Background: Ribozyme evolution experiments in cells can help our understanding of natural RNA evolution.
Results: During an experimental RNA evolution, the emergence of four cooperative mutations was more efficient under low than under high selection pressure.
Conclusion: Low selective pressure can facilitate the evolution of cooperative RNA mutations.
Significance: Understanding RNA evolution in biology requires understanding the effects of evolutionary parameters in cells.
Keywords: Cooperativity, Escherichia coli, Evolution, Ribozyme, RNA Catalysis
Abstract
Understanding the evolution of functional RNA molecules is important for our molecular understanding of biology. Here we tested experimentally how two evolutionary parameters, selection pressure and recombination, influenced the evolution of an evolving RNA population. This was done using four parallel evolution experiments that employed low or gradually increasing selection pressure, and recombination events either at the end or dispersed throughout the evolution. As model system, a trans-splicing group I intron ribozyme was evolved in Escherichia coli cells over 12 rounds of selection and amplification, including mutagenesis and recombination. The low selection pressure resulted in higher efficiency of the evolved ribozyme populations, whereas differences in recombination did not have a strong effect. Five mutations were responsible for the highest efficiency. The first mutation swept quickly through all four evolving populations, whereas the remaining four mutations accumulated later and more efficiently under low selection pressure. To determine why low selection pressure aided this evolution, all evolutionary intermediates between the wild type and the 5-mutation variant were constructed, and their activities at three different selection pressures were determined. The resulting fitness profiles showed a high cooperativity among the four late mutations, which can explain why high selection pressure led to inefficient evolution. These results show experimentally how low selection pressure can benefit the evolution of cooperative mutations in functional RNAs.
Introduction
Catalytic RNAs (ribozymes) are essential in all life forms, playing important roles in gene expression and regulation. Natural ribozymes include the ribosome (1, 2), RNase P (3), the spliceosome (4, 5), group I (6) and group II introns (7), and six small, self-cleaving ribozymes (8–13), most of which are variants of ancestral ribozymes that originated more than a billion years ago. The spliceosome, for example, appears to share a common ancestor with self-splicing group II intrions (14, 15); RNase P evolved into RNA-protein particles with different sets of proteins in bacteria, archaea and eukarya (16); and group I introns appear to follow an evolutionary round of invasion, degeneration, and loss (17). The correlation of sequences for group I intron ribozymes that were vertically inherited over hundreds of millions of years has provided insight into their loss of secondary structure elements and the concomitant loss of splicing activity (18). The comparison of biological sequences, however, cannot recapitulate their evolution because the evolutionary parameters, and evolutionary intermediates, are lost to history. In contrast, controlled evolution experiments in the laboratory are a powerful tool to understand the parameters that shape RNA evolution.
Previous experimental studies have elucidated several central features of RNA evolution in vitro. These studies showed that genetic diversity of a starting population increases the rate of adaptive evolution (19), that recombination can benefit an evolving population by reducing mutational load (20), and that two distinct, coevolving ribozymes can diversify such that each ribozyme dominates a different niche (21). In contrast to these RNA evolution studies in vitro and RNA selection experiments in cells, according to our knowledge, no experimental studies have addressed RNA evolution in cells. Evolution differs from selection by the repeated application of mutagenesis between multiple selection steps, an important difference that facilitates the successive optimization of sequences. Because nature optimizes RNA sequences by evolution it is desirable to understand the parameters affecting RNA evolution in cells. The dynamics of RNA evolution in cells are likely affected by the cellular environment, in which RNAs may be able to recruit and utilize cellular factors. This study utilizes an in vivo evolution system that allows studying the evolution of functional RNAs in E. coli cells.2
As a model system for RNA evolution in cells we utilize group I intron ribozymes, which are able to excise themselves from pre-mRNAs in vitro and in vivo (6). To do this, the introns fold into three-dimensional structures and catalyze two transesterification reactions. This results in the excision of the intron sequence and the joining of the flanking exons. These group I intron ribozymes can be converted to a trans-splicing format by removing the 5′-exon and adding a short substrate recognition sequence to the new ribozyme 5′ terminus (22, 23) (Fig. 1A). In this new format the ribozyme can specifically recognize a target site on a substrate RNA and replace the substrate 3′-portion with its own 3′ terminus. Group I intron ribozymes show trans-splicing activity in vitro (22, 23), in bacterial cells (23, 24), and in mammalian cells (25, 26).
FIGURE 1.
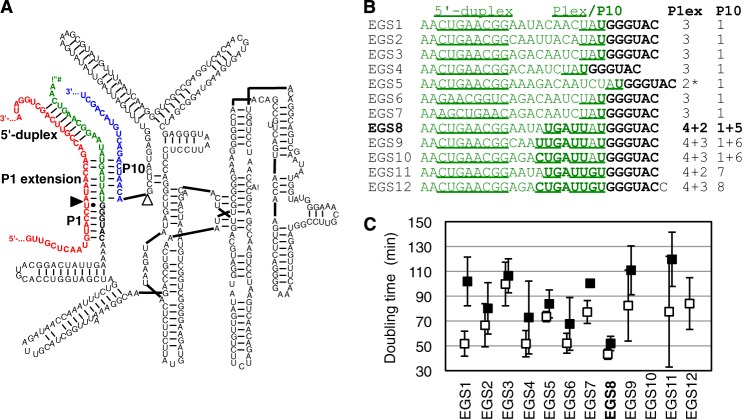
Trans-splicing ribozyme variant of the Tetrahymena group I intron that was used as parent construct for the evolutions. A, secondary structure of the ribozyme (black) with its 5′-terminal EGS (green) base-paired to the target site on the substrate (red). The ribozyme 3′-exon is in blue. The 5′-splice site is marked by a filled triangle, the 3′-splice site by an open triangle. The 5′-duplex, P1 helix, the P1 extension helix, and the P10 helix are labeled. The EGS used in this secondary structure is sequence 8 from subfigure (B). B, sequences of 12 designed EGSs that were tested on the ribozyme 5′ terminus. For each sequence, the predicted number of base pairs in the P1 helix and the P10 helix are given on the right. The underlined portion corresponds to the predicted 5′-duplex and the P1 extension helix, the bold portion to the P10 helix. The asterisk denotes a sequence where 3 bp could be formed in the P1 extension helix but are not predicted to form due to a self-structure of the EGS. C, ability of the 12 different EGSs to increase E. coli growth rates in the presence of chloramphenicol due to the repair of CAT pre-mRNA in the cells. The doubling time of E. coli cells is given for all 12 constructs, in LB medium containing 2 μg/ml chloramphenicol (open squares) and 6 μg/ml chloramphenicol (filled squares). Note that the symbols are slightly offset to clarify the error bars. EGS variant 10 did not mediate measurable growth. Note that low doubling times correspond to high ribozyme efficiencies. EGS variant 8 was chosen for the parent ribozyme of the evolution.
Trans-splicing ribozymes could be useful for therapeutic applications (for review, see Ref. 27) to correct genetic mutations on the mRNA level (25, 26, 28, 29) and specifically kill virus-infected cells (30, 31) or cancerous cells (32). Therapeutic applications are presently limited by inefficient delivery of the ribozymes into cells and by the low trans-splicing efficiency of the ribozymes in cells. The trans-splicing efficiency in cells has exceeded 10% only in exceptional cases (26). Efficiency is dependent upon on the choice of the splice site (33), the design of a 5′-terminal extension of the ribozyme sequence, the extended guide sequence (EGS3) (24, 26, 34), and, importantly, on the sequence of the ribozyme itself. Whereas the ribozyme sequence evolved in nature for cis-splicing as opposed to trans-splicing, experimental evolution could be used to adapt it to trans-splicing. Therefore, the evolution of trans-splicing ribozymes in cells serves two purposes. First, the evolution itself allows determining and quantifying the factors that guide RNA evolution in cells, through controlled experiments in the laboratory. Second, the products of the evolution experiments, more efficient trans-splicing ribozymes, could be useful tools in research and therapy.
In this study we analyzed the effects of selection pressure and recombination on the evolution of trans-splicing group I intron ribozymes in Escherichia coli cells. Four lines of evolution were conducted that differed in the application of selection pressure and recombination. The evolution of the fittest phenotype, which relied on five mutations, was most efficient under low selection pressure. Analysis of the sequences during evolution and of the evolutionary intermediates between parent ribozyme and the most efficient ribozyme found that four highly cooperative mutations resulted in two disadvantages for evolution under high selection pressure.
EXPERIMENTAL PROCEDURES
Library Plasmid
The library plasmid expressed a trans-splicing variant of the group I intron ribozyme from Tetrahymena, and the chloramphenicol acetyltransferase (CAT) pre-mRNA as described previously (24). The expression of the ribozyme was driven by a down-regulated version of the IPTG-inducible trc promoter (35), and the ribozyme carried a 3′-terminal hairpin transcription terminator. The CAT pre-mRNA was encoded in the opposite direction, and its expression was driven by the constitutive promoter derived from its parent plasmid pLysS (Novagen). It carried the frameshift mutation ΔG322 (counted with the A of the start codon as position 1), which abolished CAT activity.
In Vivo Evolution
The in vivo evolution was done essentially as described2 but with careful control of the population sizes during the selective step, the rate of mutagenesis, and the selection pressure. Briefly, electrocompetent E. coli DH5a cells (NEB) were transformed with library plasmids containing pools of ribozyme sequences, plated on LB plates containing 100 μg/ml ampicillin, and incubated at 37 °C for ∼17 h. The complexity of viable cells in each pool was estimated using dilution series plated on medium containing 100 μg/ml ampicillin. These E. coli libraries were then washed from the plates with LB medium, the A600 of the resulting cell suspension was measured, and the suspension was diluted to an A600 of 0.015. Each cell suspension was induced with a final concentration of 1 mm IPTG and shaken for 1 h at 37 °C. One million cells that contained plasmids with ribozyme inserts (as determined by colony PCR) were then plated on medium containing the appropriate chloramphenicol concentration. Chloramphenicol concentrations for evolutionary lines I and III were determined by plating each selection on plates containing three different concentrations of chloramphenicol. The concentration corresponding to ∼10% of plated clones forming visible colonies was then used for selection. After incubation at 37 °C for ∼17 h, plasmids were isolated from the grown colonies, then the ribozyme genes were subjected to mutagenic PCR (36) or recombination using the staggered extension process (37) and re-cloned into new library plasmids. The rate of mutagenesis was ∼4.3 mutations per ribozyme corresponding to 10 rounds of mutagenic PCR (36). Five to 10 plasmids were randomly selected from each population after the selective step of evolution to determine ribozyme sequences and monitor the progress of the evolution.
Construction of Ribozyme Variants
The design of variants in EGS at the ribozyme 5′ termini (see Fig. 1) used the mfold algorithm (38) to predict the structures formed by the EGS with the substrate and the ribozyme 3′ terminus. Ribozyme variants with modifications in the 5′-terminal EGS were generated by PCR with 5′-PCR primers that inserted the sequence mutations using PrimeSTAR DNA polymerase (TaKaRa). Ribozyme variants with internal mutations were constructed by site-directed mutagenesis (39). Briefly, each 50-μl reaction contained 1.25 pmol of forward primer, 1.25 pmol of reverse primer, 2.5 nmol of each dNTP, 10 μl of 5× PrimeSTAR GXL buffer, 2.5 units of PrimeSTAR GXL polymerase, and 2 ng of template plasmid. After PCR (5 min/95 °C, then 18 rounds of 50 s/95 °C, 50 s/60 °C, 4 min 45 s/68 °C and a final 7 min/68 °C), 20 units of DpnI were added, and the mixture was incubated at 37 °C for 1 h. Reactions were purified using the DNA Clean and Concentrate kit (Zymogen) and transformed into E. coli DH5α cells by electroporation.
Measurement of Cell Growth in Liquid Medium
The measurement of doubling times in liquid culture was done essentially as described (24). Briefly, fresh overnight cultures of E. coli DH5α cells with the library plasmid and the respective ribozyme variant in LB medium containing 100 μg/ml ampicillin were induced with 1 mm IPTG and shaken for 1 h at 37 °C. Each culture was diluted to an A600 of 0.05 with LB medium containing 1 mm IPTG and the indicated concentration of chloramphenicol and shaken at 37 °C. The A600 of each culture was measured every 30 min until it had exceeded 1.0. Growth rates were determined by least squares fitting.
Measurement of Cell Growth on LB Agar Plates
Fresh overnight cultures in LB medium containing 100 μg/ml ampicillin were diluted to an A600 of 0.0025, induced with a final concentration of 1 mm IPTG, and shaken at 37 °C for 1 h. Of each culture, 0.1 ml (∼50,000 viable cells) was plated on one LB agar plate containing 100 μg/ml ampicillin and one LB agar plate containing the noted concentration of chloramphenicol and IPTG at a concentration of 1 mm. Cells were grown at 37 °C for 16 h, and then each plate was washed with 1.6 ml of PBS. The A600 of each suspension from plates containing chloramphenicol was measured and normalized by the A600 from plates containing ampicillin. For most experiments the growth was measured on plates because the selection was done on plates, and the aim of measuring the growth of individual clones was to draw conclusions about their behavior during the evolution.
RESULTS
To analyze the effect of evolutionary parameters on ribozyme evolution in cells we used an in vivo selection system developed for the evolution of trans-splicing group I intron ribozymes (24). The core of this system is a plasmid encoding a gene for a trans-splicing variant of the Tetrahymena group I intron ribozyme and an inactivated CAT gene. When both the inactivated CAT mRNA and the ribozyme are expressed, the ribozyme is able to repair the CAT mRNA using trans-splicing, allowing the translation of functional CAT enzyme. The CAT enzyme uses acetyl-CoA to acetylate the antibiotic chloramphenicol (40), which abolishes binding of chloramphenicol to the ribosome (41) and mediates resistance to chloramphenicol. Repair of the inactivated CAT mRNA was facilitated by the trans-splicing ribozyme, containing a 3′-exon designed to repair the 3′ terminus of the mutated CAT mRNA (Fig. 1A). Repair of CAT mRNA by the trans-splicing group I intron ribozyme facilitated growth of E. coli cells on medium containing chloramphenicol, allowing the selection of efficient trans-splicing ribozymes in E. coli cells. The number of cells from the E. coli library that was plated on selection medium in each round of the evolution was maintained at 106. Increasing the concentration of chloramphenicol in the selection medium raised the selection pressure, to select for increased ribozyme efficiency. Mutations were introduced into the ribozyme genes by mutagenic PCR (42), and recombination events were generated by the PCR-based staggered extension process (37). Repeated rounds of mutagenesis (or recombination) and selection allowed evolving successively more active ribozyme variants.2 This in vivo evolution system was used to study the influence of selection pressure and recombination on the efficiency of the resulting ribozyme population, while keeping other evolutionary parameters constant including the rate of mutagenesis, the rate of recombination, and the population size.
As a starting point for the evolution, a trans-splicing ribozyme was designed that recognizes splice site 97 on the CAT mRNA, differing from a previous evolution on splice site 177.2 Because each splice site requires its own optimized ribozyme 5′ terminus (the EGS (24, 26, 30, 34)) 12 different EGS sequences were designed and tested to identify a 5′ terminus that could mediate efficient growth of E. coli cells in medium containing chloramphenicol (Fig. 1, B and C). The constructs differed in several aspects of the EGS design: the absence and presence of a P10 helix, the length of the P1 extension (3 or 4 bp), three different registers of the internal loop, and the specific sequence of the internal loop. The sequences of these constructs were designed such that the predicted back-folding energy of the EGS was weaker than −0.9 kcal/mol because self-structure formation of the EGS would have prevented the EGS from base pairing with the target site on the CAT pre-mRNA. The 12 constructs were measured for their ability to mediate chloramphenicol resistance due to the repair of CAT mRNA (Fig. 1C). Construct 8, which was predicted to form a 4-bp P1 extension helix and a strong P10 helix, showed the best growth at two different chloramphenicol concentrations and was chosen as the starting point for the evolutionary study.
To investigate the influence of selection pressure and recombination on the evolving ribozymes, four lines of evolution were performed in parallel, systematically varying the application of selection pressure and recombination (Fig. 2A). The first three rounds of evolution were performed as one single population, after which the population was divided into four lines, I–IV. In two of the four lines, I and III, high selection pressure was applied by successively raising the concentration of chloramphenicol from a level of 10 μg/ml in round 3 to 240 μg/ml in round 12 of the evolution (Fig. 2B). The chloramphenicol concentration was chosen in each cycle such that ∼10% of the ribozyme population formed visible colonies. In the two other lines, II and IV, a low selection pressure was applied by maintaining the chloramphenicol concentration at 10 μg/ml. Under this low selection pressure, ∼30% of the population formed visible colonies (average for evolution rounds 8–12). Recombination events were applied in three of the first 12 rounds of evolution, for all four lines (Fig. 2A). In lines I and II, the recombination events were clustered in the last three rounds, whereas in lines III and IV they were evenly distributed throughout the 12 rounds. The progress of the evolution was followed by sequencing 5–10 individual clones of the library for each line of evolution, during each round. The average number of mutations per ribozyme increased steadily, by about one mutation per ribozyme during each round of evolution (Fig. 2C).
FIGURE 2.
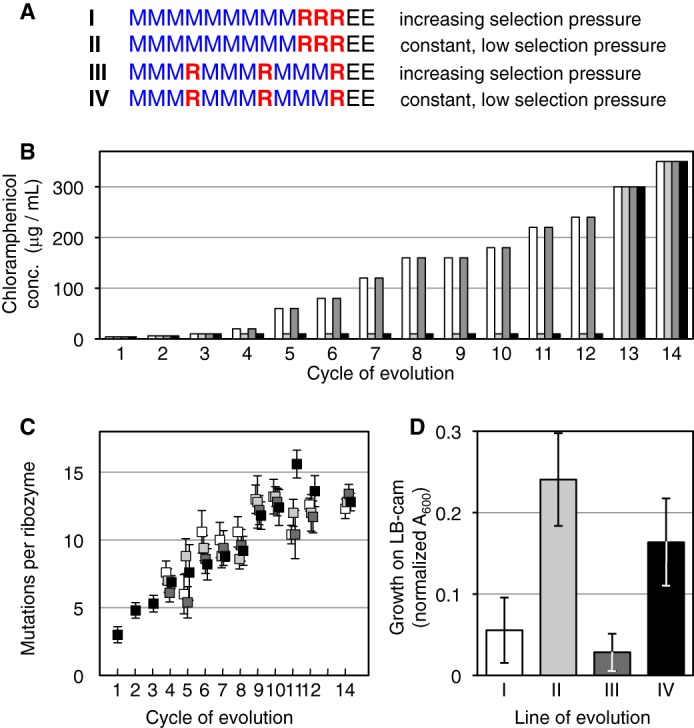
Evolution of the trans-splicing ribozyme under four different conditions (I–IV), which differed in selection pressure and recombination. A, schematic for the 12 rounds of evolution for the four separate lines. Nine rounds with mutagenesis (M, blue) and three rounds with recombination (R, red) were followed by two rounds without mutagenesis or recombination (E, black). B, selection pressure over the course of the evolution, given as the concentration of chloramphenicol in selection medium. The selection pressure for the lines I (white) and III (dark gray) increased with the activity of the pool, whereas the selection pressure for the lines II (light gray) and IV (black) was kept low, never exceeding 10 μg/ml chloramphenicol concentration. The selection pressures in rounds 13 and 14 were at the same high levels for all four lines, to select the most active ribozymes from each population. C, average number of mutations per ribozyme plotted as a function of the evolution rounds, for line I (white), II (light gray), III (dark gray), and IV (black). Each value is the average from 5–10 sequences, with error bars denoting the S.E. Note that the symbols are slightly offset to clarify the error bars. D, activities of ribozyme pools after evolution round 14, measured as cell growth on plates containing 350 μg/ml chloramphenicol. The cell growth was normalized for growth on medium with ampicillin. Error bars are S.D. of three experiments.
To identify the lines of evolution that generated the most efficient ribozymes, all four lines of evolution were subjected to two rounds of evolution under high selection pressure (rounds 13 and 14), at chloramphenicol concentrations of 300 and 350 μg/ml (Fig. 2, A and B). These two rounds did not contain mutagenesis or recombination events so that only sequences were enriched that evolved in rounds 1–12. After these two rounds of enrichment the activity of the four populations was quantified by determining the fraction of cells that grew on plates with 350 μg/ml chloramphenicol (Fig. 2D). Surprisingly, the two populations that had evolved under the lowest selection pressure showed the highest activities. The differences in recombination did not cause a significant difference among the four lines of evolution.
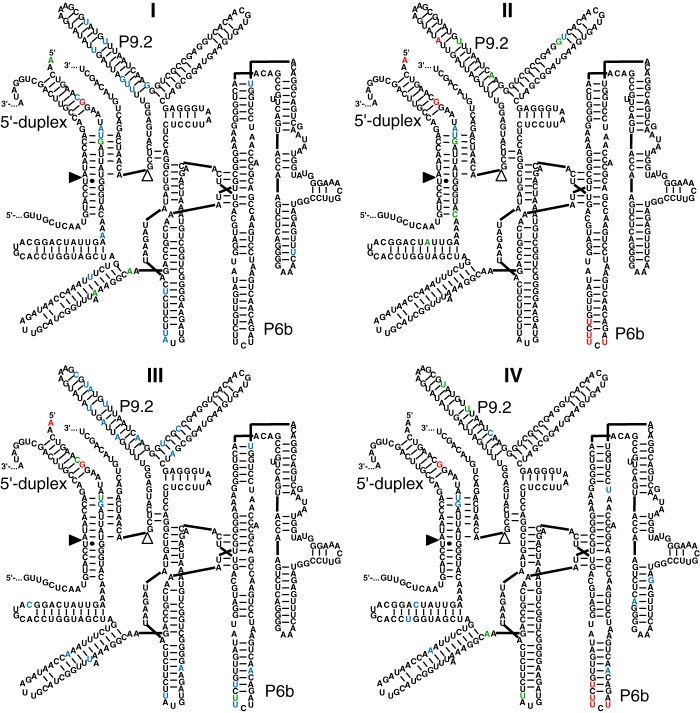
The mutations that generated the advantage for the two populations evolved under low selection pressure were identified by comparing sequences from lines II and IV (low selection pressure) with lines I and III (high selection pressure) (Fig. 3). Ten ribozymes were sequenced from each of the four populations, after evolution round 14. Mutations near the ribozyme 5′ terminus were common, with the mutation G9U dominating all four lines. Similarly, all four lines showed several mutations in the P9.2 helix. In contrast, four mutations in the P6b stem-loop clearly differentiated lines I and III from lines II and IV: U236C, U238C, U239C, and U241A. All four mutations were present in 18 of 20 sequences from populations II and IV, but in 0 of 20 in lines I and III. Three of the four mutations were in 20 of 20 clones from lines II and IV and in 3 of 20 clones from lines I and III. Conversely, 17 of 20 clones from lines I and III had none or one of the four mutations. Therefore, the increased fitness in evolutionary lines II and IV was correlated with the appearance of the four mutations in the P6b loop.
FIGURE 3.
Secondary structure representations of the mutations identified after 14 rounds of evolution in each of the four lines. Note that these mutations reflect the most active ribozymes after two rounds of enrichment at high selection pressure (rounds 13 and 14). The line (I–IV) is given for each secondary structure. For each structure, 10 sequences were analyzed. The color of the nucleotide corresponds to the frequency with which the nucleotide was found mutated: red, 8–10 mutations; green, 5–7 mutations; blue, 2–4 mutations; black, 0–1 mutation. See Fig. 1 for explanations on the secondary structure. The positions of the P6b stem-loop, P9.2 stem-loop, and the 5′-duplex are indicated. Note that the mutations in the P6b loop are highly enriched in lines II and IV but not in lines I and III.
To test whether the emergence of the four clustered mutations in the P6b loop might have been helped by an uneven coverage of mutations over the length of the ribozyme we analyzed 128 mutations in 30 sequences generated by the mutagenic PCR protocol used during the evolution (Fig. 4). No clustering of mutations and no significant deviation from random distributions were detected. As expected, the types of mutations showed a significant bias among the six possible mutations (Fig. 4C). The two types of mutations that were necessary to generate the clustered P6b mutations (U to C and U to A) were the two most frequent types of mutations obsered. Therefore, the emergence of the P6b loop mutations was likely not aided by an uneven distribution of mutations over the ribozyme but benefitted from a mutational bias in the mutagenic PCR method.
FIGURE 4.
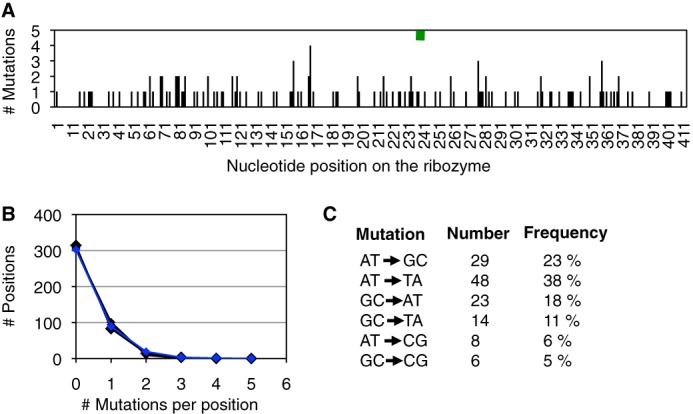
PCR analysis of mutations generated. A, distribution of 128 mutations generated by mutagenic PCR (blue) over the 414 nucleotide positions of the Tetrahymena ribozyme. The green rectangle shows the position of the P6b stem-loop mutations (positions 236–241). B, frequency of multiple mutation occurrences at the same position. Of the 414 positions, the number of positions is shown with 0, 1, 2, 3, and 4 mutations (blue). Five data sets with a random distribution are shown as comparison (black). C, mutational bias in the type of mutations. For each of the six types of mutations, their number of occurrences (total = 128) and their relative frequencies are shown. One insertion and 12 deletions were also detected (data not included).
As a more rigorous way to identify the individual mutations that gave rise to high activity, the 40 clones isolated after round 14 of the evolution were screened for chloramphenicol resistance. In a semiquantitative assay for growth on medium containing 100 μg/ml chloramphenicol, four ribozyme clones were found to mediate the most efficient growth, all of which contained the four mutations in the P6b stem-loop (data not shown). One of these clones, clone IV-12-10, was chosen for further analysis. Using site-directed mutagenesis, each of the 11 point mutations present in ribozyme IV-12-10 was individually reverted to the wild-type sequence, and the resulting ribozymes were tested in growth assays (Fig. 5A). Four mutations were necessary for maximum growth of clone IV-12-10 (G9U, U236C, U238C, and U239C). To test whether these four mutations alone were sufficient to mediate the same chloramphenicol resistance as clone IV-12-10, they were inserted into the sequence of the parent ribozyme, generating clone M4. Although the four mutations in clone M4 alone were not sufficient, a fifth mutation (U241A) completed the motif, generating clone M5, which was able to mediate growth at the level of IV-12-10 (Fig. 5B). These five mutations were identical to the mutations identified from the comparison of 40 sequences between evolutionary lines I-IV (see above), suggesting that the emergence of these five mutations in lines II and IV gave the fitness advantage to these lines.
FIGURE 5.
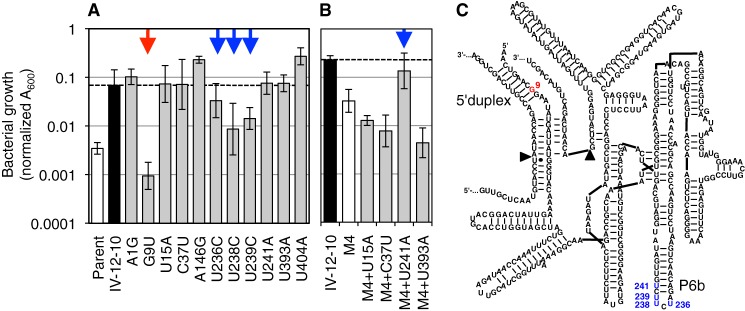
Identification of mutations that increased ribozyme activity, among the 11 mutations in clone IV-12-10. A, activity of clone IV-12-10 variants that carried single reversion mutations toward the parent ribozyme. Activity was measured as growth on LB agar containing 200 μg/ml chloramphenicol. Note the logarithmic scale that shows the growth as measured as A600. Growth of the parent ribozyme is shown as comparison, and a horizontal dashed line is shown for comparison with clone IV-12-10. The red arrow indicates a mutation reversion with >10-fold effect (G9U); the blue arrows indicate mutation reversions with <10-fold effect. B, five mutations were necessary to mediate full activity. A ribozyme containing only the four mutations identified in subfigure A (ribozyme variant M4) did not mediate full activity compared with the IV-12-10 variant. Three additional candidate mutations were added to ribozyme M4 and measured for growth in the presence of 200 μg/ml chloramphenicol. Mutation U241A, in combination with the four mutations identified in subfigure A, was found to be necessary and sufficient for full activity observed in the IV-12-10 variant. Secondary structure of the ribozyme with the positions of beneficial mutations indicated. Colorcoding is as in A. The helices containing these mutations are labeled as 5′-duplex and P6b.
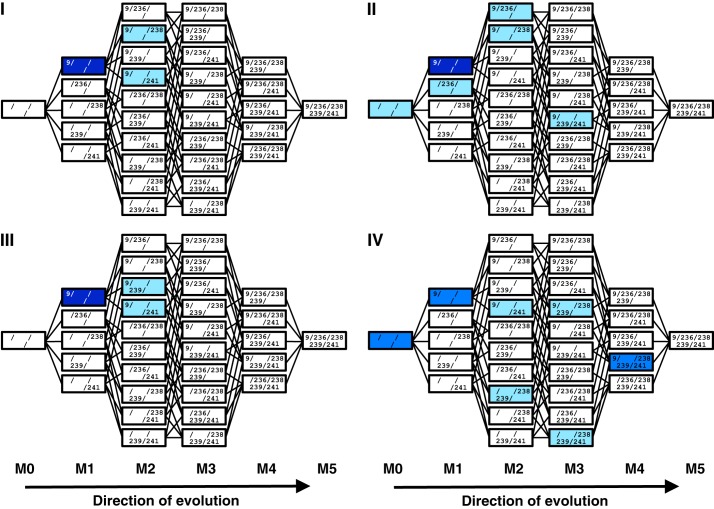
If the five M5 mutations were responsible for the difference in fitness between the four evolutionary lines, it might be possible to detect their evolutionary precursors in lines II and IV before the enrichment for the most efficient ribozymes, i.e. before round 13. To do this, the co-occurrence of M5 mutations in 80 sequences of evolution rounds 10–12 was used to identify specific evolutionary intermediates (Fig. 6). The majority of sequences in all four lines contained the G9U mutation, consistent with its dominance after round 14. The difference between the lines of evolution with regard to the M5 mutations was again correlated with high and low selection pressure: in lines I and III (high selection pressure), only single mutations were detected in addition to G9U (M2). In contrast, in lines II and IV (low selection pressure) ribozyme sequences also with two and three additional mutations (M3 and M4) were found. At this point in the evolution, M5 mutants did not dominate their populations likely because the evolutionary lines had not experienced the strong selection pressure to enrich for the most efficient ribozymes, which was applied in evolution rounds 13 and 14. The increased frequency of M3 and M4 evolutionary intermediates during rounds 10–12 suggested that low selection pressure (lines II and IV) allowed a more efficient exploration of M5 mutations. A more detailed analysis of the accumulation of M5 mutations during evolution is given in supplemental Fig. S1.
FIGURE 6.
Accumulation of evolutionary intermediates of the M5 ribozyme in rounds 10–12 of the evolution. For each of the four lines of evolution (I, II, III, and IV), all 32 evolutionary intermediates are shown, with each intermediate represented as one box. The individual mutations are listed inside. Parent ribozymes (M0) are shown on the left, and the 5-mutant ribozymes (M5) on the right. Colors illustrate the frequency with which the specific evolutionary intermediates were identified among 20 clones for each line of evolution. Light blue, 1–2 clones; blue, 3–10 clones; dark blue, 11–20 clones.
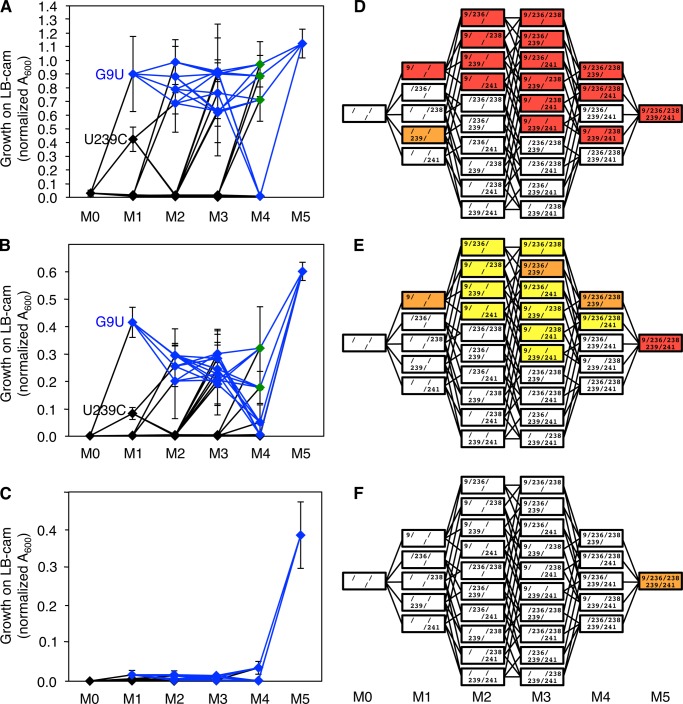
To understand how low selection pressure could have aided the evolution from the parent ribozyme to the M5 ribozyme we constructed all 30 evolutionary intermediates between the wild type ribozyme and the M5 ribozyme (see Fig. 6). The activities of these evolutionary intermediates were quantified using the same conditions as were used during the evolution (Fig. 7). Bacterial growth was measured on plates containing three different concentrations of chloramphenicol, corresponding to low selection pressure (10 μg/ml; Fig. 7A), medium selection pressure (100 μg/ml; Fig. 7B), and high selection pressure (200 μg/ml; Fig. 7C). The resulting fitness profiles confirmed that the first step of evolution, from parent ribozyme (M0) to the G9U mutant, was of large fitness benefit at low and medium selection pressure. This was consistent with the observation that mutation G9U quickly swept through all four evolving populations (supplemental Fig. S1).
FIGURE 7.
Fitness profile of all evolutionary intermediates from the parent ribozyme (M0) to the M5 ribozyme (M5), at three selection pressures. A–C, intermediates are labeled according the their number of M5 mutations (M1, M2, M3, and M4). The selection pressures correspond to chloramphenicol concentrations of 10 μg/ml (A), 100 μg/ml (B), and 200 μg/ml (C). The fitness was measured as growth on medium containing the respective chloramphenicol concentration. The two M1 ribozymes that display significant growth are labeled (G9U and U239C). All evolutionary intermediates with the G9U mutation are shown in blue or green and connected by blue lines. Green symbols highlight the gatekeeper intermediates (see the “Results” section concerning Fig. 7, and see the third paragraph of the “Discussion”). The values for all intermediates at all selection pressures are given in supplemental Fig. S2. Error bars are S.D. from three biological experiments. D–F, corresponding heat maps: 10 μg/ml (D), 100 μg/ml (E), and 200 μg/ml (F). The arrangement of evolutionary intermediates is the same as in Fig. 6. Colors denote fitness values >0.6 (red), 0.6–0.3 (orange), 0.3–0.1 (yellow), and <0.1 (white).
In contrast, the remaining four mutations in the P6b stem-loop acted highly cooperatively because when they were added to the G9U mutation (blue lines in Fig. 7) they did not mediate a fitness increase for any one, two, or three additional mutations (M2, M3, and M4, respectively). Only when all four mutations were combined in the M5 ribozyme was there a strong increase in activity. The M2, M3, and M4 intermediates even appeared to show a slight reduction in fitness relative to the G9U single mutant (Fig. 7B). In addition, several of the M4 evolutionary intermediates mediated only weak growth or no growth, such that at low and medium selection pressure, the remaining three and two evolutionary intermediates acted as “gatekeepers” on the path from the M3 intermediates to the M5 ribozyme (green symbols in Fig. 7, A and B). Finally, the high cooperativity of the mutations resulted in very low activity of all evolutionary intermediates at high selection pressure (Fig. 7C). This means that the M1–M4 evolutionary intermediates would not allow access to the M5 ribozyme at high selection pressure, which corresponded to the final 2–5 rounds of evolution for the evolutionary lines I and III (200 μg/ml; Fig. 2B).
The fitness of all evolutionary intermediates (Fig. 7, D–F) outlines the evolutionary paths that were accessible at different selection pressures. They can be used to explain why specific evolutionary intermediates were enriched during the evolution of the four strains (Fig. 6). Only two evolutionary intermediates in line IV did not appear to follow this pattern: 239/239 and 238/239/241. Both could be explained by recombination events. Intermediate 238/239 could have been generated by recombinational loss of mutation at position 9 from the detected intermediate 9/238/239. Intermediate 238/239/241 could have been generated similarly from intermediate 9/238/239/241. In both cases, the large distance to position 9 made these transitions likely. In summary, the high cooperativity of the four mutations in the P6b stem-loop generated several constraints on the emergence on this motif that were exacerbated under high selection pressure, giving an advantage to their evolutionary emergence under low selection pressure.
DISCUSSION
To study the effect of evolutionary parameters on RNA evolution in vivo we evolved four ribozyme populations, systematically varying the selection pressure and the pattern of recombination events. Lower selection pressure led to ribozyme populations with higher fitness. To determine the cause for this behavior, the ribozyme sequences were compared among the evolutionary lines. Five mutations (M5) mediating high activity were identified, and all 30 evolutionary intermediates between the parent ribozyme and the M5 ribozyme were constructed and tested for activity. Four mutations in the P6b loop of the ribozyme were highly cooperative, suggesting an explanation for the success of the evolutionary lines under low selection pressure.
Our study identified two factors that appear to have contributed to low selection pressure aiding the evolution of the four highly cooperative P6b stem-loop mutations. First and most importantly, increased selection pressure strongly reduced the survival of all evolutionary intermediates (Fig. 7). High selection pressure (200 μg/ml chloramphenicol) dramatically reduced the growth of all evolutionary intermediates, with exception of the M5 ribozyme (Fig. 7, C and F). This made the M5 ribozyme evolutionarily inaccessible at high selection pressure. At intermediate selection pressure (100 μg/ml chloramphenicol), the fitness profile generated a slight valley in the fitness landscape for the evolutionary intermediates M2, M3, and M4 relative to the M1 intermediate G9U (Fig. 7, B and E). Such a valley generates a disincentive for the population to enter these pathways, and larger drops in activity can even prevent populations from crossing the fitness valleys (43). In other words, if the evolving ribozyme populations I and III had not reached the M5 ribozyme in evolution round 7, it would have become unlikely to traverse to the M5 ribozyme, and at the evolutionary round 11 the evolution of the M5 ribozyme would have become nearly impossible (Fig. 2B).
The second, but probably less important advantage of low selection pressure was in the larger population sizes at low selection pressure. Under high selection pressure, only ∼10% of the cells formed visible colonies, whereas this fraction was ∼30% under low selection pressure. This generated ∼3-fold larger effective population sizes at low selection pressure. Large population sizes were important to evolve the four clustered mutations due to their high cooperativity, which required sampling the four-mutation sequence space without fitness benefit for any three of the four mutations. Because the ribozyme had a length of 414 nucleotides, this sequence space (414 · 413 · 412 · 411 ∼ 1010) was significantly larger than the population size of 106 cells that were plated in each selection step. The mutations, therefore, had to successively accumulate over multiple rounds of evolution, and the effective population sizes became limiting factors in the accumulation of the mutations. This constraint was strengthened by the low activity of several M4 intermediates under low and intermediate selection pressure, which reduced the number of possible evolutionary pathways to only two or three M4 gatekeeper intermediates that could lead from M3 intermediates to the M5 ribozyme (green symbols, Fig. 7). Therefore, the characteristics of the fitness profiles under different selection pressures generated a requirement for large population sizes, which was satisfied better at low than at high selection pressure.
Why did the four P6b stem-loop mutations act cooperatively? The same four mutations were identified in a previous study to increase the ribozyme efficiency in E. coli cells.2 These four mutations did not increase the in vitro trans-splicing efficiency of the ribozymes. Instead, they specifically bound the transcription termination factor Rho (in E. coli cell lysate) and increased the assembly of polysomes and the translation of the trans-spliced mRNA. Because Rho regulates the expression of many RNAs (44) these data suggested that the four P6b stem-loop mutations evolved to modulate the expression of its splicing product, the CAT mRNA. The mutations in the P6b stem-loop could have recruited Rho because Rho binds (C)7 and (C)8 sequences with micromolar affinity (45), poly(C) acts inhibitory to Rho function (46), and three of the four P6b mutations in the M5 ribozyme (U236C, U238C, and U239C) generated a (C)5 oligomer (Fig. 5). The fourth mutation (U241A) may make the (C)5 sequence more accessible for interaction with Rho because it is predicted to increase the size of the P6b loop.2 This model is consistent with a cooperative behavior of the four P6b stem-loop mutations because the lack of any of these mutations could reduce the accessibility of the loop or reduce the length of the oligo(C) sequence and thereby drastically reduce the affinity to Rho due to the length dependence of Rho binding to oligo(C) sequences (45).
Recombination did not show a strong effect on the outcome of the evolution. After enriching for the most active ribozyme variants in rounds 13 and 14 of the evolution, the activities of lines I and III were not significantly different, as were the activities between lines II and IV (Fig. 2D). Similarly, the mutations of the M5 motif that were explored in line I were almost identical to those of line III (Figs. 3 and 6). The lack of an effect by recombination on the ribozymes with the M5 mutations can be explained by the omnipresence of the G9U mutation (if all sequences contain the same mutation then recombination cannot generate a difference) and by the clustering of the four mutations in the P6b loop within six nucleotides (recombination is unlikely between closely spaced mutations).
The 12 designed 5′-terminal EGSs showed very different activities in cells, highlighting that the design principles for an EGS of trans-splicing ribozymes are not yet fully understood (24, 26, 34). EGS8, which showed the highest activity and was chosen as the starting point for the evolution, was predicted to form a P10 helix with 6 bp, which was consistent with earlier studies (26, 34) and supports the interpretation that the benefit of a P10 helix is dependent on the splice site (24). Interestingly, the evolved, strongly beneficial mutation G9U truncates the 5′-duplex from 8 to 6 or 7 bp and may increase the size of the adjacent internal loop. These results suggested that at present the most reliable way to identify the optimal EGS for a given splice site is by a combined approach between design and an in vivo selection procedure (24).
Previous in vitro evolution studies used high selection pressure to generate more efficient catalytic RNAs, which stands in contrast to the central finding of our study. High selection pressure is expected to help enrich the most efficient phenotype because it efficiently removes less active phenotypes from the population. This is especially clear for selection experiments, where all sequence diversity is contained in the starting population, and the task is to identify the most efficient sequence of that population, with selection steps as stringent and as few as possible (47–49). In contrast, evolution experiments usually do not contain the fittest phenotype in the starting population, and it is necessary to accumulate multiple mutations over successive cycles of mutagenesis, selection, amplification to access the fittest individual (50–53). Therefore, in evolution studies the fitness of evolutionary intermediates and the roughness of the fitness landscape become important. In a smooth fitness landscape, when each successive mutation increases fitness until the fitness peak is reached, high selection stringency helps a fast climb to the peak. However, at least some RNA fitness landscapes are rough (54), where high selection pressure would doom a population with low diversity to extinction (55). A larger genetic diversity speeds up the evolution of ribozymes (19), but the mutational load that is used to generate this diversity can lead to the extinction of populations, especially at small population sizes (20). This illustrates that a combination of several evolutionary parameters determines the benefit of low or high selection pressure. Our evolution in E. coli cells combined several factors that could present an obstacle for evolution at high selection pressures: the effective population sizes (∼100,000–300,000) were several orders of magnitude lower than those of most in vitro experiments, the mutagenic rate was only 3-fold below a level that previously led to extinction,2 and the cooperativity of the four P6b stem-loop mutations required the exploration of a four-mutation sequence space without gain in activity for the ribozymes with one to four mutations. Future experiments in different evolution model systems and under varying conditions are necessary to determine more generally when low selection pressure benefits the evolution of fitter phenotypes.
Previous studies selected RNAs in cells (56–60) but did not include the multiple rounds of mutagenesis and selection. In contrast to selection experiments, evolution experiments require multiple rounds of mutagenesis, selection, and amplification, such as in the present study. The experimental system employed in this study evolved a single RNA molecule inside cells, in a constant genetic background. This setup allowed a stringent control of the evolutionary parameters, the application of high mutation rates, and a relatively simple analysis and interpretation of the results. In contrast, the sequencing of complete genomes made it possible to analyze mutations in E. coli populations that evolved under experimental conditions for 2,000 generations in the laboratory (61, 62). Here, the identification of the mutations that cause the improved phenotype is quite laborious (63), creating an obstacle for experiments that evolve the complete genome. The simpler approach described in this study allows answering specific questions that can be addressed by following the evolution of specific macromolecules in cells.
Trans-splicing group I intron ribozymes were originally developed for the possible use in therapeutic applications (23). These ribozymes could be employed for the treatment of genetic disorders by repairing the mutations on the level of mRNAs (25, 26, 28, 29) and for the selective killing of cancerous or virally infected cells by splicing toxin-eoncoding RNA sequences into hTert mRNA or viral RNAs, respectively (30–32). The evolution experiments described here did not directly generate ribozymes that could be used in therapy because the most efficient ribozymes appear to rely on interactions with the bacterial protein Rho,2 which does not exist in human cells. However, similar evolution experiments can now be carried out in human cell lines. The results of the present study suggested that this would be done most efficiently by evolution experiments under low selection pressure, to allow for the enrichment of highly cooperative mutations.
Studies of protein evolution have resulted in several findings that can be compared with our study of RNA evolution. One study focused on the evolution of bacterial β-lactamase, in which a five-mutation variant mediated resistance to the antibiotic cefatoxin (64). By testing all 32 evolutionary intermediates between the wild type and the 5-mutation variant, it was found that only a fraction of the evolutionary paths were accessible under the used conditions, mirroring the results in our study (Fig. 7). However, the epigenetic interactions that generated these paths were pleiotropic effects such as increased protein aggregation and reduced thermodynamic stability, in contrast to the cooperativity of RNA mutations in our study, which was probably caused by a single factor, binding to the Rho protein.2 Other studies found that catalytic promiscuity can help in the evolution of a new function both in proteins (65, 66) and in RNAs (67). At present, the most powerful experimental system to study macromolecular evolution appears to be phage-assisted continuous evolution (68). Here the evolving molecule, usually T7 RNA polymerase, is repeatedly selected for high activity inside E. coli cells, allowing for the completion of hundreds of evolution rounds within a few days. With this technique, populations that were evolved under low selection stringency followed by high selection stringency reproducibly arrived at different sets of mutations than when evolution was done at high stringency alone, similar to the results of our study (69). Additionally, low selection stringency generated larger genetic diversity, which appeared to be the case in our study as well. Future studies will show to what extent the different chemistries of RNAs and proteins cause different evolutionary behavior.
Acknowledgments
We thank Karen Olson and Gregory Dolan for helpful discussions and William Sinko for computational help in the identification of mutations.
This work was supported, in whole or in part, by National Institutes of Health Molecular Biophysics Training Grant T32 GM008326 (to Susan Taylor). This work was also supported by Grants 2011/12 and 2012/13 from the Hellman Family Foundation (to U. F. M.).

This article contains supplemental Figs. S1 and S2.
K. E. Olson, G. F. Dolan, and U. F. Müller, unpublished results.
- EGS
- extended guide sequence
- CAT
- chloramphenicol acetyltransferase
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside
- A600
- attenuation at 600 nm (equivalent to the commonly used OD600).
REFERENCES
- 1. Noller H. F., Hoffarth V., Zimniak L. (1992) Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256, 1416–1419 [DOI] [PubMed] [Google Scholar]
- 2. Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 [DOI] [PubMed] [Google Scholar]
- 3. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 [DOI] [PubMed] [Google Scholar]
- 4. Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. (1986) Splicing of messenger RNA precursors. Annu. Rev. Biochem. 55, 1119–1150 [DOI] [PubMed] [Google Scholar]
- 5. Valadkhan S. (2005) snRNAs as the catalysts of pre-mRNA splicing. Curr. Opin. Chem. Biol. 9, 603–608 [DOI] [PubMed] [Google Scholar]
- 6. Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. (1982) Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31, 147–157 [DOI] [PubMed] [Google Scholar]
- 7. van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44, 225–234 [DOI] [PubMed] [Google Scholar]
- 8. Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. (1986) Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14, 3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buzayan J. M., Gerlach W. L., Bruening G. (1986) Satellite tobacco ringspot virus RNA: a subset of the RNA sequence is sufficient for autolytic processing. Proc. Natl. Acad. Sci. U.S.A. 83, 8859–8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharmeen L., Kuo M. Y., Dinter-Gottlieb G., Taylor J. (1988) Antigenomic RNA of human hepatitis δ virus can undergo self-cleavage. J. Virol. 62, 2674–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saville B. J., Collins R. A. (1990) A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61, 685–696 [DOI] [PubMed] [Google Scholar]
- 12. Winkler W. C., Nahvi A., Roth A., Collins J. A., Breaker R. R. (2004) Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428, 281–286 [DOI] [PubMed] [Google Scholar]
- 13. Roth A., Weinberg Z., Chen A. G. Y., Kim P. B., Amer T. D. A., Breaker R. R. (2013) A novel class of self-cleaving ribozymes is prevalent in many species of bacteria and eukarya. Nat. Chem. Biol., in press [Google Scholar]
- 14. Dayie K. T., Padgett R. A. (2008) A glimpse into the active site of a group II intron and maybe the spliceosome, too. RNA 14, 1697–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koonin E. V. (2006) The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol. Direct. 1, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walker S. C., Engelke D. R. (2006) Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 41, 77–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goddard M. R., Burt A. (1999) Recurrent invasion and extinction of a selfish gene. Proc. Natl. Acad. Sci. U.S.A. 96, 13880–13885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haugen P., Runge H. J., Bhattacharya D. (2004) Long-term evolution of the S788 fungal nuclear small subunit rRNA group I introns. RNA 10, 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hayden E. J., Ferrada E., Wagner A. (2011) Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature 474, 92–95 [DOI] [PubMed] [Google Scholar]
- 20. Soll S. J., Díaz Arenas C., Lehman N. (2007) Accumulation of deleterious mutations in small abiotic populations of RNA. Genetics 175, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Voytek S. B., Joyce G. F. (2009) Niche partitioning in the coevolution of two distinct RNA enzymes. Proc. Natl. Acad. Sci. U.S.A. 106, 7780–7785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inoue T., Cech T. R. (1985) Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 82, 648–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sullenger B. A., Cech T. R. (1994) Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing. Nature 371, 619–622 [DOI] [PubMed] [Google Scholar]
- 24. Olson K. E., Müller U. F. (2012) An in vivo selection method to optimize trans-splicing ribozymes. RNA 18, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rogers C. S., Vanoye C. G., Sullenger B. A., George A. L., Jr. (2002) Functional repair of a mutant chloride channel using a trans-splicing ribozyme. J. Clin. Invest. 110, 1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Byun J., Lan N., Long M., Sullenger B. A. (2003) Efficient and specific repair of sickle β-globin RNA by trans-splicing ribozymes. RNA 9, 1254–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fiskaa T., Birgisdottir A. B. (2010) RNA reprogramming and repair based on trans-splicing group I ribozymes. N. Biotechnol. 27, 194–203 [DOI] [PubMed] [Google Scholar]
- 28. Watanabe T., Sullenger B. A. (2000) Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc. Natl. Acad. Sci. U.S.A. 97, 8490–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kastanos E., Hjiantoniou E., Phylactou L. A. (2004) Restoration of protein synthesis in pancreatic cancer cells by trans-splicing ribozymes. Biochem. Biophys. Res. Commun. 322, 930–934 [DOI] [PubMed] [Google Scholar]
- 30. Ayre B. G., Köhler U., Goodman H. M., Haseloff J. (1999) Design of highly specific cytotoxins by using trans-splicing ribozymes. Proc. Natl. Acad. Sci. U.S.A. 96, 3507–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ryu K. J., Kim J. H., Lee S. W. (2003) Ribozyme-mediated selective induction of new gene activity in hepatitis C virus internal ribosome entry site-expressing cells by targeted trans-splicing. Mol. Ther. 7, 386–395 [DOI] [PubMed] [Google Scholar]
- 32. Song M. S., Lee S. W. (2006) Cancer-selective induction of cytotoxicity by tissue-specific expression of targeted trans-splicing ribozyme. FEBS Lett. 580, 5033–5043 [DOI] [PubMed] [Google Scholar]
- 33. Meluzzi D., Olson K. E., Dolan G. F., Arya G., Müller U. F. (2012) Computational prediction of efficient splice sites for trans-splicing ribozymes. RNA 18, 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Köhler U., Ayre B. G., Goodman H. M., Haseloff J. (1999) Trans-splicing ribozymes for targeted gene delivery. J. Mol. Biol. 285, 1935–1950 [DOI] [PubMed] [Google Scholar]
- 35. Weiss D. S., Chen J. C., Ghigo J. M., Boyd D., Beckwith J. (1999) Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181, 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cadwell R. C., Joyce G. F. (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2, 28–33 [DOI] [PubMed] [Google Scholar]
- 37. Zhao H., Giver L., Shao Z., Affholter J. A., Arnold F. H. (1998) Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 16, 258–261 [DOI] [PubMed] [Google Scholar]
- 38. Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiner M. P., Costa G. L. (1994) Rapid PCR site-directed mutagenesis. PCR Methods Appl. 4, S131–136 [DOI] [PubMed] [Google Scholar]
- 40. Shaw W. V. (1967) The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J. Biol. Chem. 242, 687–693 [PubMed] [Google Scholar]
- 41. Tritton T. R. (1979) Ribosome-chloramphenicol interactions: a nuclear magnetic resonance study. Arch. Biochem. Biophys. 197, 10–17 [DOI] [PubMed] [Google Scholar]
- 42. Cadwell R. C., Joyce G. F. (1994) Mutagenic PCR. PCR Methods Appl. 3, S136–140 [DOI] [PubMed] [Google Scholar]
- 43. Weissman D. B., Desai M. M., Fisher D. S., Feldman M. W. (2009) The rate at which asexual populations cross fitness valleys. Theor. Popul. Biol. 75, 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cardinale C. J., Washburn R. S., Tadigotla V. R., Brown L. M., Gottesman M. E., Nudler E. (2008) Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320, 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y., von Hippel P. H. (1993) Escherichia coli transcription termination factor Rho. II. Binding of oligonucleotide cofactors. J. Biol. Chem. 268, 13947–13955 [PubMed] [Google Scholar]
- 46. Walstrom K. M., Dozono J. M., von Hippel P. H. (1997) Kinetics of the RNA-DNA helicase activity of Escherichia coli transcription termination factor Rho. 2. Processivity, ATP consumption, and RNA binding. Biochemistry 36, 7993–8004 [DOI] [PubMed] [Google Scholar]
- 47. Tuerk C., Gold L. (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 [DOI] [PubMed] [Google Scholar]
- 48. Ellington A. D., Szostak J. W. (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 [DOI] [PubMed] [Google Scholar]
- 49. Bartel D. P., Szostak J. W. (1993) Isolation of new ribozymes from a large pool of random sequences. Science 261, 1411–1418 [DOI] [PubMed] [Google Scholar]
- 50. Lehman N., Joyce G. F. (1993) Evolution in vitro of an RNA enzyme with altered metal dependence. Nature 361, 182–185 [DOI] [PubMed] [Google Scholar]
- 51. Wright M. C., Joyce G. F. (1997) Continuous in vitro evolution of catalytic function. Science 276, 614–617 [DOI] [PubMed] [Google Scholar]
- 52. Ordoukhanian P., Joyce G. F. (1999) A molecular description of the evolution of resistance. Chem. Biol. 6, 881–889 [DOI] [PubMed] [Google Scholar]
- 53. Schmitt T., Lehman N. (1999) Non-unity molecular heritability demonstrated by continuous evolution in vitro. Chem. Biol. 6, 857–869 [DOI] [PubMed] [Google Scholar]
- 54. Lehman N., Donne M. D., West M., Dewey T. G. (2000) The genotypic landscape during in vitro evolution of a catalytic RNA: implications for phenotypic buffering. J. Mol. Evol. 50, 481–490 [DOI] [PubMed] [Google Scholar]
- 55. Wright S. (1932) The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proc. Sixth Int. Congress Genet. 1, 356–366 [Google Scholar]
- 56. Guo F., Cech T. R. (2002) In vivo selection of better self-splicing introns in Escherichia coli: the role of the P1 extension helix of the Tetrahymena intron. RNA 8, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ayre B. G., Köhler U., Turgeon R., Haseloff J. (2002) Optimization of trans-splicing ribozyme efficiency and specificity by in vivo genetic selection. Nucleic Acids Res. 30, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Unwalla H. J., Li H., Li S. Y., Abad D., Rossi J. J. (2008) Use of a U16 snoRNA-containing ribozyme library to identify ribozyme targets in HIV-1. Mol. Ther. 16, 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wieland M., Hartig J. S. (2008) Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. Engl. 47, 2604–2607 [DOI] [PubMed] [Google Scholar]
- 60. Chen X., Denison L., Levy M., Ellington A. D. (2009) Direct selection for ribozyme cleavage activity in cells. RNA 15, 2035–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Khan A. I., Dinh D. M., Schneider D., Lenski R. E., Cooper T. F. (2011) Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332, 1193–1196 [DOI] [PubMed] [Google Scholar]
- 62. Cooper T. F., Lenski R. E. (2010) Experimental evolution with E. coli in diverse resource environments. I. Fluctuating environments promote divergence of replicate populations. BMC Evol. Biol. 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chou H. H., Chiu H. C., Delaney N. F., Segrè D., Marx C. J. (2011) Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Weinreich D. M., Delaney N. F., Depristo M. A., Hartl D. L. (2006) Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 [DOI] [PubMed] [Google Scholar]
- 65. O'Brien P. J., Herschlag D. (1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–105 [DOI] [PubMed] [Google Scholar]
- 66. Aharoni A., Gaidukov L., Khersonsky O., McQ. Gould S., Roodveldt C., Tawfik D. S. (2005) The “evolvability” of promiscuous protein functions. Nat. Genet. 37, 73–76 [DOI] [PubMed] [Google Scholar]
- 67. Lau M. W., Unrau P. J. (2009) A promiscuous ribozyme promotes nucleotide synthesis in addition to ribose chemistry. Chem. Biol. 16, 815–825 [DOI] [PubMed] [Google Scholar]
- 68. Esvelt K. M., Carlson J. C., Liu D. R. (2011) A system for the continuous directed evolution of biomolecules. Nature 472, 499–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Leconte A. M., Dickinson B. C., Yang D. D., Chen I. A., Allen B., Liu D. R. (2013) A population-based experimental model for protein evolution: effects of mutation rate and selection stringency on evolutionary outcomes. Biochemistry 52, 1490–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]