Summary
Background
Animals can act as a reservoir and source for the emergence of novel meticillin-resistant Staphylococcus aureus (MRSA) clones in human beings. Here, we report the discovery of a strain of S aureus (LGA251) isolated from bulk milk that was phenotypically resistant to meticillin but tested negative for the mecA gene and a preliminary investigation of the extent to which such strains are present in bovine and human populations.
Methods
Isolates of bovine MRSA were obtained from the Veterinary Laboratories Agency in the UK, and isolates of human MRSA were obtained from diagnostic or reference laboratories (two in the UK and one in Denmark). From these collections, we searched for mecA PCR-negative bovine and human S aureus isolates showing phenotypic meticillin resistance. We used whole-genome sequencing to establish the genetic basis for the observed antibiotic resistance.
Findings
A divergent mecA homologue (mecALGA251) was discovered in the LGA251 genome located in a novel staphylococcal cassette chromosome mec element, designated type-XI SCCmec. The mecALGA251 was 70% identical to S aureus mecA homologues and was initially detected in 15 S aureus isolates from dairy cattle in England. These isolates were from three different multilocus sequence type lineages (CC130, CC705, and ST425); spa type t843 (associated with CC130) was identified in 60% of bovine isolates. When human mecA-negative MRSA isolates were tested, the mecALGA251 homologue was identified in 12 of 16 isolates from Scotland, 15 of 26 from England, and 24 of 32 from Denmark. As in cows, t843 was the most common spa type detected in human beings.
Interpretation
Although routine culture and antimicrobial susceptibility testing will identify S aureus isolates with this novel mecA homologue as meticillin resistant, present confirmatory methods will not identify them as MRSA. New diagnostic guidelines for the detection of MRSA should consider the inclusion of tests for mecALGA251.
Funding
Department for Environment, Food and Rural Affairs, Higher Education Funding Council for England, Isaac Newton Trust (University of Cambridge), and the Wellcome Trust.
Introduction
Staphylococcus aureus causes a wide range of diseases in human beings, from minor skin infections to severe illnesses such as septicaemia, toxic shock, endocarditis, and pneumonia.1 First described in 1961, the increasing incidence of meticillin-resistant S aureus (MRSA), and its spread in hospitals and the community, has posed a major challenge for infectious disease medicine. The evolution of meticillin resistance in S aureus is, in part, conferred by the acquisition of one of several staphylococcal cassette chromosome mec elements (SCCmec),2 which carry a gene (mecA) that encodes a penicillin binding protein (PBP2a) with low affinity for β-lactam antibiotics.3
Most isolates of MRSA in the UK are identified with antimicrobial susceptibility testing by measuring zones of growth inhibition around antibiotic-impregnated discs on agar plates or measurement of minimum inhibitory concentrations (MIC), as recommended by the British Society for Antimicrobial Chemotherapy guidelines.4 Meticillin resistance breakpoints are zone diameters of 14 mm or less around 1 μg oxacillin-impregnated discs, zone diameters of 21 mm or less around 10 μg cefoxitin-impregnated discs, an oxacillin MIC of more than 2 mg/L, or a cefoxitin MIC of more than 4 mg/L. Detection of the mecA gene by PCR or the detection of PBP2a in a slide agglutination assay5 can be used to confirm a diagnosis of MRSA when antimicrobial susceptibility results are borderline.
Before 2003, most MRSA identified belonged to multilocus sequence type clonal complexes (CC) associated with human carriage and infection. The emergence of MRSA CC398 (known as livestock-associated MRSA) in farm animals and human beings has shown that some S aureus lineages might not be strongly host-species restricted.6 A survey of slaughter pigs in the Netherlands7 showed that 39% harboured MRSA sequence type (ST)398 and another survey8 showed that 27% of people working at, or living on, a livestock farm in the Netherlands carried livestock-associated MRSA. MRSA ST398 can cause infection in people, with close animal contact being the main risk factor,9 suggesting that farm animals could provide a reservoir of MRSA.
Here we report MRSA strains obtained from cattle and human beings, which carry a new SCCmec that is undetectable by molecular diagnostic tests used for identification of MRSA.
Methods
Bacterial isolates
S aureus LGA251 and S aureus LGA254 were isolated from a bulk milk sample from a farm in southwest England in May, 2007.10 24 MRSA were obtained from a collection of 940 S aureus isolates, submitted from 465 different herds, after antibiotic susceptibility testing of all milk samples from cows with mastitis. The samples had been submitted to one of 14 regional Veterinary Laboratories Agency centres between April, 2006, and September, 2007, for bacteriological characterisation.11 None of these isolates tested positive after standard PCR tests for mecA.11 MRSA isolated from human beings, as detailed in the webappendix (p 11), were provided by the Health Protection Agency (HPA; Addenbrooke's Hospital, Cambridge, UK); the Staphylococcal Reference and Antibiotic Resistance Monitoring Laboratories, HPA (Colindale, London, UK); the Scottish MRSA Reference Laboratory (Glasgow, UK); and the National MRSA Reference Laboratory, Statens Serum Institut (Copenhagen, Denmark). Every centre identified likely candidate isolates for PCR testing for the mecA homologue, as detailed in the webappendix (p 11). Submission of all Danish MRSA to the Statens Serum Institut has been mandatory since November, 2006, and all MRSA and meticillin-susceptible S aureus submitted since 2007 have been spa typed; 678 isolates in 2007, 857 in 2008, 817 in 2009, and 1090 in 2010 were MRSA.
Procedures
Species identification of selected isolates was confirmed by PCR with primers based on the 16S–23S rRNA spacer region for S aureus.12 Testing of strains for the presence of PBP2a was done with the Mastalex test (Mast Group, Bootle, UK), which is a slide agglutination assay that detects PBP2a in MRSA by use of latex sensitised with a monoclonal antibody directed against PBP2a.5 Molecular detection of mecA, femB, and the SCCmec–orfX junction was done with PCR, as described previously.13, 14, 15, 16, 17, 18 A comparison of the primer sequences used to test for mecA and the target sequences in mecA and mecALGA251 are shown in the webappendix (p 2). Isolates were genotyped for multilocus sequence type and spa type, as described previously.19, 20
For all test isolates, the MIC of oxacillin and cefoxitin were measured by either Etest (AB Biodisk, Solna, Sweden) or agar dilution,21 depending on which laboratory did the test. The disc diffusion technique was used to establish susceptibility of LGA251 and LGA254 to penicillin, oxacillin, cefoxitin, gentamicin, neomycin, ciprofloxacin, tetracycline, erythromycin, clindamycin, fusidic acid, chloramphenicol, teicoplanin, rifampicin, trimethoprim, linezolid, and mupirocin.22 To establish whether β-lactam resistance was a result of hyperproduction of β-lactamase, tests were done with and without adjacent discs impregnated with both amoxicillin and clavulanic acid.22 The S aureus NCTC 12493 strain was used as a control for MRSA and the S aureus NCTC 6571 strain was used as a control for meticillin-susceptible S aureus (both strains from the National Collection of Type Cultures, HPA, Salisbury, UK). Growth on chromogenic MRSA screening agar, MRSA ID (bioMérieux, Basingstoke, UK), was measured by standard plating and incubation for 18 h at 35°C.
We developed a PCR assay to amplify a region of mecA and the novel homologue described here, mecALGA251. Primers were based on conserved regions of the mecA sequences of previously described S aureus strains,23 S aureus LGA251, and other Staphylococcus species (Staphylococcus epidermidis, Staphylococcus sciuri, Staphylococcus vitulinus, Staphylococcus capitis, Staphylococcus kloosii, and Staphylococcus pseudintermedius). All the mecA sequences were aligned with Bioedit (Ibis Therapeutics, Carlsbad, USA). Primers were chosen from conserved regions with a GC proportion of 40%. The chosen sequences were checked with Primer3 (version 1.1.4) for melting temperatures and self-complementarity,24 and pDraw32 (version 1.1.101) was used to confirm the amplicon size and melting temperatures. Primers were as follows: Fw, 5′ TCACCAGGTTCAAC[Y]CAAAA 3′; and Rv, 5′ CCTGAATC[W]GCTAATAATATTTC 3′. Primers for the amplification of the femB control gene were obtained from a previously described protocol.13 A 25 μL PCR reaction contained final concentrations of 2·5 units of Taq DNA polymerase (Qiagen, Crawley, UK); 1xQ solution (Qiagen); 1xQiagen CoralLoad PCR buffer (Tris-Cl, KCl, [NH4]2SO4, 15 mmol/L MgCl2, gel-loading reagent, orange dye, and red dye; pH 8·7; Qiagen); 4 mmol/L of MgCl2; 0·8 mmol/L of each dNTP (GeneAmp, Applied Biosystems, Warrington, UK); 0·4 μmol/L of each primer (Operon, Cologne, Germany); and 50 ng of DNA template. A negative control, with no target DNA, was included in the PCR run in the GeneAmp PCR System 9700 (Applied Biosystems). The amplification programme consisted of an initial denaturation step at 94°C for 5 min; 30 cycles of denaturing at 94°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 2 min; and a final extension at 72°C for 5 min. PCR products were analysed by electrophoresis on a 1% agarose gel, previously stained with ethidium bromide at 0·14 μg/mL (Sigma, Gillingham, UK), and run at 5 V/cm for 45 min. The molecular marker used was a 100 bp ladder (Promega, Southampton, UK). The sizes of the PCR products sequenced after PCR were 356 bp for the mecA gene, and 651 bp for the femB gene.
We designed a duplex-PCR to detect the mecA regulatory genes (primers: mecIF, 5′ GACACGTGAAGGCTATGATATAT 3′; mecIR, 5′ ATTCTTCAATATCATCTTCGGAC 3′; mecR1F, 5′ GGCTCAGTTAAATCATAAAGTTTG 3′; mecR1R, 5′ AAATTGCCTTACCATAGCTTGTGT 3′), a duplex-PCR to identify the two cassette recombinase genes (primers: ccrAF, 5′ GCAATAGGTTATCTACGTCAAAG 3′; ccrAR, 5′ TCTAATGATTGTGCGTTGATTCC 3′; ccrBF, 5′ TTCGTGTATCGACAGAAATGCAG 3′; ccrBR, 5′ CATCTTTACGAATATCAATACGG 3′), and a single target PCR to amplify the β-lactamase gene blaZ (primers: blaZF, 5′ AGTCGTGTTAGCGTTGATATTAA 3′; blaZR, 5′ CAATTTCAGCAACCTCACTTACTA 3′). The sizes of the expected PCR products were 344 bp for mecI, 710 bp for mecR1, 932 bp for ccrA, 1449 bp for ccrB, and 809 bp for blaZ. Except for use of an annealing temperature of 58°C instead of 55°C, we used the same method as for the other PCR assay described above.
The whole genome of the S aureus isolate LGA251 was sequenced with both capillary sequencing (on ABI 3730xl analysers; Applied Biosystems) and pyrosequencing (on 454 instruments; Life Sciences, Roche Diagnostics Corporation, Branford, CT, USA). A total of 29 300 high quality capillary reads were produced mostly from two subclone libraries (a 2–3 kb insert library and a 3–4 kb library, both with the vector pOTW12). The average read length was 650 bp and these reads represented 6·8 times coverage. The 454 sequencing produced 59·07 Mb data in reads with an average length of 225 bp. The assembly of these reads with Newbler 1.1.03.24 gave 81 contigs greater than 500 bp with a combined length of 2 699 627 bp in six scaffolds.
A combined assembly of the capillary reads, with Phrap (Version 17.0), and the consensus sequences from the 454 assembly (which were converted into overlapping 500 bp sequences) produced 26 contigs (overlapping sequences or clones from which a sequence can be obtained) greater than 2 kb with an N50 of 532 kb. A further 2310 high quality reads were produced to close gaps and to improve the quality of the sequence to finished standard. The sequence was finished and annotated as described previously.25 The sequences and annotations of the S aureus strain LGA251 genome have been entered in the EMBL database (accession numbers FR821779 for the chromosome and FR821780 for the plasmid).
Statistical analysis
Temporal trends for annual incidence of detection of mecALGA251 in Denmark for the years 2007–10, with all MRSA as the denominator, were assessed with the Cochran-Armitage trend test. Statistical analysis was done with StatXact version 9.
Role of the funding sources
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
LGA251 was resistant to penicillin, oxacillin, and cefoxitin, but susceptible to gentamicin, neomycin, ciprofloxacin, tetracycline, erythromycin, clindamycin, fusidic acid, chloramphenicol, teicoplanin, rifampicin, trimethoprim, linezolid, and mupirocin. LGA251 gave negative reactions in the latex agglutination test for PBP2a, a PCR assay with primers for mecA,13 and a PCR that amplifies a region of the SCCmec (SCCmec-orfX).18 Culture of LGA251 on agar plates with and without adjacent discs impregnated with both amoxicillin and clavulanic acid indicated that resistance was not mediated by β-lactamase hyperproduction (webappendix p 4). LGA254 had cefoxitin MICs of 24 mg/L and oxacillin Etest (AB Biodisk NA, Culver City, CA, USA) MICs of 12 mg/L, and although resistant to penicillin, was susceptible to gentamicin, neomycin, ciprofloxacin, tetracycline, erythromycin, clindamycin, fusidic acid, chloramphenicol, teicoplanin, rifampicin, trimethoprim, linezolid, and mupirocin. Both LGA251 and LGA254 were sequence type 425 by multilocus sequence type but the spa type of LGA251 was t6300 (spa repeat: 14-44-12-17-23-18-110-17-17-23-24) and the spa type of LGA254 was t6292 (spa repeat: 14-44-12-17-23-18-110-17-17-17-23-24).
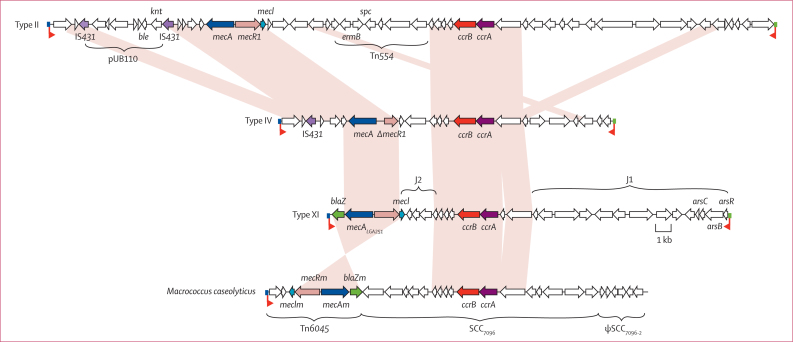
The genetic basis of the meticillin resistance in LGA251 was investigated by whole-genome sequencing. Comparative analysis of the 2·8 Mb genome revealed a 29·4 kb SCCmec element inserted in the chromosome at the same locus (3′ region of orfX) as other SCCmec elements, flanked by sequences that match the SCC integration site sequence.26 The element contained 29 coding sequences, including homologues of the site-specific recombinase genes ccrA and ccrB, and the mecA gene encoding the penicillin-binding protein PBP2a (figure 1).
Figure 1.
Comparison of the type XI SCCmec of LGA251 with other SCC elements
Schematic diagram of type II SCCmec of meticillin-resistant Staphylococcus aureus MRSA252 (top), type IV SCCmec of MW2 (upper middle), type XI SCCmec of LGA251 (lower middle), and the SCCmec-like element of Macrococcus caseolyticus JCSC7096 (bottom). The names of the coding sequences associated with drug and metal resistance are provided. Coding sequences are marked in the direction of transcription as arrows. Coding sequences belonging to the mec and ccr complexes, and IS431 are coloured, with homologues given the same colour; the individual genes belonging to these groups are named. Light pink shading joins regions that are conserved between elements. The coloured boxes at the end of the SCC elements mark the integration site sequence (blue, attL; green attR).
The site-specific recombinases CcrA and CcrB were divergent by comparison with those encoded by other SCCmec elements; CcrA was classified as ccrA1 group and CcrB was classified as ccrB3 group (webappendix p 13). The combination of ccrA1 and ccrB3 is novel and has been designated as type-8 ccr.
Comparison of the PBP2a encoded by the LGA251 mecA (MecALGA251) with proteins in the public sequence databases (Universal Protein Resource; UniProt) indicated that it is divergent in comparison with all other MecA homologues; MecALGA251 was most similar to MecA from the type III SCCmec (accession number Q93IC2), having 63% identity at the aminoacid level (70% at the DNA level; webappendix p 13). Phylogenetic analysis of MecALGA251 showed that it belongs to the PBP2a family (data not shown) and is found on a branch outside the clade that includes S aureus homologues and homologues from other Staphylococcus species (webappendix p 6).
The mecALGA251 gene was part of a complex (mecI–mecR1–mecA–blaZ) that has a different organisation to other SCCmec elements, and has been designated as class E mec gene complex. A complex with the same organisation has been identified in a plasmid,27 and an SCCmec-like element carried by Macrococcus caseolyticus,28 although the sequence conservation of the two complexes is low (<60% identity at the DNA level).
The LGA251 SCCmec element was novel and holds only two of the three joining regions normally seen (J1 and J2; figure 1). The blaZ gene was located at the left hand end of the element in a J3 region, but has been designated as part of the class E mec gene complex. The J2 region contained genes encoding hypothetical, exported, and membrane proteins, and the J1 region contained hypothetical proteins, an exported protein, membrane proteins, a lipase gene fragment, ABC transporter proteins, and genes associated with arsenic resistance. The sequence was submitted to the International Working Group on the Classification of SCC29 and has been given the designation type XI SCCmec.
Details of the geographical distribution, year of isolation, and test results for the bovine isolates are included in the table and figure 2. More than half (13 of 24; 54%) of the MRSA isolates from the survey done by the Veterinary Laboratories Agency were positive by PCR for the mecALGA251 gene and for the mecI, mecR1, ccrA, ccrB, and blaZ genes of type XI SCCmec. Sequencing of the mecA PCR amplicons from the 13 positive S aureus showed that these isolates had an identical sequence to that of mecALGA251. The 13 mecALGA251-positive S aureus isolates were genotyped by multilocus sequence type and spa type. One strain was the same sequence type as LGA251 (ST 425), the remainder resolved into four different sequence types.
Table.
Data for all bovine and human meticillin-resistant Staphylococcus aureus strains with the mecALGA251 gene, by isolate
| Host | Location | Sample | Year | Multilocus sequence type | Clonal complex | spa type | Oxacillin MIC (mg/L) | Cefoxitin MIC (mg/L) | |
|---|---|---|---|---|---|---|---|---|---|
| LGA251 | Bovine | Somerset, England | Bulk milk | 2007 | 425 | 425 | t6300 | 16 | 32 |
| LGA254 | Bovine | Somerset, England | Bulk milk | 2007 | 425 | 425 | t6292 | 12 | 24 |
| C02 467 | Bovine | Bury St Edmunds, England | Milk | 2006–07 | 130 | 130 | t6220 | 32 | 12 |
| C02 937 | Bovine | Langford, England | Milk | 2006–07 | 425 | 425 | t6292 | 16 | 12 |
| C03 125 | Bovine | Bury St Edmunds, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 32 |
| C03 362 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 2 | 12 |
| C03 363 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 24 |
| C03 364 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 12 |
| C03 365 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 24 |
| C03 366 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 12 |
| C03 367 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 12 |
| C03 370 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 32 |
| C03 371 | Bovine | Sutton Bonington, England | Milk | 2006–07 | 1245 | 130 | t843 | 16 | 24 |
| C04 288 | Bovine | Thirsk, England | Milk | 2006–07 | 1526 | 130 | t6293 | 8 | 32 |
| C04 831 | Bovine | Truro, England | Milk | 2006–07 | 151 | 705 | t529 | 16 | 4 |
| SA227 | Human | Cambridge, England | Unknown | 2008 | 130 | 130 | t1736 | 16 | 32 |
| 02.5099.D | Human | Tayside, Scotland | Finger | 2002 | 1944 | 130 | t843 | 24 | 32 |
| 08.6601.A | Human | Lothian, Scotland | Nose | 2008 | 1245 | 130 | t1535 | 32 | 24 |
| 09.2364.C | Human | Ayrshire and Arran, Scotland | Nose | 2009 | 1245 | 130 | t7947 | 24 | 32 |
| 09.3741.Aa | Human | Western Isles, Scotland | Wound | 2009 | 130 | 130 | t843 | 8 | 16 |
| 09.4633.Ca | Human | Highland, Scotland | Nose | 2009 | 1764 | 130 | t7485 | 32 | 16 |
| 09.4657.D | Human | Grampian, Scotland | Wound | 2009 | 1764 | 130 | t7485 | 16 | 24 |
| 09.7342.H | Human | Grampian, Scotland | Blood | 2009 | 130 | 130 | t843 | 16 | 24 |
| 09.8549.Q | Human | Highland, Scotland | Wound | 2009 | 1764 | 130 | t7485 | 32 | 32 |
| 10.1799.W | Human | Tayside, Scotland | Screen | 2010 | 1245 | 130 | t7946 | 16 | 16 |
| 10.3514.K | Human | Greater Glasgow and Clyde, Scotland | PEG site | 2010 | 1943 | 1943 | t7945 | 32 | 48 |
| 10.4264.V | Human | Ayrshire and Arran, Scotland | Toe | 2010 | 130 | 130 | t843 | 32 | 32 |
| 10.7365.L | Human | Greater Glasgow and Clyde, Scotland | Hand | 2010 | 130 | 130 | t843 | 16 | 16 |
| H052960441 | Human | East England | Not known | 2005 | 130 | 130 | t843 | 16 | 64 |
| H061860351 | Human | Northwest England | Sputum | 2006 | 130 | 130 | t843 | 16 | 16 |
| H074300388 | Human | Northwest of england | Skin swab | 2007 | 130 | 130 | t843 | 16 | 64 |
| H091100226 | Human | East England | Skin swab | 2009 | 425 | 425 | t6386 | 16 | 32 |
| H092840495 | Human | Northeast England | Nose screen | 2009 | 1526 | 130 | t6293 | 8 | 8 |
| H093880936 | Human | Southeast England | Ear swab | 2009 | 425 | 425 | t742 | 16 | 64 |
| H101740629 | Human | Northwest England | Skin swab | 2010 | 130 | 130 | t7734 | 16 | 16 |
| H102840444 | Human | Southwest England | Nose screen | 2010 | 425 | 425 | t742 | 8 | 32 |
| H103360314 | Human | Northwest England | Nose screen | 2010 | 1945 | 130 | t1535 | 16 | 16 |
| H104700070 | Human | Yorkshire and Humber, England | Nose screen | 2010 | 130 | 130 | t843 | 16 | 4 |
| H104720074 | Human | Yorkshire and Humber, England | Nose screen | 2010 | 130 | 130 | t843 | 16 | 4 |
| H105020135 | Human | East Midlands, England | Nose screen | 2010 | 1945 | 130 | t843 | 8 | 4 |
| H105060288 | Human | West Midlands, England | Nose screen | 2010 | 1945 | 130 | t843 | 8 | 4 |
| H105140339 | Human | Southwest England | Nose screen | 2010 | 1946 | 1943 | t978 | 16 | 4 |
| 43066 | Human | Næstved, Denmark | Wound | 2004 | 130 | 130 | t843 | 2 | 16 |
| 43067 | Human | Næstved, Denmark | Wound | 2004 | 130 | 130 | t843 | 16 | 32 |
| 45454 | Human | Herlev, Denmark | Skin | 2005 | 130 | 130 | t843 | 6 | 16 |
| 47034 | Human | Herlev, Denmark | Unknown | 2005 | 130 | 130 | t843 | 6 | 32 |
| 61425 | Human | Viborg, Denmark | Blood | 2008 | 130 | 130 | t843 | 3 | 12 |
| 65727 | Human | Sønderborg, Denmark | Nose/mouth | 2009 | n/d | 130 | t843 | 16 | 64 |
| 66519 | Human | SSI, Denmark | SSTI | 2009 | n/d | 130 | t843 | 8 | 8 |
| 66712 | Human | Aalborg, Denmark | SSTI | 2009 | 130 | 130 | t843 | 4 | 16 |
| 67909 | Human | Vejle, Denmark | SSTI | 2009 | n/d | 130 | t843 | 0·75 | 8 |
| 70355 | Human | Hillerød, Denmark | Nose/mouth | 2010 | 130 | 130 | t843 | 4 | 24 |
| 70782 | Human | Slagelse, Denmark | Eye/ear | 2010 | n/d | 130 | t843 | 6 | 32 |
| 70956 | Human | Slagelse, Denmark | SSTI | 2010 | n/d | 130 | t843 | 4 | 48 |
| 71274 | Human | Slagelse, Denmark | Blood | 2010 | n/d | 130 | t843 | 12 | 32 |
| 71795 | Human | Slagelse, Denmark | SSTI | 2010 | n/d | 130 | t843 | 1·5 | 8 |
| 71950 | Human | Aalborg, Denmark | Eye/ear | 2010 | n/d | 130 | t843 | 16 | 64 |
| 71957 | Human | Slagelse, Denmark | Unknown | 2010 | n/d | 130 | t843 | 1·5 | 16 |
| 72270 | Human | Nykbøbing F, Denmark | Screening | 2010 | n/d | 130 | t843 | 6 | 24 |
| 72890 | Human | Aalborg, Denmark | SSTI | 2010 | n/d | 130 | t843 | 24 | 48 |
| 72976 | Human | Vejle, Denmark | Fluid | 2010 | n/d | 130 | t843 | 4 | 16 |
| 73532 | Human | Herning, Denmark | SSTI | 2010 | n/d | 130 | t843 | 4 | 16 |
| 73760 | Human | Hillerød, Denmark | Blood | 2010 | n/d | 130 | t843 | 16 | 64 |
| 73829 | Human | Vejle, Denmark | Unknown | 2010 | n/d | 130 | t843 | 8 | 16 |
| 75022 | Human | Slagelse, Denmark | Unknown | 2011 | n/d | 130 | t1535 | 12 | 32 |
| Enr.7594/75 | Human | SSI, Denmark | Blood | 1975 | n/d | 130 | t843 | 3 | 16 |
PEG=percutaneous endoscopic gastrostomy. SSI=Statens Serum Institut. SSTI=skin and soft tissue infection. MIC=minimum inhibitory concentration. n/d=not done (clonal complexes for these isolates are predictions based on spa type).
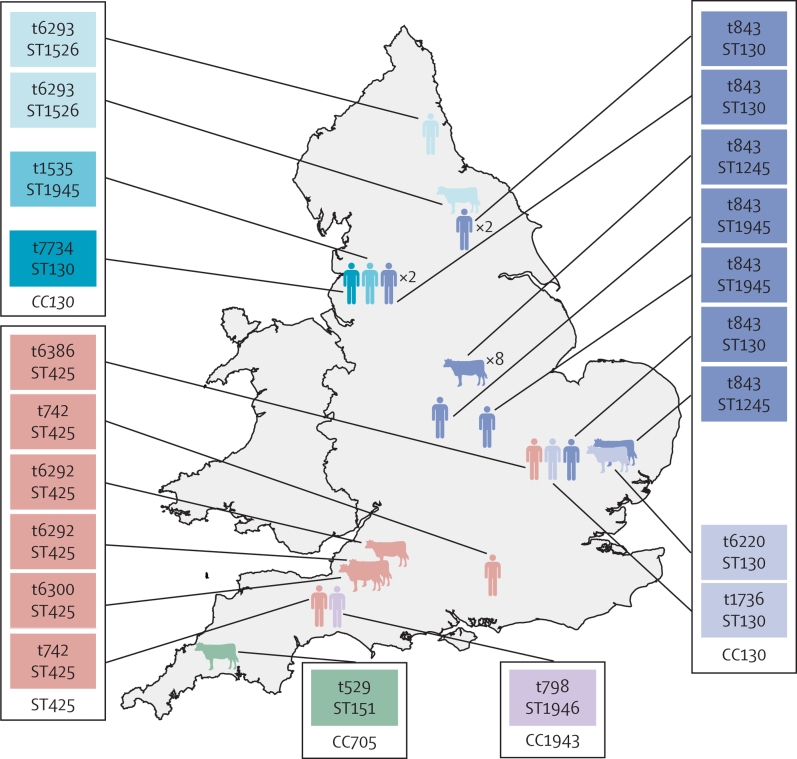
Figure 2.
Geographical distribution of the bovine and human meticillin-resistant Staphylococcus aureus strains carrying the mecALGA251 gene in England
The colouring of the symbols and labels indicates common lineage, defined on the basis of spa typing, multilocus sequence typing, or both. spa types and multilocus sequence types are indicated in the labels.
Searches for human MRSA isolates yielded 51 isolates that tested positive by PCR for mecALGA251 from likely candidates of about 120 000 clinical isolates: the only one tested from Cambridge, 14 of 25 tested from London, 12 of 16 tested from Glasgow, and 24 of 32 tested from Copenhagen. The results of tests are listed in the table and the geographical distribution of the mecALGA251 isolates is shown in figure 2 for England and in the webappendix for Scotland (p 7) and Denmark (p 8). The oxacillin MICs ranged from 0·75 mg/L to 32 mg/L, with an MIC50 of 4 mg/L and MIC90 of 16 mg/L. The cefoxitin MICs ranged from 4 mg/L to 64 mg/L, with an MIC50 of 8 mg/L and MIC90 of 24 mg/L. All the British isolates were multilocus sequence typed together with seven of 24 Danish isolates. In the Danish isolates mecALGA251 was detected in none of 678 isolates in 2007, one of 857 isolates in 2008 (0·12%, 95% CI 0–0·35%), four of 817 isolates in 2009 (0·49%, 0·01–0·97), and 13 of 1090 isolates (0·67%, 0·61–1·95) in 2010 (trend for increase: p=0·0002).
The evolutionary relation of the MRSA sequence types that carry mecALGA251 was compared within this population and with the S aureus population as a whole, considering the most frequently reported sequence types (ten or more isolates reported per sequence type).30 Analysis of the population framework shows that mecALGA251 homologues are present in phylogenetically distinct S aureus, indicating that multiple independent acquisitions of the gene have occurred (webappendix p 9).
Discussion
A novel mecA homologue, mecALGA251, associated with resistance to β-lactam antibiotics was present in clinical MRSA isolates from the UK and Denmark, and bovine milk samples from the UK. Our search for mecALGA251 MRSA has been limited to existing collections comprising isolates collected for various reasons and for which incomplete clinical data or strain typing were available. Therefore, interpretations made from our results are tentative and need to be confirmed by more systematic studies with appropriate methodology.
The detection of this gene in MRSA isolates cultured from blood and infected wound sites is strong circumstantial evidence that these organisms are capable of causing clinical disease. Whether disease caused by mecALGA251 S aureus is identical to that caused by conventional MRSA is not clear without evidence from formal epidemiological and virulence studies. Isolate collections were systematically searched for isolates that seemed to be β-lactam resistant, but tested negative by PCR for mecA or by slide agglutination test for PBP2a, and from a search for MRSA isolates that had spa types associated with mecALGA251 MRSA (webappendix p 11). The Danish isolate archive is a complete national collection. These results reveal that detection rates of mecALGA251 MRSA increased substantially between 2007 and 2010. These detection rates are good incidence estimates for those years in Denmark because the archive for 2007–10 contains all Danish MRSA, and because all MRSA isolates were spa-typed.
In this study, we provide evidence of an association between SCCmecLGA251 and β-lactam resistance—which includes meticillin resistance—in S aureus. Genetic manipulation of S aureus strains to show that the insertion or removal of the mecA homologue leads to acquisition or loss of β-lactam resistance will need to be done to show direct linkage, and such studies are underway. However, we were able to search the entire genome of LGA251 for evidence of other genes that might confer resistance. Although a β-lactamase gene (blaZ) is present in SCCmecLGA251 the inability of clavulanate (a β-lactamase inhibitor) to ablate resistance indicates that resistance is unlikely to be caused by β-lactamase. This finding is consistent with the fact that penicillinase hyperproducers do not show heteroresistance,31 as was seen with LGA251 (panel). The expected complement of genes that encode PBP is present (PBP1, PBP2, PBP3, and PBP4) in a highly conserved form compared with other sequences from other S aureus isolates (data not shown), and we recorded no additional PBPs. The most likely explanation for the β-lactam resistance seen in LGA251 is that it is mediated by the mecA homologue present in the type XI SCCmec.
Panel. Research in context.
Systematic review
The discovery of a novel mecA homologue prompted a search of public sequence databases (UniProt) with the predicted aminoacid sequence for similar mecA homologues. No identical sequence was found and the closest match had 63% identity at the aminoacid level. We searched PubMed with the terms “cattle”, “MRSA”, “livestock”, and “MRSA” for any evidence of livestock-associated meticillin-resistant S aureus (MRSA) that were not accounted for by known SCCmec types. None were found.
Interpretation
Advice on the detection and treatment of infections with mecALGA251 MRSA by antimicrobial susceptibility testing is no different to the detection and treatment of infections with other MRSA strains. However, clinicians should be aware that molecular techniques of detection of MRSA with PCR or slide agglutination tests do not detect mecALGA251 MRSA. This means that when these tests are used either for primary detection, or for confirmation of MRSA, a small chance exists that a false-negative result is obtained. The data presented in this paper suggest that the prevalence of mecALGA251 MRSA is likely to be in the range of one in 100 to one in 500 of total MRSA in the UK and Denmark. The discovery of the same mecALGA251 MRSA in dairy cows suggests that these animals might provide a reservoir of infection and close links with farms or contact with dairy cattle could be risk factors that increase the likelihood of mecALGA251 MRSA carriage or infection in patients. Until better evidence is generated from appropriate observational or experimental studies, this study provides the best evidence to inform clinical decisions concerning this new discovery.
The discovery of this new mecA homologue raises issues about the detection and confirmation of MRSA. Irrespective of whether an infection is caused by mecALGA251 MRSA or conventional MRSA, after culture and antimicrobial susceptibility testing, appropriate decisions about care of patients can be made. However, when existing PCR or monoclonal antibody methods are used as the only method to detect MRSA, or when these methods are used to confirm provisional detection of MRSA, then mecALGA251 S aureus will be wrongly diagnosed as meticillin susceptible. The use of new PCR primers such as those described in this paper, and the production of new monoclonal antibodies, would address this problem.
Another important question concerns the potential role of cattle in the epidemiology of human MRSA infections caused by mecALGA251 MRSA. Data from the Veterinary Laboratories Agency MRSA survey suggest that mecALGA251 could be present in up to 1·4% (95% CI 0·6–2·1%) of S aureus bovine mastitis isolates, and is present in up to 2·8 % (1·3–4·3) of herds. The data described in this paper do not provide direct evidence of transmission between cattle and people. However, four pieces of circumstantial evidence suggest that cattle could be the epidemiological source. First, the human isolate strain types were either CC130 (48 of 66)—a lineage that is thought to be unique to animals32—or other strain types detected in cattle during this study and not previously reported in human beings. Second, none of the strain types or spa-types carrying mecALGA251 come from lineages previously associated with human MRSA carriage or infection. Third, in England, where data from both cattle and human beings are available, evidence of geographical association in both human and bovine isolates exists (ie, human and bovine ST425 in the southwest, human and bovine ST130 in the east, and human and bovine ST1526 in the northeast; figure 2). Fourth, the Veterinary Laboratories Agency survey looking for MRSA in dairy cattle noted no other (ie, human) MRSA,11 although there are infrequent reports of human-associated lineages of mecA-positive MRSA causing mastitis in cattle in other countries.33 Such evidence suggests that a bovine reservoir exists, from which mecALGA251 MRSA is transmitted to people. Pasteurisation of milk will prevent any risk of infection via the food chain but individuals in close contact with cattle could be at higher risk of carriage. Further research is needed to test this hypothesis.
The absence of diversity between the mecALGA251 S aureus isolates from Denmark and those from the UK could be a result of a searching bias (ie, that spa-type t843 was used as one of the searching criteria, and by testing for known spa types only we might have missed mecALGA251 MRSA that had different spa types). The discovery of an isolate from a sample obtained from a patient in 1975 possessing the same spa-type shows that the mecA homologue has been in S aureus for at least 36 years, and that the Danish lineage could have changed very little during this period. This early Danish isolate also suggests that mecALGA251 S aureus might have originally been a human strain. The presence of type XI SCCmec in four separate multilocus sequence type lineages, and the fact that it is bounded by integration site sequence repeats and has intact site-specific recombination components, suggests that it has the potential to be transferred to other S aureus lineages in the future.
The search for mecALGA251 MRSA also yielded several isolates that had an MRSA phenotype but no mecA gene that could be detected by PCR. These isolates might possess other mecA homologues or other mechanisms leading to β-lactam resistance. The discovery of this previously undetected mecA homologue is potentially of public health importance. Diagnostic protocols, whether for clinical or epidemiological purposes, should consider the ramifications of not detecting S aureus strains that carry this new mecA homologue.
Acknowledgments
Acknowledgments
During the publication process we became aware that Shore and colleagues had submitted a paper for publication describing human MRSA isolates from Ireland from the clonal complex 130, which contains the type XI SCCmec (Shore AC, Deasy EC, Slickers P. Detection of staphylococcal cassette chromosome mec type XI encoding highly divergent mecA, mecI, mecR1, blaZ and ccr genes in human clinical clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother [in press]).
We are grateful to the Cambridge Infectious Diseases Consortium (CIDC; Department of Veterinary Medicine, University of Cambridge, UK) for its support during this project. We thank the Sanger Institute's Pathogen Production Group for shotgun and finishing sequencing, and the core Informatics Group for support. We thank members of the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) for their assistance and advice in naming the SCCmec element in LGA251. We are grateful to all the dairy farmers who provided samples for this study. This work was part of a PhD project supported by the Cambridge Infectious Diseases Consortium (CIDC), based at the Department of Veterinary Medicine, University of Cambridge. The CIDC was funded by the Department for Environment, Food and Rural Affairs (Defra) and the Higher Education Funding Council for England as part of the Veterinary Training and Research Initiative. Other work was directly co-funded by Defra and the Isaac Newton Trust (Trinity College, University of Cambridge, UK). The Sanger Institute is core-funded by the Wellcome Trust. SP was supported by the NIHR Cambridge Biomedical Research Centre. We would like to thank the many laboratories that submitted isolates to the reference laboratories for testing.
Contributors
LG-A and MAH collected the original bovine isolates. LG-A and HL did most of the laboratory work. DFJB, MDC, and EW repeated PCR and antimicrobial susceptibility tests and initial identification of the first human isolate. Sequencing and genetic analysis were done by MTGH, SDB, JP, KB, and DJP. CT was responsible for collection and initial characterisation of bovine isolates from the Veterinary Laboratories Agency. GFE and EKG were responsible for identification and initial characterisation of the Scottish isolates. AMK, RLRH, and BP were responsible for the identification and characterisation of the English isolates. RLS and ARL were responsible for the identification of the Danish isolates. Supervision and management of the study were done by MAH, CRW, SJP, and DJM. All authors were involved in compiling the report and approved the final version.
Conflicts of interest
All authors declare that they have no conflicts of interest.
Web Extra Material
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985;28:397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews JM, Testing BWPoS. BSAC standardized disc susceptibility testing method (version 8) J Antimicrob Chemother. 2009;64:454–489. doi: 10.1093/jac/dkp244. [DOI] [PubMed] [Google Scholar]
- 5.Brown DF, Walpole E. Evaluation of the Mastalex latex agglutination test for methicillin resistance in Staphylococcus aureus grown on different screening media. J Antimicrob Chemother. 2001;47:187–189. doi: 10.1093/jac/47.2.187. [DOI] [PubMed] [Google Scholar]
- 6.van Loo I, Huijsdens X, Tiemersma E. Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13:1834–1839. doi: 10.3201/eid1312.070384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Neeling AJ, van den Broek MJ, Spalburg EC. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2007;122:366–372. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 8.van Cleef BA, Verkade EJ, Wulf MW. Prevalence of livestock-associated MRSA in communities with high pig-densities in The Netherlands. PLoS One. 2010;5:e9385. doi: 10.1371/journal.pone.0009385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulders MN, Haenen AP, Geenen PL. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in The Netherlands. Epidemiol Infect. 2010;138:743–755. doi: 10.1017/S0950268810000075. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Alvarez L. Assessment of the role of cattle movements and other risk contacts on the spread of Staphylococcus aureus strain types between UK dairy farms. University of Cambridge; Cambridge: 2009. [Google Scholar]
- 11.Veterinary Laboratories Agency (VLA) MRSA in cattle—an investigation into selected properties of isolates recovered from clinical veterinary diagnostic samples: Department for Environment Food and Rural Affairs (Defra), UK. 2008. http://randd.defra.gov.uk/Default.aspx?Menu=Menu&Module=More&Location=None&Completed=0&ProjectID=13867 (accessed May 25, 2011).
- 12.Forsman P, Tilsala-Timisjarvi A, Alatossava T. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology. 1997;143:3491–3500. doi: 10.1099/00221287-143-11-3491. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Roth E, Claverie-Martin F, Villar J, Mendez-Alvarez S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol. 2001;39:4037–4041. doi: 10.1128/JCM.39.11.4037-4041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poulsen AB, Skov R, Pallesen LV. Detection of methicillin resistance in coagulase-negative staphylococci and in staphylococci directly from simulated blood cultures using the EVIGENE MRSA Detection Kit. J Antimicrob Chemother. 2003;51:419–421. doi: 10.1093/jac/dkg084. [DOI] [PubMed] [Google Scholar]
- 15.Kondo Y, Ito T, Ma XX. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–274. doi: 10.1128/AAC.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald RR, Antonishyn NA, Hansen T. Development of a triplex real-time PCR assay for detection of Panton-Valentine leukocidin toxin genes in clinical isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:6147–6149. doi: 10.1128/JCM.43.12.6147-6149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huletsky A, Giroux R, Rossbach V. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol. 2004;42:1875–1884. doi: 10.1128/JCM.42.5.1875-1884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shopsin B, Gomez M, Montgomery SO. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37:3556–3563. doi: 10.1128/jcm.37.11.3556-3563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown DF, Edwards DI, Hawkey PM. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA) J Antimicrob Chemother. 2005;56:1000–1018. doi: 10.1093/jac/dki372. [DOI] [PubMed] [Google Scholar]
- 22.Andrews JM. BSAC standardized disc susceptibility testing method (version 7) J Antimicrob Chemother. 2008;62:256–278. doi: 10.1093/jac/dkn194. [DOI] [PubMed] [Google Scholar]
- 23.National Center for Biotechnology Information (NCBI) The Entrez Nucleotide database. Bethesda, USA: The National Library of Medicine. http://www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore (accessed Feb 14, 2009).
- 24.Rozen S, Skaletsky HJ. In: Bioinformatics methods and protocols: methods in molecular biology. Krawetz S, Misener S, editors. Humana Press; Totowa, NJ: 2000. Primer3 on the WWW for general users and for biologist programmers; pp. 365–386. [DOI] [PubMed] [Google Scholar]
- 25.Holden MT, Hauser H, Sanders M. Rapid evolution of virulence and drug resistance in the emerging zoonotic pathogen Streptococcus suis. PLoS One. 2009;4:e6072. doi: 10.1371/journal.pone.0006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiramatsu K, Kuroda M, Baba T, Ito T, Okuma K. In: Staphylococcus aureus: molecular and clinical aspects. Ala'Aldeen D, Hiramatsu K, editors. Horwood Publishing; Chichester: 2004. The Staphylococcus aureus genome. [Google Scholar]
- 27.Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, [corrected] reflecting the ancestral genome of the human-pathogenic staphylococci. J Bacteriol. 2009;191:1180–1190. doi: 10.1128/JB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob Agents Chemother. 2010;54:1469–1475. doi: 10.1128/AAC.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International Working Group on the Classification of Staphylococcal Cassette Chromosome E Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aanensen DM. Multi locus sequence typing. http://www.mlst.net/misc/further.asp (accessed May 25, 2011).
- 31.Montanari MP, Tonin E, Biavasco F, Varaldo PE. Further characterisation of borderline methicillin-resistant Staphylococcus aureus and analysis of penicillin-binding proteins. Antimicrob Agents Chemother. 1990;34:911–913. doi: 10.1128/aac.34.5.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung JM, Lloyd DH, Lindsay JA. Staphylococcus aureus host specificity: comparative genomics of human versus animal isolates by multi-strain microarray. Microbiology. 2008;154:1949–1959. doi: 10.1099/mic.0.2007/015289-0. [DOI] [PubMed] [Google Scholar]
- 33.Vanderhaeghen W, Hermans K, Haesebrouck F, Butaye P. Methicillin-resistant Staphylococcus aureus (MRSA) in food production animals. Epidemiol Infect. 2010;138:606–625. doi: 10.1017/S0950268809991567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.