Background: The Sgs1/Top3/Rmi1 (STR) complex resolves DNA linkages formed during replication of methyl methanesulfonate (MMS)-damaged templates.
Results: Even without Sgs1 and Rmi1, Top3 can provide some resistance to MMS and resolve linkages that have recombination-dependent X-molecule (Rec-X) topology.
Conclusion: Rec-Xs form during replication of MMS-damaged DNA.
Significance: The findings provide novel insight into replication, STR function, and genome maintenance.
Keywords: DNA Damage, DNA Replication, DNA Topoisomerase, DNA Topology, Homologous Recombination, Holliday Junction, Rec-X, Sgs1, Template Switch Recombination, Top3
Abstract
The evolutionarily conserved Sgs1/Top3/Rmi1 (STR) complex plays vital roles in DNA replication and repair. One crucial activity of the complex is dissolution of toxic X-shaped recombination intermediates that accumulate during replication of damaged DNA. However, despite several years of study the nature of these X-shaped molecules remains debated. Here we use genetic approaches and two-dimensional gel electrophoresis of genomic DNA to show that Top3, unassisted by Sgs1 and Rmi1, has modest capacities to provide resistance to MMS and to resolve recombination-dependent X-shaped molecules. The X-shaped molecules have structural properties consistent with hemicatenane-related template switch recombination intermediates (Rec-Xs) but not Holliday junction (HJ) intermediates. Consistent with these findings, we demonstrate that purified Top3 can resolve a synthetic Rec-X but not a synthetic double HJ in vitro. We also find that unassisted Top3 does not affect crossing over during double strand break repair, which is known to involve double HJ intermediates, confirming that unassisted Top3 activities are restricted to substrates that are distinct from HJs. These data help illuminate the nature of the X-shaped molecules that accumulate during replication of damaged DNA templates, and also clarify the roles played by Top3 and the STR complex as a whole during the resolution of replication-associated recombination intermediates.
Introduction
The Saccharomyces cerevisiae STR complex, consisting of the DNA helicase Sgs1, the type 1A topoisomerase Top3, and the OB-fold containing protein Rmi1, functions in multiple facets of genome maintenance (1). Loss of function mutations in STR complex members cause sensitivity to a variety of DNA damaging agents, increased rates of chromosome loss, and chromosomal rearrangements (2–6). Similarly, mice and humans lacking homologs of STR complex members, such as the Werner and Bloom syndrome DNA helicases, have phenotypes ranging from strong cancer predisposition to early embryonic lethality, highlighting the evolutionarily conserved importance of the complex in maintaining genomic integrity (7, 8).
How the STR complex functions to maintain genomic stability is an active subject of investigation with evidence pointing to roles in DNA damage checkpoint responses, replication fork stability, exonucleolytic processing of DNA ends, and the resolution of homologous recombination (HR)4 intermediates (9). Of note, emerging in vitro and in vivo evidence supports roles for the STR complex in the resolution of at least two different types of HR-dependent linkages: Holliday junctions (HJ) formed, for example, during double strand break repair (DSBR), and so-called Rec-X structures (also sometimes called sister chromatid junctions) formed during template switch recombination arising from the perturbation of DNA replication (10–15). In one type of HR-based DSBR, both broken ends invade the target, and following repair synthesis, each extended end is ligated to yield a double Holliday junction (dHJ). The dHJ can be processed by a classical HJ resolvase, which can generate either crossover or non-crossover products. Alternatively, the STR complex can branch migrate the two HJs into one another to achieve dissolution without crossing over (16). In the case of template switch recombination, when a replicating polymerase encounters a block to DNA synthesis, it is thought to switch to using the newly replicated strand of the sister chromatid as a template, and then eventually return to the original template once the stall-inducing lesion has been bypassed. Alternatively, the switch could occur at a gap left behind the advancing replication fork. A Rec-X DNA intermediate is thus formed that links the sister chromatids and is characterized by entwined newly replicated strands and apparently unpaired original template strands. The Rec-X intermediate can then be dissolved by STR-catalyzed branch migration and dissolution (Fig. 1) (17).
FIGURE 1.
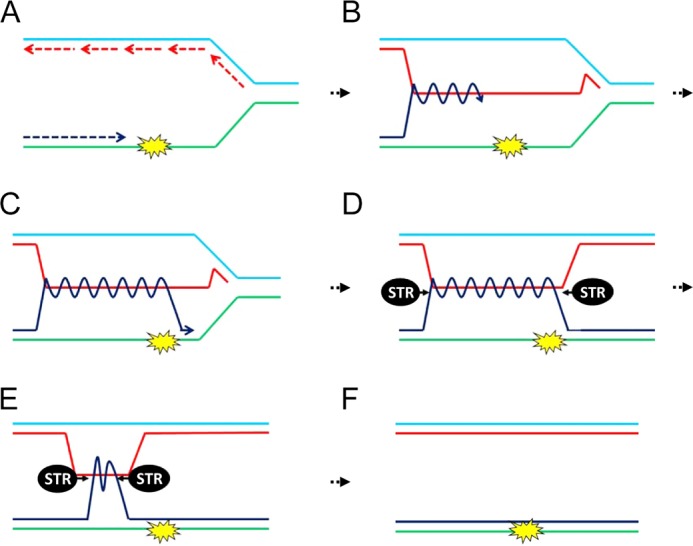
Bypass of a replication-stalling lesion through the use of Rec-X-based template switch recombination (after Liberi et al. (13)). A, a replication fork is shown with the nascent leading strand (dark blue) encountering a stall-inducing lesion. B, the leading strand switches to use of the nascent strand of the sister chromatid as a template, bypassing the lesion. C, the leading strand returns to its original template. D, a Rec-X intermediate, which is resolved by the STR complex (E and F). A Rec-X could also form during the repair of gapped DNA left behind an advancing replication fork (not shown).
The supposition that the STR complex resolves HJs is supported by robust in vitro and in vivo evidence (10, 14, 15). In contrast, evidence that the STR complex resolves Rec-X intermediates is based mostly on visualization and characterization of DNA intermediates using two-dimensional gel electrophoresis (13). The initial studies examining MMS-induced recombination intermediates from sgs1Δ cells demonstrated a prominent near-vertical “spike” at the end of the replication arc, representing joint X-shaped linkages between sister chromatids. Further analysis revealed that these sister chromatid linkages are recombination-dependent, but unlike HJs can branch migrate in a fashion unimpeded by magnesium, are refractory to cleavage by the RuvC HJ resolvase, and can be cleaved by mung bean nuclease (MBN), an endonuclease specific for regions of single-stranded DNA. These unique properties indicate that the intermediates that accumulate within MMS-treated sgs1Δ mutants are not HJs, but rather are Rec-Xs (13, 18–20). Furthermore, restoration of Sgs1 expression caused a rapid loss of Rec-X intermediates in sgs1Δ mutants, indicating Sgs1 is highly suited to resolve these structures. Similar to the results with sgs1Δ cells, other loss-of-function mutations within the STR complex have been shown to accumulate recombination dependent X-shaped molecules, consistent with involvement of the whole complex for optimal resolution of Rec-X or dHJ intermediates (21, 22).
S. cerevisiae contains three topoisomerases, Top1, Top2, and Top3, which provide relief of topological stresses generated during DNA replication, repair, and transcription. Unlike Top1 or Top2, which can relax both positive and negative DNA twist and supercoils in several contexts, Top3 is most active on negatively twisted (i.e. unwound) DNA (23). To relieve negative twist, Top3 induces a single strand nick into the phosphodiester backbone by forming a covalent linkage between its catalytically essential tyrosine residue 356 and a phosphorus present in the DNA backbone. This reaction generates a transient gate through which the intact complementary strand of DNA can pass, thus relieving superhelical tension, and is followed by restoration of the phosphodiester backbone and release of Top3 (24–26). Unlike Top1 and Top2, which provide the majority of in vivo topological relaxation, Top3 appears to function mainly in disentangling intermediates associated with DNA replication and repair (24–27). Indeed, in support of such roles, top3Δ mutants display slowed chromosome replication, chromosome missegregation, inability to undergo meiosis, and a strong sensitivity to genotoxic stressors (5, 28, 29). Because Top3 interacts physically with Sgs1 and Rmi1, and mutations that disrupt these interactions compromise DNA repair activities, Top3 is generally thought to work only within the context of the STR complex (24, 30, 32–35). However, sgs1Δ top3Δ mutants are more sensitive to MMS than sgs1Δ mutants (34, 36, 37), suggesting that Top3 might have residual Sgs1-independent activities, but this possibility remains largely unexplored.
An essential requirement for resolution of both dHJs and Rec-Xs is that DNA strands pass through transient DNA breaks, and we reasoned that Top3 may thus be the most critical component of the STR complex for resolution of these X-shaped intermediates. Although Sgs1 and Rmi1 stimulate Top3 catalysis, in principle, Top3 should have some intrinsic capacity to dissolve entangled DNA strands. Here we present evidence that Top3 indeed plays a more critical role than Sgs1 or Rmi1 in the resistance to MMS-induced DNA damage, and that in the absence of Sgs1 and Rmi1, Top3 has some capacity to resolve recombination intermediates. We utilize this Top3-dependent rescue to characterize the properties of the recombination intermediates that accumulate during replication of damaged DNA templates. Using genetic and biochemical assays, we show that the intermediates resolved by Top3 during bypass of stall-inducing lesions caused by MMS have characteristics of Rec-X structures rather than HJs. These findings clarify how the conserved STR complex promotes genome stability, and provide support for the role of Rec-X structures in DNA replication.
EXPERIMENTAL PROCEDURES
Yeast Strains
All yeast strains are derived from the BY4741/2 background. Experiments using α-factor were carried out in bar1Δ strains. The following strains were constructed: YAC174, sgs1Δ::HIS3; YAC173, sgs1Δ::HIS3, top3Δ::KanMX; YAC833, sgs1Δ::HIS3, rad52Δ::HygMX; YAC1177, sgs1Δ::HIS3, top3Δ::KanMX, rad18Δ::HygMX; YAC1637, sgs1Δ::HIS3, top3Δ::CaURA3; YAC1640:sgs1Δ::HIS3, top3Δ::CaURA3, mph1Δ::KanMX, shu1Δ::NatMX; YAC1644, sgs1Δ::HIS3, mph1Δ::KanMX, shu1Δ::NatMX; YAC1646, rmi1Δ::KanMX; YAC1887, sgs1Δ::HIS3, rad18Δ::HygMX; YAC2005, sgs1Δ::HIS3, top3Δ::NatMX, Δrad52::HygMX; YAC2341, rmi1Δ::KanMX, top3Δ::NatMX; YBB26, sgs1Δ::KanMX, top3Δ::NatMX, bar1Δ::HygMX.
Plasmids
All plasmids were based on Gateway-compatible pAG CEN/ARS vectors (38). The constitutive GPD or NOP1 promoters, which drive high or moderate levels of transcription, respectively, were each tested but gave indistinguishable results for TOP3-mediated rescue (data not shown). Plasmids used and constructed were as follows: pAG413GPD-ccdb; pAG415GPD-ccdb; pAG416GPD-ccdb; AC207, pDONR221-TOP2; AC246, pDONR221-TOP1; AC273, pAG416NOP1-ccdb; AC739, pDONR221-TOP3; AC1314, pAG416TOP3-MYC-ccdb (i.e. a TOP3 promoter-driven vector); AC1332, pDONR221-hTOP3α; AC1337, pDONR221-top3-Y356.
Analysis of X-shaped Recombination Intermediates by Two-dimensional Gel Electrophoresis
Preparation of cell cultures, DNA, two-dimensional gel electrophoresis, and Southern blots for analyzing the ARS305 region has been previously described (39), and we employed the following modifications. Briefly, α-factor was added at a final concentration of 50 nm to logarithmically growing cultures for 4 h. The synchronized cells were collected by centrifugation and inoculated at 1.2 × 107 cells/ml into fresh YPAD containing a final concentration of 0.033% MMS. Samples were either harvested 1.5 h later or the cells were washed at this point and inoculated into fresh YPAD (without MMS) at 4 × 106 cells/ml and harvested 2 and 3 h later. All comparisons were made between samples grown, prepared, and run in parallel to remove inter-experimental variation. For two-dimensional gel electrophoresis of the ribosomal DNA (rDNA) and the region adjacent to the ARS305 region, DNA was digested with BglII, and run under identical conditions as for the ARS305 region. The rDNA probe was made using PCR amplification of genomic DNA with primers rDNA-1053 and rDNA-447 as previously described (40). The ARS305-adjacent region probe was made using PCR amplification of genomic DNA with primers 5′-GTAGGAACAAAGGTTTGGCAGG-3′ and 5′-CTTCGAGATAAGGCATGGGG-3′. Gels were quantified using ImageQuant analyses of PhosphorImager scans. p values for spike:arc ratios were calculated using one-tailed t tests. MBN sensitivity of X-shaped molecules was performed by digesting 1 μg of samples with BglII in New England Biolabs Buffer 2 followed by addition of 45 units of MBN (New England Biolab) at 30 °C for 1 h. To confirm the specificity of MBN under these conditions, 0.5 fmol of a 5′-32P-end labeled oligonucleotide (oligo 1*) by itself, or annealed into a synthetic HJ, was mixed with 1 μg of BglII-digested genomic DNA and treated with MBN as above. The synthetic HJ was based on J-11, built from oligos 1, 2, 3, and 4 (41), with two modifications: an error in the reported sequence of oligo 1 (41) was corrected to generate oligo 1* (GGCGACGTGATCACCAGATGATTGCTAGGCATGCTTTCCGCAAGAGAAGC), and a C was added to the 3′ end of oligo 4 to generate oligo 4* (ACCGTTAGCAGTTCGCCTTGAGCCTAGCAATCATCTGGTGATCACGTCGCC) so that the HJ arm formed by oligos 1* and 4* was blunt ended and thus not subject to cleavage by MBN. Branch migration assays were performed as previously described (39), with the exception that the 65 °C incubation of samples was for 9 h.
Spot Assays
Yeast were grown overnight at 23 °C in YPAD or, when required for plasmid maintenance, in selective media. 105 cells, and serial 10-fold dilutions, were then spotted to YPAD plates with or without MMS. Plates were incubated at either 30 or 23 °C and imaged 2–3 days after spotting. For higher concentrations of MMS (0.02% and 0.03%) cells were spotted at 106 cells, with serial 10-fold dilutions.
Construction and Characterization of dHJ and Rec-X Substrates
Substrates were made using a two-step protocol modified from earlier constructions of dHJ from the dHJ1 and dHJ2 oligonucleotides (42, 43). Oligonucleotide Rec-X1 (5′-CCACGTTTTCGTGGCGCTGGACTAACGCTCGACACCGACCAATGCTTTTGCATTGGTCGGACCTTCAGAACCGACCAGCG-3′) was derived from dHJ1. The first step generated a covalently closed intramolecular circle from the Rec-X1 oligo (which self-anneals to form a structure containing a ligatable nick in one of its hairpinned duplex arms), and the second step generated the full Rec-X and dHJ products; this approach allowed for higher yield and purity of the Rec-X substrate. To ensure consistency between substrates, their components were processed identically and in parallel. For the first step, the dHJ1 and Rec-X1 oligonucleotides (12.5 pmol each) were phosphorylated using T4 polynucleotide kinase and a molar excess of [γ-32P]ATP, followed by purification using Centri-Spin 20 columns (Princeton Separations) containing 10 mm Tris-HCl, pH 7.5, 0.1 mm EDTA. The oligos were then subjected separately to annealing and ligation conditions (to which only Rec-X1 responded, by forming a closed circle) as follows: 6.25 pmol of oligonucleotide was suspended in 35 μl containing 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2; denatured at 99 °C for 1 min; and cooled rapidly to 4 °C. Samples were then ligated by addition of 1 mm ATP, 5 mm DTT, and 280 units of T4 DNA ligase (New England Biolabs) and incubated at 8 °C for 1 h, 10 °C for 1 h, 12 °C for 4 h, 16 °C for 12 h, and 20 °C for 12 h. The products were mixed 2:1 with formamide containing 0.1% bromphenol blue and xylene cyanol, and resolved via electrophoresis through an 8% polyacrylamide gel (19:1 acrylamide:bisacrylamide) under denaturing conditions (7.8 m urea in 0.5× TBE). The gel was exposed to a PhosphorImager screen for ∼15 min, and the image was printed out on a clear plastic sheet and placed over the gel to guide excision of the bands corresponding to the circularized Rec-X1 oligo and the linear dHJ1 oligo. The oligos were electroeluted from the gel slices using D-Tube Dialyzer Midi tubes (Novagen) with a 6–8-kDa cutoff at 6 V/cm for 1 h in 0.5× TBE, followed by dialysis overnight at 4 °C versus 1 liter of 10 mm Tris-Cl, pH 7.5, 0.1 mm EDTA. In the morning the dialysis buffer was exchanged with fresh buffer and allowed to equilibrate for an additional 2 h (this step was repeated twice). Samples were precipitated by adding 0.1 volume of NaOAc, pH 5.2, and 2.5 volumes of 100% ethanol, and washed with 70% ethanol.
For the second step, the pellets were resuspended in 20 μl of 10 mm Tris-Cl, pH 7.5, 0.1 mm EDTA containing 5′-phosphorylated dHJ2 (due to differences in substrate synthesis efficiency, dHJ2 was added at 100-fold molar excess to the Rec-X1; and at 15-fold molar excess to the dHJ1). Samples were annealed in buffer conditions as described above by denaturing at 95 °C for 3 min, shifted to 80 °C, and cooled to 5 °C at a rate of 0.125 °C/min. Ligation, electrophoresis, gel extraction, electroelution, dialysis, and precipitation were performed as above. The final products were resuspended in 10 mm Tris, pH 7.5, 0.1 mm EDTA at 0.5 fmol/μl. Substrate concentrations were estimated based on radioactive counts, measured using Image Quant analyses of PhosphorImager scans, compared with originally end-labeled oligos.
Confirmation of the Rec-X structure involved incubation of 0.5–1 fmol of substrate with restriction enzymes (New England Biolabs) and exonucleases in 50 mm KAc, 20 mm Tris acetate, 10 mm MgAc, 1 mm DTT, pH 7.9. RsaI (5 units), and HhaI (20 units) digests were carried out at 37 °C for 3 h. TaqaI (20 units) digests were carried out at 47 °C for 3 h. Escherichia coli ExoI (2.5 units; U. S. Biochemical) and ExoIII (50 units; Promega) digests were carried out at 37 °C for 30 min. After digests, enzymes were heat inactivated at 80 °C for 20 min. Electrophoresis and image analysis were performed as above.
Top3 Decatenation Reactions
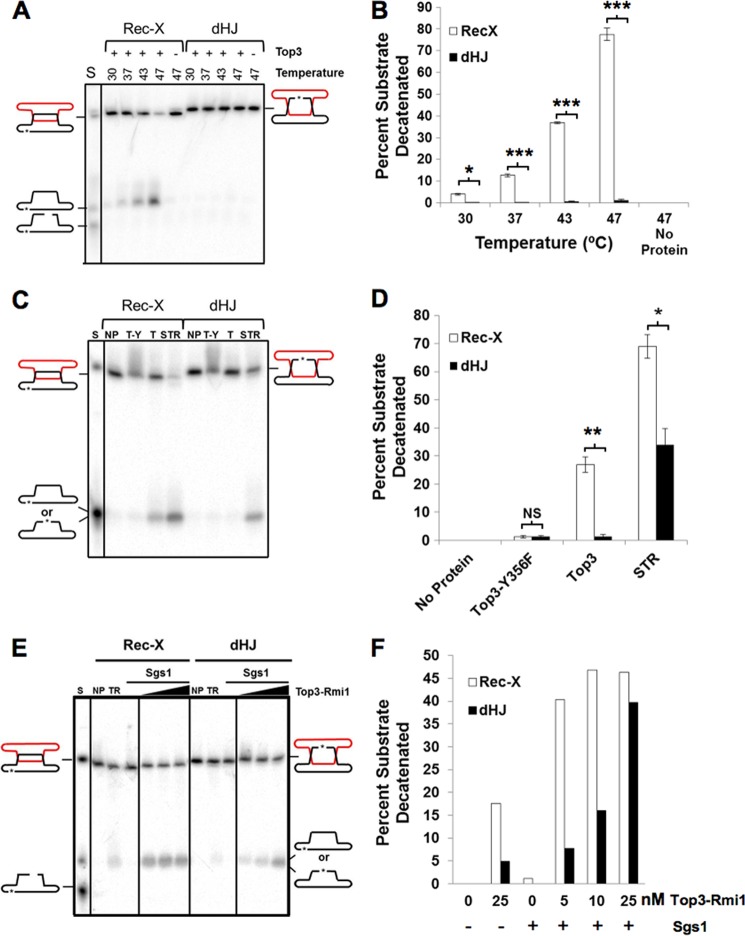
Top3 protein was prepared as described previously (44). Reactions shown in Fig. 8, A and B, were carried out using the indicated amounts of protein, and 50 pm DNA substrate in 40 mm Hepes, pH 7.0, 42% glycerol, 5 mm sodium acetate, 65 mm NaCl, 60 mm KCl, 10 μg/ml of BSA, 2 mm MgCl2, 0.2 mm EDTA, 0.2 mm DTT, 0.002% Nonidet P-40, 0.02 mm PMSF, and 1 mm spermidine, at the indicated temperatures, for 2 h. Reactions shown in Fig. 8, C and D and E and F, were carried out as above but in 7% glycerol buffer, 5 mm ATP, no or 20 mm KCl, and 4 or 6 mm MgCl2, respectively, at 37 °C. All reactions were stopped by the addition of 0.5% SDS and 0.05 mg/ml of proteinase K, incubated at 37 °C for 20 min, and electrophoresis and image analysis were performed as above. p values were calculated using two-tailed t tests.
FIGURE 8.
Top3 decatenates a Rec-X but not a dHJ substrate. A, C, and E, samples were run on denaturing polyacrylamide gels. A, decatenation of Rec-X but not dHJ by Top3. DNA substrates were incubated alone or with 40 nm Top3 at the indicated temperatures. The products of Top3-mediated decatenation migrate with the closed circular form. C, decatenation of a Rec-X by Top3 alone, but not by catalytically inactive Top3, and decatenation of a Rec-X and a dHJ by the STR complex. DNA substrates were incubated alone (NP), with 40 nm Top3-Y356F (T-Y) or Top3 (T), or 40 nm Top3-Rmi1 together with 0.6 nm Sgs1 (STR) at 37 °C. E, decatenation of a Rec-X and a dHJ by the STR complex. DNA substrates were incubated alone (NP), or with 1.2 nm Sgs1 and 0, 5, 10, or 25 nm Top3-Rmi1. B, D, and F, quantification of the data in A–C, respectively. The mobility standard (S) for all gels was made by mixing equal amounts of duplex substrate (Rec-X or dHJ), and circular and linear forms of the labeled strand. Percentages were corrected for background by subtracting the value of the no protein control for each substrate (n = 3 for B and D), *, p < 0.05; **, p < 0.01; ***, p < 0.0002.
Western Blot
Strains were grown to log phase and 108 cells were harvested by centrifugation and frozen for subsequent processing by mechanical disruption with glass beads in a 20% TCA solution. Samples were electrophoresed in 4–15% gradient acrylamide gels (Bio-Rad) and transferred to nitrocellulose membranes for probing with mouse anti-MYC antibody (Abcam #AB32) at a 1:2000 dilution, followed by HRP/chemiluminescence detection.
ADE2 Crossover Assay
Similar to a previously described crossover assay (45), a URA3 marked ARS209-containing plasmid with a ∼600-bp internal fragment of the ADE2 locus was linearized at the HpaI site within the ADE2 fragment. The resulting linear DNA was transformed into the desired yeast strains, cells were selected on SC-URA media and the percentage of red colonies was determined. Red colonies represent crossover repair events involving the formation of a dHJ that is resolved to disrupt the endogenous ADE2 locus. White colonies represent non-crossover events, e.g. repair events where the dHJs were convergently migrated and dissolved. p values were calculated using two-tailed t-tests.
RESULTS
Top3 Promotes DNA Damage Tolerance in sgs1Δ Mutants
As sgs1Δ top3Δ mutants are more MMS-sensitive than sgs1Δ mutants (34, 36, 37), we hypothesized that Top3 might promote DNA damage tolerance, in an Sgs1-independent fashion, through the resolution of toxic recombination intermediates. The alternative explanation, that Top3 impacts checkpoint responses or cell cycle kinetics in sgs1Δ mutant cells, has been ruled out previously (28). To first confirm that Top3 confers DNA damage tolerance in our genetic background, we compared the growth of MMS-treated sgs1Δ and sgs1Δ top3Δ mutants and found a 10-fold increase in DNA damage sensitivity when TOP3 was deleted from sgs1Δ cells (Fig. 2A). To provide further evidence for autonomous Top3 activity, we tested if overexpression of Top3 might further promote DNA damage tolerance in cells lacking Sgs1. A small (5–10-fold) but repeatable increase in MMS resistance was observed upon Top3 overexpression in sgs1Δ but not in wild-type cells, suggesting that the beneficial activity of Top3 is limiting, at least when Sgs1 is absent (Fig. 2B and data not shown). These data prompted us to further investigate the mechanism of Top3-dependent DNA damage resistance.
FIGURE 2.
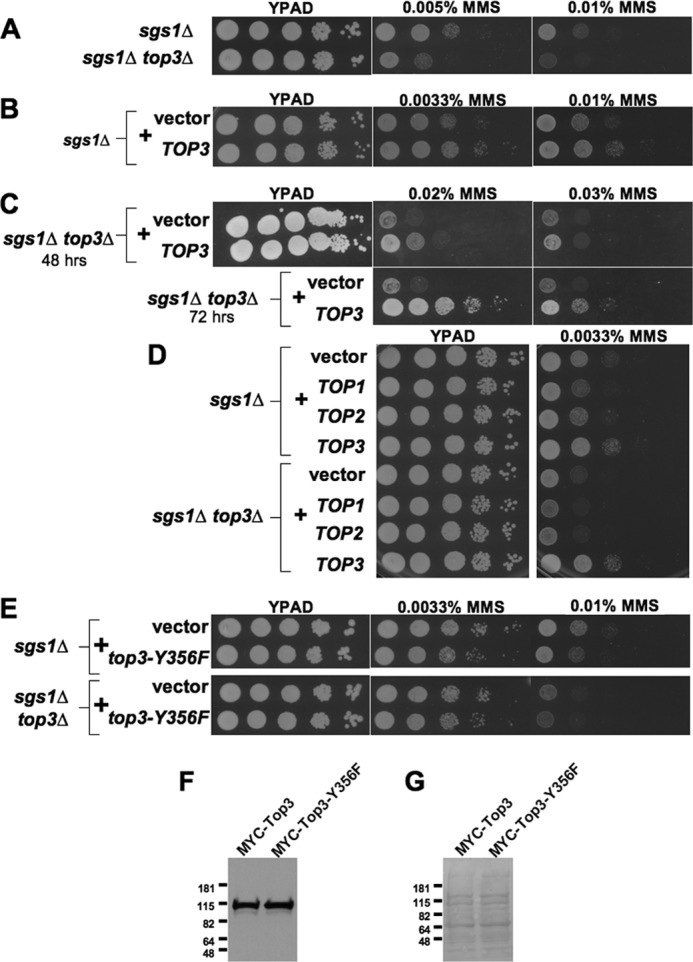
Top3 provides DNA damage resistance in sgs1Δ mutants. Spot assays comparing the effects of MMS on the growth of: A, sgs1Δ and sgs1Δ top3Δ mutants; B, sgs1Δ mutants without or with a TOP3 overexpression plasmid; C, sgs1Δ top3Δ mutants without or with a TOP3 overexpression plasmid showing resistance at higher levels of MMS; D, sgs1Δ and sgs1Δ top3Δ mutants with control vector or plasmids overexpressing TOP1, TOP2, or TOP3; and E, sgs1Δ or sgs1Δ top3Δ cells with control vector or plasmid expressing the catalytically inactive top3-Y356F allele. 10-Fold dilutions of strains were spotted onto YPAD alone or containing the indicated concentrations of MMS and grown as indicated for 48 (and for C up to 72) h. F, Western blot from log-phase sgs1Δ top3Δ cells containing plasmids expressing TOP3 promoter-driven MYC-TOP3 or MYC-top3-Y356F and probed with an anti-Myc tag antibody. G, Ponceau S staining of the same membrane as in F to confirm equal loading of protein. Molecular mass markers are indicated (kDa).
Rescue of sgs1Δ Mutants by Topoisomerase Activity Is Specific to Top3, and Can Be Conferred by Its Human Ortholog Top3α
To test if the identified role for Top3 in promoting DNA damage resistance within sgs1Δ cells is specific to Top3 or might be a general property of topoisomerases, we compared the capacity of Top1, Top2, or Top3 overexpression to rescue the MMS sensitivity of sgs1Δ and sgs1Δ top3Δ mutants. Unlike Top3, overexpression of Top1 or Top2 provided no increased DNA damage resistance in either context (Fig. 2D). The catalytic activity of Top3 was required for rescue, because overexpression of the top3-Y356F mutant, which is defective in the formation of the 5′ phosphotyrosine covalent bond between DNA and Top3, was unable to rescue DNA damage sensitivity in sgs1Δ or sgs1Δ top3Δ mutants, despite equal accumulation of wild-type and Y356F proteins (Fig. 2, E–G) (24, 36).
The selective role for Top3 in the rescue of MMS sensitivity prompted us to ask if this role might be conserved and thus shared by the human Top3 ortholog TOP3α (46). Remarkably, expression of TOP3α, which might be expected to interact poorly with endogenous yeast proteins, provided robust rescue of sgs1Δ top3Δ DNA damage sensitivity (Fig. 3A). This finding supports the idea that Top3 can function independently to confer resistance to MMS, and raises the possibility that this function is conserved between yeast and human cells.
FIGURE 3.
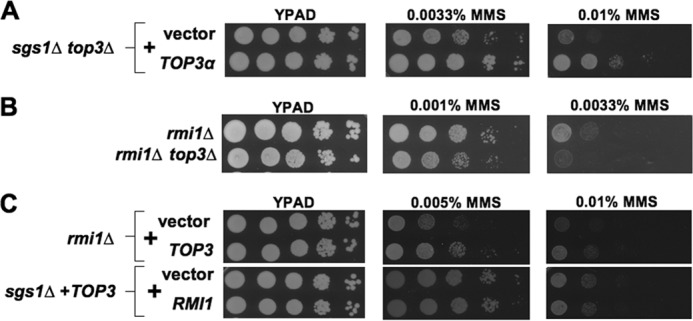
MMS resistance provided by unassisted Top3 is evolutionarily conserved and can occur without RMI1. Spot assays comparing effects of MMS on the growth of: A, sgs1Δ top3Δ cells containing either control vector or human TOP3α expression plasmid; B, rmi1Δ or rmi1Δ top3Δ mutants; and C, rmi1Δ cells with vector or TOP3 overexpression plasmid and sgs1Δ strains containing a TOP3 overexpression plasmid together with an additional control vector or plasmid overexpressing RMI1.
Top3 Promotes DNA Damage Tolerance Independent of Other STR Complex Members
In addition to its association with Sgs1, Top3 binds the OB-fold containing protein Rmi1 (2, 3). Because Rmi1 binds DNA and is reported to stimulate Top3 reaction kinetics in vitro, we asked if DNA damage tolerance provided by Top3 depends upon Rmi1 (11, 35, 44). No role for Rmi1 in Top3-mediated rescue was found as rmi1Δ top3Δ mutants were more sensitive to MMS than rmi1Δ controls, and Top3 overexpression in rmi1Δ mutants provided MMS resistance (Fig. 3, B and C). Furthermore, Rmi1 overexpression did not augment MMS resistance provided by Top3 overexpression in sgs1Δ cells (Fig. 3C). Altogether, our findings indicate that even without assistance by other STR complex members Top3 can provide some resistance to MMS-induced DNA damage.
Recombination Intermediate Resolution by Unassisted Top3 Provides DNA Damage Tolerance
We sought to determine whether the same DNA repair factors that enable the accumulation of HR intermediates during replication of MMS-treated sgs1Δ mutants are also required for the MMS resistance provided by unassisted Top3. Previous biochemical and biophysical analyses employing two-dimensional gel electrophoresis indicated that these intermediates are Rec-Xs (13, 18, 19). On two-dimensional gels, Rec-Xs run as a prominent near-vertical spike originating at the end of the replication arc (e.g. see Fig. 5A, below), as would be expected for joint linkages arising from template switch recombination between sister chromatids. Importantly, not all molecules running within the X-spike are HR-dependent, as low levels of X-shaped species are still observed in homologous recombination-deficient rad51Δ or rad52Δ strains, particularly near origins of replication (47). However, the elevated levels of Rec-Xs that accumulate when sgs1Δ mutants replicate through damaged DNA templates are entirely HR-dependent (13).
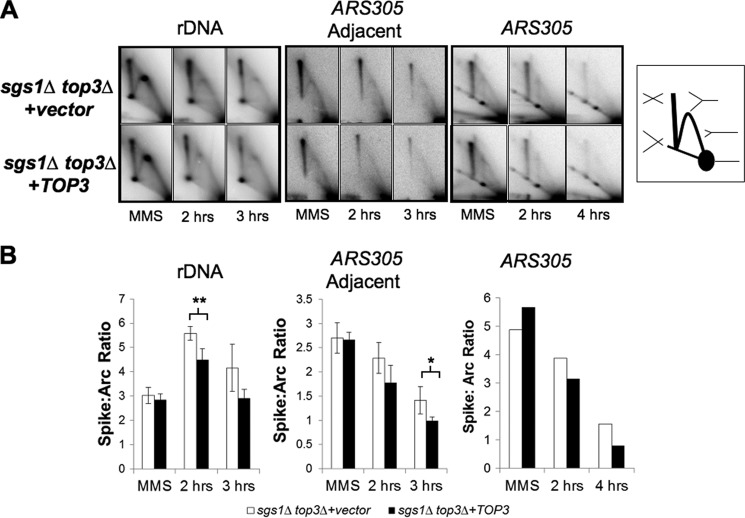
FIGURE 5.
Top3 overexpression reduces the level of X-shaped molecules within sgs1Δ top3Δ cells. A, representative two-dimensional gel electrophoresis Southern blots examining replication intermediates that accumulate in sgs1Δ top3Δ cells containing either vector or TOP3 overexpression plasmid during exposure to MMS and following release into MMS-free medium. An rDNA fragment containing the replication fork block, a fragment adjacent to an ARS305-containing fragment, as well as the ARS305-containing fragment, are shown on the left, middle, and right sets of panels, respectively. MMS indicates cells grown for 1.5 h in 0.033% MMS, and 2 h, 3 h, and 4 h indicate cells 2, 3, and 4 h after release from MMS. Right, schematic showing the location of DNA structures visualized by two-dimensional gel electrophoresis, including the 1 n spot containing non-replicating duplex, the Y-arc containing single replication forks, and the X-spike containing two duplexes interlinked at different positions along their lengths. B, quantification of the ratio of X-shaped molecules to those running within the replication arc (n = 3 biological replicates for the rDNA fragment and ARS305 adjacent fragment, and error bars represent the S.E.; *, p = 0.034; **, p = 0.009).
Several factors are necessary for Rec-X formation in sgs1Δ mutants, including the HR factor Rad52 and the post-replicative repair protein Rad18 (13, 18, 20). Furthermore, it was shown recently that cells defective in another protein complex critical for Rec-X resolution, the Smc5/6 complex, accumulate Rec-X intermediates in a fashion dependent on the DNA helicase Mph1 and the pro-recombination Shu protein complex (consisting of Shu1, Shu2, Psy3, and Csm2) (22, 48–50). Combined deletion of MPH1 and SHU1 completely suppresses Rec-X formation in smc5/6 mutants, and given the biochemical similarity between the Rec-X molecules in smc5/6 and sgs1 mutants, we predicted and confirmed similar suppression of X-shaped molecule formation in sgs1Δ top3Δ by combined deletion of MPH1 and SHU1 (data not shown). Therefore, MPH1 or SHU1, like RAD52 and RAD18, are required for accumulation of X-shaped structures in sgs1Δ top3Δ mutant cells.
If the Sgs1-independent functions of Top3 involve resolution of Rec-X intermediates, Top3 should not impact MMS resistance in backgrounds lacking Sgs1 and factors required for Rec-X formation. As predicted, sgs1Δ rad52Δ, sgs1Δ shu1Δ mph1Δ, and sgs1Δ rad18Δ mutants showed little to no effect of top3 deletion (Fig. 4A). Similarly, Top3 overexpression did not improve the MMS sensitivity of these mutants (Fig. 4B).
FIGURE 4.
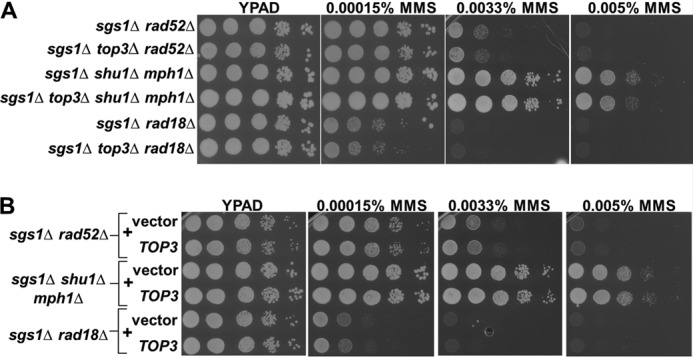
Rescue by unassisted Top3 requires factors that enable the accumulation of unresolved recombination intermediates within sgs1Δ mutants. A, sgs1Δ cells lacking additional factors that render them unable to accumulate recombination intermediates, with or without endogenous TOP3, were spotted onto YPAD containing the indicated concentrations of MMS. B, identical to A except comparing the effects of TOP3 overexpression.
To test directly for effects of Top3 on replication intermediates, we employed two-dimensional gel electrophoresis followed by Southern blotting to visualize events at different genomic regions. As is standard (13, 51–53), higher levels of MMS (0.01–0.033%) were used in two-dimensional gel electrophoresis assays to enable visualization of enhanced X-structure levels under conditions where cells are exposed to MMS for only a single S-phase and where replication must be perturbed in any given genomic fragment that is visualized; importantly, we confirmed that Top3 overexpression improves growth even when cells are exposed to these higher levels of MMS (Fig. 2C).
Logarithmically growing cells were synchronized with α-factor, and released into media containing MMS. Samples were taken at 1.5 h after treatment, and also at 2 and 3 h after release from MMS into fresh YPAD. In agreement with our MMS-resistance analyses, Top3 overexpression within sgs1Δ top3Δ mutants decreased X-shaped intermediates during recovery from MMS treatment (Fig. 5). Whereas there was a trend toward suppression by Top3 overexpression of X-shaped molecule accumulation in a genomic fragment containing ARS305, this suppression did not achieve statistical significance in our assays (Fig. 5 and data not shown). We reasoned that the background of origin-dependent X-structures may have obscured a difference in the MMS-dependent X-structures. To address this, we examined genomic fragments replicated by forks emanating from origins outside the fragments, including an rDNA region containing the replication fork block and a region adjacent and telomeric to the fragment containing ARS305. Significant and reproducible suppression of X-shaped molecules by Top3 overexpression was observed within both of these fragments (Fig. 5). Overall, our findings indicate that Top3 is capable of providing MMS resistance and promoting the resolution of recombination intermediates independently of other STR members.
Unassisted Top3 Does Not Resolve HJ Intermediates Efficiently
Recently, HJ-cleaving enzymes (RusA, Gen1, Mus81/Mms4) were shown to diminish levels of MMS-induced X-shaped molecules within STR complex mutants, leading to the suggestion that the X-structures may be HJs rather than Rec-Xs (see “Discussion”) (52, 53). Because Top3 confers resistance to DNA damage and decreases the level of X-shaped molecules that accumulate in STR mutant cells, we used Top3 manipulation to further dissect the nature of the X-shaped molecules. The X-shaped molecules that accumulate during DSBR are known to be HJs (10), and we therefore tested whether Top3, independent of Sgs1, could affect the outcomes of DSBR. We employed a previously characterized ADE2 recombination repair assay (45) in which the rate of crossover and non-crossover events are reflected by the frequencies of red and white colonies, respectively. As demonstrated previously, sgs1Δ mutants show a statistically significant increase in the ratio of crossover events as compared with wild-type strains (Fig. 6A) (12). However, in contrast to its effects on MMS resistance and replication-associated X-shaped molecules, and consistent with earlier findings, the deletion of TOP3 from sgs1Δ mutants causes no additional change in crossover events (12, 54). Similarly, overexpression of Top3 in the sgs1Δ top3Δ background did not alter the frequency of crossover events (Fig. 6B). These findings are consistent with the idea that Top3 cannot by itself resolve HJ intermediates, and support the idea that the X-shaped intermediates that are resolved by such unassisted Top3 during replication are likely not HJ intermediates.
FIGURE 6.
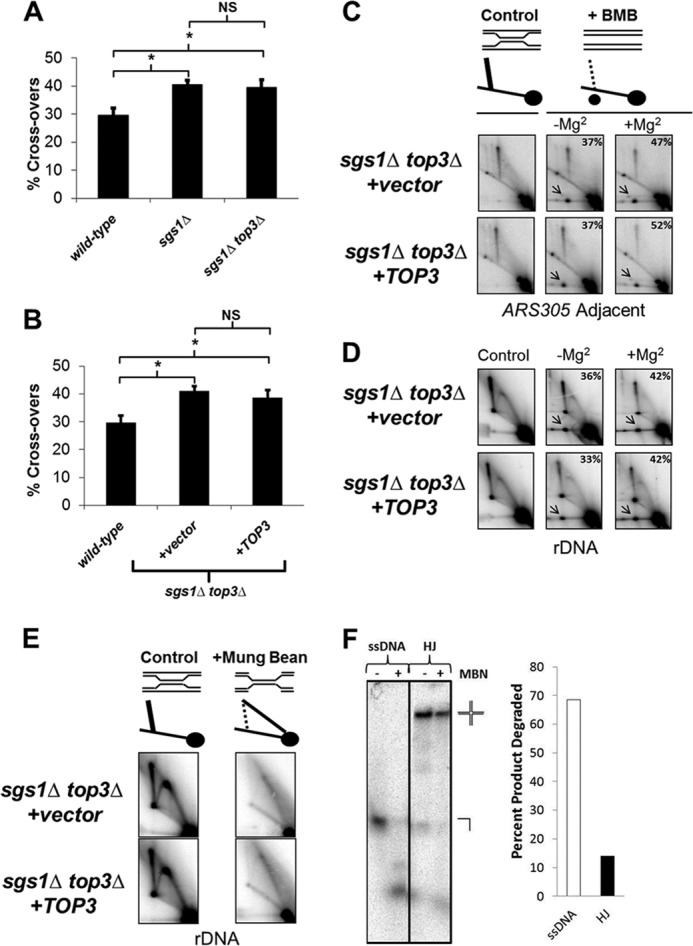
Unassisted Top3 promotes Rec-X resolution but not HJ resolution. A, wild-type, sgs1Δ, and sgs1Δ top3Δ cells were transformed with the ADE2 cassette, and the ratios of crossover-containing to total (crossover + non-crossover) colonies are plotted on the y axis. Data are from three independent experiments and error bars represent the S.E.; *, p < 0.05; NS, not significant. B, identical to A except testing the effects of Top3 overexpression. C and D, schematic: resolved products are indicated by the new spot in the right panel. Southern blot of genomic DNA extracted from sgs1Δ top3Δ cells containing either vector or TOP3 overexpression plasmid, showing resolution of X-shaped molecules into branch migrated products (arrows) at the ARS305 adjacent fragment (C) and rDNA fragment (D). Samples were incubated at 65 °C in branch migration buffer (BMB) ± Mg2+ between the first and second dimensions of electrophoresis. Percentages of branch migrated products are indicated in the top right corner of panels. E, MBN treatment of genomic DNA extracted from sgs1Δ top3Δ cells containing either vector or TOP3 overexpression plasmid, showing alteration in rDNA migration, consistent with Rec-X structures as indicated in the schematic. F, 8% native polyacrylamide gel showing MBN digestion products of a 5′-32P-end labeled 50-base long oligonucleotide alone (ssDNA) or annealed into a synthetic HJ substrate that was added into genomic DNA MBN digests under the same conditions as in E. The percentages of substrates digested are shown on the right. Note that the samples for C and E are from cells grown for 1.5 h in 0.033% MMS, prior to Top3-induced differences in X-structure resolution, whereas samples for D are from cells 2 h after release from MMS.
The X-shaped Molecules That Accumulate without Top3 Are Rec-X Structures
Previous analyses of the MMS-induced recombination intermediates that accumulate within sgs1Δ mutants revealed that they are Rec-X structures which, unlike HJs, can branch migrate unhindered by magnesium and are susceptible to cleavage by MBN (13). Although we expected a similar identity for the X-structures in sgs1Δ top3Δ, it was conceivable that they were instead HJs or regressed forks (i.e. “chicken foot” structures). We examined samples from cells grown for 1.5 h in 0.033% MMS, prior to Top3-induced differences in X-structure resolution. As predicted, the MMS-induced X-shaped molecules in sgs1Δ top3Δ mutants showed an equivalent capacity to be branch migrated to resolution in the presence or absence of Mg2+ (Fig. 6, C and D). Furthermore, they demonstrated sensitivity to cleavage by MBN as well as a migration pattern consistent with the digested products of Rec-X structures (Fig. 6E). MBN specificity under these conditions was confirmed by demonstrating efficient cleavage of trace quantities of 5′-32P-end labeled ssDNA but not HJ substrates added into genomic DNA digestion reactions that were otherwise identical to those above (Fig. 6F).
Unassisted Top3 Decatenates Rec-X but Not dHJ Substrates in Vitro
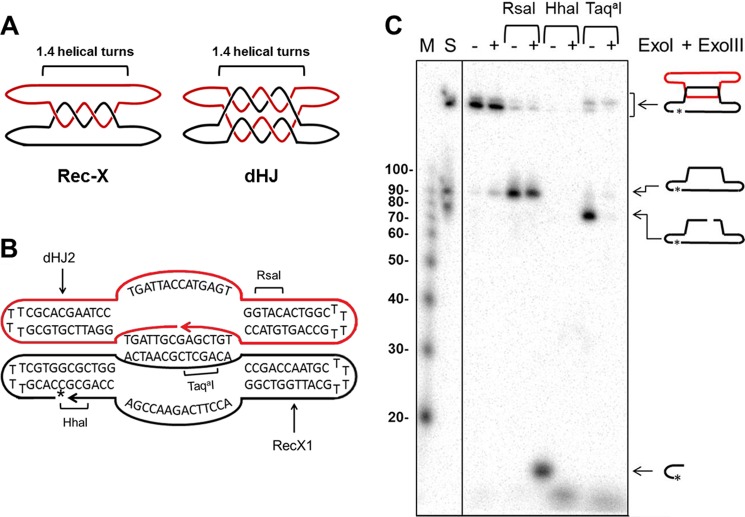
To confirm that unassisted Top3 can directly and selectively resolve Rec-Xs, we generated synthetic Rec-X and dHJ substrates, and tested their resolution by purified Top3 in vitro. The dHJ substrate has been described previously (15, 42, 43) and was assembled from two 80-mers, each forming intrastrand hairpinned duplex arms and two interstrand plectonemically coiled duplexes, each ∼1.4 helical turns in length (Fig. 7A). The Rec-X was derived from the dHJ 80-mers, with one strand modified such that the two strands formed only one of the duplexes, and had unpaired “outside” strands (Fig. 7, A and B; see “Experimental Procedures”). The Rec-X structure was confirmed using site-specific endonucleases together with exonucleases (Fig. 7C). Each substrate contained a single 32P-labeled strand and a total of two interlinks between strands.
FIGURE 7.
Characterization of synthetic Rec-X structure. A, schematic representations of the Rec-X and dHJ substrates, showing where the two oligonucleotides are topologically interlinked. For simplicity, the helical turns are not drawn in subsequent figures. B, Rec-X map showing the sequences of the RecX1 and dHJ2 oligos that compose it, regions of double-stranded DNA, and RsaI, HhaI, and TaqaI cut sites. The 32P-labeled phosphate is indicated by an asterisk. C, 8% polyacrylamide denaturing gel confirming the structure of Rec-X. First lane (M), 5′-32P-end labeled 10-bp DNA Step ladder (Promega); second lane 2 (S) standard comprising Rec-X, and ligated and unligated forms of the 80-nt long RecX1 oligo; third and fourth lanes, Rec-X without restriction enzyme treatment; fifth and sixth lanes, digested with RsaI; seventh and eighth lanes, digested with HhaI; ninth and tenth lanes, digested with TaqaI. Fourth, sixth, eighth, and tenth lanes, samples were further treated with E. coli ExoI and ExoIII, which degrade single-stranded and double-stranded DNA, respectively, in a 3′ → 5′ direction. Note that the TaqaI digest was performed at 47 °C, indicating that the inter-strand duplex region remained at least partially double stranded during the Top3 decatenation experiments, even at an elevated temperature. In addition we independently confirmed the structure of dHJ in a fashion similar to the characterization of the Rec-X (data not shown), as described previously (43).
Top3 was incubated with each substrate and strand decatenation was assessed using denaturing PAGE to separate the substrates from products (Fig. 8). A range of temperatures was examined, because Top3 topoisomerase activity is enhanced at elevated temperatures (25, 44, 55). Consistent with previous studies demonstrating that the dHJ substrate is resistant to Top3 alone but can be completely dissolved by Top3 combined with Sgs1 (11), Top3 was unable to decatenate the dHJ substrate even at high temperatures. However, Top3 displayed robust decatenation activity on the Rec-X substrate (up to 76%), in a temperature-dependent fashion (Fig. 8, A and B). Increased temperature alone did not result in decatenation of the Rec-X substrate, as evidenced by the no protein control, which was also treated at the highest temperature (47 °C). We suspect that the increased temperature could allow the Rec-X to adopt partial ssDNA character, making it a better substrate for Top3. However, we note that this does not appear to explain the specificity of Top3 for the Rec-X substrate because elevated temperatures should affect dHJ substrate similarly. It is also clear that the Rec-X retains significant duplex character at elevated temperatures because it is cleaved efficiently by the TaqaI endonuclease at 47 °C (Fig. 7). Decatenation of the Rec-X was dependent on the catalytic activity of Top3, as the catalytically inactive Top3-Y356F was incapable of resolving the structure (Fig. 8, C and D). As expected, incubation with the entire STR complex showed robust decatenation of both the dHJ and the Rec-X (Fig. 8, C and D). The fact that the STR complex is more active than Top3 in the resolution of the Rec-X is consistent with our in vivo data showing that although unassisted Top3 has some capacity to resolve X-shaped replication intermediates and provide some resistance to MMS, it does not do so as robustly as the full STR complex. These findings are also consistent with the capacity of purified human TOP3α to decatenate entwined single-stranded circles, in a fashion stimulated by BLM and RMI1 (35). Interestingly, the STR complex as a whole was almost twice as active on the Rec-X than on the dHJ at lower temperatures, and showed considerably more activity on the Rec-X than the dHJ at low protein concentrations, consistent with the idea that the STR complex processes Rec-X as well as dHJ substrates in vivo (Fig. 8, C–F). We note that Rmi1 does enable weak resolution by Top3 of the dHJ, as well as slightly stimulate the ability of Top3 to resolve the Rec-X (Fig. 8, E and F, and see Fig. 8, A and B; and data not shown). This contrasts with the activity of TOP3 unassisted by SGS1 in vivo, for which RMI1 does not enable crossover inhibition or stimulate resistance to MMS. A possible explanation for these apparent discrepancies is that the two interlinks between the entwined strands of the synthetic Rec-X and dHJ substrates are fewer in number than the interlinks between X-structures in vivo. Rmi1 has been shown to stimulate Top3 activity particularly at the final decatenation step (11), and thus might provide greater assistance to Top3 on the synthetic substrates than on highly interlinked substrates in vivo.
DISCUSSION
Here we show that the type 1A topoisomerase Top3 can confer significant resistance to MMS-induced DNA damage in the absence of its STR complex partners, Sgs1 and Rmi1. This function of Top3 requires its catalytic activity and also involves HR pathways, as strains defective in the accumulation of X-shaped recombination intermediates (sgs1Δ rad52Δ, sgs1Δ shu1Δ mph1Δ, or sgs1Δ rad18Δ mutants) failed to show altered DNA damage sensitivity upon TOP3 deletion or overexpression. Consistent with these findings, Top3 overexpression diminished X-structures visualized by two-dimensional gel electrophoresis and Southern blotting, indicating that Top3 promotes resolution of X-structures.
Although we have no reason to believe that Top3 functions apart from the STR complex under natural conditions, we took advantage of its unassisted activity to investigate the MMS-induced X-shaped replication intermediates, whose structure has been a matter of debate (56). In particular, recent investigations of factors (e.g. HJ resolvases) capable of resolving recombination intermediates that accumulate within STR mutants have concluded that the X-shaped intermediates previously characterized as Rec-Xs instead represent unresolved HJs (52, 53). However, to our knowledge, in all cases where X-structures induced by replication in the presence of MMS have been examined at a biochemical level, they have been found to have features most consistent with Rec-X species rather than HJs (13, 18, 20). These features include branch migration unimpeded by Mg2+, susceptibility of the outside single-stranded regions to cleavage by MBN, and relative resistance to cleavage by nucleases that cleave HJs, including T4 endonuclease VII and RuvC. On the other hand, it is clear that X-shaped intermediates formed during DSBR such as during meiotic crossover events are HJs (57). During DSBR, the STR complex promotes the branch migration of double HJs and ultimately their mutual removal by a dissolution mechanism. Consistent with a role for unassisted Top3 in the resolution of Rec-X intermediates but not HJs, we found that Top3 overexpression did not impact crossover frequencies in a DSBR assay, and that sgs1Δ top3Δ mutants had crossover levels no greater than sgs1Δ mutants. These observations are in agreement with previous studies (12, 54). We also note that although conversion of a stalled replication fork to a DSB is presumably followed by formation of a HJ during HR-dependent resumption of replication, MMS does not actually cause significant levels of DSBs, consistent with MMS-induced X-structures not being HJs (58).
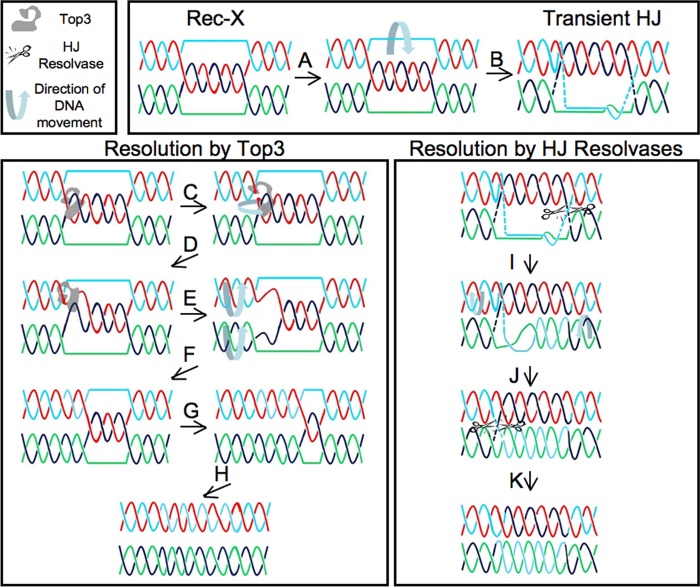
How then, can one reconcile the apparently conflicting viewpoints concerning the nature of replication-related X-structures? We suggest two non-mutually exclusive possibilities. First, the HJ resolvases identified as assisting in the resolution of X-shaped molecules within STR complex mutants (Mus81/Mms4, RusA, and Gen1(1–527)) may process Rec-X substrates, although perhaps inefficiently. These nucleases clearly process HJs, but when carefully tested in vitro, each of these nucleases also binds and cleaves additional substrates (59–64). Because the discovery of Rec-X intermediates is still rather new, none of these identified HJ resolvases have been tested to see if they possess in vitro activity against Rec-X molecules. Indeed, to our knowledge, our in vitro studies with Top3 represent the first construction of a synthetic Rec-X substrate. We speculate that because Rec-X molecules have properties in common with HJs and replication forks, both of which Mus81/Mms4, RusA, and Gen1 all bind and act upon, it would not be surprising if each of these enzymes can process Rec-X molecules. Mus81/Mms4 in particular is a good candidate Rec-X resolvase as it cleaves HJs very inefficiently in vitro (65), but nonetheless has been demonstrated to be the enzyme responsible for resolution of MMS-induced X-spikes in the absence of the STR complex (31, 53). The second possibility is that Rec-Xs might transiently assume a more HJ-like character, potentially allowing processing by HJ resolvases. This conversion of a Rec-X into a HJ might take place by base-pairing the outside strands at a Rec-X junction (via paranemic coiling) to generate a HJ (Fig. 9). The very slow rate at which X-spikes are removed by overexpressed HJ resolvases is consistent with both of these models (52, 53). Ultimately, to settle this issue it will be important to test the activity of HJ resolvases on model Rec-X structures and also to perform more detailed structural studies on the X-shaped molecules that accumulate during replication of damaged templates.
FIGURE 9.
Models for Rec-X-HJ isomerization, and for Rec-X resolution via Top3 versus a HJ resolvase. Top: model for how a Rec-X can isomerize into a HJ. The outside strands are capable of base pairing (A), which would be limited by stress induced by the positive twist in the region of base pairing and paranemic coiling. This stress could be absorbed by compensatory negative twist in the adjacent stretch(es) of the outside strands (not shown), and so base pairs should be able to form at least transiently. Here such base pairing is shown adjacent to the right junction, thus forming a HJ (B). Lower left, model for how a Rec-X can be resolved by Top3. Top3 binds the Rec-X and cleaves one of the plectonemically coiled strands (red; C), enables passage of the intact strand (black), then religates the strand (D). The resulting negative twist is relieved by duplex rotation (E), leading to a Rec-X with one less linkage (F). G and H, the products expected after steps C–F are repeated one and two more times, respectively. Lower right, model for how a Rec-X can be resolved by a HJ resolvase. The transient HJ described above is cleaved by the HJ resolvase (I), allowing the original outside strands to coil plectonemically around each other (J). This enables the linkage on the left to form a HJ that is subsequently cleaved by the HJ resolvase (K). Note that crossing-over does not occur with resolution by Top3, but can occur with resolution via HJ resolvases.
Acknowledgments
We thank Amy Tsou, Anthony Chi, Martin Kupiec, Lorraine Symington, Xiaolan Zhao, and members of the Johnson lab, including Jay Johnson and Jen Wanat, for thoughtful discussions and/or comments on the manuscript. We thank Jesse Platt for help with experiments related to an earlier version of the manuscript. Aaron Gitler is acknowledged for the generous gifts of yeast strains and plasmids.
This work was supported, in whole or in part, by National Institutes of Health Grant R25 CA101871 from the NCI (to M. R. G.) and Grants sT32-AG000255 (to A. C.), R01-GM071268 (to S. J. B.), and R01-AG021521 and P01-AG031862 (to F. B. J.).
- HR
- homologous recombination
- STR
- Sgs1/Top3/Rmi1
- HJ
- Holliday junctions
- DSBR
- double strand break repair
- MBN
- mung bean nuclease
- MMS
- methyl methanesulfonate.
REFERENCES
- 1. Larsen N. B., Hickson I. D. (2013) RecQ Helicases. Conserved guardians of genomic integrity. Adv. Exp. Med. Biol. 767, 161–184 [DOI] [PubMed] [Google Scholar]
- 2. Chang M., Bellaoui M., Zhang C., Desai R., Morozov P., Delgado-Cruzata L., Rothstein R., Freyer G. A., Boone C., Brown G. W. (2005) RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J. 24, 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mullen J. R., Nallaseth F. S., Lan Y. Q., Slagle C. E., Brill S. J. (2005) Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol. Cell. Biol. 25, 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Myung K., Datta A., Chen C., Kolodner R. D. (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27, 113–116 [DOI] [PubMed] [Google Scholar]
- 5. Wallis J. W., Chrebet G., Brodsky G., Rolfe M., Rothstein R. (1989) A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58, 409–419 [DOI] [PubMed] [Google Scholar]
- 6. Watt P. M., Louis E. J., Borts R. H., Hickson I. D. (1995) Sgs1. A eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81, 253–260 [DOI] [PubMed] [Google Scholar]
- 7. Bohr V. A. (2008) Rising from the RecQ-age. The role of human RecQ helicases in genome maintenance. Trends Biochem. Sci. 33, 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monnat R. J., Jr. (2010) Human RECQ helicases. Roles in DNA metabolism, mutagenesis and cancer biology. Semin. Cancer Biol. 20, 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ashton T. M., Hickson I. D. (2010) Yeast as a model system to study RecQ helicase function. DNA Repair 9, 303–314 [DOI] [PubMed] [Google Scholar]
- 10. Bzymek M., Thayer N. H., Oh S. D., Kleckner N., Hunter N. (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cejka P., Plank J. L., Bachrati C. Z., Hickson I. D., Kowalczykowski S. C. (2010) Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 17, 1377–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ira G., Malkova A., Liberi G., Foiani M., Haber J. E. (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oh S. D., Lao J. P., Hwang P. Y., Taylor A. F., Smith G. R., Hunter N. (2007) BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu L., Hickson I. D. (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 16. Kass E. M., Jasin M. (2010) Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 584, 3703–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Branzei D. (2011) Ubiquitin family modifications and template switching. FEBS Lett. 585, 2810–2817 [DOI] [PubMed] [Google Scholar]
- 18. Chavez A., George V., Agrawal V., Johnson F. B. (2010) Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution. J. Biol. Chem. 285, 11922–11930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee J. Y., Kozak M., Martin J. D., Pennock E., Johnson F. B. (2007) Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLOS Biol. 5, e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Branzei D., Vanoli F., Foiani M. (2008) SUMOylation regulates Rad18-mediated template switch. Nature 456, 915–920 [DOI] [PubMed] [Google Scholar]
- 21. Mankouri H. W., Hickson I. D. (2006) Top3 processes recombination intermediates and modulates checkpoint activity after DNA damage. Mol. Biol. Cell 17, 4473–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mankouri H. W., Ngo H. P., Hickson I. D. (2007) Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell 18, 4062–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koster D. A., Crut A., Shuman S., Bjornsti M. A., Dekker N. H. (2010) Cellular strategies for regulating DNA supercoiling. A single-molecule perspective. Cell 142, 519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bennett R. J., Wang J. C. (2001) Association of yeast DNA topoisomerase III and Sgs1 DNA helicase. Studies of fusion proteins. Proc. Natl. Acad. Sci. U.S.A. 98, 11108–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim R. A., Wang J. C. (1992) Identification of the yeast TOP3 gene product as a single strand-specific DNA topoisomerase. J. Biol. Chem. 267, 17178–17185 [PubMed] [Google Scholar]
- 26. Wang J. C. (2002) Cellular roles of DNA topoisomerases. A molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 27. Cejka P., Plank J. L., Dombrowski C. C., Kowalczykowski S. C. (2012) Decatenation of DNA by the S. cerevisiae Sgs1-Top3-Rmi1 and RPA complex. A mechanism for disentangling chromosomes. Mol. Cell 47, 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chakraverty R. K., Kearsey J. M., Oakley T. J., Grenon M., de La Torre Ruiz M. A., Lowndes N. F., Hickson I. D. (2001) Topoisomerase III acts upstream of Rad53p in the S-phase DNA damage checkpoint. Mol. Cell. Biol. 21, 7150–7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gangloff S., de Massy B., Arthur L., Rothstein R., Fabre F. (1999) The essential role of yeast topoisomerase III in meiosis depends on recombination. EMBO J. 18, 1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bennett R. J., Noirot-Gros M. F., Wang J. C. (2000) Interaction between yeast sgs1 helicase and DNA topoisomerase III. J. Biol. Chem. 275, 26898–26905 [DOI] [PubMed] [Google Scholar]
- 31. Szakal B., Branzei D. (2013) Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 32, 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gangloff S., McDonald J. P., Bendixen C., Arthur L., Rothstein R. (1994) The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog. A potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14, 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ui A., Satoh Y., Onoda F., Miyajima A., Seki M., Enomoto T. (2001) The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol. Genet. Genomics 265, 837–850 [DOI] [PubMed] [Google Scholar]
- 34. Fricke W. M., Kaliraman V., Brill S. J. (2001) Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 276, 8848–8855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J., Bachrati C. Z., Ou J., Hickson I. D., Brown G. W. (2010) Human topoisomerase IIIα is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J. Biol. Chem. 285, 21426–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Onodera R., Seki M., Ui A., Satoh Y., Miyajima A., Onoda F., Enomoto T. (2002) Functional and physical interaction between Sgs1 and Top3 and Sgs1-independent function of Top3 in DNA recombination repair. Genes Genet. Syst. 77, 11–21 [DOI] [PubMed] [Google Scholar]
- 37. Wagner M., Price G., Rothstein R. (2006) The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174, 555–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alberti S., Gitler A. D., Lindquist S. (2007) A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liberi G., Cotta-Ramusino C., Lopes M., Sogo J., Conti C., Bensimon A., Foiani M. (2006) Methods to study replication fork collapse in budding yeast. Methods Enzymol. 409, 442–462 [DOI] [PubMed] [Google Scholar]
- 40. Fritsch O., Burkhalter M. D., Kais S., Sogo J. M., Schär P. (2010) DNA ligase 4 stabilizes the ribosomal DNA array upon fork collapse at the replication fork barrier. DNA Repair 9, 879–888 [DOI] [PubMed] [Google Scholar]
- 41. Whitby M. C., Bolt E. L., Chan S. N., Lloyd R. G. (1996) Interactions between RuvA and RuvC at Holliday junctions. Inhibition of junction cleavage and formation of a RuvA-RuvC-DNA complex. J. Mol. Biol. 264, 878–890 [DOI] [PubMed] [Google Scholar]
- 42. Bachrati C. Z., Hickson I. D. (2009) Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIα. Methods Mol. Biol. 582, 91–102 [DOI] [PubMed] [Google Scholar]
- 43. Fu T. J., Kemper B., Seeman N. C. (1994) Cleavage of double-crossover molecules by T4 endonuclease VII. Biochemistry 33, 3896–3905 [DOI] [PubMed] [Google Scholar]
- 44. Chen C. F., Brill S. J. (2007) Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J. Biol. Chem. 282, 28971–28979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tay Y. D., Sidebotham J. M., Wu L. (2010) Mph1 requires mismatch repair-independent and -dependent functions of MutSα to regulate crossover formation during homologous recombination repair. Nucleic Acids Res. 38, 1889–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hanai R., Caron P. R., Wang J. C. (1996) Human TOP3. A single-copy gene encoding DNA topoisomerase III. Proc. Natl. Acad. Sci. U.S.A. 93, 3653–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lopes M., Cotta-Ramusino C., Liberi G., Foiani M. (2003) Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell 12, 1499–1510 [DOI] [PubMed] [Google Scholar]
- 48. Chavez A., Agrawal V., Johnson F. B. (2011) Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J. Biol. Chem. 286, 5119–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen Y. H., Choi K., Szakal B., Arenz J., Duan X., Ye H., Branzei D., Zhao X. (2009) Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. U.S.A. 106, 21252–21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choi K., Szakal B., Chen Y. H., Branzei D., Zhao X. (2010) The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2306–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vanoli F., Fumasoni M., Szakal B., Maloisel L., Branzei D. (2010) Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 6, e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mankouri H. W., Ashton T. M., Hickson I. D. (2011) Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 108, 4944–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ashton T. M., Mankouri H. W., Heidenblut A., McHugh P. J., Hickson I. D. (2011) Pathways for Holliday junction processing during homologous recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 31, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robert T., Dervins D., Fabre F., Gangloff S. (2006) Mrc1 and Srs2 are major actors in the regulation of spontaneous crossover. EMBO J. 25, 2837–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wilson T. M., Chen A. D., Hsieh T. (2000) Cloning and characterization of Drosophila topoisomerase IIIβ. Relaxation of hypernegatively supercoiled DNA. J. Biol. Chem. 275, 1533–1540 [DOI] [PubMed] [Google Scholar]
- 56. Liberi G., Foiani M. (2010) The double life of Holliday junctions. Cell Res. 20, 611–613 [DOI] [PubMed] [Google Scholar]
- 57. Schwacha A., Kleckner N. (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83, 783–791 [DOI] [PubMed] [Google Scholar]
- 58. Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A. S., Helleday T. (2005) Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33, 3799–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boddy M. N., Gaillard P. H., McDonald W. H., Shanahan P., Yates J. R., 3rd, Russell P. (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548 [DOI] [PubMed] [Google Scholar]
- 60. Bolt E. L., Lloyd R. G. (2002) Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 10, 187–198 [DOI] [PubMed] [Google Scholar]
- 61. Chan S. N., Harris L., Bolt E. L., Whitby M. C., Lloyd R. G. (1997) Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem. 272, 14873–14882 [DOI] [PubMed] [Google Scholar]
- 62. Ip S. C., Rass U., Blanco M. G., Flynn H. R., Skehel J. M., West S. C. (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456, 357–361 [DOI] [PubMed] [Google Scholar]
- 63. Kaliraman V., Mullen J. R., Fricke W. M., Bastin-Shanower S. A., Brill S. J. (2001) Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rass U., Compton S. A., Matos J., Singleton M. R., Ip S. C., Blanco M. G., Griffith J. D., West S. C. (2010) Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 24, 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fricke W. M., Bastin-Shanower S. A., Brill S. J. (2005) Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair 4, 243–251 [DOI] [PubMed] [Google Scholar]