miR-146a-SHH signaling. A–H, the expression levels of proinflammatory genes, Il-12, Tnf-α, Il-6, Ccl5, and Cxcl9 (A–D) and SHH signaling markers expression (E–H) were measured along ascending colon (AC), transverse colon (TC), descending colon (DC), and small intestine (SI) in WT and iNOS−/− animals (n = 6 each) using quantitative real time RT-PCR. *, p < 0.05 versus DSS-treated WT mice (Student's t test). I–L, quantitative real time RT-PCR analysis of miR-146a after DSS treatment of WT or iNOS null animals (n = 6) in ascending colon, transverse colon, descending colon, and small intestine is shown. *, p < 0.05 versus WT control; **, p < 0.05 versus WT DSS (one-way ANOVA). WT, wild type; iNOS−/−, iNOS knock-out.
Recent studies have implicated a crucial role of SHH signaling in intestinal inflammation (19). As represented in Fig. 2, E–G, SHH signaling activation is dependent on iNOS activity. Thus, we explored the possible role for iNOS/NO in activation of SHH signaling in the current in vivo model for IBDs. In accordance with the above observations (Fig. 7, A–D), DSS-administered WT mice showed significant increase in SHH signaling activation. However, iNOS null mice did not exhibit SHH activation even with DSS administration (Fig. 7, E–H). Corroborating these results, DSS-administered iNOS null mice failed to induce miR-146a expression with DSS administration along the ascending colon, transverse colon, descending colon, and small intestine as compared with results in WT mice (Fig. 7, I–L). Together, our observations suggest that iNOS/NO-miR-146a-mediated SHH signaling activation presents a new mechanism for modulations of inflammatory diseases such as IBD.
DISCUSSION
NOD2 is a cytoplasmic surveillance sensor that governs homeostasis through regulation of immune responses, inflammation, and apoptosis (33–35). Several studies implicate NOD2 polymorphism in CD, the major form of IBDs. Strikingly, these variants of NOD2 have been documented not only in CD patients but also in healthy individuals, underscoring the presence of predisposition for development of CD (36). Although surplus NOD2 signaling during certain kinds of CD is evidenced, the role of the NOD2 downstream signaling network under such conditions is imprecisely understood. The deeper insight into the NOD2-activated molecular signaling is crucial for understanding NOD2-driven inflammation and designing target-specific pharmacological agents to treat inflammation. In this perspective, our current investigation identifies NO-triggered miR-146a as a novel downstream messenger of NOD2 signaling in modulating SHH signaling and inflammatory responses.
NO often acts as a key signaling molecule that defines critical rate-limiting steps for pathogenesis of chronic or acute inflammation across varying cellular contexts (37). Significantly, excessive production of NO in response to a variety of pathologic or homeostatic stimuli leads to modulation of several cellular processes (38–40). Although NO is implicated in IBD responses, its role in inflammatory modulation remains inadequately understood. On the other hand, SHH signaling is a fundamental pathway that directs gut development and maintains gut homeostasis through epithelial signaling (16). Studies have also implicated elevated SHH signaling during inflammatory diseases (19, 41). In accordance with these observations, we explored the possible link between NO and SHH signaling. NOD2-responsive NO was found to induce SHH signaling in macrophages. Importantly, the in vivo DSS model revealed a possible NOD2 gain-of-function scenario of IBD as excessive NO-dependent SHH responses were observed in mice.
Unrestrained SHH signaling during inflammatory disorders recurrently entails the high risk of neoplastic transformations (42). In this context, the system of checks and balances that diligently titers the SHH signaling by several negative regulators such as NUMB assumes central importance. Corroborating our previous results, we found diminished levels of NUMB protein due to post-transcriptional regulation by miR-146a, which in itself is a consequence of NOD2/NO signaling. Supporting this, characterized inflammatory miRNAs such as miR-146a are implicated in various inflammatory diseases such as IBD, systemic lupus erythematosus, Sjögren syndrome, and rheumatoid arthritis (43). Furthermore, miR-146a expression correlates with iNOS expression as documented in one study (29). Interestingly, NO is known to mediate regulatory effects by modulating the activity of several miRNAs, suggesting a critical contribution of reactive nitrogen intermediates in modulating post-transcriptional modifications (44, 45).
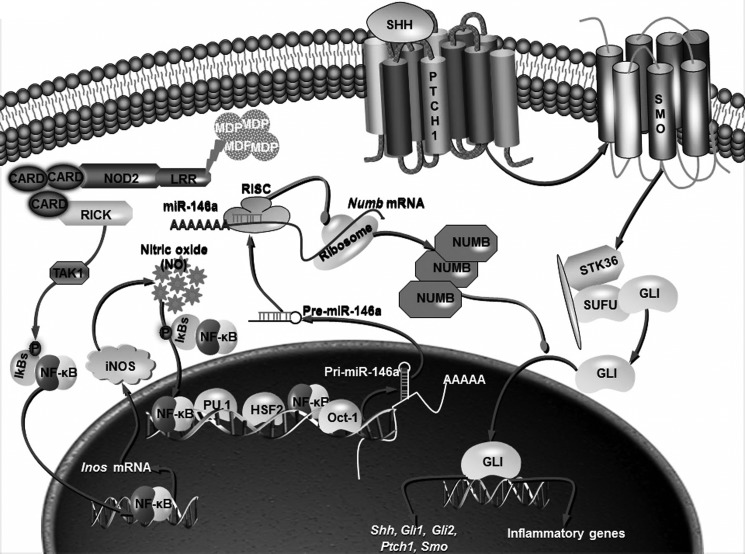
In summary, our work demonstrates that NOD2-driven inflammation is critically regulated by NO-responsive miR-146a that facilitates activation of SHH signaling by targeting NUMB expression. These molecular events contribute to the expression of inflammatory genes such as IL-12, TNF-α, IL-6, CCL-5, and CXCL-9 (Fig. 8). The in vivo DSS-induced colitis murine model presented unique roles for NO-dependent miR-146a and SHH signaling during IBD especially similar to that exemplified by the NOD2 gain-of-function variant. We believe that various novel molecular regulators identified in the current study would thus serve as potential therapeutic targets.
FIGURE 8.
Molecular mechanism of DSS-induced colitis in murine model. NOD2-triggered iNOS/NO regulates miR-146a expression through transcription factors, NF-κB, PU.1, HSF2, and Oct-1. miR-146a thus produced targets Numb mRNA, leading to reduced expression of NUMB and activation of SHH signaling. Overall, NOD2/NO pathway establishes cross-talk with SHH pathway via miR-146a-mediated degradation of NUMB to control inflammatory responses.
Acknowledgments
We thank the Central Animal facility, Indian Institute of Science (IISc) for providing mice for experimentation. We acknowledge Dr. Kushagra Bansal and Shambhuprasad TK for critical comments and timely help. We sincerely thank Dr. Andrew McMahon, Harvard University, for pCDNA3 vector, pCDNA3 SHH vector; Dr. Hideyuki Okano, Keio University, Tokyo, Japan, for pCDNA3 NUMB vector, pGVP2-NUMB 3′-UTR luciferase reporter construct, and pGVP2-NUMB 3′-UTR reverse luciferase reporter construct; and Dr. Erik Flemington, Tulane University, New Orleans, LA for WT miR-146a promoter luciferase construct and mutant miR-146a promoter luciferase for binding sites of transcription factors NF-κB, c-ETS, PU.1, c-Myc, HSF2, and Oct-1 constructs.
This study was supported by funds from the Department of Biotechnology (DBT), Department of Science and Technology (DST), Council for Scientific and Industrial Research (CSIR), Indian Council of Medical Research (ICMR), Government of India, and the Indo-French Center for Promotion of Advanced Research (IFCPAR/CEFIPRA) and by infrastructure support from the ICMR (Center for advanced study in Molecular Medicine), DST (FIST), and UGC (special assistance) (to K. N. B.).
- NOD
- nucleotide-binding oligomerization domain
- CD
- Crohn disease
- DSS
- dextran sulfate sodium
- iNOS
- inducible nitric-oxide synthase
- IBD
- inflammatory bowel disease
- MDP
- muramyl dipeptide
- SHH
- Sonic hedgehog
- miRNA/miR
- microRNA
- ANOVA
- analysis of variance
- GLI
- glioma-associated oncogene family zinc finger
- PTCH1
- Patched 1
- SMO
- Smoothened.
REFERENCES
- 1. Kufer T. A. (2008) Signal transduction pathways used by NLR-type innate immune receptors. Mol. Biosyst. 4, 380–386 [DOI] [PubMed] [Google Scholar]
- 2. Newton K., Dixit V. M. (2012) Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 4, a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Proell M., Riedl S. J., Fritz J. H., Rojas A. M., Schwarzenbacher R. (2008) The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One 3, e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cho J. H., Abraham C. (2007) Inflammatory bowel disease genetics: Nod2. Annu. Rev. Med. 58, 401–416 [DOI] [PubMed] [Google Scholar]
- 5. Wang Z. W., Ji F., Teng W. J., Yuan X. G., Ye X. M. (2011) Risk factors and gene polymorphisms of inflammatory bowel disease in population of Zhejiang, China. World J. Gastroenterol. 17, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanauer S. B. (2006) Inflammatory bowel disease: epidemiology, pathogenesis, and therapeutic opportunities. Inflamm. Bowel Dis. 12, Suppl. 1, S3–S9 [DOI] [PubMed] [Google Scholar]
- 7. Thoreson R., Cullen J. J. (2007) Pathophysiology of inflammatory bowel disease: an overview. Surg. Clin. North Am. 87, 575–585 [DOI] [PubMed] [Google Scholar]
- 8. Heliö T., Halme L., Lappalainen M., Fodstad H., Paavola-Sakki P., Turunen U., Färkkilä M., Krusius T., Kontula K. (2003) CARD15/NOD2 gene variants are associated with familially occurring and complicated forms of Crohn's disease. Gut 52, 558–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eckmann L., Karin M. (2005) NOD2 and Crohn's disease: loss or gain of function? Immunity 22, 661–667 [DOI] [PubMed] [Google Scholar]
- 10. Netea M. G., Kullberg B. J., de Jong D. J., Franke B., Sprong T., Naber T. H., Drenth J. P., Van der Meer J. W. (2004) NOD2 mediates anti-inflammatory signals induced by TLR2 ligands: implications for Crohn's disease. Eur. J. Immunol. 34, 2052–2059 [DOI] [PubMed] [Google Scholar]
- 11. Rumbo M., Courjault-Gautier F., Sierro F., Sirard J. C., Felley-Bosco E. (2005) Polarized distribution of inducible nitric oxide synthase regulates activity in intestinal epithelial cells. FEBS J. 272, 444–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kolios G., Valatas V., Ward S. G. (2004) Nitric oxide in inflammatory bowel disease: a universal messenger in an unsolved puzzle. Immunology 113, 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Beck P. L., Li Y., Wong J., Chen C. W., Keenan C. M., Sharkey K. A., McCafferty D. M. (2007) Inducible nitric oxide synthase from bone marrow-derived cells plays a critical role in regulating colonic inflammation. Gastroenterology 132, 1778–1790 [DOI] [PubMed] [Google Scholar]
- 14. Ozturk H., Tuncer M. C., Ozturk H., Buyukbayram H. (2007) Nitric oxide regulates expression of Sonic hedgehog and hypoxia-inducible factor-1α in an experimental model of kidney ischemia-reperfusion. Ren. Fail. 29, 249–256 [DOI] [PubMed] [Google Scholar]
- 15. de Santa Barbara P., van den Brink G. R., Roberts D. J. (2003) Development and differentiation of the intestinal epithelium. Cell. Mol. Life Sci. 60, 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van den Brink G. R. (2007) Hedgehog signaling in development and homeostasis of the gastrointestinal tract. Physiol. Rev. 87, 1343–1375 [DOI] [PubMed] [Google Scholar]
- 17. Ghorpade D. S., Holla S., Kaveri S. V., Bayry J., Patil S. A., Balaji K. N. (2013) Sonic hedgehog-dependent induction of MicroRNA 31 and MicroRNA 150 regulates Mycobacterium bovis BCG-driven Toll-like receptor 2 signaling. Mol. Cell. Biol. 33, 543–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhou X., Liu Z., Jang F., Xiang C., Li Y., He Y. (2012) Autocrine Sonic hedgehog attenuates inflammation in cerulein-induced acute pancreatitis in mice via upregulation of IL-10. PLoS One 7, e44121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lees C. W., Zacharias W. J., Tremelling M., Noble C. L., Nimmo E. R., Tenesa A., Cornelius J., Torkvist L., Kao J., Farrington S., Drummond H. E., Ho G. T., Arnott I. D., Appelman H. D., Diehl L., Campbell H., Dunlop M. G., Parkes M., Howie S. E., Gumucio D. L., Satsangi J. (2008) Analysis of germline GLI1 variation implicates hedgehog signalling in the regulation of intestinal inflammatory pathways. PLoS Med. 5, e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varjosalo M., Taipale J. (2008) Hedgehog: functions and mechanisms. Genes Dev. 22, 2454–2472 [DOI] [PubMed] [Google Scholar]
- 21. Bansal K., Balaji K. N. (2011) Intracellular pathogen sensor NOD2 programs macrophages to trigger Notch1 activation. J. Biol. Chem. 286, 5823–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brooks M. N., Rajaram M. V., Azad A. K., Amer A. O., Valdivia-Arenas M. A., Park J. H., Núñez G., Schlesinger L. S. (2011) NOD2 controls the nature of the inflammatory response and subsequent fate of Mycobacterium tuberculosis and M. bovis BCG in human macrophages. Cell. Microbiol. 13, 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bansal K., Trinath J., Chakravortty D., Patil S. A., Balaji K. N. (2011) Pathogen-specific TLR2 protein activation programs macrophages to induce Wnt-β-catenin signaling. J. Biol. Chem. 286, 37032–37044 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Pan Q., Kravchenko V., Katz A., Huang S., Ii M., Mathison J. C., Kobayashi K., Flavell R. A., Schreiber R. D., Goeddel D., Ulevitch R. J. (2006) NF-κB-inducing kinase regulates selected gene expression in the Nod2 signaling pathway. Infect. Immun. 74, 2121–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Po A., Sico M. A., Alimandi M., Giannini G., Maroder M., Screpanti I., Gulino A. (2006) Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 8, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 26. Stecca B., Ruiz i Altaba A. (2010) Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J. Mol. Cell Biol. 2, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuang W., Tan J., Duan Y., Duan J., Wang W., Jin F., Jin Z., Yuan X., Liu Y. (2009) Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem. Biophys. Res. Commun. 378, 259–263 [DOI] [PubMed] [Google Scholar]
- 28. Schaefer J. S., Montufar-Solis D., Vigneswaran N., Klein J. R. (2011) Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. J. Immunol. 187, 5834–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sohn J. J., Schetter A. J., Yfantis H. G., Ridnour L. A., Horikawa I., Khan M. A., Robles A. I., Hussain S. P., Goto A., Bowman E. D., Hofseth L. J., Bartkova J., Bartek J., Wogan G. N., Wink D. A., Harris C. C. (2012) Macrophages, nitric oxide and microRNAs are associated with DNA damage response pathway and senescence in inflammatory bowel disease. PLoS One 7, e44156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perě M., Cerar A. (2012) Dextran sodium sulphate colitis mouse model: traps and tricks. J. Biomed. Biotechnol. 2012, 718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waldner M. J., Neurath M. F. (2009) Chemically induced mouse models of colitis. in Current Protocols in Pharmacol, Chapter 5, Unit 5 55, John Wiley & Sons, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 32. Pilichos C. J., Kouerinis I. A., Zografos G. C., Korkolis D. P., Preza A. A., Gazouli M., Menenakos E. I., Loutsidis A. E., Zagouri F., Gorgoulis V. G., Fotiadis C. I. (2004) The effect of nitric oxide synthases inhibitors on inflammatory bowel disease in a rat model. In Vivo 18, 513–516 [PubMed] [Google Scholar]
- 33. Ferrero-Miliani L., Nielsen O. H., Andersen P. S., Girardin S. E. (2007) Chronic inflammation: importance of NOD2 and NALP3 in interleukin-1β generation. Clin. Exp. Immunol. 147, 227–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kufer T. A., Banks D. J., Philpott D. J. (2006) Innate immune sensing of microbes by Nod proteins. Ann. N.Y. Acad. Sci. 1072, 19–27 [DOI] [PubMed] [Google Scholar]
- 35. Richardson W. M., Sodhi C. P., Russo A., Siggers R. H., Afrazi A., Gribar S. C., Neal M. D., Dai S., Prindle T., Jr., Branca M., Ma C., Ozolek J., Hackam D. J. (2010) Nucleotide-binding oligomerization domain-2 inhibits Toll-like receptor-4 signaling in the intestinal epithelium. Gastroenterology 139, 904–917, 917.e901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Economou M., Trikalinos T. A., Loizou K. T., Tsianos E. V., Ioannidis J. P. (2004) Differential effects of NOD2 variants on Crohn's disease risk and phenotype in diverse populations: a metaanalysis. Am. J. Gastroenterol. 99, 2393–2404 [DOI] [PubMed] [Google Scholar]
- 37. Laroux F. S., Lefer D. J., Kawachi S., Scalia R., Cockrell A. S., Gray L., Van der Heyde H., Hoffman J. M., Grisham M. B. (2000) Role of nitric oxide in the regulation of acute and chronic inflammation. Antioxid. Redox Signal. 2, 391–396 [DOI] [PubMed] [Google Scholar]
- 38. Blantz R. C., Munger K. (2002) Role of nitric oxide in inflammatory conditions. Nephron 90, 373–378 [DOI] [PubMed] [Google Scholar]
- 39. Cooke J. P., Dzau V. J. (1997) Nitric oxide synthase: role in the genesis of vascular disease. Annu. Rev. Med. 48, 489–509 [DOI] [PubMed] [Google Scholar]
- 40. Reiter C. D., Gladwin M. T. (2003) An emerging role for nitric oxide in sickle cell disease vascular homeostasis and therapy. Curr. Opin. Hematol. 10, 99–107 [DOI] [PubMed] [Google Scholar]
- 41. Nielsen C. M., Williams J., van den Brink G. R., Lauwers G. Y., Roberts D. J. (2004) Hh pathway expression in human gut tissues and in inflammatory gut diseases. Lab. Invest. 84, 1631–1642 [DOI] [PubMed] [Google Scholar]
- 42. El-Zaatari M., Saqui-Salces M., Waghray M., Todisco A., Merchant J. L. (2009) Sonic hedgehog in gastric physiology and neoplastic transformation: friend or foe? Curr. Opin. Endocrinol. Diabetes Obes. 16, 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Iborra M., Bernuzzi F., Invernizzi P., Danese S. (2012) MicroRNAs in autoimmunity and inflammatory bowel disease: crucial regulators in immune response. Autoimmun. Rev. 11, 305–314 [DOI] [PubMed] [Google Scholar]
- 44. Bauersachs J., Thum T. (2011) Biogenesis and regulation of cardiovascular microRNAs. Circ. Res. 109, 334–347 [DOI] [PubMed] [Google Scholar]
- 45. Mathé E., Nguyen G. H., Funamizu N., He P., Moake M., Croce C. M., Hussain S. P. (2012) Inflammation regulates microRNA expression in cooperation with p53 and nitric oxide. Int. J. Cancer 131, 760–765 [DOI] [PMC free article] [PubMed] [Google Scholar]