Background: The subtilisin-like SUB1 is involved in Plasmodium egress from erythrocytes.
Results: Using conditional mutagenesis, we show that SUB1 plays an essential role during Plasmodium hepatic stages.
Conclusion: SUB1 has a dual pivotal role in parasite egress from host hepatocytes and erythrocytes.
Significance: Its critical involvement in hepatic and erythrocytic parasite development qualifies SUB1 as a multistage drug target.
Keywords: Hepatocyte, Malaria, Molecular Genetics, Plasmodium, Protease, Conditional Mutagenesis, Egress, Subtilisin
Abstract
In their mammalian host, Plasmodium parasites have two obligatory intracellular development phases, first in hepatocytes and subsequently in erythrocytes. Both involve an orchestrated process of invasion into and egress from host cells. The Plasmodium SUB1 protease plays a dual role at the blood stage by enabling egress of the progeny merozoites from the infected erythrocyte and priming merozoites for subsequent erythrocyte invasion. Here, using conditional mutagenesis in P. berghei, we show that SUB1 plays an essential role at the hepatic stage. Stage-specific sub1 invalidation during prehepatocytic development showed that SUB1-deficient parasites failed to rupture the parasitophorous vacuole membrane and to egress from hepatocytes. Furthermore, mechanically released parasites were not adequately primed and failed to establish a blood stage infection in vivo. The critical involvement of SUB1 in both pre-erythrocytic and erythrocytic developmental phases qualifies SUB1 as an attractive multistage target for prophylactic and therapeutic anti-Plasmodium intervention strategies.
Introduction
Malaria is a widespread parasitic disease caused by the Plasmodium parasite. In 2011, ∼600,000 deaths due to malaria were reported by the World Health Organization, whereas half of the world's population is exposed to the parasites (1). Parasite resistance to multiple antimalarials is increasing, and resistance of Plasmodium falciparum to artemisinin derivatives has already been described (2–5). This strengthens the urgent need for new chemotherapeutic approaches targeting vital parasite processes.
Plasmodium is an obligate intracellular protozoan of the Apicomplexa phylum. Infection starts when the parasite form called sporozoite is injected into the skin of the host during the Anopheles mosquito bite. Sporozoites rapidly reach the liver and invade hepatocytes, into which they multiply and differentiate into erythrocyte-infecting parasite forms, called merozoites. Once released into the bloodstream, merozoites invade erythrocytes, initiating the intraerythrocytic cycles that cause the symptoms of the disease.
Plasmodium sporozoites and merozoites, like most invasive stages of Apicomplexa, invade their respective host cell by actively penetrating inside a parasitophorous vacuole (PV),5 which is mainly derived from the invaginated host cell membrane around the internalized parasite. After multiplication by schizogony inside the PV, the progeny merozoites egress from erythrocytes following a sequential quick lysis of both the PV and host cell membranes (6–8). In contrast, the way merozoites egress from hepatocytes involves a more distinct two-step event; they first rupture the PV membrane (PVM) but not the hepatocyte plasma membrane (HPM), inducing the formation of merosomes, which have not been observed during the egress of erythrocytic merozoites (9, 10). Merosomes are HPM-bound vesicles that bud into the liver sinusoid and act as shuttles for hepatic merozoites (9, 11), avoiding their phagocytosis by macrophages in the liver. The merosome/HPM membrane appears to rupture only in the lung microvasculature, releasing hepatic merozoites in a safer environment (10, 12).
Although little is known regarding the egress of hepatic merozoites, several actors have been shown to participate in Plasmodium erythrocytic merozoite egress, including host enzymes (13, 14) and parasite perforin-like proteins (15–17). Importantly, a cascade of parasite proteases plays a pivotal role during the egress of erythrocytic merozoites and subsequent invasion into erythrocytes (6, 9, 16, 18–20). The subtilisin-like protease SUB1, a bacterial like serine protease, is a Plasmodium merozoite product essential for the parasite erythrocytic cycle (18). In Plasmodium falciparum, SUB1 was shown to have a dual function. It is crucial for merozoite egress from erythrocytes and renders released merozoites competent for a new round of erythrocyte invasion (18, 21). In the process of P. falciparum merozoite egress from erythrocytes, SUB1 carries out the maturation of the family of papain-like proteases called serine repeat antigens (SERAs) (18, 19, 22). It remains unknown whether the SERAs have a direct role in membrane disruption or whether they in turn maturate other effectors, but it is established that their SUB1-dependent maturation is essential for the egress of merozoite (18). The reported role of SUB1 in P. falciparum merozoite invasion is to undertake the processing of merozoite surface proteins (MSPs), including MSP1, the major merozoite surface protein and a leading malaria vaccine candidate (21). After SUB1 maturation, the membrane-anchored C-terminal 42-kDa fragment of MSP1 remains associated with the other MSP1 fragments and additional partners, including MSP6 and MSP7 (21, 23), which are also processed by SUB1 (21). Although functional studies suggest that MSPs promote initial binding of the merozoite to the erythrocyte surface, their exact function remains unclear. However, MSP processing by SUB1 is necessary for efficient erythrocyte invasion.
Whether SUB1 also plays a role at the hepatic stages of Plasmodium is unknown. Although SUB1 has so far only been reported as being expressed in erythrocytic stages, the SERAs are expressed in both erythrocytic and liver stages. The P. falciparum genome contains nine SERA-encoding genes, all transcribed during the parasite erythrocytic cycle (20). The genome of the rodent malaria parasite Plasmodium berghei encodes five SERAs (24), and four of these are known to be expressed in liver stages (25, 26). Moreover, hepatic merozoites express MSPs and are able to invade erythrocytes. In this work, we show that SUB1 is expressed by hepatic merozoites, and, by generating sub1-null liver stages using FLP/flippase recognition target (FRT)-mediated conditional mutagenesis in P. berghei (27, 28), we show that SUB1-deficient hepatic merozoites are unable to egress from infected hepatocytes and to establish a blood infection. Together, these data show that SUB1 plays a key role during the last phase of Plasmodium biological development into host hepatocytes, qualifying this enzyme as an attractive target of intervention strategies aiming at preventing as well as curing infection in humans.
EXPERIMENTAL PROCEDURES
Parasites, Mosquitoes, and Mice
The wild-type and GFP-expressing P. berghei ANKA clones were used to study PbSUB1 in erythrocytic and liver stages, respectively (29, 30). The P. berghei NK65 UIS4/FLP and GFP-expressing clone, which expresses the FLP under the control of the uis4 promoter (27), was transfected to obtain the SUB1/Cond conditional clones. Anopheles stephensi (Sda500 strain) mosquitoes were fed on infected mice 3–5 days after emergence and kept at 21 °C and 70% humidity as described (11). Sporozoites were collected from salivary glands dissected on day 18–21 postfeeding. To evaluate the parasite infectivity in vivo, 30,000 sporozoites were injected intravenously into 4-week-old C57BL/6 mice (Janvier). Blood stage parasitemia was assessed daily by flow cytometry as described previously (31). The study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Institut Pasteur and complied with the European Union guidelines for the handling of laboratory animals. The procedures were approved by the Institut Pasteur animal care and use committee. Animal care and handling was approved by the Ministère de l'Agriculture et de la Pêche (Report 107503056792, clearance number 75-273), and the protocols and procedures used were approved by the Direction Départementale des Services Vétérinaires de Paris (Ref. RL-07031395-30701147). All experiments were planned in order to minimize animal suffering.
P. berghei Transfection and Cloning
The targeting pSUB1/FRT plasmid was constructed by cloning three PCR products into a plasmid that carries two FRT sites and the human dihydrofolate reductase expression cassette (27, 28): (i) a 736-bp fragment upstream from the Pbsub1 locus; (ii) a 3479-bp DNA fragment encompassing the 5′ FRT site, 1177 bp corresponding to the 5′-untranslated region (UTR) of Pbsub1, the complete PbSUB1 open reading frame (1799 bp), and 503 bp corresponding to the Pbsub1 3′-UTR; and (iii) a 673-bp fragment downstream from the Pbsub1 locus. The latter was amplified using the oligonucleotides 5′-CCCGGGCATTTGGTGTATATTATTGTTTTTCTTG-3′ and 5′-GGTACCTCAATGTGCATTTATTGTTATGC-3′ and was cloned first in the plasmid using SmaI and KpnI restriction sites (underlined sequences). To the resulting plasmid was added the 736-bp fragment upstream of the Pbsub1 locus amplified using the primers 5′-AAGCTTTAAATAAATCACCAAATATTAAATTGG-3′ and 5′-GCGGCCGCATGTGTGTTATATAAGTGATTATTCTTTAAGC-3′ using HindIII and NotI restriction sites (underlined sequences). Finally, the entire Pbsub1 locus was amplified using the 5′-GATATCTAAATCGAAGAAAATATAAACATTCG-3′ and 5′-GGGCCCAAATATATGTCGGTGTAATGCACC-3′ primers and cloned using the EcoRV and ApaI restriction sites. The sequence of all DNA fragments has been controlled by sequencing (Beckman Coulter Genomics). Prior to transfection, the final pSUB1/Cond plasmid was linearized using BmrI and KpnI (Biolabs) restriction enzymes.
P. berghei erythrocytic stages were transfected as described previously (29). Briefly, P. berghei blood stages collected from an infected rat were cultured in vitro overnight at 37 °C, in complete RPMI1640 medium supplemented with 20% FBS and in an atmosphere composed of 5% O2, 5% CO2, and 90% N2. The resulting mature schizonts were purified by Nycodenz gradient, and 107 to 108 merozoites resuspended in 40 μl of complete medium were electroporated using the U33 program in the presence of 100 μl of human T cell Nucleofactor solution (Amaxa) and 5 μg of linearized plasmid DNA. The transfection mix was injected intravenously into two 3-week-old female Swiss mice that were treated with WR99210 (16 mg/kg) injected subcutaneously from day 1 to 3 post-transfection. At day 5, the emerging parasite population was transferred from each mouse to two naive mice that were treated again with WR99210 for 3 days. When parasitemia in initial and transferred mice was >2%, blood was collected to prepare cryostabilates and genomic DNA. Clones of the selected mutants were obtained by limiting dilution via the infection of at least 30 mice by 0.5 parasites.
Screening for the correct integration of the 5′ FRT was performed by PCR using primers P4 and P5. The P4-P5 amplified product (2 kb) was digested by XbaI (Biolabs), and two fragments of 1.2 and 0.8 kb were obtained when the XbaI-containing 5′ FRT site was present. Excision or non-excision of the Pbsub1 locus was assessed by PCR using primers P1-P2 and P3-P2, respectively.
Southern Blot, PCR, and RT-PCR Analysis
Southern blot analyses were carried out as described (27, 32). Genomic DNA from P. berghei parental and SUB1/Cond clones 5 and 22 was extracted using the QIAamp DNA blood kit (Qiagen); digested with NcoI, XbaI, or AccI; separated by gel electrophoresis; and transferred overnight to Hybond N+ membrane (GE Healthcare). The PCR DIG probe synthesis kit (Roche Applied Science) and primers 5′-ATGATAAAAAATTAGGTAGATTAGG-3′ and 5′-TTAATTTCTTTTTTTACTTTTCCACATGCG-3′ were used to amplify a 600-bp fragment that corresponds to the 3′-end of the PbSUB1 open reading frame. Hybridization was performed overnight at 42 °C, and washing and signal detection were carried according to the manufacturer's instructions.
Excision or non-excision of the Pbsub1 locus was assessed by PCR using primers P1-P2 and P3-P2, respectively. Primers sequences were as follows: P1, 5′-ATATTTGGTCACTCTTGATGG-3′; P2, 5′-ACATTAAAATTTTCGAGCATGC-3′; P3, 5′-TCTCTTCAATGATTCATAAATAGTTGG-3′. PCR was performed using Taq polymerase (Invitrogen) with one cycle at 95 °C for 2 min followed by 35 cycles of 95 °C for 30 s, 58 °C for 1 min, and 68 °C for 1 min and 1 cycle of 68 °C for 10 min. Screening for the correct integration of the 5′ FRT was performed by PCR using primers P4 and P5, the sequences of which are 5′-TTGTTATGTACATTTTGTACCTTTATGG-3′ and 5′-TACAAGCATAGATAAAAACTGTTCG-3′, respectively. The P4-P5 amplified product (2 kb) was digested by XbaI (Biolabs), and two fragments of 1.2 and 0.8 kb were obtained when the 5′ FRT site, which contains the XbaI site, was present. For RT analysis, total RNA was purified from erythrocytic schizonts, non-infected HepG2 cells, and parasitized HepG2 cells collected 24, 48, and 62 h postinfection with sporozoites using TRIzol (Ambion). Reverse transcription was performed using SuperScript II reverse transcriptase (Invitrogen). cDNA was used as a template for PCR using primers specific for P. berghei Pbsub1 and Pbtub, leading to the amplification of fragments of 515 and 421 bp, respectively. The two primer pairs 5′-AGCTTTAGATGATAAAAAATTAGGTAGATTAGG-3′ plus 5′-CAACTTGTTTATACGTTAAATCTGGATTAATTGA-3′ and 5′-TGGAGCAGGAAATAACTGGG-3′ plus 5′-ACCTGACATAGCGGCTGAAA-3′ were used to amplify Pbsub1 and Pbtub, respectively.
Western Blot and Immunofluorescence Analysis
Protein extracts from P. berghei were prepared as described previously (33), whereas liver stages were obtained following enrichment by a cell sorting procedure. Briefly, P. berghei GFP+-infected HepG2 cells (ratio 1:0.5) cultured for 21, 48, and 62 h postinfection with P. berghei GFP+ sporozoites were sorted using the FACSAria III cell sorter piloted with FACSDiva version 6.1.3 software. Western blot analyses were carried out as described previously (34), except that cells were lysed in XT sample buffer (Bio-Rad) supplemented with 10 mm dithiothreitol. Following separation on 4–12% BisTris Criterion precast gel (Bio-Rad) and transfer onto nitrocellulose (Amersham Biosciences), membranes were probed with respective antiserum diluted in Tween-buffered saline (10 mm Tris, pH 8 (Sigma), 150 mm NaCl (Sigma), 0.2% Tween (Sigma), and 5% (v/v) milk). Primary antibodies used were the 1D11 anti-PbSUB1 anti-peptide mAb obtained following immunization of mice with the KNSYKKSFFNDEYRC PbSUB1 peptide coupled to keyhole limpet hemocyanin (Genscript), the anti-PbMSP1–42 rabbit polyclonal serum (35), and the anti-PbSERA3 rat polyclonal serum (26). Following washing and incubation with horseradish peroxidase (HRP)-conjugated anti-species IgG antibodies (Promega), signals were revealed using the SuperSignal (ThermoScientific) or the ECL Prime (Amersham Biosciences) luminescent substrate. Alternatively, membranes were probed with an HRP-conjugated rabbit anti-GAPDH (Santa Cruz Biotechnology, Inc.) or a rabbit anti-GFP serum (Clontech). Relative protein proportions were evaluated using Quantity One software (Bio-Rad).
Before incubation with antibodies of interest, erythrocytic or liver schizonts and free merozoites were fixed on 10-well slides with cold acetone/methanol (4:1) for 10 min at −20 °C. Following extensive washing, the presence of IgGs was revealed following incubation with anti-species IgG coupled to Alexa Fluor 488 or 594 (Invitrogen). Slides were mounted with Vectashield, and images were taken using an Axiovert II fluorescence microscope (Zeiss) and treated using ImageJ software.
Culture of P. berghei Liver Stages in HepG2 Cells and Preparation of Hepatic Merozoites
HepG2 cells were grown in DMEM+ Glutamax-1 media (Invitrogen) supplemented with 10% FCS (PAA Laboratories GmbH) and penicillin/streptomycin/neomycin solution stabilized (P4083, Sigma) at 37 °C in the presence of 5% CO2 and 10% O2. The day before infection, 4 × 104 cells were plated per well in 8-well chamber slides (Nalge Nunc International, Rochester, NY), and 24 h later, 4 × 104 CONT or SUB1/Cond sporozoites were added. The medium was changed daily. The quantification of infection was carried out at 24 and 48 h postinfection by flow cytometry using the MACSQUANT cytometer, allowing counting of the P. berghei GFP-infected HepG2. At 65 h postinfection, the culture supernatant was collected, and cells were scraped and lysed using an insulin syringe. Merozoites contained in both fractions were counted using KOVA slides (Hycor) and an Axiovert II fluorescence microscope (Zeiss) with ×40 objective. The time lapse slides have been extracted from the movies accessible as supplemental material, which were recorded using an Axiovert II fluorescence microscope (Zeiss) with 10× objective using ImageJ software. Hepatic merozoites were prepared from parasitized HepG2 cells cultivated 62 h postinfection with sporozoites. The culture supernatant was collected, the cells were scraped and mechanically lysed by pipetting, and merozoites were prepared by filtration using a 5-μm filter (Sartorius Stedium Biotech-Minisart) and counted using KOVA slides (Hycor) and an Axiovert II fluorescence microscope (Zeiss) with ×40 objective. 50,000 merozoites of each fraction of CONT or SUB1/Cond were injected intravenously into three 4-week-old C57BL/6 mice.
Transmission Electron Microscopy
Infected HepG2 cells collected 59 or 65 h postinfection (hpi) were fixed in 2.5% glutaraldehyde in 0.1 m cacodylate buffer at 4 °C for 24 h. After several washes in 0.1 m cacodylate buffer, samples were postfixed for 1 h in the same buffer containing 1% osmium tetroxide (Merck). After dehydration in a graded ethanol series, the samples were embedded in Epon resin. Contrasted ultrathin sections (60 nm) were observed in a JEM 1010 transmission electron microscope (Jeol).
RESULTS
SUB1 Is Expressed in P. berghei Liver Stage Parasites
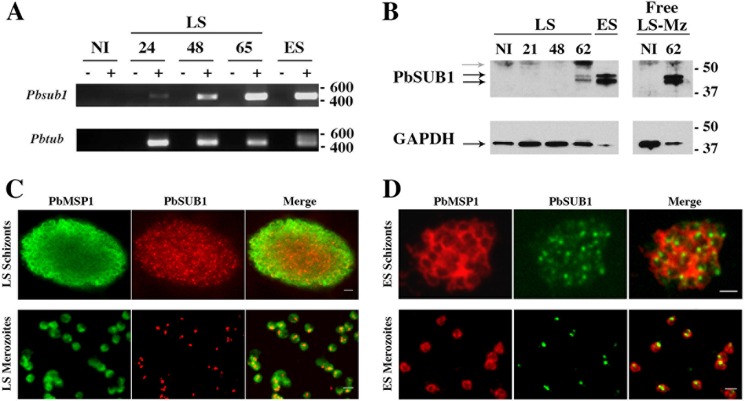
Expression of sub1 was examined in P. berghei parasites developing inside cultured hepatoma HepG2 cells (Fig. 1). RT-PCR analysis indicated that sub1 transcription by P. berghei-containing HepG2 cells increased from 24 to 65 hpi by sporozoites (Fig. 1A). SUB1 protein was barely detected 48 hpi (data not shown) and abundantly present at 62 hpi (Fig. 1B). Immunofluorescence assays confirmed the presence of SUB1 in mature hepatic schizonts as well as in free merozoites released from infected hepatocytes (Fig. 1C), similar to the SUB1 staining in erythrocytic parasite stages (Fig. 1D). In both intrahepatocytic and intraerythrocytic schizonts, SUB1 staining displayed both punctate and diffuse patterns, the latter possibly reflecting the release of SUB1 into the PV.
FIGURE 1.
PbSUB1 is a protein of blood stage and hepatic merozoites. A, mRNA of Pbsub1 was detected in erythrocytic segmented schizonts (ES) and in HepG2 cells 24, 48, and 65 h postinfection with P. berghei sporozoites. mRNA of Pbtub was amplified as a control of RNA preparation. mRNA extracted from non-infected HepG2 (ni) cells was amplified as a control. Molecular mass markers are in base pairs. B, Western blot using the 1D11 anti-PbSUB1 monoclonal antibody. PbSUB1 mature forms (black arrows) are detected in protein extracts prepared from erythrocytic (ES), exoerythrocytic schizonts in HepG2 cells 62 h postinfection with P. berghei sporozoites and in liver stages merosomes and merozoites (LS-Mz) collected in the culture supernatant of infected HepG2 62 h postinfection. Protein extracts from non-infected (NI) erythrocytes, HepG2 cells, and culture supernatants of HepG2 cells have been probed as a control, allowing the identification of a host cell-specific signal (gray arrow). For a loading control, the same membranes have been probed with anti-GAPDH antibodies. Molecular mass markers are in kilodaltons. Shown is immunofluorescence analysis of liver stages (LS) (C) and erythrocytic stages (ES) (D) of P. berghei schizonts and free merozoites. Fixed liver stage schizonts and merozoites were incubated with anti-PbSUB1 mAb (red) and anti-PbMSP1 (green). Fixed erythrocytic stage schizonts and merozoites were incubated with anti-PbSUB1 mAb (green) or anti-PbMSP1 (red). Scale bars, 2 μm.
Generation of P. berghei SUB1 Conditional Lines
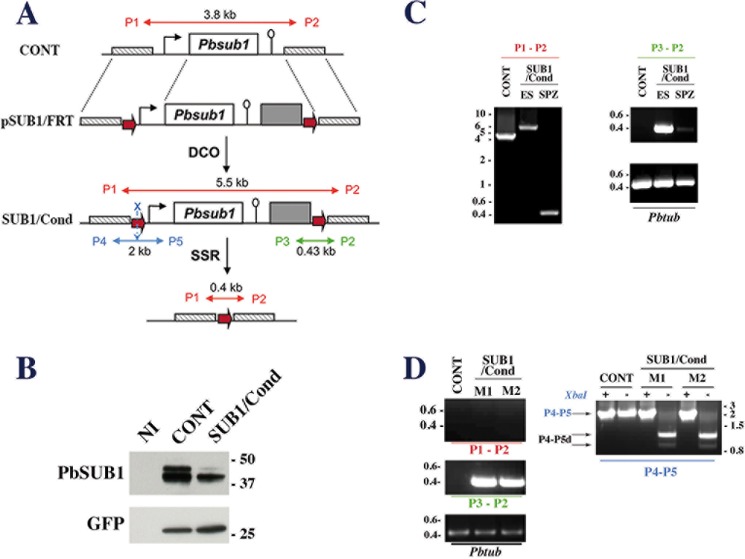
To decipher SUB1 biological function in liver stage parasites and hepatic merozoites, we used a conditional inactivation strategy because SUB1 is essential in blood stage parasites (18). We deleted the sub1 coding sequence in sporozoites using the FLP/FRT site-specific recombination system (27, 28). The targeting plasmid pSUB1/FRT (Fig. 2A) was transfected into UIS4/FLP(−)F parasites, which constitutively express GFP and express the FLP recombinase specifically in salivary glands sporozoites (27). The plasmid pSUB1/FRT was designed to place, following double crossover recombination, a 5′ FRT site 736 bp upstream from the sub1 start codon and a 3′ FRT site downstream from the 3′-UTR of sub1 and the selectable marker (Fig. 2A).
FIGURE 2.
Generation of the P. berghei conditional SUB1/Cond mutant. A, schematic representations of the wild-type Pbsub1 locus in control (CONT) parental parasites, the targeting plasmid pSUB1/FRT (top), the expected Pbsub1 recombinant locus in the SUB1/Cond clone obtained following a double crossover event (DCO; middle), and the recombinant Pbsub1-null locus after the FLP-recombinase mediated site-specific recombination (SSR; bottom). The linearized pSUB1/FRT plasmid carries the following from 5′ to 3′: a 736-bp fragment located upstream of the sub1 locus (cross-hatched box), an FRT site (red arrow), the Pbsub1 locus composed of a 5′-untranslated regulatory sequence (1177 bp; black arrow), the Pbsub1 coding sequence (1799 bp; open box), a 3′-untranslated regulatory sequence (503 bp; open lollipop), the human dihydrofolate reductase cassette (gray box), the second FRT site, and a 673-bp fragment located downstream of the Pbsub1 locus. The P1 and P2 primers amplify a 3.8- or 5.5-kb fragment on CONT or SUB1/Cond loci, respectively, and a 0.4-kb fragment following FLP-specific SSR excision of Pbsub1. P3/P2 and P4/P5 couples of primers are specific for the SUB1/Cond recombinant locus; P3 and P2 amplify a 0.43-kb fragment, whereas P4 and P5 amplify a 2-kb fragment surrounding the 5′ FRT site, which contains an XbaI restriction site (blue X). B, Western blot analysis of SUB1/Cond clone5 and CONT erythrocytic segmented schizonts probed for PbSUB1. Protein extracts corresponding to 5 × 105 infected red blood cells and an equal number of non-infected erythrocytes (NI) were probed by 1D11 anti-PbSUB1 and anti-GFP antibodies; the latter was used as a loading control. C, PCR analysis of the Pbsub1 locus of the recombinant SUB1/Cond clone 5 compared with the parental (CONT) parasites. P3-P2 and P1-P2 primers were used to analyze the SUB1/Cond gDNA prepared from erythrocytic stages (ES) and sporozoites (SPZ) collected on day 20 postinfection. Primers P1-P2 amplify both the excised (SPZ; 0.4 kb) and non-excised Pbsub1 loci (ES; 5.5 kb) from SUB1/Cond and the parental Pbsub1 locus (CONT; 3.8 kb). Primers P3-P2 amplify only non-excised Pbsub1 locus (0.43 kb) from gDNA prepared from sporozoites and erythrocytic stages of SUB1/Cond parasites. The P. berghei tubulin coding gene (Pbtub) was amplified as a control of gDNA. D, PCR analysis of genomic DNA of CONT and SUB1/Cond erythrocytic stages following infection of mice by sporozoites. Genomic DNA was prepared from the two parasitized mice (M1 and M2) corresponding to experiment 6 (see Table 1) and analyzed using P1-P2, P3-P2, and P4-P5 primers as indicated in C. As shown by the amplification of a 0.43-kb fragment with the P3-P4 primers and a 2-kb fragment with the P4-P5 primers, erythrocytic parasites collected from both mice display a SUB1/Cond genotype. XbaI-digested (+) or undigested (−) P4-P5 fragments (P4-P5, blue characters) are shown. In the presence of a complete 5′ FRT site containing the XbaI site, two fragments of 1.2 and 0.8 kb are generated (P4-P5d; black arrows) and are the signature of a complete SUB1/Cond recombinant locus. Importantly, no fragment of 0.4 kb corresponding to a Pbsub1 excised locus could be amplified with P1-P2 primers. Molecular mass markers are in kilobase pairs and in kilodaltons.
After transfection of pSUB1/FRT into UIS4/FLP(−)F GFP-expressing P. berghei parasites (hereafter called CONT, for control parental parasites), WR99210-resistant parasites were readily selected. PCR genotyping identified several recombinant clones, including SUB1/Cond-5 and SUB1/Cond-22, originating from independent transfection experiments. Their genotype was confirmed by Southern bot analysis to contain the expected sub1 recombinant locus (data not shown). Both SUB1/Cond-5 (Fig. 2B) and SUB1/Cond-226 clones expressed the expected forms of PbSUB1 but in a >50% reduced amount as compared with the parental CONT clone, and they were able to cycle through the Anopheles vector (Table 1).
TABLE 1.
SUB1/Cond sporozoites are unable to establish a blood parasitemia
Groups of 4–5 mice were infected intravenously by SUB1/Cond or CONT sporozoites to determine the prepatent period before the establishment of a blood parasitemia. The number of animals in each group that were positive for erythrocytic stages is shown. Asterisks indicate that the analysis of genomic DNA prepared from blood stage parasites collected from these mice has shown that only the non-excised (NE) genotype for the Pbsub1 locus could be detected. PCR genotyping of the resulting erythrocytic SUB1/Cond parasites has been performed on parasites prepared from infected mice labeled with an asterisk. A representative PCR analysis showing that only the SUB1/Cond non-excised genotype could be detected and corresponding to the erythrocytic parasites prepared from the two infected mice (Experiment 6) is shown in Fig. 2F.
| Experiment | Parasite clone | No. of injected sporozoites | No. of positive mice/no. of injected mice (blood parasitemia) | Prepatent period (days) | Genotype of recovered parasites |
|---|---|---|---|---|---|
| 1 | SUB1/Cond-22 | 30,000 | 2*/10 | 6 | NE Pbsub1 |
| CONT | 10/10 | 4 | WT | ||
| 2 | SUB1/Cond-22 | 30,000 | 0/3 | ||
| CONT | 3/3 | 5 | WT | ||
| 3 | SUB1/Cond-22 | 30,000 | 2*/4 | 6 | NE Pbsub1 |
| CONT | 4/4 | 6 | WT | ||
| 4 | SUB1/Cond-5 | 30,000 | 1*/5 | 7 | NE Pbsub1 |
| CONT | 5/5 | 6 | WT | ||
| 5 | SUB1/Cond-5 | 30,000 | 2*/4 | 4 | NE Pbsub1 |
| CONT | 4/4 | 4 | WT | ||
| 6 | SUB1/Cond-5 | 30,000 | 2*/5 | 4 | NE Pbsub1 |
| CONT | 5/5 | 4 | WT |
SUB1 Is Essential for Parasite Infectivity
Clone SUB1/Cond-5, hereafter called SUB1/Cond, was selected to analyze the role of SUB1 in pre-erythrocytic stages of the parasite. A. stephensi mosquitoes were allowed to feed on SUB1/Cond- or CONT-infected mice. At day 20 post-transmission, similar numbers of SUB1/Cond and CONT salivary gland sporozoites were obtained, and site-specific recombination efficiency was evaluated by PCR analysis of the sub1 locus in SUB1/Cond sporozoites; P1-P2 primers (Fig. 2A), which amplified both the non-excised and excised loci, albeit with different efficiencies, generated a band of 0.4 kb corresponding to the excised locus with gDNA prepared from sporozoites but not erythrocytic stages of the SUB1/Cond clone (Fig. 2C, left). However, P3-P2 primers, designed to amplify a small fragment of 0.43 kb specifically from the non-excised locus of SUB1/Cond, revealed the presence of some non-excised parasites in both erythrocytic stages and sporozoites of the SUB1/Cond clone (Fig. 2C, right). This indicated that SUB1/Cond sporozoites contained a mixture of excised and non-excised parasites for the sub1 locus. By normalizing the gDNA amount to the parasite tubulin PCR product (Fig. 2C) and scoring excision efficiency using PCR band intensity, the proportion of excised parasites was evaluated to be >90%, in agreement with previous uses of the UIS4/FLP(−)F system (27, 28, 36, 37).
To evaluate SUB1/Cond parasite infectivity, mice were infected with day 20 sporozoites, and the emergence of blood stage parasites was monitored. In all experiments and using either SUB1/Cond clone, fewer animals developed a blood parasitemia after inoculation of mutant compared with CONT sporozoites (Table 1). The average prepatent period (i.e. the delay before the apparition of a blood parasitemia following the injection of sporozoites) induced by SUB1/Cond clones was similar to or prolonged compared with CONT parasites, with an average increase of 1–2 days in experiments 1 and 4, respectively (Table 1), corresponding to a 10–100-fold decrease in SUB1/Cond parasite infectivity. Importantly, in all experiments, animals that developed blood stage infection after injection of SUB1/Cond sporozoites only contained recombinant SUB1/Cond non-excised parasites, as shown by PCR analysis using the P3-P2 and P4-P5 primers (Fig. 2D) and the specific failure of P1-P2 primers to amplify the small 0.4-kb fragment corresponding to the excised locus (Table 1 and Fig. 2D). This counterselection of excised parasites demonstrated a crucial role for SUB1 in parasite infectivity, which might be required during the pre-erythrocytic phase of the parasite development in addition to its pivotal role during Plasmodium intraerythrocytic cycle.
SUB1 Is Important for Hepatic Merozoite Egress from the Parasitophorous Vacuole
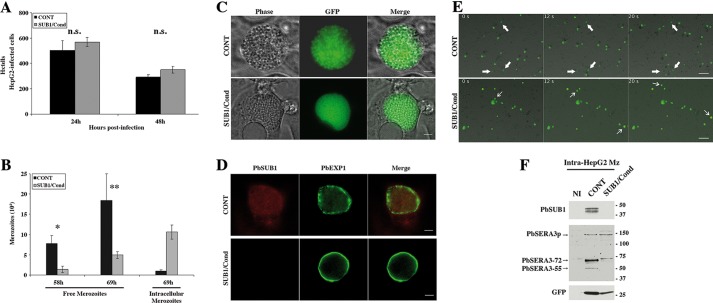
To explore whether SUB1 was essential in the hepatic phase, we compared SUB1/Cond and CONT parasite development inside hepatocytes in vitro. At 24 and 48 hpi with sporozoites, similar numbers of normal average size liver stages of both parasite clones were counted by flow cytometry (Fig. 3A). This confirmed, in agreement with the PbSUB1 expression pattern described above, that SUB1 was not important for sporozoite invasion of hepatocytes or parasite intrahepatocytic development. To assess merozoite egress from hepatocytes, we counted free merozoites released in the culture supernatant by fluorescence microscopy (Fig. 3B). Merosomes normally bud off from infected HepG2 cells starting at ∼58–60 hpi, occasionally rupturing and releasing merozoites in the medium. At 58 and 69 hpi, ∼6- and ∼4-fold more CONT than SUB1/Cond merozoites, respectively, were counted. This difference was observed for five independent experiments and was shown to be statistically significant (p < 0.001) using an exact one-sided Wilcoxon rank sum test and combined using the meta-analytical approach of Fisher. Conversely, at 69 hpi, ∼10-fold more SUB1/Cond than CONT intracellular merozoites were counted following their mechanical release from infected HepG2 (Fig. 3B). These data suggested that SUB1/Cond merozoites were impaired in their egress from hepatocytes.
FIGURE 3.
SUB1/Cond parasites invade and develop normally into the hepatocyte HepG2 cells but are defective for the release and maturation of hepatic merozoites. A, HepG2 cells were infected with SUB1/Cond or CONT GFP+-P. berghei sporozoites. GFP+ infected HepG2 cells collected 24 and 48 hpi were counted by flow cytometry. Results from a typical experiment are shown and correspond to the mean and S.D. (error bars) of internal triplicates. n.s., no significant difference, as determined by a Wilcoxon rank sum test. B, free merozoites released in the supernatant of SUB1/Cond or CONT HepG2-infected cells were collected 58 and 69 hpi and quantified by fluorescence microscopy. To quantify intracellular merozoites, HepG2-infected cells were collected 69 hpi and mechanically lysed to obtain released merozoites. Results from a typical experiment are shown and correspond to the mean and S.D. of internal triplicates. * and **, statistically significant differences (p < 0.001); the p values of five independent experiments were computed using an exact one-sided Wilcoxon rank sum test and combined using the meta-analytical approach of Fisher. C, light (phase) and fluorescent (GFP) microscopy analysis of SUB1/Cond and CONT schizonts (65 h postinfection) into HepG2 cells are shown and are representative of five independent experiments. Scale bar, 20 μm. D, immunofluorescence analysis of SUB1/Cond and CONT LS schizonts. Fixed LS schizonts became GFP− following fixation and were incubated with anti-PbSUB1 mAb (red) and anti-EXP1 (green). Scale bar, 20 μm. E, time lapse fluorescent video microscopy analysis of SUB1/Cond and CONT schizont-infected HepG2 cells 60 hpi. Budding from the ruptured infected HepG2 cells, CONT merosomes are floating in the supernatant, whereas only a few parasites remain intracellular and are immobile (thick white arrows). In contrast, only a few SUB1/Cond merosomes float in the supernatant (thin white arrows), whereas most of the SUB1/Cond schizonts remain intracellular and are immobile. The slides shown correspond to pictures captured at 0, 12, and 20 s from supplemental Movies 1 and 2. Scale bar, 20 μm. F, Western blot analysis of CONT and SUB1/Cond schizont-infected HepG2 cells collected by cell sorting at 58 hpi. Protein extracts corresponding to non-infected (NI) and CONT or SUB1/Cond merozoite-infected HepG2 were probed by the 1D11 anti-PbSUB1 mAb and the anti-PbSERA3 serum. In contrast to PbSERA3 precursor (PbSERA3p), mature PbSUB1 and PbSERA3 maturation products (PbSERA3-72 and PbSERA3-55) could only be detected in CONT intracellular merozoites. Anti-GFP antibodies were used as parasite loading control. Molecular mass markers are in kilodaltons.
Liver stages developing inside HepG2 cells at 65 hpi were then imaged by light and fluorescence microscopy. SUB1/Cond parasites appeared slightly smaller than the CONT counterparts (Fig. 3C). This size difference correlated with an intact labeling of PVM around SUB1/Cond liver stages using anti-EXP1 antibodies (Fig. 3D). In contrast, the EXP1 labeling enclosing CONT liver stages, when visible, was discontinuous (Fig. 3D), as expected from PVM rupture at this time point. Hepatic merozoite egress and merosome formation were then analyzed by video microscopy. Merosomes are typically seen moving in the culture supernatant as budding, free floating structures from ∼60 hpi (9). At 60 hpi, 85% of the CONT parasites analyzed appeared as “moving” (i.e. giving rise to drifting merosomes), whereas the remainder appeared as “immobile” parasites (n = 20; Fig. 3E and supplemental Movie 1). In contrast, the proportion of SUB1/Cond parasites generating merosomes dropped to <12% (n = 17; Fig. 3E and supplemental Movie 2). This difference in the proportion of PVM containing SUB1/Cond and CONT schizonts was shown to be statistically significant (Pearson's χ2 test with simulated p value based on 105 replicates; χ2 = 14.854, p < 0.0001).
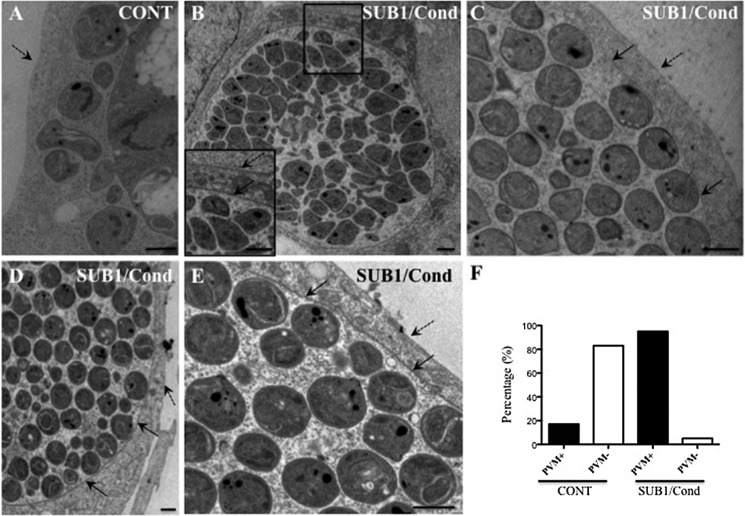
Next, merozoite capacity to escape from the PVM was investigated by transmission electron microscopy of infected HepG2 cells at ∼59 or 65 hpi (Fig. 4). At ∼59 h, numbers and shapes of intracellular merozoites were similar for the SUB1/Cond and CONT clones, confirming unimpaired merozoite formation in the SUB1/Cond liver stages. At this time point, only 2 of 12 analyzed CONT liver stages displayed a visible PVM, whereas the remainder were surrounded only by the HPM (Fig. 4, A and F). In contrast, 15 of 17 SUB1/Cond liver stages were still surrounded by a visible PVM in addition to the HPM (Fig. 4, B, C, and F). At 65 hpi, when CONT merozoites were no longer present inside hepatocytes, SUB1/Cond merozoites were still enclosed in a PVM and an HPM (Fig. 4, D and E). These observations provide direct evidence for a role of SUB1 in rupturing the PVM within the hepatocyte and hence in merozoite egress.
FIGURE 4.
SUB1/Cond hepatic merozoites are defective for the parasitophorous vacuole membrane rupture. Transmission electron microscopy analysis of intra-HepG2 SUB1/Cond and CONT schizonts prepared 59 h (A–C) or 65 h (D and E) postinfection with their respective sporozoites. A, representative example of a hepatic CONT schizont without PVM. B–E, representative examples of SUB1/Cond hepatic schizonts with intact PVM, shown by black arrows; the HepG2 plasma membrane is indicated by dotted black arrows. Differentiated merozoites appeared morphologically normal but remained inside the parasitophorous vacuole. Scale bars, 2 μm. F, histogram of the percentage of 17 SUB1/Cond and 12 CONT schizonts with (PVM+) or without (PVM−) a visible PVM at 59 h postinfection. The different proportion of PVM-containing SUB1/Cond and CONT schizonts is statistically significant (Pearson's χ2 test with simulated p value based on 105 replicates; χ2 = 14.854, p < 0.0001).
SUB1 Processes SERA3 in Parasite Liver Stages
Generation of SUB1-deficient parasites provided for the first time the opportunity to directly test the role of SUB1 in processing its protein targets. In P. berghei, SERA3 is expressed in mature liver stages, before the egress of hepatic merozoites, and its 130-kDa precursor undergoes specific processing (26). It is noteworthy that PfSERA6, the PbSERA3 ortholog in P. falciparum, is maturated by SUB1 at the blood stages (22). To investigate PbSERA3 processing in PbSUB1-deficient merozoites, infected HepG2 cells were isolated by cell sorting at 58 hpi, immediately prior to merozoite egress, and protein extracts were analyzed by Western blot. Although present in CONT liver stages, SUB1 protein remained undetectable in SUB1/Cond liver stages, confirming that the vast majority of SUB1/Cond merozoites were excised individuals lacking sub1. Results also showed that SERA maturation products, known as PbSERA3-72 and PbSERA3-55 (26), were also undetectable in SUB1/Cond liver stages, although their precursor of 130 kDa was detectable (PbSERA3p; Fig. 3F). This indicates that SERA3 processing in liver stages is SUB1-dependent.
SUB1-deficient Hepatic Merozoites Do Not Process MSP1 and Are Unable to Establish a Blood Infection
A primary target of SUB1 in erythrocytic merozoites is MSP1, which is cleaved into four fragments of 83, 30, 38, and 42 kDa that remain at the merozoite surface (21). To investigate whether SUB1 carries out MSP1 processing at the hepatic stage, SUB1-dependent MSP1 processing was analyzed in CONT and SUB1/Cond hepatic merozoites collected at 59 hpi by mechanical rupture of infected HepG2 cells. At the protein level, whereas SUB1 was detected in CONT hepatic merozoites, this was not the case in SUB1/Cond hepatic merozoites (Fig. 5A). In parallel, the processed MSP1 42-kDa form accumulated in CONT merozoites (4.2 times more than in SUB1/Cond merozoites) but was barely detectable in SUB1/Cond merozoites in which the 230-kDa MSP1 precursor accumulated (3.13 times more than in CONT merozoites; Fig. 5A). Because PCR analysis confirmed that most SUB1/Cond merozoites had an excised sub1 locus (Fig. 5B), this indicated that SUB1 was involved in the primary processing of MSP1 in hepatic merozoites. To test the capacity of SUB1/Cond merozoites to trigger a blood infection, mice were infected by CONT or SUB1/Cond hepatic merozoites, and the emergence of blood stage parasites was monitored. In three independent experiments, few or no animals developed a blood parasitemia after inoculation of SUB1/Cond hepatic merozoites (Table 2) compared with CONT merozoites. In addition, genotyping of the blood stage parasites collected from the positive mice infected with SUB1/Cond merozoites showed that all mice harbored only non-excised Pbsub1 parasites (Fig. 5B). In line with the known essential role played by SUB1 during the erythrocytic cycle of P. falciparum in vitro (18), these observations provide direct evidence that SUB1 is essential to establish a blood stage infection in vivo.
FIGURE 5.
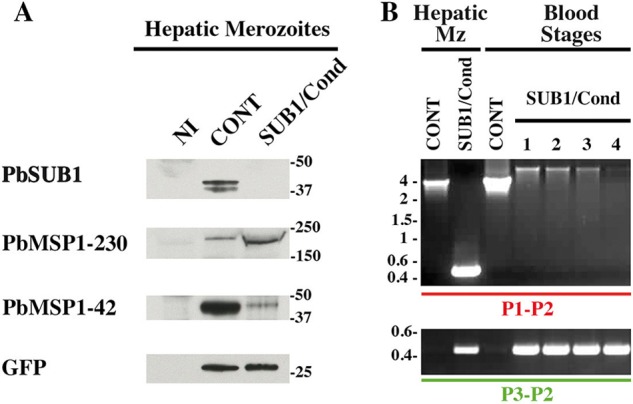
SUB1/Cond hepatic merozoites display a default of MSP1 maturation and are unable to establish a blood parasitemia. A, protein extracts of CONT and SUB1/Cond hepatic merozoites mechanically released from infected HepG2 cultures 60 hpi were probed with the 1D11 anti-PbSUB1 mAb and the rabbit anti-PbMSP1 polyclonal serum. Anti-GFP antibodies were used as a parasite loading control. Molecular mass markers are in kilodaltons. B, PCR genotyping of the sub1 locus using the P1-P2 and P3-P2 primers (Fig. 2A). gDNA was prepared from SUB1/Cond or CONT hepatic merozoites (Mz) and blood stage parasites collected from mice infected by 50,000 hepatic CONT or SUB1/Cond merozoites, corresponding to experiment 3 presented in Table 2. PCR analysis of one representative CONT gDNA is shown, whereas lanes 1–4 correspond to the analysis of the gDNA prepared from the four mice that developed a blood parasitemia following the injection of SUB1/Cond hepatic merozoites (Table 2). The SUB1/Cond genotype of these parasites was confirmed by amplification of the 0.43-kb fragment by the P3-P2 primers. However, no parasites with an excised Pbsub1 locus could be detected, as shown by the failure to amplify the Pbsub1 excised specific fragment of 0.4 kb using the P1-P2 primers, whereas the full-length Pbsub1 non-excised locus is detected, as shown by the amplification of the 5.5-kb fragment (Fig. 2A). Molecular mass markers are in kilobase pairs.
TABLE 2.
SUB1/Cond hepatic merozoites are unable to establish a blood parasitemia
Groups of 6–7 mice were infected intravenously by SUB1/Cond or CONT hepatic merozoites. The number of animals in each group that developed a parasitemia is shown. Asterisks indicate that only the non-excised (NE) Pbsub1 locus could be detected by PCR genotyping of genomic DNA prepared from blood stage parasites collected from these mice. A representative PCR analysis of the four infected mice of Experiment 3 is shown in Fig. 5B.
| Experiment | Parasite clone | No. of injected merozoites | No. of positive mice/no. of injected mice (blood parasitemia) | Genotype of recovered blood stage parasites |
|---|---|---|---|---|
| 1 | SUB1/Cond-5 | 50,000 | 3*/6 | NE Pbsub1 |
| CONT | 4/6 | WT | ||
| 2 | SUB1/Cond-5 | 50,000 | 0/6 | |
| CONT | 4/6 | WT | ||
| 3 | SUB1/Cond-5 | 50,000 | 4*/7 | NE Pbsub1 |
| CONT | 7/7 | WT |
DISCUSSION
This study provides direct genetic evidence for successive roles of SUB1 at different steps of the P. berghei life cycle. Previous studies have shown that P. falciparum SUB1 was important in vitro for merozoite egress from erythrocytes (18) as well as for priming merozoites for erythrocyte invasion (21). These conclusions were based on the use of SUB1 inhibitors, including chemical compounds (18, 38) or peptidic derivatives (19, 39). The issue of inhibitor specificity is a possible confounder in such an approach. Here, using genetically modified SUB1-deficient parasites, we show that SUB1 plays a central role during both the pre-erythrocytic and erythrocytic phases of Plasmodium life cycle, being critically involved in merozoite egress from host hepatocytes and in the establishment of a subsequent intraerythrocytic cycle.
In pre-erythrocytic stages of P. berghei, we show that SUB1, while being dispensable for sporozoite invasion of hepatocytes, is important for merozoite egress from hepatocytes. SUB1-deficient hepatic merozoites remained enclosed inside two clearly discernable membranes, namely the PVM and the HPM, demonstrating that SUB1 is crucial for rupturing at least the PVM. The egress defect of SUB1-deficient hepatic merozoites was associated with the absence of merosome formation and the accumulation of the cysteine-protease PbSERA3 precursor, known to be maturated in P. berghei hepatic schizonts (26). PbSERA3 involvement in the PVM rupture was suggested by the inhibition of PVM degradation in the presence of the cysteine inhibitor E64 (9), which correlates with a defect of PbSERA3 secretion to the cytoplasm of the schizont-infected hepatocytes (26). In P. falciparum blood stages, PfSERA6, the P. berghei SERA3 ortholog, is maturated by PfSUB1, which is thought to activate the PfSERA6 protease involved in the rupture of the erythrocytic schizont PVM (22). Taken together, these observations and our data show that SUB1 and SERA form a conserved kit to trigger the egress of Plasmodium merozoites from both erythrocytes and hepatocytes.
In addition, the P. berghei cGMP-dependent protein kinase (PbPKG) has been shown to play a key role for the egress of hepatic merozoites (40). Its P. falciparum orthologue (PfPKG) has recently been shown to participate in an intracellular shift in calcium concentration that triggers the secretion of the active PfSUB1 from merozoite exonemes into the PV (41, 42). It is tempting to propose that, together with SUB1/SERA, the parasite PKG composes a multipartner-regulated pathway that plays equivalent roles in Plasmodium egress from hepatocytes and red blood cells.
There are, however, important differences in the way hepatic and erythrocytic merozoites egress from their respective host cells. As already mentioned, the egress of erythrocytic merozoites occurs after the rapidly successive ruptures of the PVM and the HPM (7, 8), whereas the ruptures of these membranes by hepatic merozoites are separated in time (10). Therefore, whereas merozoite egress from both cell types involves common molecular basis, such as the SUB1/SERA kit, merozoite egress from hepatocytes requires yet unknown specific factors to prevent the premature rupture of the HPM and allow merosome formation. Even the PVM rupture has a distinct molecular basis at both stages because, for instance, it is dependent on and independent of the liver stage-specific protein 1 (LISP1) in hepatocytes and erythrocytes, respectively (43). Whether a link exists between SUB1/SERA- and LISP1-mediated steps during PVM rupture inside hepatocytes remains to be investigated.
The contributions of the various members of the SERA family to membrane disruption and whether they function as effectors of the process remain unclear. In P. falciparum, two of the nine sera genes, Pfsera5 and Pfsera6, cannot be inactivated in blood stages. In contrast, Pfsera4-null parasites complete the blood cycle normally, whereas overexpressing Pfsera5 (20, 44). P. berghei produces five PbSERAs. PbSERA5, also called ECP1, is essential for sporozoite egress from the oocyst (45), the parasite stage that develops in the mosquito midgut and in which one zygote transforms into thousands of sporozoites. Interestingly, unlike the erythrocytic or liver stages in the mammalian host, the oocyst does not reside intracellularly inside a PV but instead extracellularly, although it is enclosed in a thick wall. The other four PbSERAs are produced after 50 h in infected HepG2 cells (26). Gene inactivation experiments showed that PbSERA1 and PbSERA2 are dispensable for the parasite life cycle, including in combination, but mutants increase production of PbSERA3 in liver stages, suggesting a mechanism of biological compensation (25). Therefore, as in erythrocytic stages of P. falciparum, the SERAs expressed by P. berghei liver stages might be subject to functional hierarchy and to some redundancy. In particular, the fact that the SUB1/SERA kit is involved only in the rupture of the PVM, without affecting the HPM, suggests that other liver stage-specific protein(s) might regulate or prevent a SERA-mediated rupture of the hepatocyte PM to permit merosome formation. Clearly, although evidence suggests a role of SERAs in PVM rupture, determination of the SERAs that are important and their exact functions awaits further exploration. It also remains unknown whether SUB1 might process other factors involved in parasite egress.
Little is known about hepatic merozoites, which critically bridge the parasite pre-erythrocytic phase to the erythrocytic cycle. Our data indicate that SUB1-deficient hepatic merozoites, when mechanically released from hepatocytes, fail to mature MSP1 and are unable to establish a blood stage infection. In P. falciparum, PfMSP1 interacts with other MSPs, PfMSP6 and -7, forming a complex on the merozoite surface (46–48). PfSUB1 matures the three components of this complex (21), which remains associated at the surface of the merozoite and is critical for merozoite invasion, probably via a merozoite-erythrocyte attachment step, prior to parasite reorientation and internalization inside the host cell (23, 49, 50). More work is needed to unravel the complex basis of merozoite attachment to the erythrocyte and protein maturation events involved in the process.
This work establishes that SUB1, in addition to its pivotal role in the erythrocytic stage, is also involved in the pre-erythrocytic phase of the malaria parasite life cycle. This suggests that anti-SUB1 compounds should have both prophylactic and therapeutic properties. SUB1 might play additional roles at other steps of the parasite life cycle (e.g. during the egress of gametes, which are also located inside a PV inside erythrocytes). SUB1 thus emerges as a promising multistage drug target, the inhibitor of which could avoid the establishment of Plasmodium blood parasitemia, therefore preventing the occurrence of symptoms of malaria.
Acknowledgments
We thank Christiane Rondeau (Université de Montréal Dpt de Pathologie et Biologie Cellulaire, Canada) and Marie-Christine Prévost (Plate-forme de Microscopie Ultrastructurale, Institut Pasteur) for help with transmission electron microscopy, Joana Tavares and Daniel Y. Bargieri for helpful discussion, and Emmanuel Bischoff for statistical analysis. We thank Volker Heussler, Tania De Koning-Ward, and Patricia Baldacci for the gift of the anti-SERA3, anti-MSP1, and anti-EXP1 antibodies, respectively, and members of the Centre de Production et d'Infection des Anophèles (CEPIA, Institut Pasteur) for rearing A. stephensi mosquitoes.
This work was supported by grants from the “Institut Carnot-Pasteur Maladies Infectieuses” and “Agence Nationale pour la Recherche” Grant ANR-11-RPIB-002 (to J. C. B.).

This article contains supplemental Movies 1 and 2.
L. Tawk, C. Lacroix, P. Gueirard, R. Kent, O. Gorgette, S. Thiberge, O. Mercereau-Puijalon, R. Ménard, and J.-C. Barale, unpublished observation.
- PV
- parasitophorous vacuole
- PVM
- PV membrane
- SERA
- serine repeat antigen
- MSP
- merozoite surface protein
- FRT
- flippase recognition target
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- hpi
- hours postinfection
- gDNA
- genomic DNA.
REFERENCES
- 1. WHO Global Malaria Programme (2011) World Malaria Report, World Health Organization, Geneva [Google Scholar]
- 2. Dondorp A. M., Nosten F., Yi P., Das D., Phyo A. P., Tarning J., Lwin K. M., Ariey F., Hanpithakpong W., Lee S. J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S. S., Yeung S., Singhasivanon P., Day N. P., Lindegardh N., Socheat D., White N. J. (2009) Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Witkowski B., Lelièvre J., Barragán M. J., Laurent V., Su X. Z., Berry A., Benoit-Vical F. (2010) Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother. 54, 1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Witkowski B., Khim N., Chim P., Kim S., Ke S., Kloeung N., Chy S., Duong S., Leang R., Ringwald P., Dondorp A. M., Tripura R., Benoit-Vical F., Berry A., Gorgette O., Ariey F., Barale J. C., Mercereau-Puijalon O., Menard D. (2013) Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother. 57, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Noedl H., Se Y., Schaecher K., Smith B. L., Socheat D., Fukuda M. M. (2008) Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 359, 2619–2620 [DOI] [PubMed] [Google Scholar]
- 6. Salmon B. L., Oksman A., Goldberg D. E. (2001) Malaria parasite exit from the host erythrocyte. A two-step process requiring extraerythrocytic proteolysis. Proc. Natl. Acad. Sci. U.S.A. 98, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wickham M. E., Culvenor J. G., Cowman A. F. (2003) Selective inhibition of a two-step egress of malaria parasites from the host erythrocyte. J. Biol. Chem. 278, 37658–37663 [DOI] [PubMed] [Google Scholar]
- 8. Abkarian M., Massiera G., Berry L., Roques M., Braun-Breton C. (2011) A novel mechanism for egress of malarial parasites from red blood cells. Blood 117, 4118–4124 [DOI] [PubMed] [Google Scholar]
- 9. Sturm A., Amino R., van de Sand C., Regen T., Retzlaff S., Rennenberg A., Krueger A., Pollok J. M., Menard R., Heussler V. T. (2006) Manipulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science 313, 1287–1290 [DOI] [PubMed] [Google Scholar]
- 10. Sturm A., Heussler V. (2007) Live and let die. Manipulation of host hepatocytes by exoerythrocytic Plasmodium parasites. Med. Microbiol. Immunol. 196, 127–133 [DOI] [PubMed] [Google Scholar]
- 11. Thiberge S., Blazquez S., Baldacci P., Renaud O., Shorte S., Ménard R., Amino R. (2007) In vivo imaging of malaria parasites in the murine liver. Nat. Protoc. 2, 1811–1818 [DOI] [PubMed] [Google Scholar]
- 12. Baer K., Klotz C., Kappe S. H., Schnieder T., Frevert U. (2007) Release of hepatic Plasmodium yoelii merozoites into the pulmonary microvasculature. PLoS Pathog. 3, e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roggwiller E., Fricaud A.-C., Blisnick T., Braun-Breton C. (1997) Host urokinase-type plasminogen activator participates in the release of malaria merozoites from infected erythrocytes. Mol. Biochem. Parasitol. 86, 49–59 [DOI] [PubMed] [Google Scholar]
- 14. Chandramohanadas R., Davis P. H., Beiting D. P., Harbut M. B., Darling C., Velmourougane G., Lee M. Y., Greer P. A., Roos D. S., Greenbaum D. C. (2009) Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science 324, 794–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roiko M. S., Carruthers V. B. (2009) New roles for perforins and proteases in apicomplexan egress. Cell. Microbiol. 11, 1444–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blackman M. J. (2008) Malarial proteases and host cell egress. An “emerging” cascade. Cell Microbiol. 10, 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kafsack B. F., Carruthers V. B. (2010) Apicomplexan perforin-like proteins. Commun. Integr. Biol. 3, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yeoh S., O'Donnell R. A., Koussis K., Dluzewski A. R., Ansell K. H., Osborne S. A., Hackett F., Withers-Martinez C., Mitchell G. H., Bannister L. H., Bryans J. S., Kettleborough C. A., Blackman M. J. (2007) Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell 131, 1072–1083 [DOI] [PubMed] [Google Scholar]
- 19. Arastu-Kapur S., Ponder E. L., Fonović U. P., Yeoh S., Yuan F., Fonović M., Grainger M., Phillips C. I., Powers J. C., Bogyo M. (2008) Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 4, 203–213 [DOI] [PubMed] [Google Scholar]
- 20. Miller S. K., Good R. T., Drew D. R., Delorenzi M., Sanders P. R., Hodder A. N., Speed T. P., Cowman A. F., de Koning-Ward T. F., Crabb B. S. (2002) A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J. Biol. Chem. 277, 47524–47532 [DOI] [PubMed] [Google Scholar]
- 21. Koussis K., Withers-Martinez C., Yeoh S., Child M., Hackett F., Knuepfer E., Juliano L., Woehlbier U., Bujard H., Blackman M. J. (2009) A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 28, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruecker A., Shea M., Hackett F., Suarez C., Hirst E. M., Milutinovic K., Withers-Martinez C., Blackman M. J. (2012) Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte. J. Biol. Chem. 287, 37949–37963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kauth C. W., Woehlbier U., Kern M., Mekonnen Z., Lutz R., Mücke N., Langowski J., Bujard H. (2006) Interactions between merozoite surface proteins 1, 6, and 7 of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 281, 31517–31527 [DOI] [PubMed] [Google Scholar]
- 24. Kooij T. W., Carlton J. M., Bidwell S. L., Hall N., Ramesar J., Janse C. J., Waters A. P. (2005) A Plasmodium whole-genome synteny map. Indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 1, e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Putrianti E. D., Schmidt-Christensen A., Arnold I., Heussler V. T., Matuschewski K., Silvie O. (2010) The Plasmodium serine-type SERA proteases display distinct expression patterns and non-essential in vivo roles during life cycle progression of the malaria parasite. Cell. Microbiol. 12, 725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schmidt-Christensen A., Sturm A., Horstmann S., Heussler V. T. (2008) Expression and processing of Plasmodium berghei SERA3 during liver stages. Cell. Microbiol. 10, 1723–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lacroix C., Giovannini D., Combe A., Bargieri D. Y., Späth S., Panchal D., Tawk L., Thiberge S., Carvalho T. G., Barale J. C., Bhanot P., Ménard R. (2011) FLP/FRT-mediated conditional mutagenesis in pre-erythrocytic stages of Plasmodium berghei. Nat. Protoc. 6, 1412–1428 [DOI] [PubMed] [Google Scholar]
- 28. Combe A., Giovannini D., Carvalho T. G., Spath S., Boisson B., Loussert C., Thiberge S., Lacroix C., Gueirard P., Ménard R. (2009) Clonal conditional mutagenesis in malaria parasites. Cell Host Microbe 5, 386–396 [DOI] [PubMed] [Google Scholar]
- 29. Janse C. J., Ramesar J., Waters A. P. (2006) High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1, 346–356 [DOI] [PubMed] [Google Scholar]
- 30. Ishino T., Orito Y., Chinzei Y., Yuda M. (2006) A calcium-dependent protein kinase regulates Plasmodium ookinete access to the midgut epithelial cell. Mol. Microbiol. 59, 1175–1184 [DOI] [PubMed] [Google Scholar]
- 31. Sultan A. A., Thathy V., Nussenzweig V., Ménard R. (1999) Green fluorescent protein as a marker in Plasmodium berghei transformation. Infect. Immun. 67, 2602–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barale J. C., Attal-Bonnefoy G., Brahimi K., Pereira da Silva L., Langsley G. (1997) Plasmodium falciparum asparagine and aspartate rich protein 2 is an evolutionary conserved protein whose repeats identify a new family of parasite antigens. Mol. Biochem. Parasitol. 87, 169–181 [DOI] [PubMed] [Google Scholar]
- 33. Uzureau P., Barale J. C., Janse C. J., Waters A. P., Breton C. B. (2004) Gene targeting demonstrates that the Plasmodium berghei subtilisin PbSUB2 is essential for red cell invasion and reveals spontaneous genetic recombination events. Cell. Microbiol. 6, 65–78 [DOI] [PubMed] [Google Scholar]
- 34. Girardin S. E., Benjannet S., Barale J. C., Chrétien M., Seidah N. G. (1998) The LIM homeobox protein mLIM3/Lhx3 induces expression of the prolactin gene by a Pit-1/GHF-1-independent pathway in corticotroph AtT20 cells. FEBS Lett. 431, 333–338 [DOI] [PubMed] [Google Scholar]
- 35. de Koning-Ward T. F., O'Donnell R. A., Drew D. R., Thomson R., Speed T. P., Crabb B. S. (2003) A new rodent model to assess blood stage immunity to the Plasmodium falciparum antigen merozoite surface protein 119 reveals a protective role for invasion inhibitory antibodies. J. Exp. Med. 198, 869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Giovannini D., Späth S., Lacroix C., Perazzi A., Bargieri D., Lagal V., Lebugle C., Combe A., Thiberge S., Baldacci P., Tardieux I., Ménard R. (2011) Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa. Cell Host Microbe 10, 591–602 [DOI] [PubMed] [Google Scholar]
- 37. Zhang M., Mishra S., Sakthivel R., Rojas M., Ranjan R., Sullivan W. J., Jr., Fontoura B. M., Ménard R., Dever T. E., Nussenzweig V. (2012) PK4, a eukaryotic initiation factor 2α(eIF2α) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc. Natl. Acad. Sci. U.S.A. 109, 3956–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bouillon A., Giganti D., Benedet C., Gorgette O., Pêtres S., Crublet E., Girard-Blanc C., Witkowski B., Ménard D., Nilges M., Mercereau-Puijalon O., Stoven V., Barale J. C. (2013) In silico screening on the three-dimensional model of the Plasmodium vivax SUB1 protease leads to the validation of a novel anti-parasite compound. J. Biol. Chem. 288, 18561–18573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bastianelli G., Bouillon A., Nguyen C., Crublet E., Pêtres S., Gorgette O., Le-Nguyen D., Barale J. C., Nilges M. (2011) Computational reverse-engineering of a spider-venom derived peptide active against Plasmodium falciparum SUB1. PLoS One 6, e21812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Falae A., Combe A., Amaladoss A., Carvalho T., Menard R., Bhanot P. (2010) Role of Plasmodium berghei cGMP-dependent protein kinase in late liver stage development. J. Biol. Chem. 285, 3282–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Agarwal S., Singh M. K., Garg S., Chitnis C. E., Singh S. (2013) Ca(2+) -mediated exocytosis of subtilisin-like protease 1. A key step in egress of Plasmodium falciparum merozoites. Cell. Microbiol. 15, 910–921 [DOI] [PubMed] [Google Scholar]
- 42. Collins C. R., Hackett F., Strath M., Penzo M., Withers-Martinez C., Baker D. A., Blackman M. J. (2013) Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog. 9, e1003344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ishino T., Boisson B., Orito Y., Lacroix C., Bischoff E., Loussert C., Janse C., Ménard R., Yuda M., Baldacci P. (2009) LISP1 is important for the egress of Plasmodium berghei parasites from liver cells. Cell. Microbiol. 11, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCoubrie J. E., Miller S. K., Sargeant T., Good R. T., Hodder A. N., Speed T. P., de Koning-Ward T. F., Crabb B. S. (2007) Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases. Implications for vaccine and drug design. Infect. Immun. 75, 5565–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aly A. S., Matuschewski K. (2005) A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202, 225–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stafford W. H., Blackman M. J., Harris A., Shai S., Grainger M., Holder A. A. (1994) N-terminal amino acid sequence of the Plasmodium falciparum merozoite surface protein-1 polypeptides. Mol. Biochem. Parasitol. 66, 157–160 [DOI] [PubMed] [Google Scholar]
- 47. Pachebat J. A., Ling I. T., Grainger M., Trucco C., Howell S., Fernandez-Reyes D., Gunaratne R., Holder A. A. (2001) The 22 kDa component of the protein complex on the surface of Plasmodium falciparum merozoites is derived from a larger precursor, merozoite surface protein 7. Mol. Biochem. Parasitol. 117, 83–89 [DOI] [PubMed] [Google Scholar]
- 48. Trucco C., Fernandez-Reyes D., Howell S., Stafford W. H., Scott-Finnigan T. J., Grainger M., Ogun S. A., Taylor W. R., Holder A. A. (2001) The merozoite surface protein 6 gene codes for a 36 kDa protein associated with the Plasmodium falciparum merozoite surface protein-1 complex. Mol. Biochem. Parasitol. 112, 91–101 [DOI] [PubMed] [Google Scholar]
- 49. Goel V. K., Li X., Chen H., Liu S. C., Chishti A. H., Oh S. S. (2003) Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc. Natl. Acad. Sci. U.S.A. 100, 5164–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Holder A. A. (2009) The carboxy-terminus of merozoite surface protein 1. Structure, specific antibodies and immunity to malaria. Parasitology 136, 1445–1456 [DOI] [PubMed] [Google Scholar]