Background: miR-29 is a positive regulator of osteoblastogenesis, but its role in osteoclastogenesis is undefined.
Results: Expression of all miR-29 family members increased during osteoclastic differentiation. miR-29 knockdown impaired migration, osteoclast commitment, and formation. Six novel miR-29 targets were identified.
Conclusion: miR-29 promotes osteoclastogenesis.
Significance: These data expand our understanding of osteoclastogenesis, providing insight into miR-29 function in hematopoietic cells and other lineages.
Keywords: Bone, Cell Differentiation, Cell Migration, MicroRNA, Osteoclast, 3′-UTR, miRNA, Post-transcriptional Regulation
Abstract
Osteoclast differentiation is regulated by transcriptional, post-transcriptional, and post-translational mechanisms. MicroRNAs are fundamental post-transcriptional regulators of gene expression. The function of the miR-29 (a/b/c) family in cells of the osteoclast lineage is not well understood. In primary cultures of mouse bone marrow-derived macrophages, inhibition of miR-29a, -29b, or -29c diminished formation of TRAP (tartrate-resistant acid phosphatase-positive) multinucleated osteoclasts, and the osteoclasts were smaller. Quantitative RT-PCR showed that all miR-29 family members increased during osteoclast differentiation, in concert with mRNAs for the osteoclast markers Trap (Acp5) and cathepsin K. Similar regulation was observed in the monocytic cell line RAW264.7. In stably transduced RAW264.7 cell lines expressing an inducible miR-29 competitive inhibitor (sponge construct), miR-29 knockdown impaired osteoclastic commitment and migration of pre-osteoclasts. However, miR-29 knockdown did not affect cell viability, actin ring formation, or apoptosis in mature osteoclasts. To better understand how miR-29 regulates osteoclast function, we validated miR-29 target genes using Luciferase 3′-UTR reporter assays and specific miR-29 inhibitors. We demonstrated that miR-29 negatively regulates RNAs critical for cytoskeletal organization, including Cdc42 (cell division control protein 42) and Srgap2 (SLIT-ROBO Rho GTPase-activating protein 2). Moreover, miR-29 targets RNAs associated with the macrophage lineage: Gpr85 (G protein-coupled receptor 85), Nfia (nuclear factor I/A), and Cd93. In addition, Calcr (calcitonin receptor), which regulates osteoclast survival and resorption, is a novel miR-29 target. Thus, miR-29 is a positive regulator of osteoclast formation and targets RNAs important for cytoskeletal organization, commitment, and osteoclast function. We hypothesize that miR-29 controls the tempo and amplitude of osteoclast differentiation.
Introduction
Osteoclasts are the only cells able to resorb mineralized matrix. The activity of these cells is critical for bone growth, normal bone remodeling, and fracture repair. A fine balance between the number and activity of osteoblasts and osteoclasts is necessary for bone homeostasis (1). Pathologies associated with abnormal osteoclast number or function include osteopetrosis, osteoporosis, and inflammatory osteolysis. Osteopetrosis is caused by impaired resorption, caused by insufficient osteoclast formation or activity, and results in augmented bone density. In this disorder, changes in bone morphology are often accompanied by immunodeficiency and anemia, caused by narrowing of the bone marrow cavity and reduced expansion of the hematopoietic cell populations (2, 3). Osteoporosis is caused by excessive bone resorption coupled with insufficient bone formation. Systemic loss of bone mass can be triggered and supported by hormonal imbalance, such as estrogen deficiency. In inflammatory osteolysis, signaling from immune cells in conditions such as rheumatoid arthritis and periodontal disease determines bone loss localized at the joints or in the oral cavity. Thus, abnormal osteoclastic activity can result in higher predisposition to fractures, impaired joint mechanics, and loss of teeth (4).
The differentiation of osteoclasts from hematopoietic precursors is a complex multistep process (1). It begins with the commitment of multipotent precursors to differentiation along the osteoclast lineage. These committed monocytic cells subsequently migrate and fuse together to form multinucleated mature osteoclasts (5). Bone resorption is initiated when the osteoclast polarizes and organizes the cytoskeletal structures that form the sealing zone and ruffled border. These dynamic structures, which in vitro appear as an actin-rich ring, mediate the degradation of the bone surface, creating an acidic environment and secreting proteolytic enzymes, to degrade the inorganic and organic components of bone matrix, respectively (6). Tight control of the complex osteoclast differentiation process is accomplished by the regulation of gene expression at multiple transcriptional, post-transcriptional, and post-translational levels (7).
Substantial progress has been made in describing the mechanisms of macrophage colony-stimulating factor (M-CSF)-driven2 and receptor activator of nuclear factor κB ligand (RANKL)-driven osteoclastogenesis and bone resorption, and key transcription factors involved include c-FOS, NFATc1, and NFκB. In addition, several studies highlight the role of post-translational modifications, mainly phosphorylation, in regulating the activity of receptors and kinases important for transducing intracellular signals, such as the M-CSF receptor (c-FMS), SRC, and JNK (7, 8). However, in the last decade, the importance of an additional level of gene regulation has emerged: post-transcriptional control by microRNAs (miRNAs).
miRNAs are short sequences of noncoding, single-stranded RNA that can bind target mRNAs based on sequence complementarity. This process involves the RNA-induced silencing complex, which, for the most part, mediates the inhibition of gene expression by decreasing translation and/or by decreasing mRNA stability (9). Often, miRNAs regulate biological functions by modulating the expression of multiple genes that participate in the same or correlated pathways (10). miRNA levels are rapidly altered during embryonic development, as well as in adulthood, resulting in prompt and efficient post-transcriptional control (11, 12).
The overall importance of the miRNA processing pathway in the osteoclast lineage was reported. In vitro silencing of key factors involved in miRNA processing, including DGCR8 (DiGeorge syndrome critical region 8 gene), AGO2 (Argonaute2), and DICER1, suppressed osteoclast differentiation and activity (13). In vivo, deletion of Dicer in the monocyte/macrophage lineage, using a Cd11b promoter driven-cre recombinase, as well as in mature osteoclasts using a cathepsin K promoter driven-cre, resulted in the development of a mild osteopetrotic phenotype (13, 14).
Recent studies identified specific miRNAs and miRNA targets involved in osteoclast commitment and differentiation. For example, miR-223 promotes osteoclast formation, at least in part, through the inhibition of NFIA (nuclear factor 1/A) (13, 15). Decreased NFIA expression is necessary for the terminal differentiation of osteoclasts (13), as well as granulocytes and monocytes (16, 17). Further, miR-21 promotes osteoclast differentiation, and it was shown to target Pdcd4 (programmed cell death domain 4) mRNA. PDCD4 represses AP-1 (activator protein 1)-dependent transcription, and the AP-1 family member c-FOS is essential for osteoclastogenesis (7). Therefore, by suppressing AP-1 function, PDCD4 may exert a negative effect on osteoclast differentiation. Another report demonstrated a negative effect of miR-155 on osteoclastogenesis. miR-155 promotes the commitment of progenitor cells to the macrophage lineage, through repression of Mitf (microphthalmia-associated transcription factor) mRNA (18). MITF is required in the later phases of osteoclast formation, where it promotes the expression of genes crucial for osteoclast maturation and function, like Trap, Oscar (osteoclast-associated immunoglobulin-like receptor), and cathepsin K (19).
We and others have studied the role of the miR-29 family in cells of the osteoblast lineage. Although miR-29 family members target several critical extracellular matrix mRNAs and limit their expression, this miRNA family promotes osteoblastic differentiation by targeting negative regulators of this process (20–22). We considered that miR-29 family members may also play a role in osteoclastogenesis, given that altered miR-29 levels were associated with hematopoietic malignancies. For example, diminished miR-29 levels were found in patients with chronic lymphocytic leukemia and correlate with advanced clinical features and poor prognosis in acute myeloid leukemia (23–26).
In this study, we characterized the expression of miR-29 family members during the differentiation of murine bone marrow-derived osteoclast cultures and an osteoclast precursor cell line. We show that miR-29 is important for cell migration, osteoclast commitment, and differentiation, and we identified six novel miR-29 targets in osteoclastic cells.
EXPERIMENTAL PROCEDURES
Cell Culture
Primary osteoclast precursor cultures were established from bone marrow of 6–8-week-old C57BL/6 male mice that had been depleted of B220/CD45R-positive and CD3-positive cells (B and T lymphocytes, respectively). Briefly, bone marrow was isolated from femurs, tibias, and humeri (27). Cells were incubated with phycoerythrin-conjugated primary antibodies for B220 and CD3 (eBioscience, San Diego, CA), and with magnetically labeled anti-phycoerythrin microbeads (Miltenyi Biotec, Auburn, CA). Magnetic activated cell sorting (MACS®) column technology (Miltenyi Biotec) was used to capture B220- and CD3-positive cells in the column, and the flow-through contained a population of cells enriched for the monocytic and non-lymphoid lineage cells (28). Flow cytometric analysis confirmed that this procedure depleted 93–95% of T and B cells, thereby decreasing the heterogeneity of the marrow cells that were subsequently plated for experiments (data not shown). Cells were cultured in α-MEM (Invitrogen) supplemented with 10% FBS (Atlas Biologicals, Fort Collins, CO) and 30 ng/ml murine recombinant M-CSF (eBioscience). Bone marrow-derived osteoclast precursor cells were plated in the presence of 30 ng/ml murine recombinant M-CSF and RANKL (eBioscience) for up to 6 days.
The mouse monocytic RAW264.7 cell line was obtained from American Type Culture Collection (Manassas, VA) (TIB-71TM) and cultured in DMEM (Invitrogen) supplemented with 10% FBS. Cells were cultured in α-MEM supplemented with 10% FBS and 30 ng/ml RANKL to stimulate osteoclastic differentiation.
The human embryonic kidney (HEK293T) PhoenixTM-Eco cell line was a gift from the Nolan laboratory of Stanford University (29) and was used for retrovirus production. These cells were cultured in DMEM supplemented with 10% FBS. The HEK293FT cell line was obtained from Invitrogen and used for lentivirus production. 293FT cells were cultured in DMEM supplemented with 10% FBS.
In Vitro Osteoclast Formation Assay
Cells were fixed in 2.5% glutaraldehyde in PBS, and TRAP activity was detected according to the manufacturer's instructions, using the acid phosphatase leukocyte kit (Sigma-Aldrich). TRAP-positive multinucleated cells containing more than three nuclei were counted as osteoclasts under microscopic examination. Osteoclast area was quantified using cellSens imaging software (Olympus, Center Valley, PA).
Quantitative Real Time PCR
Primary osteoclast precursors and RAW264.7 cells were plated at 53,000 cells/cm2. Total RNA was isolated from differentiating cultures using the miRNeasy Mini kit (Qiagen). On-column DNase treatment was performed to reduce contamination with genomic DNA, and an additional treatment with RQ1 DNase (Promega, Madison, WI) was performed prior to gene expression analysis. miR-29 expression levels were analyzed with the TaqMan microRNA assay (Invitrogen). According to the manufacturer's instructions, 22.5 ng of RNA were reverse transcribed with specific primers to generate cDNA. miR-29 expression was detected by quantitation PCR in a MiQqPCR cycler (Bio-Rad) and normalized to U6 small nuclear RNA (RNUB6) levels using the absolute quantification method.
To quantify mRNA levels in total RNA, DNased RNA was reverse-transcribed using Moloney murine leukemia virus-reverse transcriptase (Invitrogen) and quantified by qPCR with iQ SYBR Green Supermix (Bio-Rad). The primer sets used are shown in supplemental Table S1. RNA levels were determined using absolute quantification and normalized to hypoxanthine-guanine phosphoribosyltransferase (Hprt) mRNA. RNA experiments were performed at least twice, and each experiment contained biological triplicates. For qRT-PCR, each sample was analyzed in duplicate.
Retroviral Constructs
To obtain miR-29 knockdown, double-stranded oligonucleotides targeting the miR-29a precursor were cloned into the retroviral vector pSilencer 5.1-H1 Retro (Ambion). The sequences of the oligonucleotides used are indicated in supplemental Table S2. The silencing construct was inserted into the pSilencer vector using BamHI and HindIII restriction enzymes. As negative control, pSilencer 5.1 Retro Scrambled was used (Ambion). Retrovirus was produced using the HEK293T PhoenixTM-Eco cell line (29).
miR-29 Knockdown and Osteoclast Formation
Whole bone marrow was isolated from 6–8-week-old C57BL/6 mice and plated overnight on a tissue culture plastic plate to limit the amount of stromal cells. The nonadherent population of cells was centrifuged on a Ficoll gradient, to enrich for macrophage precursors (30). These bone marrow-derived macrophages were seeded at 5,000 cells/well in 96-well plates, in α-MEM supplemented with 10% FBS in the presence of 30 ng/ml M-CSF. 48 h after plating, cells were transfected with 50 nm anti-miRNA inhibitors (Dharmacon) using BioT reagent (Bioland Scientific, Paramount, CA). Alternatively, cells were transduced with retroviruses harboring a miR-29a knockdown construct or a scrambled control. Osteoclast differentiation was induced with RANKL treatment (10 ng/ml), and osteoclast formation was evaluated by TRAP staining.
pSLIK Lentiviral Constructs
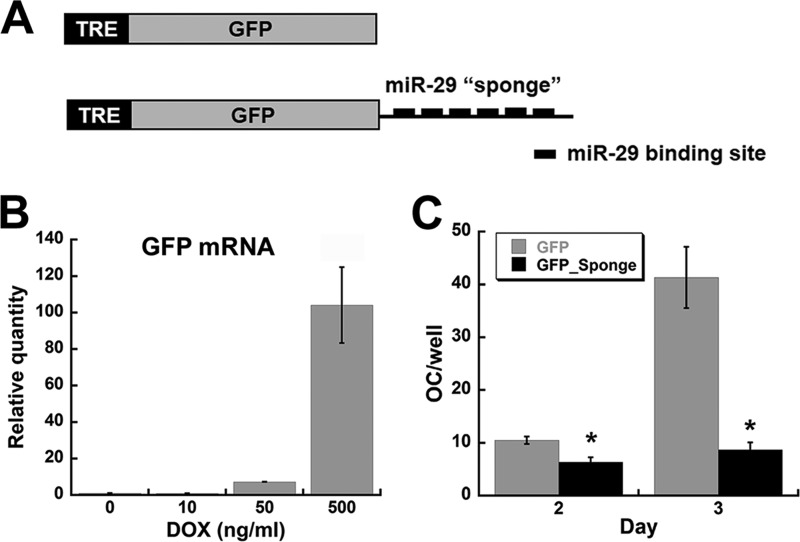
To knock down the activity of all miR-29 family members, we generated a miR-29 “sponge,” which works as a competitive target for miR-29, relieving the repression of its endogenous target mRNAs (see Fig. 3) (31). The murine osteonectin 3′-UTR contains a pair of miR-29 binding sites, within cDNA bases 1083–1149. Three copies of this tandem miR-29 binding site were cloned downstream of a GFP reporter gene to generate the miR-29 sponge. The GFP alone control or GFP_Sponge cDNAs were subcloned in the pEN_Tmcs entry vector (plasmid 25751; Addgene, Cambridge, MA) (32), which contains a tetracycline inducible promoter (tetracycline-responsive element). These constructs were subjected to Gateway recombination with the lentiviral construct pSLIK, harboring a hygromycin resistance selectable marker gene (plasmid 25737; Addgene) (32). The ubiquitin C promoter in the pSLIK vector constitutively drives the expression of the tetracycline activator (rtTA3), which, in the presence of doxycycline (DOX), promotes the expression of the GFP or GFP_Sponge transgene from the tetracycline-responsive element promoter.
FIGURE 3.
miR-29 knockdown inhibits osteoclastic differentiation of RAW264.7 cells. A, schematic representation of DOX-inducible GFP and GFP_Sponge constructs. B, GFP RAW264.7 cell line was treated for 24 h with increasing concentrations of DOX. GFP mRNA expression was quantified by qRT-PCR and normalized to HPRT. C, GFP and GFP_Sponge RAW264.7 cells were treated with 500 ng/ml of DOX. After 2 and 3 days of differentiation with RANKL (30 ng/ml), osteoclast formation was evaluated by TRAP staining (n = 6, 96-well plate). *, significantly different from GFP, p < 0.05. OC, osteoclasts; TRE, tetracycline-responsive element.
Lentivirus Production and Transduction
pSLIK constructs containing GFP alone or GFP_Sponge genes were co-transfected in the HEK293FT cell line, along with the expression vectors for the viral packaging proteins. These include the viral transactivators Tat (pHDM-tat1b) and Rev (pRC/CMV-rev1b); the viral core polyprotein and reverse transcriptase, encoded by the GAG and POL genes, respectively (pHDM-Hgpm2); and the VSV-G (vesicular stomatitis virus) envelope glycoprotein (pHDM-G) (gift from the Lee laboratory, Harvard Gene Therapy Initiative, Cambridge, MA). Culture medium containing the lentiviral particles was used to transduce RAW264.7 cells. Pools of stably transduced cells were established by culture in the presence of hygromycin (100 μg/ml).
miR-29 Sponge Expression and Osteoclast Differentiation
GFP and GFP_Sponge RAW264.7 cell lines were seeded at a cell density of 1,000 cells/well in 96-well plates, in DMEM supplemented with 10% FBS. 24 h later, osteoclast differentiation was stimulated by treatment with 30 ng/ml RANKL in α-MEM supplemented with 10% FBS. The expression of the transgene was induced by addition of 500 ng/ml DOX to the culture medium (Sigma-Aldrich). Osteoclast formation was evaluated by TRAP staining.
Cell Viability Assay
GFP and GFP_Sponge RAW264.7 cells were plated in 96-well plates at 5,000 cells/well, in DMEM supplemented with 10% FBS. 24 h later, cells were treated with 30 ng/ml RANKL, to induce osteoclastogenesis, in the presence or absence of 500 ng/ml DOX, to activate transgene expression. Cell viability was assessed over 3 days by MTS assay using the CellTiter 96 AQueous One Solution cell proliferation assay kit, as indicated by the manufacturer's instructions (Promega).
Phagocytosis Assay
GFP and GFP_Sponge RAW264.7 cells were cultured in α-MEM supplemented with 10% FBS in the presence of 500 ng/ml of DOX and 30 ng/ml of RANKL for 24 h prior to the assay. Cells were then plated at 100,000 cells/well in 96-well plates and allowed to adhere to the plate for 3 h. Culture medium was replaced with a solution of pHrodo Staphylococcus aureus bacterial particles (250 μg/ml), which will emit fluorescence when phagocytosed (Invitrogen). Cells were incubated for 1 h at 37 °C, and nuclei were stained with DAPI. Cultures were analyzed by fluorescence microscopy. Phagocyotosis of pHrodo particles was quantified by measuring fluorescence emitted at 590 nm and normalized to DAPI fluorescence at 460 nm.
Macrophage Commitment Assay
GFP and GFP_Sponge RAW264.7 cells were plated at 26,000 cells/cm2 in DMEM supplemented with 10% FBS. Subsequently, cells were cultured in α-MEM supplemented with 10% FBS in the presence of 500 ng/ml of DOX and 30 ng/ml of RANKL for 24 h. Total RNA was isolated from the cultures using the miRNeasy mini kit (Qiagen), and the expression of macrophage marker genes was analyzed by qPCR as previously described. The primer sets used are shown in supplemental Table S1.
Cell Migration Assay
GFP and GFP_Sponge RAW264.7 cells were cultured in DMEM, in the absence of FBS, and in the presence of 500 ng/ml of DOX for 24 h prior to the assay. The cells were then plated at 200,000 cells/well on 8-μm pore polycarbonate membrane inserts in 6.5-mm Transwell membranes (Corning, Tewksbury, MA). 30 ng/ml M-CSF was added to the bottom chamber as a chemotactic agent. Cells were incubated overnight, in the presence of DOX. Cells that did not migrate were removed from the top side of the Transwell membrane using a cotton swab. Upon fixation with 3.7% formaldehyde, cells on the bottom side of the Transwell membrane were stained with 0.05% crystal violet solution. Crystal violet stain was solubilized using 100% methanol, and optical density was quantified at 540 nm.
Apoptosis Assay
GFP and GFP_Sponge RAW264.7 were plated at 3,125 cells/cm2 in DMEM supplemented with 10% FBS. 24 h later, culture medium was switched to α-MEM supplemented with 10% FBS, with the addition of 30 ng/ml of RANKL. After 2 days of differentiation, expression of the transgene was induced with 500 ng/ml of DOX. 2 days later, osteoclast apoptosis was assessed by Caspase-3 colorimetric assay kit, as indicated by the manufacturer's instructions (GenScript, Piscataway, NJ).
Actin Ring Formation Assay
GFP and GFP_Sponge RAW264.7 cells were seeded on glass chamber slides at 3,125 cells/cm2, in DMEM supplemented with 10% FBS. 24 h later, culture medium was switched to α-MEM supplemented with 10% FBS, with the addition of 30 ng/ml of RANKL and 500 ng/ml of DOX. End point cultures were fixed with 3.7% formaldehyde, and F-actin was labeled with rhodamine phalloidin conjugate (Invitrogen). Nuclei were visualized by using mounting medium containing DAPI (Invitrogen). Cultures were analyzed by fluorescent microscopy.
Luciferase Constructs
Gene-specific PCR primers were used to amplify from mouse genomic DNA template the coding sequences or UTRs for calcitonin receptor, TRAP, cathepsin K, Cdc42, Srgap2, Gpr85, Cd93, and Nfia, which contained potential miR-29-binding sites (primer sequences are reported in supplemental Table S2). Using the appropriate restriction enzymes, these fragments were cloned downstream from Luciferase, in the cytomegalovirus promoter-driven Luciferase reporter vector pMIR-REPORT (Ambion, Austin, TX). Site-directed mutagenesis, to delete putative miR-29 binding sites in selected constructs, was performed by overlap extension (primer sequences are reported in supplemental Table S3). Luciferase plasmids containing the regions of interest were used as templates for mutagenesis. All constructs were verified by sequencing.
Luciferase Activity Assay
RAW264.7 cells were plated at 58,000 cells/cm2. After 24 h, cells were co-transfected with Luciferase constructs and a constitutively expressing β-galactosidase construct, as a control vector for transfection efficiency (GenBankTM accession number U02451) (Clontech) using BioT (BioT:DNA ratio 1.5 μl:1 μg). In selected experiments, 50 nm anti-miR-29-a or -c or negative control (scrambled) miRNA inhibitors were also transfected. 6 h post-transfection, cells were treated with or without RANKL (30 ng/ml) in α-MEM supplemented with 10% FBS. Following 48 h, cell lysates were harvested using reporter lysis buffer (Promega). Samples were analyzed for Luciferase activity using a Luciferase assay system (Promega) and normalized to β-galactosidase activity, which was assessed using Galacton® reagent (Applied Biosystem, Foster City, CA). Each Luciferase experiment was performed at least three times, using n = 6.
Data Analysis
The data are presented as the means ± S.E. The data were analyzed by Student's t test or one-way analysis of variance with Bonferroni post hoc test as appropriate (KaleidaGraph; Synergy Software, Reading, PA).
RESULTS
miR-29 Expression Increases during in Vitro Osteoclast Differentiation
We analyzed the expression of miR-29 family members in mouse bone marrow depleted of B220- and CD3-positive cells and cultured in the presence of M-CSF and RANKL for up to 5 days. In these cultures, osteoclasts were evident by day 3, and osteoclast number and size were highest at day 5 (Fig. 1, B–D). qRT-PCR showed that all the miR-29 family members, miR-29a, -b, and -c, were expressed at a similar level and that their expression was not increased until between days 3 and 5 of differentiation (Fig. 1A). In contrast, mRNA levels for the osteoclast markers Trap and cathepsin K were increased from days 1 to 3 and sustained at day 5 (Fig. 1E).
FIGURE 1.
miR-29 increases during osteoclast differentiation in vitro. Primary bone marrow osteoclast precursors depleted of B and T cells were differentiated by culturing with 30 ng/ml each M-CSF and RANKL. A, miR-29a, -b, and -c expression was quantified after 1, 3, and 5 days of culture and normalized to U6 (n = 4). B, number of TRAP(+) osteoclasts/well (n = 3, 96-well plate). C, osteoclast area. Box plot lines represent the 25% quartile of the data, the median, and the 75% quartile. Outliers are denoted by dots (n = 3 wells, 96-well plate). D, representative images of TRAP stained cultures after 1, 3, and 5 days of differentiation were captured using 10× magnification. E, osteoclast marker mRNAs were quantified by qRT-PCR and normalized to HPRT (n = 4). *, significantly different from day 1, p < 0.05. OCS, osteoclasts; CATK, cathepsin K.
miR-29 Is a Positive Regulator of Osteoclastogenesis
To investigate the role of miR-29 in osteoclast differentiation, we inhibited miR-29 activity in primary cultures of bone marrow-derived macrophage/osteoclast precursors (bone marrow-derived macrophages), using transiently transfected miR-29a, -b, or -c specific oligonucleotide inhibitors. Throughout the time course analyzed, miR-29 inhibition resulted in a significant reduction in the number of TRAP positive multinucleated osteoclasts compared with the scrambled, nontargeting control (Table 1). These data suggest that miR-29 inhibition does not merely delay osteoclastogenesis and that miR-29 activity is important for osteoclast formation. Consistent with these data, primary bone marrow-derived precursors transduced with a retrovirus expressing a miR-29a inhibitor also showed decreased osteoclast formation compared with control (supplemental Fig. S1).
TABLE 1.
miR-29 knockdown decreases osteoclast formation in vitro
Mature sequences of the nontargeting control (cel-miR-67) and the miR-29 family members are indicated. Divergent bases are underlined. Seed binding regions (bases 2–8) are in italics. Primary bone marrow macrophages were transfected with 50 nm anti-miR-29a, -b, or -c inhibitor or nontargeting scrambled control oligonucleotides. Cells were treated for 3–6 days with M-CSF (30 ng/ml) and RANKL (10 ng/ml). Osteoclast formation was evaluated by TRAP staining (n = 4 wells, 96-well plate).
| Inhibitor | Mature miRNA sequence | Day 3 | Day 4 | Day 5 | Day 6 |
|---|---|---|---|---|---|
| Scrambled | ucacaaccuccuagaaagaguaga | 5.2 ± 1.6a | 21.6 ± 6.3a | 46.0 ± 4.3a | 203.8 ± 17.9a |
| miR-29a | uagcaccaucugaaaucgguua | 0 | 0 | 11.0 ± 1.7 | 17.3 ± 1.4 |
| miR-29b | uagcaccauuugaaaucgguua | 0.7 ± 0.5 | 2.8 ± 1.0 | 10.2 ± 1.9 | 17.8 ± 1.9 |
| miR-29c | uagcaccauuugaaaucaguguu | 0 | 0 | 6.5 ± 2.3 | 17.8 ± 2.1 |
a Significantly different from 29a, 29b, or 29c inhibitor (p < 0.01).
miR-29 family members have identical seed binding regions (miRNA bases 2–8). miR-29a and -29c differ by only one base, whereas miR-29b is more divergent. In the transient transfection studies shown in Table 1, the miR-29b inhibitor appeared to be somewhat less efficacious at the early time points, days 3 and 4. However, the isoform-specific inhibitors had similar effects at the later time points, days 5 and 6. Overall, the miR-29 isoform inhibitors had similar activity. Given the degree of conservation among the miR-29 isoforms, it may be difficult to tease out isoform-specific effects using an inhibitor strategy.
When bone marrow-derived macrophages were plated at a higher density, the formation of large osteoclasts (>8 nuclei) was significantly diminished in cells transiently transfected with miR-29a inhibitor (large osteoclast number (per well) in scrambled 108 ± 5 versus miR-29a inhibitor 74 ± 3; p < 0.01). This indicates that miR-29 knockdown negatively affects osteoclast size, suggesting that miR-29 activity plays a positive role in osteoclast maturation.
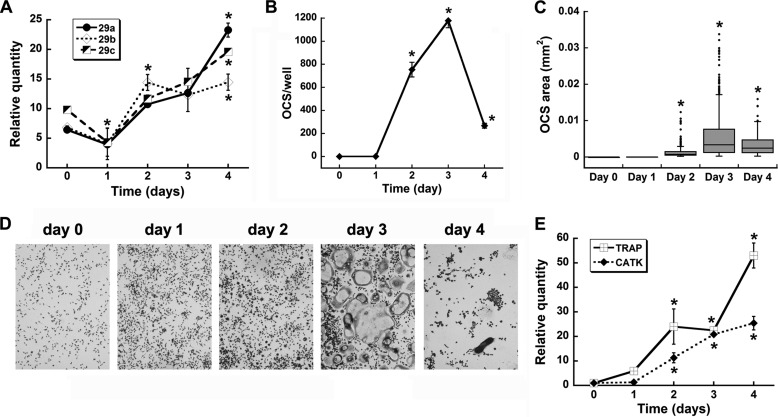
The primary cultures obtained from bone marrow are heterogeneous, even when depleted of lymphocytes (33). For this reason, we sought a more simplified and homogeneous model system for the purpose of evaluating the mechanisms by which miR-29 regulates osteoclast differentiation. Therefore, we chose to use the monocytic cell line RAW264.7. We first characterized the expression of miR-29 family members in these cells after treatment with RANKL for up to 4 days. We found that expression of all miR-29 family members was modestly but significantly decreased after 24 h of RANKL treatment (Fig. 2A). Levels of miR-29a, -b, and -c then increased as osteoclastogenesis progressed, in a trend similar to that observed in the primary cultures (compare with Fig. 1A). In these cultures, osteoclast number and size peaked on day 3 and were decreased on day 4 because of apoptosis of mature osteoclasts (Fig. 2, B–D). As observed in the primary cells, the osteoclast marker genes, Trap and cathepsin K, also increased with osteoclastic differentiation (Fig. 2E). These data suggested that RAW264.7 cells could be a valid surrogate for analyzing the mechanisms by which miR-29 regulates osteoclastogenesis.
FIGURE 2.
miR-29 expression in RAW264.7 cells recapitulates the pattern observed in primary osteoclast precursor cells. A, the expression of miR-29a, -b, and -c was analyzed over 4 days of differentiation with RANKL (30 ng/ml). B, number of osteoclasts per well (n = 3, 96-well plate). C, osteoclast area. Box plot lines represent the 25% quartile of the data, the median, and the 75% quartile. Outliers are denoted by dots (n = 3 wells, 96-well plate). D, representative images of TRAP stained cultures at days 0, 1, 2, 3, and 4 of differentiation, respectively, were captured using 10× magnification. E, gene expression levels for osteoclast markers. *, significantly different from day 0, p < 0.05. OCS, osteoclasts; CATK, cathepsin K.
Inhibition of miR-29 Activity Impairs Osteoclastic Differentiation of RAW264.7
To further define the activity of miR-29 in osteoclastogenesis, we developed an inducible lentiviral knockdown construct, based on the miRNA sponge strategy (31). The miR-29 sponge consisted of six miR-29 binding sites cloned downstream of GFP, in a DOX-inducible lentiviral vector (Fig. 3A). Expression of the GFP_Sponge RNA can work as a decoy or competitive inhibitor for all the members of the miR-29 family). RAW264.7 cells were stably transduced with lentivirus harboring the miR-29 sponge (GFP_Sponge cells) or GFP alone (GFP cells).
To confirm DOX-inducible expression of the transgene, we quantified GFP mRNA levels in GFP cells treated for 24 h with different doses of DOX. qRT-PCR verified that GFP RNA was induced in a dose-responsive fashion, in the presence of increasing concentrations of DOX (Fig. 3B). The 500 ng/ml dose of DOX was chosen for the subsequent assays because it produced a level of transgene expression similar to that of Trap mRNA, while representing a DOX dose well below that recently reported to decrease osteoclast differentiation in bone marrow cells in vitro (34).
We used the GFP_Sponge RAW264.7 cells as a relatively homogeneous cell population to study the mechanisms by which miR-29 regulates osteoclastogenesis. In cells treated with DOX and RANKL, we observed reduced formation of TRAP-positive multinucleated cells in the GFP_Sponge cultures after 2 days, in comparison with the cells expressing the GFP transgene only (Fig. 3C). After 3 days, this disparity was even more pronounced, because the formation of TRAP-positive multinucleated cells was increased only in the GFP RAW264.7 cultures. There was no difference in the ability of GFP and GFP_Sponge cells to form TRAP-positive multinucleated cells in the absence of DOX (data not shown). These results confirm the positive role of miR-29 in osteoclastogenesis, as observed in primary cells (Table 1).
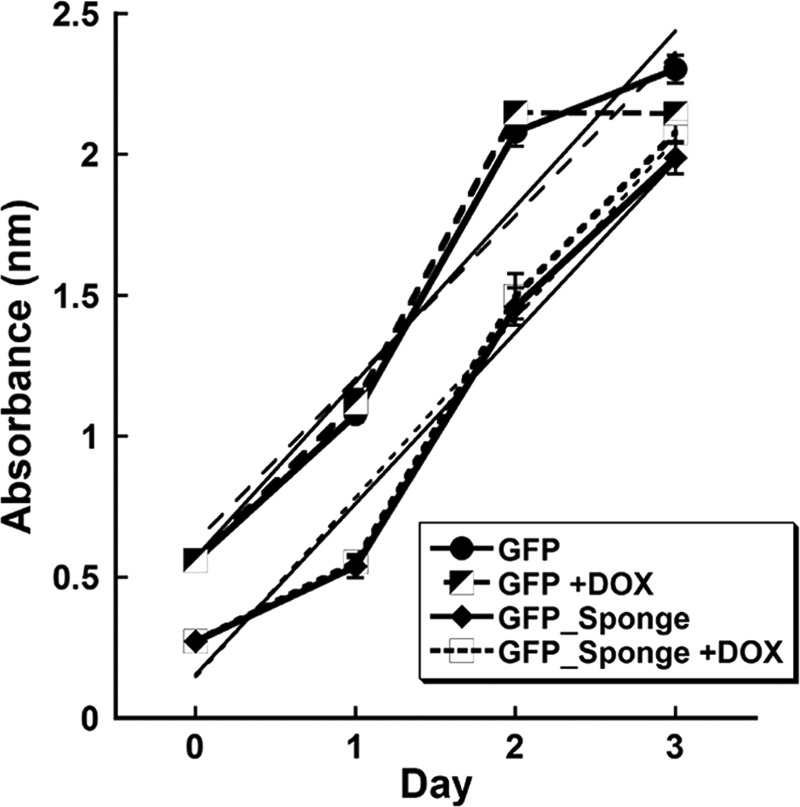
To determine whether miR-29 knockdown in the GFP_Sponge cultures impaired osteoclast formation because of altered cell proliferation or viability, GFP or GFP_Sponge cells were cultured in the presence or absence of DOX for up to 3 days. Viable cells were quantified by MTS assay. All cultures displayed similar growth curves, suggesting that miR-29 did not promote osteoclast formation by regulating cell growth (Fig. 4).
FIGURE 4.
Inhibition of miR-29 does not affect cell viability. GFP and GFP_Sponge RAW264.7 cells were treated with RANKL (30 ng/ml) in the presence or absence of DOX (500 ng/ml). Cell viability was measured over 3 days by MTS assay (n = 6). Growth curves in the presence or absence of DOX were superimposable. The lines without symbols illustrate linear growth curves with similar slopes.
Inhibition of miR-29 Activity Impairs RAW264.7 Cell Commitment to the Osteoclast Lineage
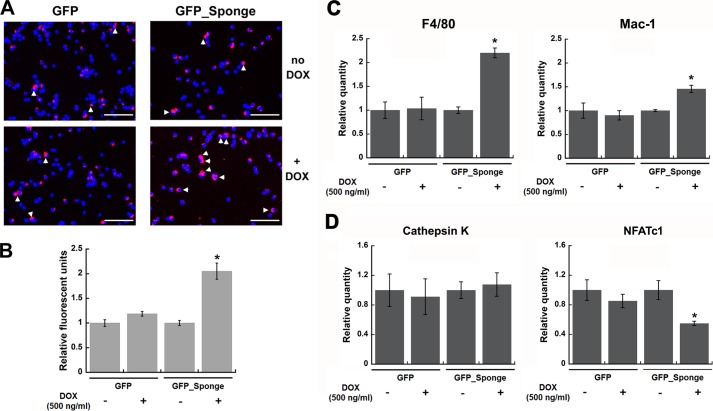
Some miRNAs regulate differentiation by promoting commitment to one cell fate at the expense of another. Because RAW264.7 cells have the potential to differentiate into osteoclasts or macrophages, we evaluated the effect of miR-29 knockdown on their lineage commitment. RAW264.7 cells have phagocytic properties, which are increased when committed to the macrophage fate. Therefore, we quantified phagocytosis in GFP and GFP_Sponge cells that had been induced toward osteoclastic differentiation by culture in the presence of RANKL for 24 h. Phagocytosis was not altered in the GFP cells treated with DOX. However, when miR-29 activity was knocked down in GFP_Sponge cultures treated with DOX, we observed increased phagocytic activity (Fig. 5, A and B).
FIGURE 5.
miR-29 knockdown supports RAW264.7 cell commitment to the macrophage lineage. GFP and GFP_Sponge RAW264.7 cells were induced toward osteoclastic differentiation by 24-h RANKL treatment (30 ng/ml) in the presence or absence of DOX (500 ng/ml). A, lineage commitment was assessed by phagocytosis assay. The cells were incubated for 1 h with pHrodo bacterial particles (250 μg/ml). Cells that have phagocytosed the particles display red fluorescence (arrowheads). Nuclei were stained using DAPI reagent. Representative images of the cultures were captured using 10× magnification. Scale bar, 100 μm. B, phagocytosis was quantified by measuring rhodamine fluorescence and normalized to DAPI fluorescence (n = 6). C and D, macrophage marker mRNAs (C) and osteoclast marker mRNAs (D) were quantified by qRT-PCR and normalized to Hprt mRNA (n = 3). *, significantly different from no DOX, p < 0.05.
In addition, we analyzed the expression of genes associated with macrophage and osteoclast differentiation. DOX treatment of GFP_Sponge cells increased mRNA for macrophage markers F4/80 and Mac-1/Cd11b and decreased mRNA for the early osteoclast marker Nfatc1. In contrast, mRNA for the more mature osteoclast marker, cathepsin K, was not affected (Fig. 5, C and D). These data suggest that miR-29 knockdown promotes the commitment of the RAW264.7 cell line to the macrophage lineage, at the expense of osteoclastogenesis.
Inhibition of miR-29 Activity Impairs RAW264.7 Migration
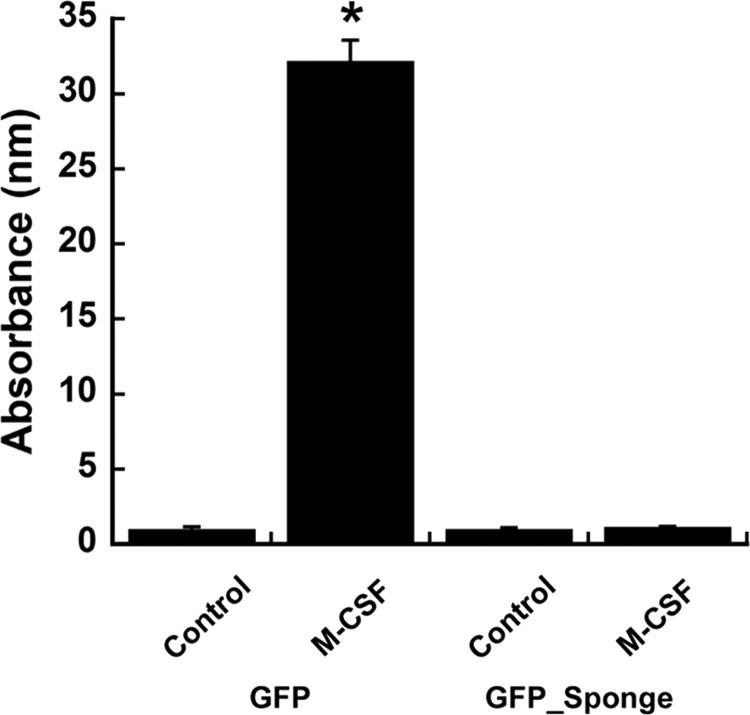
To form osteoclasts, the migration of precursor cells is critical. We analyzed the ability of GFP and GFP_Sponge cells to migrate in response to a chemotactic stimulus using a modified Boyden chamber (Transwell) assay. We found that GFP-expressing RAW264.7 cells displayed robust migration toward M-CSF supplemented culture medium (Fig. 6). However, expression of the miR-29 sponge completely abrogated the capability of these osteoclast precursor cells to respond to the chemotactic agent, strongly indicating that miR-29 expression supports cell migration (Fig. 6).
FIGURE 6.
miR-29 promotes the migration of RAW264.7 cells. GFP and GFP_Sponge cells were cultured for 24 h with DOX (500 ng/ml). Cells were plated on Transwell membranes with 8-μm pores and allowed to migrate overnight toward M-CSF (30 ng/ml). The cells that migrated through the membrane were stained with crystal violet. Crystal violet staining was quantified. *, significantly different from no M-CSF, p < 0.05 (n = 6).
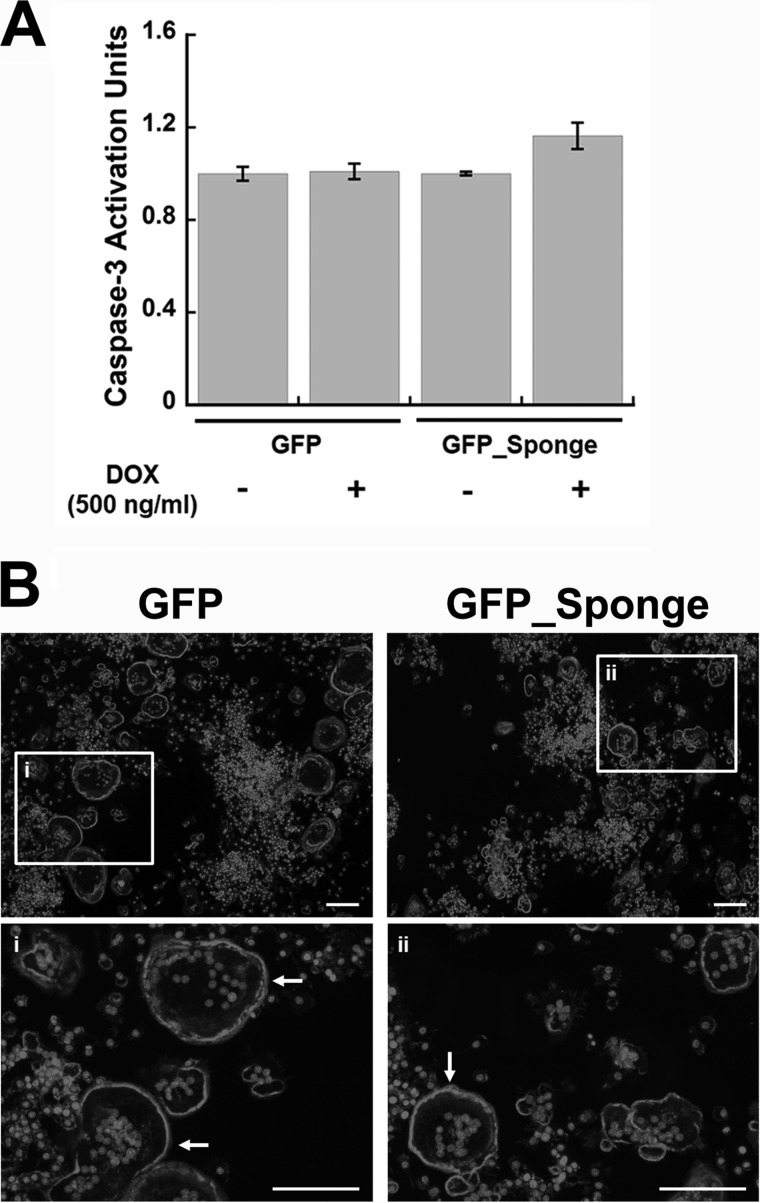
Inhibition of miR-29 Activity Does Not Affect the Apoptosis of Mature Osteoclasts or Actin Ring Formation
Osteoclasts are terminally differentiated cells, and their ultimate fate is to undergo apoptosis (i.e., Fig. 2B). It is possible that increased apoptosis could contribute to the decrease in number and size of osteoclastic cells observed in the presence of the miR-29 sponge. Therefore, we analyzed Caspase-3 activity in GFP and GFP_Sponge cells cultured for 4 days in the presence of RANKL. The miR-29 sponge was only expressed during days 3 and 4 of culture, to allow initiation of osteoclastic differentiation. We did not detect a significant difference in Caspase-3 activity in cells expressing the miR-29 sponge, compared with the other groups (Fig. 7A). This suggests that miR-29 knockdown does not affect the survival of mature osteoclasts.
FIGURE 7.
Inhibition of miR-29 does not affect apoptosis of mature osteoclasts, or actin ring formation. A, GFP and GFP_Sponge RAW264.7 cells were differentiated for 2 days with RANKL (30 ng/ml). The expression of the transgene was induced by addition of DOX (500 ng/ml) at day 3. Caspase-3 activity in mature osteoclast cultures was quantified after 4 days of differentiation (n = 6). B, GFP and GFP_Sponge RAW264.7 cells were cultured on glass chamber slides for 4 days in the presence of DOX (500 ng/ml) and RANKL (30 ng/ml). Actin ring formation was evaluated by phalloidin staining, and nuclei were visualized with DAPI reagent. Representative images were captured using 5× magnification. Boxed regions i and ii are visualized at 20× magnification. Actin rings are indicated by arrows. Scale bar, 100 μm.
The formation of actin rings is a critical step for osteoclast-mediated bone resorption. To determine whether miR-29 knockdown affects actin ring formation, the GFP and GFP_Sponge were cultured for 4 days in the presence of RANKL and subjected to phalloidin staining. Although fewer and smaller multinucleated TRAP-positive cells were identified in miR-29 sponge cultures, their actin ring structures were intact (Fig. 7B, panels i and ii).
miR-29 Targets RNAs Important for the Macrophage/Osteoclast Lineage
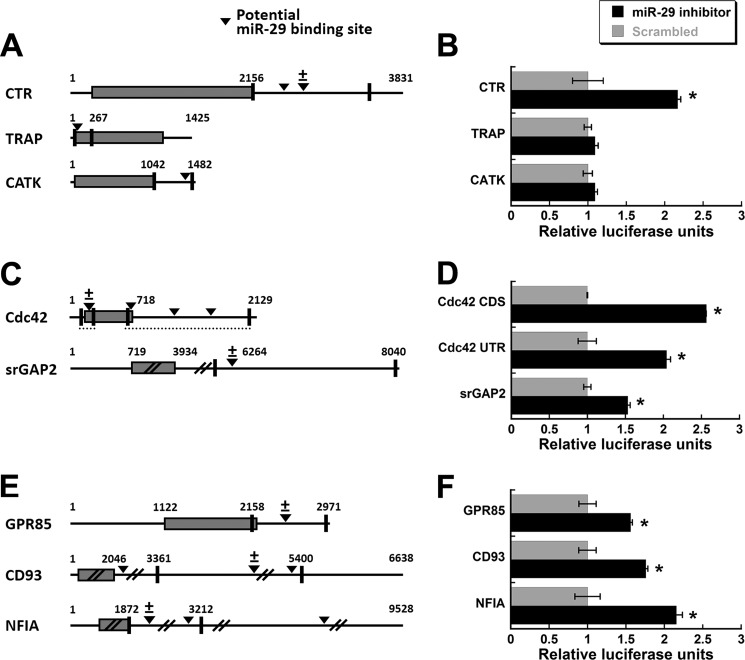
Our functional assays indicated that miR-29 is important for osteoclastogenesis and promotes cell migration and osteoclast commitment. To better understand the underlying mechanisms, we focused on identifying the mRNAs that are targeted by miR-29 and whose functions are important in the macrophage/osteoclast lineage. We analyzed a list of genes expressed in osteoclastic cells3 for potential miR-29 targets, using several different algorithms for miRNA target prediction (miRanda, DIANA-mirExTra, PicTar, and RNAhybrid). The list of potential targets was refined based on the ability of the miRNA to base pair with the target mRNA. We chose to clone and analyze eight candidate genes with a well documented role in osteoclasts (Fig. 8A), with a role in cell migration (Fig. 8C), or with a role in the macrophage lineage (Fig. 8E) (supplemental Tables S2 and S4).
FIGURE 8.
Luciferase analysis of miR-29 targets. Potential miR-29 targets were identified using a bioinformatic approach. Putative miR-29 binding sites (arrowheads) are present in the coding sequence and/or in the 3′-UTR. The corresponding regions (between the vertical lines) were cloned into the pMIR-REPORT Luciferase vector, downstream of the Luciferase gene. The ± symbol denotes the binding site deleted for the constructs shown in Fig. 9. A, putative miR-29 binding sites were identified in genes important for osteoclastogenesis. B, Luciferase activity was quantified in RAW264.7 cells co-transfected with a miR-29c inhibitor or a scrambled nontargeting control, and normalized to β-galactosidase activity. Cells were treated with RANKL (30 ng/ml) for 48 h after transfection. C, putative miR-29 binding sites were identified in genes important for cytoskeletal remodeling and cell migration. D, Luciferase activity. E, potential miR-29 target genes associated with the macrophage lineage. F, luciferase activity. *, significantly different from scrambled, p < 0.05 (n = 6). CATK, cathepsin K; CDS, coding sequence.
We cloned the sequences containing the potential binding sites for miR-29 into a Luciferase reporter vector. Most constructs contained regions of interest ≥1 kb in length (Fig. 8, A, C, and E). In these constructs, Luciferase expression was driven by a strong, constitutive promoter, with the cloned regions serving as 3′-UTR for the Luciferase gene. Therefore, Luciferase activity represents the regulatory activity of the sequence of interest. RAW264.7 cells were transiently transfected with the Luciferase reporter plasmids and miR-29c inhibitor oligonucleotides. Increased Luciferase activity in the presence of the miR-29 inhibitor would suggest that miR-29 targets that RNA region.
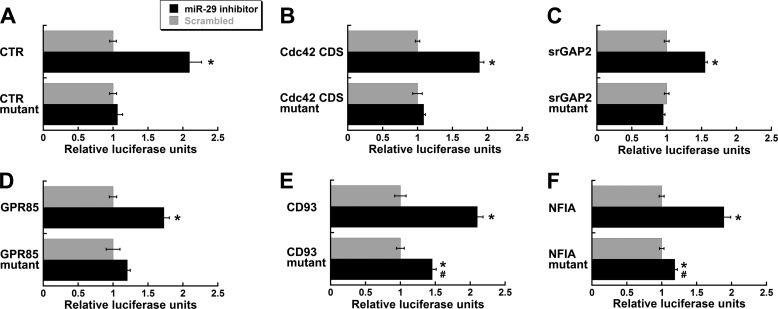
Of the RNAs with a well known function in osteoclastic cells, Calcr (calcitonin receptor), Trap, and cathepsin K (Ctsk, Catk) had potential miR-29 binding sites. The potential miR-29 binding site in the Trap RNA was in the coding region, whereas those for Calcr and Ctsk were in the 3′-UTR. There were two potential miR-29 binding sites in the Calcr 3′-UTR. miR-29c inhibitor only increased Luciferase activity from the construct containing the Calcr 3′-UTR, suggesting that Calcr RNA is targeted by miR-29, whereas Trap and Ctsk are not (Fig. 8B). The potential miR-29 binding site at base 2565 in the Calcr 3′-UTR had the most complementarity to miR-29 family members. When we deleted this miR-29 binding site, the ability of the miR-29 inhibitor to increase Luciferase activity was lost, indicating that this sequence is specific for miR-29-mediated regulation (Fig. 9A).
FIGURE 9.
Deletion mutants of miR-29 binding sites. Putative miR-29 binding sites (marked with ± in Fig. 8) in the miR-29 target genes were deleted from pMIR-REPORT Luciferase vectors. Luciferase activity was quantified in the RAW264.7 cells co-transfected with a miR-29c inhibitor or a scrambled nontargeting control and normalized to β-galactosidase activity. Cells were treated with RANKL (30 ng/ml) for 48 h after transfection. *, significantly different from scrambled; #, significantly different from 29c inhibitor in the wild-type vector, p < 0.05 (n = 6). CDS, coding sequence. A, calcitonin receptor gene; B, Cdc42 coding sequence; C, Srgap2; D, Gpr85; E, Cd93; F, Nfia.
For RNAs important for cytoskeletal organization, we analyzed Cdc42 (cell division control protein 42) and Srgap2 (SLIT-ROBO Rho GTPase-activating protein 2) (Fig. 8C). CDC42 is important for osteoclast function and migration, and srGAP2 participates in the same signaling pathway as CDC42 (35). Although expressed in osteoclasts, srGAP2 has not been studied in the osteoclast lineage. There were potential miR-29 binding sites in the coding region and in the 3′-UTR of Cdc42, and the miR-29 inhibitor increased Luciferase activity from constructs containing either region, as well as the construct carrying the Srgap2 3′-UTR (Fig. 8D). Deletion of the potential miR-29 binding site in the coding region of Cdc42 abolished the ability of the miR-29c inhibitor to relieve repression of Luciferase activity, indicating that this is a miR-29 binding site (Fig. 9B). Similarly, deletion of the potential miR-29 binding site in the Srgap2 3′-UTR construct abrogated the response of the construct to miR-29c inhibitor, indicating that the Srgap2 3′-UTR is targeted by miR-29 (Fig. 9C). Deletion mutagenesis was not performed on the mouse Cdc42 3′-UTR construct because functional miR-29 binding sites in the human 3′-UTR were previously reported (50).
With regard to genes associated with the macrophage lineage, we chose to examine GPR85 (G protein-coupled receptor 85), NFIA (nuclear factor I/A), and CD93 (Fig. 8E). NFIA expression inhibits both macrophage and osteoclast maturation, whereas GPR85 and CD93 are expressed during macrophage differentiation (13, 17, 46, 49). Nfia and Cd93 each had two potential miR-29 binding sites in the 3′-UTR segment analyzed, whereas Gpr85 had one site. The miR-29c inhibitor increased Luciferase activity for GPR85, CD93, and NFIA constructs, suggesting that these UTRs are miR-29 targets (Fig. 8F). Deletion of the potential miR-29 binding site in the Gpr85 3′-UTR eliminated the miR-29c inhibitor-mediated increase in Luciferase activity, indicating that this site is directly regulated by miR-29 (Fig. 9D). For Cd93, we deleted the potential miR-29 binding site at base 4403, and for Nfia, we deleted the miR-29 site at 2125. These sites were chosen because, of the two present in the UTR construct, they had the most complementarity to miR-29 family members. The ability of the miR-29c inhibitor to relieve repression of Luciferase activity in the Cd93 and Nfia mutant constructs was significantly decreased, but not totally abolished, likely because of the remaining functional miR-29 binding site in the construct (Fig. 9, E and F) (supplemental Table S4).
It should be noted that the transfection studies shown in Fig. 8 were performed in cells treated with RANKL, and similar trends were also noted in the absence of RANKL (supplemental Fig. S2). Moreover, similar results were obtained when miR-29a inhibitor was used instead of miR-29c inhibitor (data not shown). Overall, miR-29 may promote osteoclastogenesis by repressing RNAs important for differentiation to the alternative macrophage lineage. miR-29 targeting of RNAs important for osteoclast function and actin remodeling may allow subtle regulation of the rate of osteoclast differentiation (Table 2).
TABLE 2.
Newly validated miR-29 targets and their biology
| Gene | Biology |
|---|---|
| Calcr, Ctr (calcitonin receptor) | Decreases resorption and increases osteoclast survival (44) |
| Cdc42 (cell division cycle 42) | Promotes osteoclast survival and differentiation; regulates rate of actin ring formation (52) |
| Srgap2 (SLIT-ROBO Rho GTPase-activating protein) | Induces membrane protrusions; knockdown reduces cell adhesion and increases migration (35, 65) |
| Gpr85 (G protein-coupled receptor 85) | Transiently induced by LPS in macrophages (48, 49) |
| Cd93 (complement component 1q receptor 1) | Promotes differentiation of monocytes into macrophages (46) |
| Nfia (nuclear factor I/A) | Negatively controls c-FMS; overexpression inhibits osteoclast formation; increased in Dicer−/− null osteoclast precursors (13, 17) |
DISCUSSION
Osteoclast commitment and maturation is an intricate, multistep process, modulated by the combined activity of numerous signaling pathways. Because miRNAs can control the expression of several genes working in one or multiple pathways, it is likely that miRNAs orchestrate many of the changes in gene expression or activity necessary for osteoclast differentiation. In this study, we demonstrate that miR-29 plays a positive role in osteoclastogenesis. Its expression increases during differentiation, and miR-29 knockdown impairs migration, commitment, and osteoclastogenesis. Our study is unique in that we validated a set of six novel miR-29 targets, which will contribute to our understanding of miR-29 function in osteoclasts and in other cell types (Table 2).
The miR-29 family consists of four genes that encode three mature miRNAs. These genes are organized in genomic clusters: miR-29a and miR-29b-1 are transcribed as a single polycistronic primary transcript from mouse chromosome 6, and miR-29b-2 and miR-29c are also transcribed as a polycistronic transcript from chromosome 1 (36, 37). The three mature miRNAs of this family, miR-29a, -b, and -c, present high sequence conservation and genomic organization in mouse, rat, and human, and nucleotides in positions 2–8, seed bases that are important for target recognition and binding, are identical (Table 1) (38). Although miR-29 family members may have overlapping targets, the mature miR-29 family members can be expressed at different levels, suggesting distinct transcriptional or post-transcriptional regulation of these genes (39, 40).
The expression of miR-29 family members increased during the osteoclast differentiation process, in both primary cultures and in RAW264.7 cells, and miR-29 knockdown decreased osteoclast formation, suggesting that this miRNA family plays a positive role in differentiation. Studies from our laboratory and others demonstrate a positive role of miR-29 in the differentiation of other lineages, including osteoblastic and myogenic (20, 22, 41, 42). More recently, miR-29 was identified as one of seven miRNAs that, in concert, can restrict proliferation and promote differentiation (43). Thus, the increase in miR-29 expression seen in the later stages of osteoclast differentiation may be in response to RANKL-induced differentiation program and withdrawal from the cell cycle.
The Calcr 3′-UTR is targeted by miR-29, and CTR plays an important role in osteoclast function and cell survival. CTR is a G protein-coupled receptor that mediates the anti-apoptotic effect of calcitonin on mature osteoclasts while inhibiting their resorption activity (44). Thus, the targeting of CTR by miR-29 in mature osteoclasts could promote resorption.
We identified three new miR-29 targets that may be involved in commitment of precursor cells to the osteoclast lineage: Nfia, Cd93, and Gpr85 (Figs. 8 and 9 and Table 2). Transcript levels for these genes are decreased in primary cultures of osteoclast progenitors treated for 4 days with RANKL.3 NFIA is known to repress the differentiation of hematopoietic cells, including granulocytes and monocytes, and NFIA is a negative regulator of the M-CSF receptor in osteoclasts (16, 17). Because the M-CSF receptor is a positive regulator of osteoclastogenesis, the targeting of Nfia mRNA by miR-29, like miR-223, could contribute to differentiation (Fig. 10A) (13).
FIGURE 10.
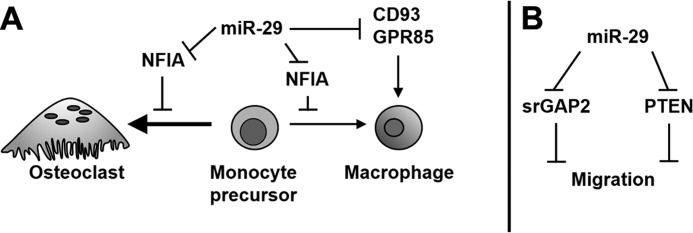
Model of miR-29 regulation of osteoclastogenesis. Schematic representations of the potential mechanisms regulated by miR-29 in the osteoclast lineage are shown. A, cell lineage commitment. B, cell migration.
In hematopoietic cells, the role of the other two miR-29 targets, Cd93 and Gpr85, is less well characterized. However, it is known that CD93 (C1qRp) is a transmembrane receptor regulating phagocytosis and cell adhesion and is present on cells of the myeloid lineage (45). CD93 expression is increased with monocyte differentiation and macrophage activation (46, 47). Targeting of Cd93 mRNA by miR-29 could promote commitment to osteoclastogenesis, preventing monocytic differentiation. GPR85, also called SREB2 (superconserved receptor expressed in brain 2) is a G protein-coupled receptor abundant in neurons and is involved in determining brain size and functionality (48). In macrophages, GPR85 expression increases upon inflammatory stimulation with LPS (49). Here, we showed that inhibition of miR-29 activity promotes the commitment of the RAW264.7 cells to the macrophage fate (Fig. 5). It is possible that down-regulation of Nfia, Cd93, and Gpr85 mRNAs by miR-29 could play a role in decreasing the potential of the cell to differentiate into the macrophage lineage, thus promoting osteoclastogenesis (Fig. 10A).
Osteoclast migration and fusion require extensive cytoskeletal reorganization, as does polarization and actin ring formation, initial steps for bone resorption. In this study, we demonstrated that knockdown of miR-29 family members, through the expression of an inducible miRNA sponge construct, suppressed the chemotactic migration of RAW264.7 cells (Fig. 6). Decreased cell motility in the presence of the miR-29 sponge likely contributed to the observed decrease in osteoclast size. However, not all functions involved in cytoskeletal remodeling, such as actin ring formation, were affected by knockdown of miR-29 activity (Fig. 7B). It is possible that the factors or pathways specific for regulating actin ring formation may be less affected by miR-29 in osteoclasts.
Cdc42 mRNA was identified as a miR-29 target in humans, and we confirmed miR-29 targeting of the mouse homolog (Figs. 8 and 9) (50). CDC42 is a small GTPase that regulates actin remodeling, as well as cell cycle control and survival (51). Although not required for actin ring formation, CDC42 regulates the rate of formation, as well as cell polarization (52). CDC42 also regulates migration by controlling podosome turnover, and it is important for the movement of hematopoietic progenitors and macrophages toward chemotactic signals (53, 54). Although CDC42 is critical for osteoclast formation and survival (52), Cdc42 transcript levels in osteoclasts are less than those found in macrophages or monocytes (55). Further, the levels of Cdc42 mRNA do not change dramatically during osteoclastic differentiation (56). It is possible that translational regulation by miR-29 family members could play a role in fine-tuning the CDC42 levels during osteoclastogenesis. However, it is also important to consider that Cdc42 transcripts are subject to alternative splicing, which can give rise to alternative 3′-UTRs. There is little known about Cdc42 splice variants in cells of the osteoclast lineage. It is possible that alternative 3′-UTR usage may be one means to vary the ability of Cdc42 to be targeted by miRNAs.
Srgap2 mRNA is one of the novel miR-29 targets identified in this study. srGAP2 is a Rho-GTPase-activating protein that participates in the same signaling pathway as CDC42 (35). Although srGAP2 function in osteoclasts has not been investigated, it has been shown to repress cell migration during neuronal development (35, 57) (Fig. 10 and Table 2). Knockdown of miR-29 activity could decrease cell motility, in part by causing an increase in srGAP2. Intriguingly, in other cell systems, miR-29 was shown to target Pten (tumor-suppressor phosphatase and tensin homolog), a lipid phosphatase involved in the phosphatidylinositol metabolism. Recent studies identified the suppression of PTEN as one mechanism by which miR-29 promotes migration in endothelial cells, breast cancer cells, and hepatocellular carcinoma cells (58). In RAW264.7 cells, activation of PTEN inhibits RANKL-mediated osteoclastogenesis and osteopontin-induced migration (59). It is possible that miR-29 knockdown could increase PTEN levels and contribute to repressed migration of RAW264.7 cells (Fig. 10).
It should be noted that our results appear to conflict with a recent report that lentivirus-mediated overexpression of miR-29b in human CD14+ peripheral blood mononuclear cells reduced osteoclast formation (60). In that paper, Rossi et al. reported that miR-29b expression decreased during osteoclast formation and that constitutive overexpression of miR-29b decreased the expression of osteoclast marker genes and impaired collagen degradation. However, the miR-29 overexpression study by Rossi et al. differs from our work in several key areas. First, Rossi et al. used semisorted circulating human osteoclast precursors from the periphery, whereas we used murine bone marrow resident osteoclast precursors, which may not circulate. Second, the study by Rossi et al. did not report on the expression of other miR-29 family members, which should be present, because miR-29b is transcribed on the same pri-miRNA as miR-29c and miR-29a. Third, Rossi et al. overexpressed miR-29b. It has been shown that superphysiological expression of a particular miRNA can alter global recruitment of miRNAs to the RNA-induced silencing complex, which can confound interpretation of a resulting cell phenotype (61, 62). Further, knockdown of components important for miRNA processing, such as DGCR8, Ago2, and Dicer1, inhibits osteoclastogenesis (13). It is possible that the miR-29b precursor overexpressed by Rossi et al. could compete for miRNA processing machinery, providing an alternative explanation for the inhibition of osteoclastogenesis observed by those investigators.
In our study, we observed similar inhibitory effects on osteoclastogenesis when miR-29a, -29b, or -29c were individually targeted by transiently transfected inhibitors (Table 1) and when their activity was inhibited by the miR-29 sponge competitive inhibitor (Fig. 3C). These data, and the rest of the data herein, strongly support the conclusion that miR-29 family members promote osteoclastogenesis by several mechanisms. Whereas some miRNAs may act as “switches” for the commitment to one cell fate or another, many miRNAs are more subtle regulators of gene expression, modulating the amplitude and tempo of a differentiation program (13, 18). miR-29 family members are likely subtle regulators of multiple osteoclast mRNA targets.
In conclusion, we demonstrated that miR-29 family members sustain migration and commitment of the precursor to osteoclastogenesis, and we validated six novel targets for this miRNA family. These data contribute to our understanding of the basic mechanisms regulating osteoclast differentiation and provide insight into the function of miR-29 family members in cells of the hematopoietic lineage and in other tissue types. Dysregulation of miR-29 family members is implicated in the pathology of multiple malignancies and in conditions such as diabetes and fibrosis, and aging (63). Additional studies, in vivo, will better define the role of miR-29 in osteoclastogenesis. It is possible that increased miR-29 levels could contribute to increased osteoclast formation with aging (64).
Acknowledgments
We thank Dr. Kristie Kapinas for making the miR-29a knockdown retroviral construct and Jeong-Shin Lee (Deputy Director, Harvard Gene Therapy Initiative, Harvard Medical School) for the gift of the packaging constructs for lentivirus production.
This work was supported, in whole or in part, by National Institutes of Health Grants AR44877 (to A. M. D.) and AR055143 (to S.-K. L.).

This article contains supplemental Tables S1–S4 and Figs. S1 and S2.
S.-K. Lee, unpublished data.
- M-CSF
- macrophage colony-stimulating factor
- qRT-PCR
- quantitative RT-PCR
- miRNA
- microRNA
- HPRT
- hypoxanthine-guanine phosphoribosyltransferase
- DOX
- doxycycline
- RANKL
- receptor activator of nuclear factor κB ligand
- TRAP
- tartrate-resistant acid phosphatase.
REFERENCES
- 1. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Osteoclast differentiation and activation. Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 2. Mughal M. Z. (2009) Miscellaneous bone disorders. Endocr. Dev. 16, 191–217 [DOI] [PubMed] [Google Scholar]
- 3. Saftig P., Hunziker E., Wehmeyer O., Jones S., Boyde A., Rommerskirch W., Moritz J. D., Schu P., von Figura K. (1998) Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 95, 13453–13458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Østergaard M., Pedersen S. J., Døhn U. M. (2008) Imaging in rheumatoid arthritis–status and recent advances for magnetic resonance imaging, ultrasonography, computed tomography and conventional radiography. Best Pract. Res. Clin. Rheumatol. 22, 1019–1044 [DOI] [PubMed] [Google Scholar]
- 5. Roodman G. D. (1999) Cell biology of the osteoclast. Exp. Hematol. 27, 1229–1241 [DOI] [PubMed] [Google Scholar]
- 6. Novack D. V., Faccio R. (2011) Osteoclast motility. Putting the brakes on bone resorption. Ageing Res. Rev. 10, 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asagiri M., Takayanagi H. (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 8. Bruzzaniti A., Baron R. (2006) Molecular regulation of osteoclast activity. Rev. Endocr. Metab. Disord. 7, 123–139 [DOI] [PubMed] [Google Scholar]
- 9. Fabian M. R., Sonenberg N. (2012) The mechanics of miRNA-mediated gene silencing. A look under the hood of miRISC. Nat. Struct. Mol. Biol. 19, 586–593 [DOI] [PubMed] [Google Scholar]
- 10. Liang M. (2009) MicroRNA. A new entrance to the broad paradigm of systems molecular medicine. Physiol. Genomics 38, 113–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Winter J., Jung S., Keller S., Gregory R. I., Diederichs S. (2009) Many roads to maturity. MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 11, 228–234 [DOI] [PubMed] [Google Scholar]
- 12. He L., Hannon G. J. (2004) MicroRNAs. Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 13. Sugatani T., Hruska K. A. (2009) Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 284, 4667–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mizoguchi F., Izu Y., Hayata T., Hemmi H., Nakashima K., Nakamura T., Kato S., Miyasaka N., Ezura Y., Noda M. (2010) Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell. Biochem. 109, 866–875 [DOI] [PubMed] [Google Scholar]
- 15. Sugatani T., Vacher J., Hruska K. A. (2011) A microRNA expression signature of osteoclastogenesis. Blood 117, 3648–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M. L., Nervi C., Bozzoni I. (2005) A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell 123, 819–831 [DOI] [PubMed] [Google Scholar]
- 17. Rosa A., Ballarino M., Sorrentino A., Sthandier O., De Angelis F. G., Marchioni M., Masella B., Guarini A., Fatica A., Peschle C., Bozzoni I. (2007) The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc. Natl. Acad. Sci. U.S.A. 104, 19849–19854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mann M., Barad O., Agami R., Geiger B., Hornstein E. (2010) miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc. Natl. Acad. Sci. U.S.A. 107, 15804–15809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hershey C. L., Fisher D. E. (2004) Mitf and Tfe3. Members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone 34, 689–696 [DOI] [PubMed] [Google Scholar]
- 20. Kapinas K., Kessler C. B., Delany A. M. (2009) miR-29 suppression of osteonectin in osteoblasts. Regulation during differentiation and by canonical Wnt signaling. J. Cell. Biochem. 108, 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kapinas K., Kessler C., Ricks T., Gronowicz G., Delany A. M. (2010) miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J. Biol. Chem. 285, 25221–25231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z., Hassan M. Q., Jafferji M., Aqeilan R. I., Garzon R., Croce C. M., van Wijnen A. J., Stein J. L., Stein G. S., Lian J. B. (2009) Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 284, 15676–15684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pekarsky Y., Santanam U., Cimmino A., Palamarchuk A., Efanov A., Maximov V., Volinia S., Alder H., Liu C. G., Rassenti L., Calin G. A., Hagan J. P., Kipps T., Croce C. M. (2006) Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 66, 11590–11593 [DOI] [PubMed] [Google Scholar]
- 24. Garzon R., Heaphy C. E., Havelange V., Fabbri M., Volinia S., Tsao T., Zanesi N., Kornblau S. M., Marcucci G., Calin G. A., Andreeff M., Croce C. M. (2009) MicroRNA 29b functions in acute myeloid leukemia. Blood 114, 5331–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu C., Wang Y., Kuai W., Sun X., Chen H., Hong Z. (2013) Prognostic value of miR-29a expression in pediatric acute myeloid leukemia. Clin. Biochem. 46, 49–53 [DOI] [PubMed] [Google Scholar]
- 26. Wang X. S., Gong J. N., Yu J., Wang F., Zhang X. H., Yin X. L., Tan Z. Q., Luo Z. M., Yang G. H., Shen C., Zhang J. W. (2012) MicroRNA-29a and microRNA-142–3p are regulators of myeloid differentiation and acute myeloid leukemia. Blood 119, 4992–5004 [DOI] [PubMed] [Google Scholar]
- 27. Dobson K. R., Reading L., Haberey M., Marine X., Scutt A. (1999) Centrifugal isolation of bone marrow from bone. An improved method for the recovery and quantitation of bone marrow osteoprogenitor cells from rat tibiae and femurae. Calcif. Tissue Int. 65, 411–413 [DOI] [PubMed] [Google Scholar]
- 28. Jacquin C., Gran D. E.., Lee S. K., Lorenzo J. A., Aguila H. L. (2006) Identification of multiple osteoclast precursor populations in murine bone marrow. J. Bone Miner. Res. 21, 67–77 [DOI] [PubMed] [Google Scholar]
- 29. Swift S., Lorens J., Achacoso P., Nolan G. P. (2001) Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Protoc. Immunol. Unit 10.17C [DOI] [PubMed] [Google Scholar]
- 30. Lee S. K., Gardner A. E., Kalinowski J. F., Jastrzebski S. L., Lorenzo J. A. (2006) RANKL-stimulated osteoclast-like cell formation in vitro is partially dependent on endogenous interleukin-1 production. Bone 38, 678–685 [DOI] [PubMed] [Google Scholar]
- 31. Ebert M. S., Neilson J. R., Sharp P. A. (2007) MicroRNA sponges. Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shin K. J., Wall E. A., Zavzavadjian J. R., Santat L. A., Liu J., Hwang J. I., Rebres R., Roach T., Seaman W., Simon M. I., Fraser I. D. (2006) A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. U.S.A. 103, 13759–13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee S. K., Kalinowski J. F., Jacquin C., Adams D. J., Gronowicz G., Lorenzo J. A. (2006) Interleukin-7 influences osteoclast function in vivo but is not a critical factor in ovariectomy-induced bone loss. J. Bone Miner. Res. 21, 695–702 [DOI] [PubMed] [Google Scholar]
- 34. Kinugawa S., Koide M., Kobayashi Y., Mizoguchi T., Ninomiya T., Muto A., Kawahara I., Nakamura M., Yasuda H., Takahashi N., Udagawa N. (2012) Tetracyclines convert the osteoclastic-differentiation pathway of progenitor cells to produce dendritic cell-like cells. J. Immunol. 188, 1772–1781 [DOI] [PubMed] [Google Scholar]
- 35. Guo S., Bao S. (2010) srGAP2 arginine methylation regulates cell migration and cell spreading through promoting dimerization. J. Biol. Chem. 285, 35133–35141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang T. C., Yu D., Lee Y. S., Wentzel E. A., Arking D. E., West K. M., Dang C. V., Thomas-Tikhonenko A., Mendell J. T. (2008) Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mott J. L., Kurita S., Cazanave S. C., Bronk S. F., Werneburg N. W., Fernandez-Zapico M. E. (2010) Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-κB. J. Cell. Biochem. 110, 1155–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brodersen P., Voinnet O. (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat. Rev. Mol. Cell Biol. 10, 141–148 [DOI] [PubMed] [Google Scholar]
- 39. Wang H., Garzon R., Sun H., Ladner K. J., Singh R., Dahlman J., Cheng A., Hall B. M., Qualman S. J., Chandler D. S., Croce C. M., Guttridge D. C. (2008) NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang Y. C., Hasegawa H., Wang S. W., Ku C. C., Lin Y. C., Chiou S. S., Hou M. F., Wu D. C., Tsai E. M., Saito S., Yamaguchi N., Yokoyama K. K. (2011) Jun dimerization protein 2 controls senescence and differentiation via regulating histone modification. J. Biomed. Biotechnol. 2011, 569034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Winbanks C. E., Wang B., Beyer C., Koh P., White L., Kantharidis P., Gregorevic P. (2011) TGF-β regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 286, 13805–13814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steiner D. F., Thomas M. F., Hu J. K., Yang Z., Babiarz J. E., Allen C. D., Matloubian M., Blelloch R., Ansel K. M. (2011) MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 35, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marzi M. J., Puggioni E. M., Dall'Olio V., Bucci G., Bernard L., Bianchi F., Crescenzi M., Di Fiore P. P., Nicassio F. (2012) Differentiation-associated microRNAs antagonize the Rb-E2F pathway to restrict proliferation. J. Cell Biol. 199, 77–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuo Y. J., Tsuang F. Y., Sun J. S., Lin C. H., Chen C. H., Li J. Y., Huang Y. C., Chen W. Y., Yeh C. B., Shyu J. F. (2012) Calcitonin inhibits SDCP-induced osteoclast apoptosis and increases its efficacy in a rat model of osteoporosis. PLoS One 7, e40272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Danet G. H., Luongo J. L., Butler G., Lu M. M., Tenner A. J., Simon M. C., Bonnet D. A. (2002) C1qRp defines a new human stem cell population with hematopoietic and hepatic potential. Proc. Natl. Acad. Sci. U.S.A. 99, 10441–10445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jeon J. W., Jung J. G., Shin E. C., Choi H. I., Kim H. Y., Cho M. L., Kim S. W., Jang Y. S., Sohn M. H., Moon J. H., Cho Y. H., Hoe K. L., Seo Y. S., Park Y. W. (2010) Soluble CD93 induces differentiation of monocytes and enhances TLR responses. J. Immunol. 185, 4921–4927 [DOI] [PubMed] [Google Scholar]
- 47. Beyer M., Mallmann M. R., Xue J., Staratschek-Jox A., Vorholt D., Krebs W., Sommer D., Sander J., Mertens C., Nino-Castro A., Schmidt S. V., Schultze J. L. (2012) High-resolution transcriptome of human macrophages. PLoS One 7, e45466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matsumoto M., Straub R. E., Marenco S., Nicodemus K. K., Matsumoto S., Fujikawa A., Miyoshi S., Shobo M., Takahashi S., Yarimizu J., Yuri M., Hiramoto M., Morita S., Yokota H., Sasayama T., Terai K., Yoshino M., Miyake A., Callicott J. H., Egan M. F., Meyer-Lindenberg A., Kempf L., Honea R., Vakkalanka R. K., Takasaki J., Kamohara M., Soga T., Hiyama H., Ishii H., Matsuo A., Nishimura S., Matsuoka N., Kobori M., Matsushime H., Katoh M., Furuichi K., Weinberger D. R. (2008) The evolutionarily conserved G protein-coupled receptor SREB2/GPR85 influences brain size, behavior, and vulnerability to schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 105, 6133–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lattin J. E., Schroder K., Su A. I., Walker J. R., Zhang J., Wiltshire T., Saijo K., Glass C. K., Hume D. A., Kellie S., Sweet M. J. (2008) Expression analysis of G protein-coupled receptors in mouse macrophages. Immunome Res. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Park S. Y., Lee J. H., Ha M., Nam J. W., Kim V. N. (2009) miR-29 miRNAs activate p53 by targeting p85 α and CDC42. Nat. Struct. Mol. Biol. 16, 23–29 [DOI] [PubMed] [Google Scholar]
- 51. Heasman S. J., Ridley A. J. (2008) Mammalian Rho GTPases. New insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 9, 690–701 [DOI] [PubMed] [Google Scholar]
- 52. Ito Y., Teitelbaum S. L., Zou W., Zheng Y., Johnson J. F., Chappel J., Ross F. P., Zhao H. (2010) Cdc42 regulates bone modeling and remodeling in mice by modulating RANKL/M-CSF signaling and osteoclast polarization. J. Clin. Invest. 120, 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Moreau V., Tatin F., Varon C., Anies G., Savona-Baron C., Génot E. (2006) Cdc42-driven podosome formation in endothelial cells. Eur. J. Cell Biol. 85, 319–325 [DOI] [PubMed] [Google Scholar]
- 54. Allen W. E., Zicha D., Ridley A. J., Jones G. E. (1998) A role for Cdc42 in macrophage chemotaxis. J. Cell Biol. 141, 1147–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Helden S. F., Anthony E. C., Dee R., Hordijk P. L. (2012) Rho GTPase expression in human myeloid cells. PLoS One 7, e42563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brazier H., Stephens S., Ory S., Fort P., Morrison N., Blangy A. (2006) Expression profile of RhoGTPases and RhoGEFs during RANKL-stimulated osteoclastogenesis. Identification of essential genes in osteoclasts. J. Bone Miner. Res. 21, 1387–1398 [DOI] [PubMed] [Google Scholar]
- 57. Guerrier S., Coutinho-Budd J., Sassa T., Gresset A., Jordan N. V., Chen K., Jin W. L., Frost A., Polleux F. (2009) The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 138, 990–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Salmena L., Carracedo A., Pandolfi P. P. (2008) Tenets of PTEN tumor suppression. Cell 133, 403–414 [DOI] [PubMed] [Google Scholar]
- 59. Sugatani T., Alvarez U., Hruska K. A. (2003) PTEN regulates RANKL- and osteopontin-stimulated signal transduction during osteoclast differentiation and cell motility. J. Biol. Chem. 278, 5001–5008 [DOI] [PubMed] [Google Scholar]
- 60. Rossi M., Pitari M. R., Amodio N., Di Martino M. T., Conforti F., Leone E., Botta C., Paolino F. M., Del Giudice T., Iuliano E., Caraglia M., Ferrarini M., Giordano A., Tagliaferri P., Tassone P. (2013) miR-29b negatively regulates human osteoclastic cell differentiation and function. Implications for the treatment of multiple myeloma-related bone disease. J. Cell. Physiol. 228, 1506–1515 [DOI] [PubMed] [Google Scholar]
- 61. Wang W. X., Wilfred B. R., Hu Y., Stromberg A. J., Nelson P. T. (2010) Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA 16, 394–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khan A. A., Betel D., Miller M. L., Sander C., Leslie C. S., Marks D. S. (2009) Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat. Biotechnol. 27, 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ugalde A. P., Ramsay A. J., de la Rosa J., Varela I., Mariño G., Cadiñanos J., Lu J., Freije J. M., López-Otín C. (2011) Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 30, 2219–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cao J. J., Wronski T. J., Iwaniec U., Phleger L., Kurimoto P., Boudignon B., Halloran B. P. (2005) Aging increases stromal/osteoblastic cell-induced osteoclastogenesis and alters the osteoclast precursor pool in the mouse. J. Bone Miner. Res. 20, 1659–1668 [DOI] [PubMed] [Google Scholar]
- 65. Mason F. M., Heimsath E. G., Higgs H. N., Soderling S. H. (2011) Bi-modal regulation of a formin by srGAP2. J. Biol. Chem. 286, 6577–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]