Background: Three genes encode human nonmuscle myosin II (NM II) heavy chains, and the proteins have different intracellular roles and localizations.
Results: NM II paralogs form bipolar filaments, but there are important differences in filament structure, enzymatic, and actin binding behavior.
Conclusion: NM II filaments show diverse interactions with actin.
Significance: NM II filaments are adapted to work in cytoskeletal networks.
Keywords: Actin, ATPases, Cytoskeleton, Electron Microscopy (EM), Molecular Motors, Myosin
Abstract
Nonmuscle myosin IIs (NM IIs) are a group of molecular motors involved in a wide variety of cellular processes including cytokinesis, migration, and control of cell morphology. There are three paralogs of the NM II heavy chain in humans (IIA, IIB, and IIC), each encoded by a separate gene. These paralogs are expressed at different levels according to cell type and have different roles and intracellular distributions in vivo. Most previous studies on NM II used tissue-purified protein or expressed fragments of the molecule, which presents potential drawbacks for characterizing individual paralogs of the intact protein in vitro. To circumvent current limitations and approach their native properties, we have successfully expressed and purified the three full-length human NM II proteins with their light chains, using the baculovirus/Sf9 system. The enzymatic and structural properties of the three paralogs were characterized. Although each NM II is capable of forming bipolar filaments, those formed by IIC tend to contain fewer constituent molecules than those of IIA and IIB. All paralogs adopt the compact conformation in the presence of ATP. Phosphorylation of the regulatory light chain leads to assembly into filaments, which bind to actin in the presence of ATP. The nature of interactions with actin filaments is shown with different paralogs exhibiting different actin binding behaviors under equivalent conditions. The data show that although NM IIA and IIB form filaments with similar properties, NM IIC forms filaments that are less well suited to roles such as tension maintenance within the cell.
Introduction
Myosins are a large superfamily of molecular motors that use the energy provided by ATP hydrolysis to generate force. The force generated by these mechanochemical enzymes is employed in a wide range of biological processes from the intracellular transport of nanometer scale cargoes through to the muscle-based movement of whole animals. The phylogenetic tree of myosins includes up to 37 distinct classes, grouped on the basis of their sequence homology (1, 2). The class II myosins, which were the first to be discovered and are therefore often referred to as “conventional myosins,” are filament-forming proteins and include those responsible for skeletal, smooth, and cardiac muscle contraction. Since the original isolation of myosin as an extract of skeletal muscle by Kühne (3) in the 1850s, there have been countless studies relating to the mechanisms underlying force generation in muscle, leading to a sophisticated understanding of the myosin motor in general. Skeletal muscle myosin has been shown to form large bipolar filaments both in vitro and in vivo, whereas smooth muscle myosin has been shown to be capable of forming either short bipolar filaments or large side polar filaments in vitro (4). In addition to the muscle myosin II paralogs, there are also nonmuscle myosin II paralogs present in all cell types that play roles in a wide range of cellular and subcellular processes. These roles include but are not limited to cell migration (5–7), cell adhesion (8–10), and cytokinesis (11, 12).
In common with all members of the myosin II class, nonmuscle myosin II (NM II)4 is a hexamer formed by dimerization of two heavy chains, each ∼2,000 amino acids long, and each heavy chain binds two light chains. The heavy chain begins with an N-terminal motor domain that is responsible for ATP hydrolysis and is the source of force generation. Following this is a lever arm region that binds the light chains. Proximal to the motor domain is the essential light chain (ELC) that acts to structurally stabilize the lever arm. The remainder of the lever is stabilized by the regulatory light chain (RLC), which plays a role in regulating the enzymatic activity of the myosin in a phosphorylation-dependent manner (13, 14). C-terminal to the lever arm is a long (∼1,100 amino acids) α-helical region responsible for dimerization of the heavy chains to form the characteristic coiled coil tails of myosin II molecules. A nonhelical region at the tip of the tail terminates the heavy chain. We will subsequently refer to this structure as the myosin monomer because it is the functional unit that undergoes intermolecular association via its tail region to form filaments.
In mammals there are three genes (MYH9, MYH10, and MYH14) encoding NM II heavy chains. The heavy chains together with the bound RLCs and ELCs are termed NM IIA, NM IIB, and NM IIC, respectively. Alternative splicing of NM II transcripts leads to a greater range of expressed paralogs in vivo (15–17). Although two sites for splicing have been identified at the protein level for NM IIB and NM IIC, each producing splice variants with inserts in the loop 1 and 2 regions, only mRNA has been reported for NM IIA (17). The NM IIB and NM IIC splicing gives rise to four heavy chain sequences per gene (and therefore eight different NM II heavy chains). Because loop 1 is important for determining the nucleotide binding properties of the motor and loop 2 is important for determining the actin binding/actin activation properties of the motor, the insertions produce molecules with different enzymatic properties (18–22).
The assembly of myosin isolated from human platelets was originally described by Niederman and Pollard (23). It was subsequently shown that platelets express exclusively the NM IIA paralog (24, 25). The study by Niederman and Pollard showed that platelet myosin formed uniform bipolar filaments that were much shorter than those formed from skeletal muscle myosin. Unlike platelets, most cell types express a mixture of NM II paralogs (26). This, combined with the difficulty of expressing intact full-length protein in bacterial systems, means that no studies to date have compared the properties of the three full-length NM II paralogs in vitro, although some aspects of NM IIA and NM IIB regulation have been investigated using recombinant protein (27, 28). Much of the information to date regarding the macromolecular assembly of NM II has been obtained by studying tail fragments of the molecule, usually expressed as recombinant proteins in Escherichia coli (29–34). These tail fragments do not form uniform discrete bipolar assemblies such as those seen with full-length myosin and therefore are not ideal as a system for comparing the properties of the native filaments. Recently, however, the use of baculovirus expression systems has allowed the properties of recombinant full-length proteins to be investigated (27, 28, 35). Here we report on the physical and biochemical properties of filaments formed from the full-length baculovirus expressed NM II paralogs and compare and contrast them to previous reports for the tissue-purified proteins.
EXPERIMENTAL PROCEDURES
Generation of Baculovirus Expression Vectors
cDNAs encoding human nonmuscle myosin II heavy chain (NMHC II) were amplified by PCR using plasmid DNA templates described previously (12, 36–38) and cloned into the baculovirus transfer vector pFastBac1. This was modified to incorporate nucleotides encoding a FLAG tag epitope (DYKDDDDK) for purification and the appropriate restriction sites for cloning. The FLAG tag was located at the N terminus of each construct. The primers for this modification were 5′-GGAATTCATGGACTACAAGGACGACGATGATAAGGCGGCCGCGTCGACGGTACCGG-3′ and the complementary antisense primer. The primers used, the plasmid DNA or cDNA template, and the references are listed in supplemental Table S1. The nucleotide sequences of the cloned DNA fragments were confirmed in all cases by sequencing. The shortest splice forms of each NMHC II (i.e., those lacking insertions in loop 1 or 2) were created, and they are referred to simply as NMHC IIA, IIB, and IIC throughout this manuscript. The splice variant of IIB, which contains an insert in the loop 2 region, was also produced, and this is referred to as NMHC IIB2 hereafter.
Expression and Purification of Full-length NM II Proteins
Full-length NMHC II, together with regulatory and essential light chains (RLC and ELC), were expressed in the baculovirus/Sf9 system (Invitrogen). 3 × 109 Sf9 cells were co-infected with full-length NMHC II virus and a virus including both light chains (bovine nonmuscle myosin RLC (MYL9) and chicken nonmuscle myosin ELC (MYL6)). The infected Sf9 cells were harvested by sedimentation following 72 h of growth and then stored at −80 °C for future use. Immunoblot analysis was used, prior to purification, to confirm the level of expression. The proteins were purified according to a previous report (39) with modification. During purification, all buffers included 0.5 m NaCl. The frozen pellet (∼10 ml in volume) was thawed in the extraction buffer including 0.5 m NaCl, 10 mm MOPS (pH 7.3), 10 mm MgCl2, 1 mm EGTA, 3 mm NaN3, 2 mm ATP, 0.1 mm phenylmethylsulfonyl fluoride, 0.1 mm dithiothreitol, 5 μg/ml leupeptin, and proteinase inhibitor mixture (Sigma) and homogenized using a ground glass homogenizer. The proteins were purified by FLAG affinity chromatography using M2 FLAG affinity gel (Sigma). The eluted protein was dialyzed in Buffer A (10 mm MOPS, pH 7.0, 0.1 mm EGTA, 2 mm MgCl2) containing 50 mm KCl. The protein pellet was concentrated by centrifuging at 2,000 × g for 5 min and dissolved in an appropriate amount of Buffer A containing 0.5 m KCl. Successful purification of NM II was confirmed by PageBlue staining as shown in Fig. 1 (Fermentas Inc., Glen Burnie, MD) following separation on a 10% Bis-Tris polyacrylamide-SDS gel (Invitrogen). Protein concentration was determined using a spectrophotometer and calculated according to the formula [myosin](mg/ml) = (A280 − A320)/0.52. RLC phosphorylation of purified protein was quantified using the PhostagTM SDS-PAGE system and gel densitometry (Wako Chemicals, Richmond, VA) (40). Phosphorylation levels were 0.2, 0.7, and 1.5% for NM IIA, NM IIB, and NM IIC, respectively, for myosin as isolated from Sf9 cells (n = 3 for NM IIA and NM IIC, n = 2 for NM IIB).
FIGURE 1.
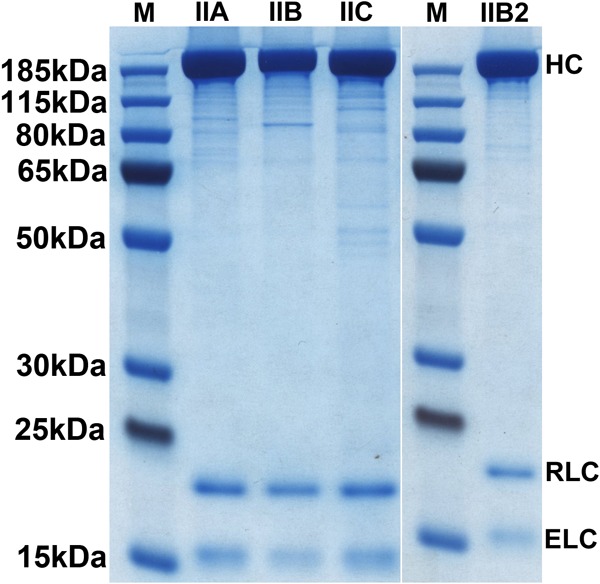
Expression and purification of FLAG-tagged full-length NM IIs from baculovirus-infected Sf9 cells. NM II proteins were purified by FLAG affinity chromatography (see “Experimental Procedures”). Purified NM II heavy chains and two light chains were separated on a 10% (Bis-Tris) polyacrylamide-SDS gel, followed by PageBlue staining. The positions of the NM II heavy chain (HC), RLC, and ELC are shown on the right. M, molecular mass marker (PageRulerTM Plus Prestained).
Preparation of Other Proteins
Skeletal muscle actin was purified from rabbit skeletal muscle (41). A clone for rat calmodulin (CaM) was expressed in E. coli. Baculovirus driving the expression of FLAG-tagged rabbit smooth muscle myosin light chain kinase (MLCK) (NP_001075775) was generated, and protein was purified from Sf9 cells using FLAG affinity chromatography.
ATPase Activity Assay
Actin-activated MgATPase activities were determined by an NADH-linked assay as described previously (42) with modifications for assaying full-length NM II. The myosin was phosphorylated for a minimum of 30 min at room temperature in a Buffer A containing 150 mm KCl, 1 mm ATP, 0.2 mm CaCl2, 1 μm CaM, 10 μg/ml MLCK. Phosphorylation under these conditions was confirmed using the PhostagTM SDS-PAGE system. Phosphorylation levels were 97, 97, and 100% (5, 5, and 1% diphosphorylated) for NM IIA, NM IIB, and NM IIC, respectively (n = 3 for NM IIA and NM IIC, n = 2 for NM IIB). The assay was performed at 25 °C in Buffer A containing 150 mm KCl, 1 mm ATP, 0.2 mm CaCl2, 1 μm CaM, 40 units/ml lactate dehydrogenase, 200 units/ml pyruvate kinase, 1 mm phosphoenolpyruvate, 0.2 mm NADH, and various concentrations of actin (1–12 μm). Actin filaments were stabilized by a 1.5-fold molar excess of phalloidin (Calbiochem, San Diego, CA) and passed through a 25-gauge needle several times to shear the actin filaments into smaller pieces. The data were corrected for the background MgATPase activity of actin. MgATPase activity of NM II at 0 μm actin concentration was subtracted from each data point. To determine the kinetic constants Vmax and KATPase, the experimental data sets were fitted to the Michaelis-Menten mathematical equation using Sigmaplot (Systat Software Inc., San Jose, CA). The reported data represent the means and S.D. from at least two separate experiments.
Electron Microscopy and Image Processing
For myosin in the absence of actin, samples were diluted to 100 nm myosin in buffer A with the appropriate concentration of KCl to achieve the stated final concentration (0.15 m KCl in most cases). In experiments using RLC-phosphorylated myosin, phosphorylation was carried out for a minimum of 30 min at room temperature in Buffer A containing 150 mm KCl, 1 mm ATP, 0.2 mm CaCl2, 1 μm CaM, 20 μg/ml MLCK. Phosphorylation under these conditions was confirmed using the PhostagTM SDS-PAGE system. Total phosphorylated RLC was 100, 99, and 99% (17, 17, and 9% diphosphorylated) for NM IIA, NM IIB, and NM IIC, respectively (n = 3 for NM IIA and NM IIC, n = 2 for NM IIB). Phosphorylated myosin was then mixed with F-actin (in Buffer A containing 150 mm KCl) to give final concentrations of 50 nm myosin, 500 nm F-actin. Samples were applied to the grid immediately after mixing. For all EM experiments, a 5-μl drop of sample was applied to a carbon-coated copper grid (pretreated with UV light to produce a hydrophilic surface) and stained with 1% uranyl acetate. Micrographs were recorded on a JEOL 1200EX II microscope operating at room temperature. Data were recorded on an AMT XR-60 CCD camera. Catalase crystals were used as a size calibration standard. Image processing was carried out using SPIDER software as described previously (43, 44). Measurements were carried out using ImageJ software. Contour lengths were measured using a freehand line, and a spline was fitted to reduce interpolation error. Individual raw images in the manuscript are displayed following application of the ImageJ FFT bandpass filter operation (between 40 and 2 pixels) and are autoscaled after filtering to allow easier visualization.
Glutaraldehyde Cross-linking
Proteins at 0.05–0.1 mg/ml in Buffer A containing 150 mm KCl were cross-linked for 1 min at room temperature using 0.1% glutaraldehyde. The reaction was quenched by adding Tris-HCl (pH 8.0) to a final concentration of 100 mm. For NM II cross-linked in the presence of ATP, 1 mm ATP was added to a suspension of filaments 5 min prior to cross-linking.
Statistical Analysis
Statistical analysis of filament measurements was carried out using Microsoft Excel and GraphPad Prism 5 software.
Light Scattering
Light scattering was measured at 90° in a PerkinElmer Life Sciences LS 55 fluorescence spectrophotometer. Excitation was performed at 365 nm (slit width, 15 nm) and detected at 365 nm (slit width, 5 nm). Relative light scattering was calculated using the intensities immediately prior to initiation of filament disassembly and reassembly as 100 and 0%, respectively. Initially, 150 μl of a suspension of unphosphorylated myosin filaments (0.15 mg/ml NM II in Buffer A containing 150 mm KCl) was measured. At the indicated time point (300 s), ATP was added to initiate filament disassembly (final concentration, 1 mm ATP). At the second indicated time point (800 s), filament reassembly was initiated by the addition of a premixed solution of MLCK, CaM, and CaCl2 (final concentrations, 10 μg/ml MLCK, 1 μm CaM, 0.2 mm CaCl2).
Modeling of Charge Periodicity in the NM II tail
Charge along the myosin tail was determined using a method based on Straussman et al. (45). Briefly, the register of each tail residue within the heptad repeat was determined using the Paircoil2 algorithm (46). Residues at positions A and D were given a value of 0. Aspartate and glutamate residues were given a value of −1, and arginine and lysine were assigned a value of +1. All other residues were given a value of 0, and positions beyond the boundaries of the coiled coil were given values of 0. The relative charge at position n along the tail was determined by summing charges in a surrounding 98-residue window (from n − 48 to n + 49). A three-dimensional model of myosin II HMM (3DTP.pdb) was fused to an idealized coiled coil to produce a model representing the overall structure of NM II. Coiled coil residues were colored according to charge (calculated as described above) using UCSF Chimera software (47).
RESULTS
Expression and Purification of Full-length FLAG-tagged NM II Heavy Chain with Bound ELCs and RLCs
All three paralogs of full-length NM II were successfully expressed using the baculovirus/Sf9 expression system and purified using FLAG affinity chromatography. For each paralog, the splice variants lacking insertions in loops 1 and 2 were expressed and purified. These are referred to as NM IIA, NM IIB, and NM IIC. The NM IIB splice isoform containing the loop 2 insert was also expressed. This is referred to as NM IIB2. In each case, expressed heavy chains bound both of the co-expressed light chains. Fig. 1 shows a PageBlue-stained SDS-polyacrylamide gel indicating each of the purified full-length human NM II heavy chains together with bound ELC and RLC. The yield of each of these proteins was ∼1–2 mg of NM II/liter of cultured cells.
Enzymatic Activity of Full-length NM II
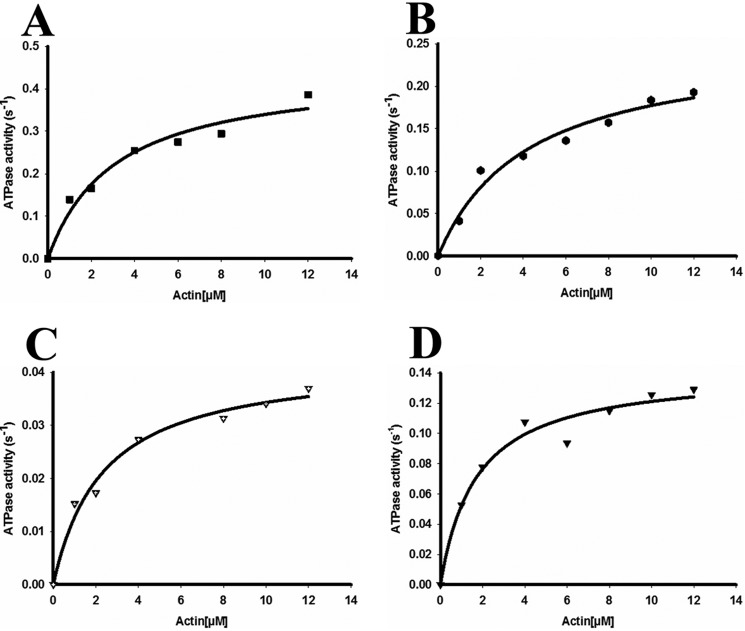
Measurement of the actin activated MgATPase activities of full-length myosin preparations initially proved problematic because of interactions of actin filaments with myosin filaments, which resulted in superprecipitation and nonlinear rates. To circumvent this as much as possible, we used 150 mm KCl and mechanically sheared the actin filaments by repeatedly pipetting the solution prior to the start of the assay (48). This allowed us to obtain linear rates of ATP hydrolysis. The actin-activated MgATPase activity of full-length NM II after phosphorylation is indicated in Fig. 2 (A–D). The Vmax of full-length NM II displayed a tendency of NM IIA > IIB > IIC > IIB2. KATPase values exhibited the tendency of NM IIB > IIA > IIB2 > IIC. The values for the kinetic constants obtained from the full-length proteins are compared with those for the corresponding HMM fragments in Table 1. For all paralogs, basal ATPase rates in the absence of actin were <0.01 s−1.
FIGURE 2.
Enzymatic activity of full-length wild-type NM II molecules. Actin-activated MgATPase activity of full-length wild-type NM IIA (A), NM IIB (B), NM IIB2 (C), and NM IIC (D) was measured at 150 mm KCl at 25 °C (details under “Experimental Procedures”). NM II was phosphorylated with MLCK at room temperature for 30 min prior to the assay. The actin concentration varied from 0 to 12 μm. MgATPase activity of NM II at 0 μm actin concentration was subtracted from each data point (< 0.01 s−1 for all paralogs). Data sets were fitted to a hyperbolic equation to determine the kinetic constants, namely, Vmax and KATPase. The data shown were from the measurement of a single preparation of NM II. Note the different y axis scales.
TABLE 1.
Actin-activated MgATPase activity of full-length NM IIs and their HMM counterparts
The actin-activated MgATPase activity was measured at 25 °C as described under “Experimental Procedures.” HMM data from previously published papers are shown for comparison.
General Assembly Properties of Full-length NM II
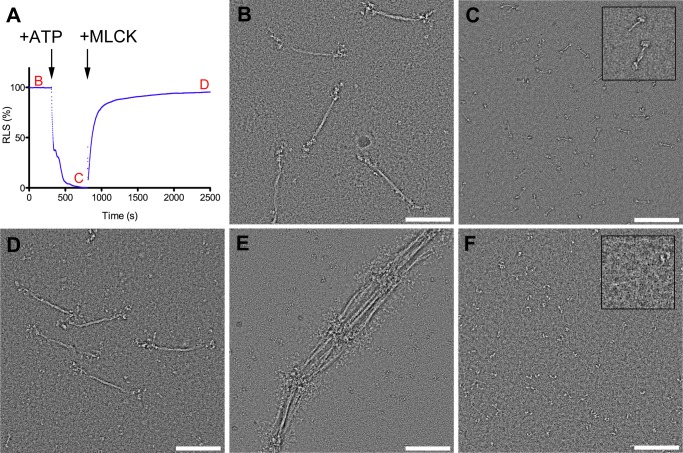
All three recombinant NM II paralogs displayed behavior similar to that demonstrated previously for tissue-purified NM II in response to changing ionic strength, presence of ATP, and RLC phosphorylation (49–51). Fig. 3 shows this behavior using NM IIB as an example. Myosin in high ionic strength buffer (10 mm MOPS, pH 7.0, 2 mm MgCl2, 0.1 mm EGTA, 500 mm KCl) was an extended molecule and showed the characteristic two heads and long tail (Fig. 3F). In buffer approximating physiological ionic strength (10 mm MOPS, pH 7.0, 2 mm MgCl2, 0.1 mm EGTA, 150 mm KCl), myosin polymerized into filaments ∼300 nm in length (Fig. 3B). In lower ionic strength buffer (10 mm MOPS, pH 7.0, 2 mm MgCl2, 0.1 mm EGTA, 100 mm KCl), filaments associated in an end to end fashion, resulting in large extended or weblike aggregates, but did not increase in overall length (Fig. 3E). Addition of 1 mm ATP to myosin filaments in the absence of RLC phosphorylation (in 150 mm KCl) resulted in rapid disassembly of filaments into compact molecules (Fig. 3, A and C) (51, 52). Phosphorylation of the RLC within these compact molecules leads to reassembly into filaments ∼300 nm in length and similar in overall appearance to RLC unphosphorylated, ATP-free filaments (Fig. 3, A and D).
FIGURE 3.
General assembly behavior of NM II as exemplified by NM IIB. A, time course of relative light scattering (RLS). The arrows indicate the times where ATP and MLCK (with Ca2+ and CaM) were added. Red letters refer to EM images in subsequent panels. B, bipolar thick filaments formed at approximately physiological ionic strength (150 mm KCl) in the absence of ATP. C, addition of 1 mm ATP to a suspension of filaments at physiological ionic strength leads to disassembly into compact individual molecules. Inset shows examples of a compact monomer and antiparallel dimer. D, compact molecules reassemble into filaments following RLC phosphorylation with MLCK. E, lowering the ionic strength to 100 mm KCl leads to lateral and serial aggregation of filaments by their head containing ends. F, in high ionic strength buffer (0.5 m KCl), myosin is extended and unpolymerized. Inset highlights one such molecule.
Comparison of NM IIA, IIB, and IIC Filaments
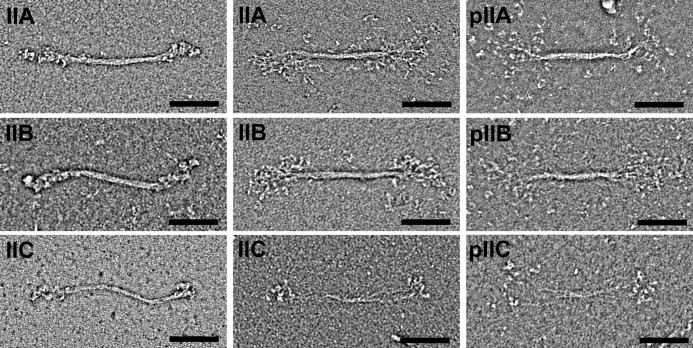
Negative stain EM of unphosphorylated NM II proteins in 0.15 m KCl, and the absence of ATP revealed the presence of disperse bipolar filaments. Examples of filaments from the three paralogs are shown in Fig. 4. In all three paralogs, filaments were seen in which the heads are tightly packed against the filament backbone as well as filaments in which the heads are splayed outwards, away from the filament backbone. The majority of filaments have a compact appearance: 70, 88, and 73% for NM IIA, NM IIB, and NM IIC, respectively. The overall appearance is similar to that observed in studies of myosin purified from human platelets, which corresponds to NM IIA (23). To compare the three paralogs, 100 filaments of each paralog were quantified in terms of the total contour length, the contour length of the bare zone (the central region that lacks projecting heads), and the filament width at the center of the bare zone. The contour length distributions of filaments formed by the three wild-type NM II paralogs were 301 (±24 nm S.D.), 323 (±24 nm S.D.), and 293 (±33 nm S.D.) for NM IIA, NM IIB, and NM IIC respectively (Fig. 5). Mann-Whitney tests demonstrate no significant difference between NM IIC and NM IIA, whereas the length of NM IIB filaments is deemed to be significantly different from NM IIA or NM IIC (A versus B, p < 0.0001; A versus C, p = 0.6139; and B versus C, p < 0.0001). The mean widths of NM IIA and NM IIB were 11.2 (±2.4 nm S.D.) and 11.5 (±2.3 nm), respectively, whereas NM IIC had a mean width of 7.9 (±2.1 nm). Mann-Whitney tests demonstrate that the difference in width between NM IIC and either NM IIA or NM IIB is statistically significant, whereas NM IIA and NM IIB are not significantly different from each other (A versus B, p = 0.2118; A versus C, p < 0.0001; B versus C, p < 0.0001). NM IIC filaments also had a much longer mean bare zone length than the other paralogs. The mean bare zone lengths were 167 (±19 nm) for NM IIA, 166 (±16 nm) for NM IIB, and 219 (±30 nm) for NM IIC. Mann-Whitney tests demonstrate that the difference in bare zone between NM IIC and either NM IIA or NM IIB is statistically significant, whereas NM IIA and NM IIB are not significantly different from each other (A versus B, p = 0.6242; A versus C, p < 0.0001; and B versus C, p < 0.0001).
FIGURE 4.
Appearance of wild-type NM II filaments. The left two columns show unphosphorylated filaments in the absence of nucleotide. The right column shows filaments in which the RLCs are phosphorylated. In the absence of nucleotide, NM IIA, B, and C form filaments with similar gross morphology. In all cases the filaments are bipolar and ∼300 nm in size. Some filaments have a compact appearance where the heads are packed closely against the backbone (left panels), whereas in others a bouquet of heads is seen with individual molecules splayed out from the filament backbone (right panels). The appearances of unphosphorylated and phosphorylated filaments are similar. Scale bar, 100 nm.
FIGURE 5.
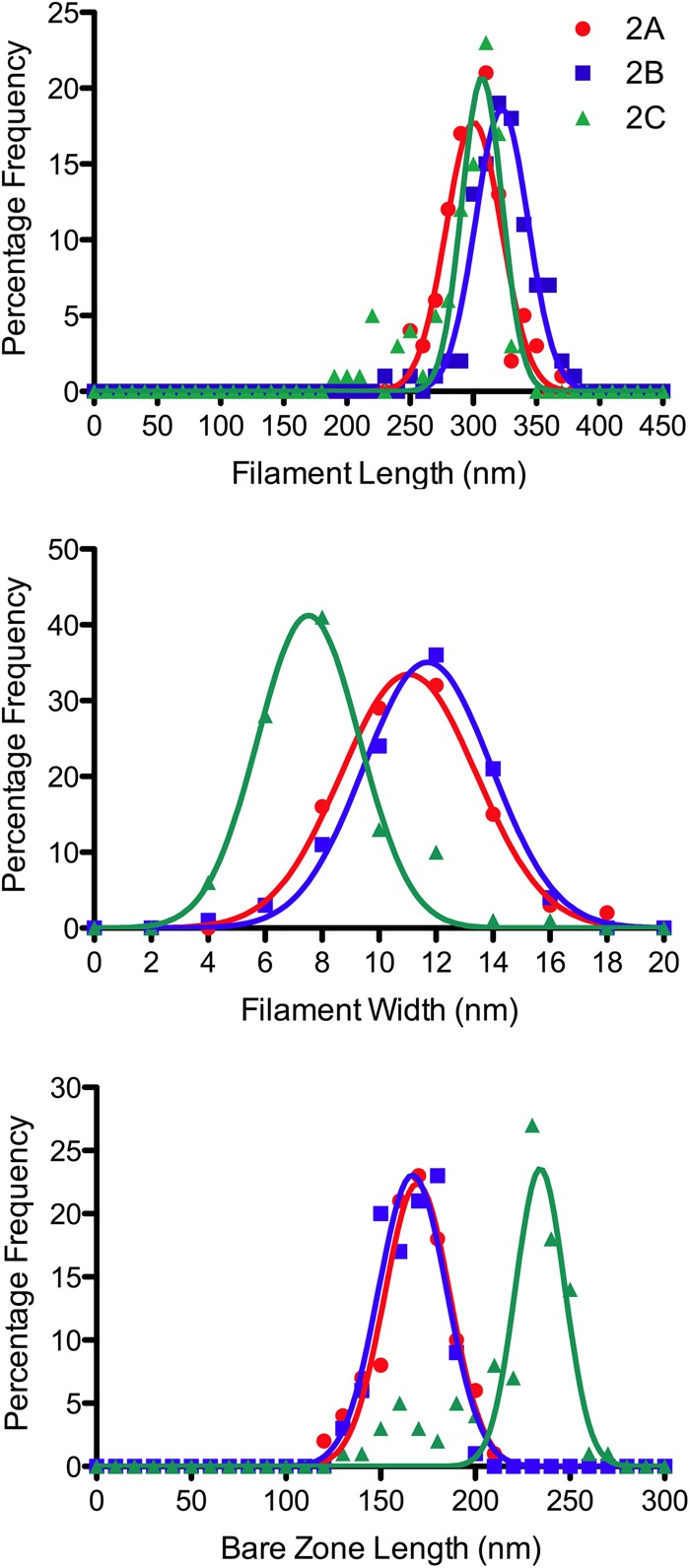
Comparison of filament dimensions for NM IIA, IIB, and IIC. Length distributions of filaments from NM II paralogs are similar. Mean contour lengths were 301 (±24 nm S.D.), 323 (±24 nm S.D.), and 293 (±33 nm S.D.) for NM IIA, NM IIB, and NM IIC respectively. NM IIC filaments tended to be narrower and have a longer bare zone than those made from NM IIA and NM IIB, indicating fewer constituent molecules in NM IIC filaments. Mean widths were 11.2 (±2.4 nm S.D.), 11.5 (±2.3 nm), and 7.9 nm (±2.1 nm) for NM IIA, NM IIB, and NM IIC, respectively. Mean bare zone lengths were 167 (±19 nm) for NM IIA, 166 (±16 nm) for NM IIB, and 219 (±30 nm) for NM IIC.
Effect of ATP on Unphosphorylated NM II
Following addition of 1 mm ATP, NM II filaments rapidly disassembled into individual molecules. The disassembly of NM IIB monitored by light scattering is shown in Fig. 3A. Filaments from all three paralogs as well as the NM IIB2 splice variant disassembled on a similar time scale. After addition of ATP, molecules of all three paralogs of NM II adopted a compact conformation resembling that seen previously in other myosins and previously demonstrated for NM IIA in which the heads interact asymmetrically to form an enzymatically inactive state of the molecule and the tail folds into three approximately equal size segments (27, 44, 53, 54) (Fig. 6, upper row). To stabilize the 10 S (folded) structure and allow it to be investigated in more detail, glutaraldehyde cross-linking was used. This has previously been shown to preserve the compact form of myosin II without changing the conformation of the molecule at the resolution of the negative stain method (44). Alignment and classification of 2938 images of NM IIB cross-linked in the presence of ATP revealed that the heads are organized in the compact conformation first shown in detail in Wendt et al. (53). Fig. 7 (top two rows) shows the global averages (left column), variances (middle column), and examples of class averages of the compact conformation (right column) in the characteristic right (top row) and left views (second row). The maximum contour length of this compact structure, measured from 40 class averages, is 50.1 nm (±1.1 nm S.D.) with the tail section beyond the heads contributing 32.8 nm (±1.1 nm S.D.) of this distance. In addition to this folded monomer, many molecules were present as antiparallel pairs of myosin monomers creating a small bipolar “dogbone” appearance (Figs. 6, lower row, and 7, bottom two rows). The maximum contour length of these dogbones, measured from 41 class averages, was 72.5 (±3.9 nm S.D.). The antiparallel overlap in these dogbones is therefore ∼28 nm. Although there were examples of structures containing more than two associated myosin monomers, they were much rarer. The most common of these structures was a trimer with two head pairs at one end and one head pair at the other. The ratio of monomers to trimers was ∼26:1. To determine whether the antiparallel dimer was a result of an incomplete disassembly of filaments, the ratio of monomers to antiparallel dimers present after 1 min of glutaraldehyde cross-linking was compared using a fixed myosin concentration (0.1 mg/ml) and found to be independent of the time after addition of ATP to a suspension of filaments. The same ratio was also seen when NM IIB was preincubated with ATP for 10 min prior to a reduction in ionic strength (i.e., the same the ratio exists when compact molecules are formed directly rather than from the disassembly of filaments). In all cases, the ratio of monomers to dimers was ∼2:1 (2.1:1 for NM IIB cross-linked in the first minute after addition of ATP, 2.2:1 for myosin cross-linked in the second minute after addition of ATP, and 2.3:1 for myosin cross-linked in the tenth minute after addition of ATP. For myosin preincubated with ATP prior to lowering the ionic strength, the ratio was 2.2:1. Note that these numbers disregard the partially disassembled filaments seen at the shorter time scales). The results demonstrate that even in the compact unphosphorylated state, NM II has a tendency to associate in an antiparallel manner.
FIGURE 6.
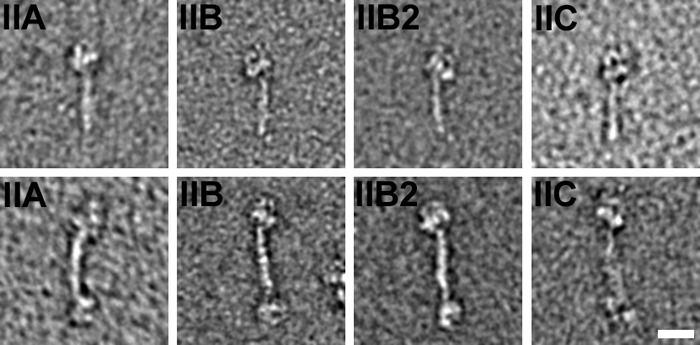
Effect of ATP on unphosphorylated RLC NM II. All three paralogs of NM II, as well as the IIB2 splice variant, form a compact structure following addition of 1 mm ATP (upper panels). In all three paralogs, pairs of compact molecules are associated in an antiparallel dogbone structure (lower panels). Scale bar, 20 nm (applies to all panels).
FIGURE 7.
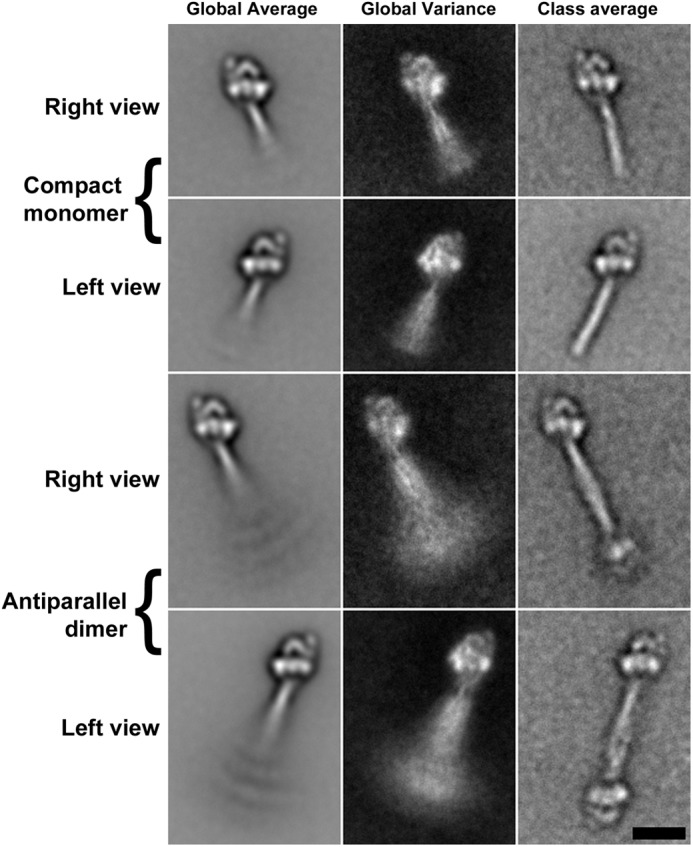
Image processing reveals the compact NM II structures in more detail. Alignment and classification of images of NM IIB in the presence of 1 mm ATP shows the structure to be very similar to that previously detailed for smooth muscle myosin in which the heads interact in an asymmetric fashion and the tail is folded and wrapped around the heads to form a compact molecule ∼50 nm in length (upper two rows) (44, 53, 54). The dogbone structure is an antiparallel pair of compact molecules in which the folded tail regions of two myosins are associated with a mean overlap of 28 nm (lower two rows). The left column shows global average images, the central column shows global variance images, and the right column shows a selected class average in each case. Scale bar, 20 nm (applies to all panels).
Effect of RLC Phosphorylation
Following phosphorylation with MLCK, NM II forms filaments at 150 mm KCl even in the presence of ATP. The assembly of filaments following addition of MLCK is shown in Fig. 3 (A and D). These filaments are similar in gross morphology and size to those seen in the absence of ATP and phosphorylation, although there is an increased tendency for the heads to be splayed away from the filament backbone, creating a wide “bouquet” of heads (Fig. 4).
Interactions of Active NM II Filaments with Actin
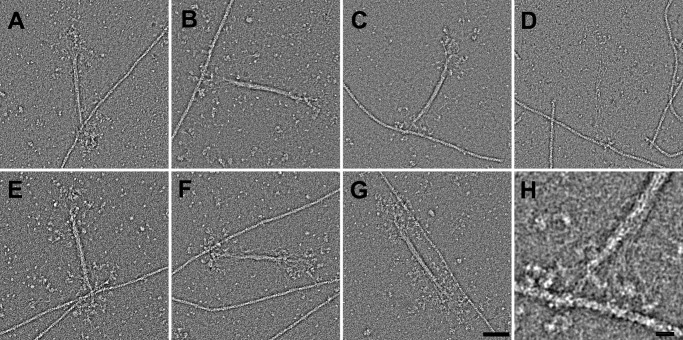
When mixed with actin, NM II filaments in which the RLCs are phosphorylated can be seen bound to actin in the presence of 1 mm ATP. Multiple types of attachment are observed (Fig. 8). To allow comparison of the actin binding behaviors of different paralogs, the proportions of bound versus unbound filaments were compared under a fixed set of conditions and from random regions of the grid (50 nm myosin, 500 nm actin) (Table 2). The types of binding were also quantified as type 1, where binding is to a single F-actin filament via one side of the bipolar NM II filament (Fig. 8, A–D); type 2, where binding is to a single F-actin by both sides of the bipolar NM II filament (Fig. 8G); type 3, where a single side of the bipolar NM II filament is bound to multiple F-actins (Fig. 8E); and type 4, where the two ends of a bipolar filament contact different actin filaments (Fig. 8F). Under the conditions tested, the NM IIB paralog had the greatest tendency to bind actin (89% bound; see Table 2) and accordingly, the greatest tendency to cross-link actin. Actin binding and cross-linking were reduced in the NM IIB2 splice variant, although the majority of NM II filaments were still bound (64%). NM IIA also showed a high tendency for filaments to bind actin (72% bound), whereas less than half of the NM IIC filaments were bound to an actin filament (47%). In all paralogs, there was an approximately equal chance of cross-linking multiple F-actins by one side of the NM II filament as by the two ends. In the case of NM IIA and NM IIB, some regions of the grid contained dense actomyosin networks caused by the strong tendency of these paralogs to bind and cross-link actin (Fig. 9). In cases where these networks were too dense to view individual filaments, no micrograph was recorded (therefore the proportion of bound filaments in these cases may be an underestimate). This kind of superprecipitating behavior was not seen with NM IIC or NM IIB2.
FIGURE 8.
Interactions of phosphorylated RLC NM II with actin in the presence of 1 mm ATP. Following phosphorylation of the RLC with MLCK, NM II from all paralogs (as well as the IIB2 splice variant) forms filaments that are capable of binding to actin. A, NM IIA; B, NM IIB; C, NM IIB2; D, NM IIC. Multiple modes of interaction with actin can be seen. E and F, in addition to the single sided attachments seen in A–D, single myosin filaments can bind multiple actin filaments via a single filament end (E) or via both filament ends (F). G, NM II filaments are also capable of binding to a single actin filament via both ends, although this is less common than the single sided attachment. H, because individual myosins splay away from the filament backbone, a large area of actin, spanning multiple helical repeats, is accessible to the bouquet of heads. E–H show NM IIB2. Scale bar in G, 50 nm (applies to A–G); scale bar in H, 10 nm.
TABLE 2.
Comparison of actin binding behavior among different paralogs
For each NM II, the percentage of filaments bound to actin or unbound is shown. Bound filaments were classified as type 1 (bound to a single actin filament via one side of the myosin filament; e.g., Fig. 8, A–D), type 2 (bound to a single actin filament by both sides of the myosin filament; e.g., Fig. 8G), type 3 (bound to multiple actin filaments by one side of the myosin filament; e.g., Fig. 8E), or type 4 (bound to multiple actin filaments by both sides of the myosin filament; e.g., Fig. 8F). Each type is quantified as a percentage of all filaments, and types 2, 3, and 4 are not mutually exclusive.
| Paralog | Bound | Unbound | Type 1 | Type 2 | Type 3 | Type 4 |
|---|---|---|---|---|---|---|
| % | % | % | % | % | % | |
| NM IIA | 72 | 28 | 15 | 15 | 40 | 43 |
| NM IIB | 89 | 11 | 14 | 24 | 66 | 68 |
| NM IIB2 | 64 | 36 | 37 | 6 | 14 | 16 |
| NM IIC | 47 | 53 | 30 | 3 | 7 | 11 |
FIGURE 9.
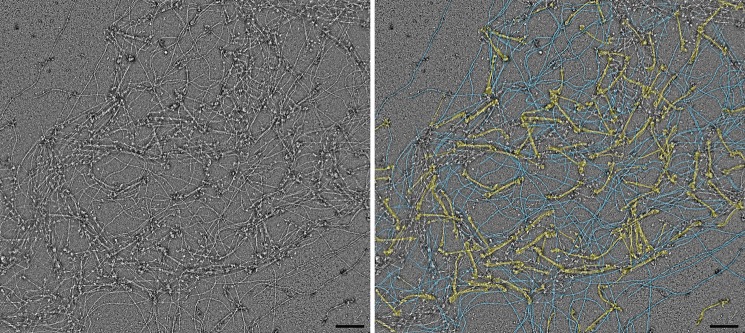
Superprecipitation of actin caused by phosphorylated RLC NM IIB filaments in the presence of ATP. The left panel shows an example of a dense actomyosin network resulting from interaction of 500 nm actin with 50 nm NM IIB. The right panel shows a false color version of the same image highlighting myosin filaments (yellow, myosin; blue, actin).
DISCUSSION
Our study has made use of the baculovirus/Sf9 system to study the filament forming properties of NM II filaments. The use of the expressed NM IIs has two major advantages compared with most of the published studies on NM II filament assembly. First, there are multiple paralogs of NM II present in most mammalian cells and tissues (15). This hampers the determination of the properties of individual NM II paralogs when using tissue-purified protein. An exception to this is platelets, which have been shown to express only NM IIA (26). Purification of NM II from brain tissue should result in myosin, which is highly enriched in NM IIB, but no cell types are known in which exclusively NM IIB or NM IIC are expressed. The extensive studies of NM II from brush border (49–52) were probably carried out with a mixture of all three NM II paralogs (15). Thus, distinguishing the properties of individual NM II paralogs is not possible using tissue-purified myosin. Second, our studies (and earlier studies using platelet NM II) show conclusively that tail fragments of NM IIs do not serve as models for the assembly of full-length NM II. NM II rod and light meromyosin fragments form large aggregates that can be amorphous to paracrystalline in nature (30–32, 34). They can obtain lengths greater than 1 μm and have variable thicknesses. The assemblies formed from NM II rods and light meromyosin are not sensitive to ATP concentrations because the motor domain is lacking. Studies looking at intracellular distribution of these fragments show large puncta, indicative of aggregation, with different localizations compared with full-length myosins (55, 56). Although some elements of the interactions involved in paracrystal formation and NM II filaments may be shared, the overall structures are very different. The appearance of filaments formed from the three paralogs of NM II is broadly similar; in all cases, filaments are bipolar and ∼300 nm long. This is particularly important to note for the case of NM IIC, which by phylogenetic analysis is no more closely related to NM IIA and NM IIB than it is to smooth muscle myosin (57). Smooth muscle myosin forms side polar filaments, whereas NM IIC only forms bipolar filaments similar to those of NM IIA and NM IIB. NM II filaments are similar in size and appearance to the filaments of platelet myosin (NM IIA) described by Niederman and Pollard (23). In that study, the authors noted that ATP had no effect on the appearance of the filaments, indicating that the purified platelet myosin was either RLC-phosphorylated or not capable of binding and/or hydrolyzing ATP, because it is clear from our study and previous ones that ATP depolymerizes unphosphorylated NM II filaments (49, 51). Niederman and Pollard used potassium iodide in the purification procedure to help depolymerize actin, which was subsequently shown to affect the enzymatic properties of some myosins (58, 59). In contrast, filaments formed from the recombinant NM II used in this study could be readily disassembled with ATP and reassembled by RLC phosphorylation, indicating that the purified myosin lacks RLC phosphorylation and is enzymatically competent.
The similarity of NM IIA and IIB filaments in terms of their overall dimensions indicates that the different properties, functions, and localization of these motors in vivo do not arise from the NM II paralogs forming different types of macromolecular assemblies. Instead, the known differences in the enzymatic activities of the motor domains, different regulatory inputs via paralog specific phosphorylation of the heavy chains, interactions with proteins such as S100A4 that regulate NM II filament assembly, and distinct expression patterns are the likely means by which the different functional properties and spatial distributions of these two paralogs emerge (60–62). In the case of NM IIC, the lower mean width and longer bare zone of filaments relative to NM IIA and NM IIB are both indicative of filaments consisting of fewer molecules relative to the other paralogs. This has clear implications for the ability of such filaments to remain bound to actin because the processivity of the filament is determined by both the number of motors and the duty ratio of the individual motors (35). Consistent with this, under conditions where the majority of NM IIA (72%) and NM IIB (89%) filaments are bound to actin, less than half of NM IIC filaments were bound (47%). The difference is even more striking when the proportions bound to multiple actin filaments are compared.
The homogenous appearance of recombinant NM II filaments suggests that there is an upper limit imposed on filament growth that results from the inherent properties of the NM II itself. This is unlike the behavior of purified skeletal muscle myosins in vitro where filaments have a relatively large length distribution (63, 64). In the case of skeletal muscle myosin, the formation of filaments has been divided into two stages, an initial stage where an antiparallel array of molecules is formed and acts as a nucleus upon which further molecules are added, via parallel interactions, to form relatively large filaments, often many microns in length (65). In the case of NM II, the stage of filament elongation appears to be absent or unfavorable, leaving only a relatively small assembly that is analogous to the core of a muscle myosin filament (66). Indeed, comparison of the NM II filaments shown here with the core of the skeletal muscle filament shows the structures to be very similar. The interactions involved in the formation of NM II filaments may therefore be a conserved feature of myosin II filament assemblies in general.
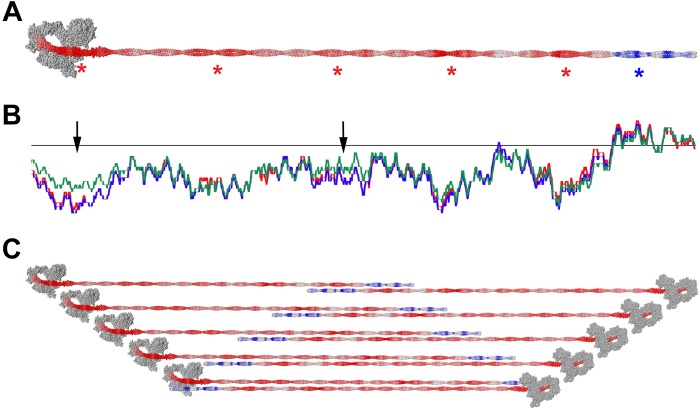
In the Niedermann and Pollard study on platelet myosin filaments (NM IIA), the number of myosin molecules per filament was estimated to be 28 (i.e., 56 heads) based on a hexagonally packed array of myosin molecules (23). Applying a similar analysis to the average filament dimensions measured here, we obtain values of 29 molecules per filament (58 heads) for NM IIA, 30 molecules for NM IIB, and 14 molecules (28 heads) for NM IIC (this calculation assumes a head spacing of 14.3 nm along the filament and a tail length of 160 nm). The coiled coil of NM II contains a charge repeat of 196 residues as described in Ref. 45. When the charge of the exposed residues of the coiled coil is summed in a 98-residue window, this results in five troughs (more negative) and five peaks (less negative). Only the C-terminal end of the coiled coil shows a region of overall positive charge. Assuming that antiparallel overlaps dominate the NM II filament assembly, there are therefore five more negatively charged regions with which the C-terminal positively charged regions can most favorably interact. A filament containing all of these overlaps therefore has 5 crowns of heads per end, corresponding to 20 molecules per filament if there are 2 molecules per crown or 30 if there are 3 (Fig. 10).
FIGURE 10.
Model of antiparallel overlaps in NM II filaments. Charge distribution of exposed residues is shown on a model of NM IIB. Charges are smoothed over a surrounding window of 98 residues. Red is negative charge, and blue is positive charge. Asterisks show the positions of strongly charged regions (A). Charge distribution of each NM II paralog along the coiled coil. Red, blue, and green represent charges of NM IIA, NM IIB, and NM IIC, respectively (B). Schematic diagram showing antiparallel overlaps, which allow the positively charged region to interact with the more negatively charged regions of the coiled coil (C). The heads in this figure are displayed in the compact, folded position for visual simplicity rather than to imply compact head packing within the filament.
The reason why NM IIC forms filaments with fewer component molecules and a longer bare zone may relate to differences in the charge periodicity within the tail region. A previous study looking at charge repeats along the NM II tail revealed some difference between NM IIC and the other two paralogs (45). In a comparison of the NM II paralogs, as well as skeletal and smooth muscle myosin, NM IIC was shown to be unique in that it contained a region of strong positive charge in the coiled coil proximal to the lever/rod junction. However, having repeated the analysis conducted in that study, we were unable to locate this positively charged sequence in NM IIC. This may relate to a frameshift error present in the early consensus human NM IIC sequence (GenBankTM accession number AAO39147.1) that has since been updated (UniProtKB number Q7Z406). Having performed the analysis using the corrected sequence, the difference between charge distribution in NM IIC relative to the other paralogs becomes much less pronounced. Despite this, two of the charge troughs (Fig. 10, arrows) are less negatively charged in NM IIC than the other two paralogs, which may lower the tendency for NM IIC molecules to form long overlaps.
The existence of the compact structure has previously been demonstrated for NM IIA as well as smooth, cardiac, and skeletal muscle myosin II (27, 67). In this study, we show the presence of this conformation in all NM II paralogs in the presence of ATP and the absence of RLC phosphorylation. A proposed role for this compact structure is as an enzymatically inactive molecule, which can diffuse throughout the cytoplasm and forms a pool of NM II, available for activation and polymerization when required. We also demonstrate the existence of antiparallel pairs of compact molecules for each of the NM II paralogs. This kind of antiparallel dimerization has previously been shown to occur in smooth muscle myosin (68, 69). It is possible that the anti-parallel dimer acts as a seed for filament formation because having an existing antiparallel interaction means that as the RLCs become phosphorylated, the molecule can extend and begin the polymerization process.
Following phosphorylation of the RLC using MLCK, NM II in the presence of ATP forms filaments. The filaments are very similar in overall appearance to those formed in the absence of ATP and RLC phosphorylation. The heads have a higher tendency to be splayed out from the filament backbone in RLC phosphorylated filaments, as has previously been observed in thick filaments from skeletal muscle (70). This may indicate that the more compact filaments seen in the absence of RLC phosphorylation are similar to the relaxed state seen in muscle myosin filaments in which the head-head interaction discussed above in relation to the compact molecules is present within a filament (71). The physiological relevance of this is unclear because it is not known that an assembled, unphosphorylated NM II filament is capable of existing in vivo. However, in vitro studies showed that the presence of some portion of phosphorylated nonmuscle myosin in filaments consisting of mixtures of phosphorylated and unphosphorylated myosin partially stabilized the unphosphorylated portion against ATP-dependent depolymerization.
RLC phosphorylated NM IIB filaments are capable of binding to and translocating along actin filaments as a processive unit (35). The types of interaction seen by EM show that NM II filaments can bind to multiple actin filaments via a single side or both sides of the bipolar structure. This leads to a tendency to cross-link actin filaments and produce large superprecipitated regions of actomyosin (Fig. 9). This effect is especially evident in the case of NM IIA and NM IIB, which contain more motors per filament than NM IIC and have a higher probability of actin attachment. In the region where the myosin heads make contact with actin, the splaying effect resulting from the N-terminal regions of the myosin tail peeling away from the filament backbone means that large numbers of actin subunits are accessible to the resulting bouquet of heads. One effect of this is to increase the probability of myosin heads binding to actin. It is clear from the images that there is high degree of flexibility of individual myosins within the filament. There is enough flexibility that in some cases both ends of the NM II filaments can be seen bound to the same actin filament. In images of filaments bound to actin, it was common to observe binding of motors from one side of the filament spread by more than 100 nm along the actin filament. In the cell, actomyosin networks are arranged in a variety of different geometries. These include sarcomeric structures such as those seen in stress fibers and purse strings as well as less ordered networks such as those seen at the peripheral region of the lamella (72–74). Because of the splaying and flexibility of individual NM II molecules from the filament, the actomyosin interactions seen here occur over a wide range of geometries, explaining how NM II filaments can maintain interactions with actin in the absence of well ordered sarcomere-like structures. Indeed, when mixed with actin, NM IIA and NM IIB filaments tended to form disordered actomyosin meshworks, which resemble those seen in some regions of the cell (Fig. 9) (72).
The NM IIC paralog has been shown to play roles in neuritogenesis and maintenance of apical cell junctions in epithelial cells within the cochlea (73, 75). NM IIC filaments contain fewer heads per filament. This has clear implications for the ability of such filaments to remain bound to actin for long periods because the processivity of the filament is determined by both the number of motors and the duty ratio of the individual motors (35). Consistent with this, under conditions where the majority of NM IIA (72%) and NM IIB (89%) filaments are bound to actin, less than half of NM IIC filaments were bound (47%). The difference is even more striking when the proportions bound to multiple actin filaments are compared with NM IIC having a 5–6-fold lower tendency to cross-link actin filaments than NM IIA and a 8–9-fold lower tendency to cross-link actin filaments than NM IIB (Table 2).
The NM IIB2 splice variant has been shown to be important in the development of Purkinje neurons, with expression beginning postnatally. NM IIB2-ablated Purkinje cells display lower numbers of dendritic spines and branches, and NM IIB2-ablated mice have impaired motor coordination. Comparing the behavior of the loop 2 inserted (NM IIB2) versus uninserted splice variants (NM IIB) reveals that NM IIB2 appears to be regulated in the same way as NM IIB in vitro. In both cases, the unphosphorylated proteins in the presence of ATP exist as compact single molecules, which then assemble into filaments following phosphorylation of the RLC. The filaments of the splice variants are indistinguishable when viewed in isolation but show very different behavior when viewed in the presence of actin. The NM IIB2 splice variant has a lower tendency to bind to actin than the uninserted splice variant (64% versus 89% bound, respectively), and NM IIB2 has a 4–5-fold lower tendency to cross-link actin filaments than NM IIB under equivalent conditions. It is reasonable to assume that in vivo, NM IIB2 will be regulated in the same way as NM IIB because the heavy chain phosphorylation sites of the two are the same. A switch from NM IIB to NM IIB2 expression may therefore result in an overall reduction in the contractile behavior of the actomyosin network of the cell while maintaining the regulatory inputs that coordinate filament assembly and disassembly on smaller temporal and spatial scales. Studies of the B2 insert in the context of a soluble HMM-like fragment suggested that the NM IIB2 HMM had a much lower actin-activated MgATPase activity than did the unspliced NM IIB HMM isoform, which led the authors to speculate that the NM IIB2 isoform may be in a permanent “off” state (i.e., equivalent to that shown in Figs. 6 and 7, in which the heads interact asymmetrically and are blocked from interacting with actin) (76). The low but measurable increase in the MgATPase activity of the full-length NM IIB2 molecules in the presence of actin and its ability to form filaments when phosphorylated argue that the earlier proposed model is incorrect and that the NM IIB2 isoform has a finite but down-regulated enzymatic activity instead.
There are surprisingly few electron microscopic images of in vivo myosin filaments in cells present in the literature. Part of this is due to the crowded nature of the cytoskeleton, which makes it difficult to distinguish the small myosin filaments from the other elements, and part of this is due to difficulties in preserving their structures. Nevertheless, the images of filaments that are available bear a strong resemblance in size and shape to the in vitro filaments presented here (72, 74, 77). Of additional interest is the resemblance of the ribbon-like structures that we find at slightly lower ionic strength where the myosin filaments interact in both a head to head manner and in a parallel stacking arrangement. Recent advances in superresolution light microscopy and correlative EM are allowing the nature of NM II filaments to be investigated in vivo, and the results obtained are consistent with the active form of NM II being the small bipolar filaments examined here (73, 78). The results shown here help us to bridge the gap between understanding the properties of the NM IIs as isolated motor units and understanding the roles and behavior of NM II as large oligomeric units in the cell.
Acknowledgments
We thank the Electron Microscopy Core Facility of the NHLBI for support, advice, and use of facilities. We also thank Mary Anne Conti for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant HL0017633.

This article contains supplemental Table S1.
- NM II
- nonmuscle myosin II
- ELC
- essential light chain
- RLC
- regulatory light chain
- NMHC II
- nonmuscle myosin II heavy chain
- CaM
- calmodulin
- MLCK
- myosin light chain kinase
- HMM
- heavy meromyosin.
REFERENCES
- 1. Odronitz F., Kollmar M. (2007) Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 8, R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Richards T. A., Cavalier-Smith T. (2005) Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118 [DOI] [PubMed] [Google Scholar]
- 3. Kühne W. (1864) Untersuchungen über das protplasma und di contractilitat, W. Engelmann, Leipzig, Germany [Google Scholar]
- 4. Craig R., Megerman J. (1977) Assembly of smooth muscle myosin into side-polar filaments. J. Cell Biol. 75, 990–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lo C. M., Buxton D. B., Chua G. C., Dembo M., Adelstein R. S., Wang Y. L. (2004) Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol. Biol. Cell 15, 982–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betapudi V., Licate L. S., Egelhoff T. T. (2006) Distinct roles of nonmuscle myosin II isoforms in the regulation of MDA-MB-231 breast cancer cell spreading and migration. Cancer Res. 66, 4725–4733 [DOI] [PubMed] [Google Scholar]
- 7. Vicente-Manzanares M., Zareno J., Whitmore L., Choi C. K., Horwitz A. F. (2007) Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176, 573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shewan A. M., Maddugoda M., Kraemer A., Stehbens S. J., Verma S., Kovacs E. M., Yap A. S. (2005) Myosin 2 is a key Rho kinase target necessary for the local concentration of E-cadherin at cell-cell contacts. Mol. Biol. Cell 16, 4531–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ivanov A. I., Samarin S. N., Bachar M., Parkos C. A., Nusrat A. (2009) Protein kinase C activation disrupts epithelial apical junctions via ROCK-II dependent stimulation of actomyosin contractility. BMC Cell Biol. 10, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Conti M. A., Even-Ram S., Liu C., Yamada K. M., Adelstein R. S. (2004) Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J. Biol. Chem. 279, 41263–41266 [DOI] [PubMed] [Google Scholar]
- 11. Pollard T. D. (2010) Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 22, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma X., Kovács M., Conti M. A., Wang A., Zhang Y., Sellers J. R., Adelstein R. S. (2012) Nonmuscle myosin II exerts tension but does not translocate actin in vertebrate cytokinesis. Proc. Natl. Acad. Sci. U.S.A. 109, 4509–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heissler S. M., Manstein D. J. (2013) Nonmuscle myosin-2. Mix and match. Cell Mol. Life Sci. 70, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vicente-Manzanares M., Ma X., Adelstein R. S., Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Golomb E., Ma X., Jana S. S., Preston Y. A., Kawamoto S., Shoham N. G., Goldin E., Conti M. A., Sellers J. R., Adelstein R. S. (2004) Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J. Biol. Chem. 279, 2800–2808 [DOI] [PubMed] [Google Scholar]
- 16. Takahashi M., Kawamoto S., Adelstein R. S. (1992) Evidence for inserted sequences in the head region of nonmuscle myosin specific to the nervous system. Cloning of the cDNA encoding the myosin heavy chain-B isoform of vertebrate nonmuscle myosin. J. Biol. Chem. 267, 17864–17871 [PubMed] [Google Scholar]
- 17. Li Y., Lalwani A. K., Mhatre A. N. (2008) Alternative splice variants of MYH9. DNA Cell Biol. 27, 117–125 [DOI] [PubMed] [Google Scholar]
- 18. Spudich J. A. (1994) How molecular motors work. Nature 372, 515–518 [DOI] [PubMed] [Google Scholar]
- 19. Pato M. D., Sellers J. R., Preston Y. A., Harvey E. V., Adelstein R. S. (1996) Baculovirus expression of chicken nonmuscle heavy meromyosin II-B. Characterization of alternatively spliced isoforms. J. Biol. Chem. 271, 2689–2695 [DOI] [PubMed] [Google Scholar]
- 20. Kim K. Y., Kovács M., Kawamoto S., Sellers J. R., Adelstein R. S. (2005) Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J. Biol. Chem. 280, 22769–22775 [DOI] [PubMed] [Google Scholar]
- 21. Jana S. S., Kim K. Y., Mao J., Kawamoto S., Sellers J. R., Adelstein R. S. (2009) An alternatively spliced isoform of non-muscle myosin II-C is not regulated by myosin light chain phosphorylation. J. Biol. Chem. 284, 11563–11571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heissler S. M., Manstein D. J. (2011) Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J. Biol. Chem. 286, 21191–21202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niederman R., Pollard T. D. (1975) Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J. Cell Biol. 67, 72–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burridge K., Bray D. (1975) Purification and structural analysis of myosins from brain and other non-muscle tissues. J. Mol. Biol. 99, 1–14 [DOI] [PubMed] [Google Scholar]
- 25. Kawamoto S., Adelstein R. S. (1991) Chicken nonmuscle myosin heavy chains. Differential expression of two mRNAs and evidence for two different polypeptides. J. Cell Biol. 112, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma X., Jana S. S., Conti M. A., Kawamoto S., Claycomb W. C., Adelstein R. S. (2010) Ablation of nonmuscle myosin II-B and II-C reveals a role for nonmuscle myosin II in cardiac myocyte karyokinesis. Mol. Biol. Cell 21, 3952–3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung H. S., Komatsu S., Ikebe M., Craig R. (2008) Head-head and head-tail interaction. A general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell 19, 3234–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kiboku T., Katoh T., Nakamura A., Kitamura A., Kinjo M., Murakami Y., Takahashi M. (2013) Nonmuscle myosin II folds into a 10S form via two portions of tail for dynamic subcellular localization. Genes Cells 18, 90–109 [DOI] [PubMed] [Google Scholar]
- 29. Rosenberg M. M., Ronen D., Lahav N., Nazirov E., Ravid S., Friedler A. (2013) High resolution characterization of myosin IIC protein tailpiece and its effect on filament assembly. J. Biol. Chem. 288, 9779–9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakasawa T., Takahashi M., Matsuzawa F., Aikawa S., Togashi Y., Saitoh T., Yamagishi A., Yazawa M. (2005) Critical regions for assembly of vertebrate nonmuscle myosin II. Biochemistry 44, 174–183 [DOI] [PubMed] [Google Scholar]
- 31. Rosenberg M., Straussman R., Ben-Ya'acov A., Ronen D., Ravid S. (2008) MHC-IIB filament assembly and cellular localization are governed by the rod net charge. PLoS One 3, e1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ricketson D., Johnston C. A., Prehoda K. E. (2010) Multiple tail domain interactions stabilize nonmuscle myosin II bipolar filaments. Proc. Natl. Acad. Sci. U.S.A. 107, 20964–20969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hodge T. P., Cross R., Kendrick-Jones J. (1992) Role of the COOH-terminal nonhelical tailpiece in the assembly of a vertebrate nonmuscle myosin rod. J. Cell Biol. 118, 1085–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Franke J. D., Dong F., Rickoll W. L., Kelley M. J., Kiehart D. P. (2005) Rod mutations associated with MYH9-related disorders disrupt nonmuscle myosin-IIA assembly. Blood 105, 161–169 [DOI] [PubMed] [Google Scholar]
- 35. Nagy A., Takagi Y., Billington N., Sun S. A., Hong D. K., Homsher E., Wang A., Sellers J. R. (2013) Kinetic characterization of nonmuscle Myosin IIb at the single molecule level. J. Biol. Chem. 288, 709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bao J., Jana S. S., Adelstein R. S. (2005) Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. J. Biol. Chem. 280, 19594–19599 [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y., Conti M. A., Malide D., Dong F., Wang A., Shmist Y. A., Liu C., Zerfas P., Daniels M. P., Chan C. C., Kozin E., Kachar B., Kelley M. J., Kopp J. B., Adelstein R. S. (2012) Mouse models of MYH9-related disease. Mutations in nonmuscle myosin II-A. Blood 119, 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang A., Ma X., Conti M. A., Liu C., Kawamoto S., Adelstein R. S. (2010) Nonmuscle myosin II isoform and domain specificity during early mouse development. Proc. Natl. Acad. Sci. U.S.A. 107, 14645–14650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang F., Harvey E. V., Conti M. A., Wei D., Sellers J. R. (2000) A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry 39, 5555–5560 [DOI] [PubMed] [Google Scholar]
- 40. Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. (2006) Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5, 749–757 [DOI] [PubMed] [Google Scholar]
- 41. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 42. Wang F., Kovacs M., Hu A., Limouze J., Harvey E. V., Sellers J. R. (2003) Kinetic mechanism of non-muscle myosin IIB. Functional adaptations for tension generation and maintenance. J. Biol. Chem. 278, 27439–27448 [DOI] [PubMed] [Google Scholar]
- 43. Burgess S. A., Walker M. L., Thirumurugan K., Trinick J., Knight P. J. (2004) Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 147, 247–258 [DOI] [PubMed] [Google Scholar]
- 44. Jung H. S., Burgess S. A., Billington N., Colegrave M., Patel H., Chalovich J. M., Chantler P. D., Knight P. J. (2008) Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc. Natl. Acad. Sci. U.S.A. 105, 6022–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Straussman R., Squire J. M., Ben-Ya'acov A., Ravid S. (2005) Skip residues and charge interactions in myosin II coiled-coils. Implications for molecular packing. J. Mol. Biol. 353, 613–628 [DOI] [PubMed] [Google Scholar]
- 46. McDonnell A. V., Jiang T., Keating A. E., Berger B. (2006) Paircoil2. Improved prediction of coiled coils from sequence. Bioinformatics 22, 356–358 [DOI] [PubMed] [Google Scholar]
- 47. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Butt T., Mufti T., Humayun A., Rosenthal P. B., Khan S., Molloy J. E. (2010) Myosin motors drive long range alignment of actin filaments. J. Biol. Chem. 285, 4964–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scholey J. M., Taylor K. A., Kendrick-Jones J. (1980) Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature 287, 233–235 [DOI] [PubMed] [Google Scholar]
- 50. Kendrick-Jones J., Taylor K. A., Scholey J. M. (1982) Phosphorylation of nonmuscle myosin and stabilization of thick filament structure. Methods Enzymol. 85, 364–370 [DOI] [PubMed] [Google Scholar]
- 51. Kendrick-Jones J., Smith R. C., Craig R., Citi S. (1987) Polymerization of vertebrate non-muscle and smooth muscle myosins. J. Mol. Biol. 198, 241–252 [DOI] [PubMed] [Google Scholar]
- 52. Scholey J. M., Smith R. C., Drenckhahn D., Groschel-Stewart U., Kendrick-Jones J. (1982) Thymus myosin. Isolation and characterization of myosin from calf thymus and thymic lymphocytes, and studies on the effect of phosphorylation of its Mr = 20,000 light chain. J. Biol. Chem. 257, 7737–7745 [PubMed] [Google Scholar]
- 53. Wendt T., Taylor D., Messier T., Trybus K. M., Taylor K. A. (1999) Visualization of head-head interactions in the inhibited state of smooth muscle myosin. J. Cell Biol. 147, 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wendt T., Taylor D., Trybus K. M., Taylor K. (2001) Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. U.S.A. 98, 4361–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ben-Ya'acov A., Ravid S. (2003) Epidermal growth factor-mediated transient phosphorylation and membrane localization of myosin II-B are required for efficient chemotaxis. J. Biol. Chem. 278, 40032–40040 [DOI] [PubMed] [Google Scholar]
- 56. Burns C. G., Reedy M., Heuser J., De Lozanne A. (1995) Expression of light meromyosin in Dictyostelium blocks normal myosin II function. J. Cell Biol. 130, 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Berg J. S., Powell B. C., Cheney R. E. (2001) A millennial myosin census. Mol. Biol. Cell 12, 780–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Collins J. H., Korn E. D. (1981) Purification and characterization of actin-activatable, Ca2+-sensitive myosin II from Acanthamoeba. J. Biol. Chem. 256, 2586–2595 [PubMed] [Google Scholar]
- 59. Pollard T. D., Thomas S. M., Niederman R. (1974) Human platelet myosin. I. Purification by a rapid method applicable to other nonmuscle cells. Anal. Biochem. 60, 258–266 [DOI] [PubMed] [Google Scholar]
- 60. Ford H. L., Silver D. L., Kachar B., Sellers J. R., Zain S. B. (1997) Effect of Mts1 on the structure and activity of nonmuscle myosin II. Biochemistry 36, 16321–16327 [DOI] [PubMed] [Google Scholar]
- 61. Dulyaninova N. G., Malashkevich V. N., Almo S. C., Bresnick A. R. (2005) Regulation of myosin-IIA assembly and Mts1 binding by heavy chain phosphorylation. Biochemistry 44, 6867–6876 [DOI] [PubMed] [Google Scholar]
- 62. Elliott P. R., Irvine A. F., Jung H. S., Tozawa K., Pastok M. W., Picone R., Badyal S. K., Basran J., Rudland P. S., Barraclough R., Lian L. Y., Bagshaw C. R., Kriajevska M., Barsukov I. L. (2012) Asymmetric mode of Ca2+-S100A4 interaction with nonmuscle myosin IIA generates nanomolar affinity required for filament remodeling. Structure 20, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Josephs R., Harrington W. F. (1966) Studies on the formation and physical chemical properties of synthetic myosin filaments. Biochemistry 5, 3474–3487 [DOI] [PubMed] [Google Scholar]
- 64. Kaminer B., Bell A. L. (1966) Myosin filamentogenesis. Effects of pH and ionic concentration. J. Mol. Biol. 20, 391–401 [DOI] [PubMed] [Google Scholar]
- 65. Davis J. S. (1986) A model for length-regulation in thick filaments of vertebrate skeletal myosin. Biophys. J. 50, 417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reisler E., Cheung P., Oriol-Audit C., Lake J. A. (1982) Growth of synthetic myosin filaments from myosin minifilaments. Biochemistry 21, 701–707 [DOI] [PubMed] [Google Scholar]
- 67. Trybus K. M., Huiatt T. W., Lowey S. (1982) A bent monomeric conformation of myosin from smooth muscle. Proc. Natl. Acad. Sci. U.S.A. 79, 6151–6155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Trybus K. M., Lowey S. (1984) Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J. Biol. Chem. 259, 8564–8571 [PubMed] [Google Scholar]
- 69. Onishi H., Wakabayashi T. (1984) Electron microscopic studies on myosin molecules from chicken gizzard muscle III. Myosin dimers. J. Biochem. 95, 903–905 [DOI] [PubMed] [Google Scholar]
- 70. Levine R. J., Kensler R. W., Yang Z., Stull J. T., Sweeney H. L. (1996) Myosin light chain phosphorylation affects the structure of rabbit skeletal muscle thick filaments. Biophys. J. 71, 898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zoghbi M. E., Woodhead J. L., Moss R. L., Craig R. (2008) Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. U.S.A. 105, 2386–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shutova M., Yang C., Vasiliev J. M., Svitkina T. (2012) Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS One 7, e40814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ebrahim S., Fujita T., Millis B. A., Kozin E., Ma X., Kawamoto S., Baird M. A., Davidson M., Yonemura S., Hisa Y., Conti M. A., Adelstein R. S., Sakaguchi H., Kachar B. (2013) NMII forms a contractile transcellular sarcomeric network to regulate apical cell junctions and tissue geometry. Curr. Biol. 23, 731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Goeckeler Z. M., Bridgman P. C., Wysolmerski R. B. (2008) Nonmuscle myosin II is responsible for maintaining endothelial cell basal tone and stress fiber integrity. Am. J. Physiol. Cell Physiol. 295, C994–C1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wylie S. R., Chantler P. D. (2008) Myosin IIC. A third molecular motor driving neuronal dynamics. Mol. Biol. Cell 19, 3956–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim K. Y., Kawamoto S., Bao J., Sellers J. R., Adelstein R. S. (2008) The B2 alternatively spliced isoform of nonmuscle myosin II-B lacks actin-activated MgATPase activity and in vitro motility. Biochem. Biophys. Res. Commun. 369, 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Verkhovsky A. B., Borisy G. G. (1993) Non-sarcomeric mode of myosin II organization in the fibroblast lamellum. J. Cell Biol. 123, 637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hoelzle M. K., Svitkina T. (2012) The cytoskeletal mechanisms of cell-cell junction formation in endothelial cells. Mol. Biol. Cell 23, 310–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kovács M., Tóth J., Nyitray L., Sellers J. R. (2004) Two-headed binding of the unphosphorylated nonmuscle heavy meromyosin. ADP complex to actin. Biochemistry 43, 4219–4226 [DOI] [PubMed] [Google Scholar]