Background: AlgX is required for the biosynthesis and export of the exopolysaccharide alginate.
Results: The structure of AlgX has been determined, and the functional characterization of AlgX and mutant variants has been performed.
Conclusion: AlgX contains an SGNH hydrolase-like domain and carbohydrate-binding module. Mutation of the Ser-His-Asp triad in vivo results in non-acetylated alginate.
Significance: This is the first structural characterization of a polysaccharide acetyltransferase.
Keywords: Biofilm, Microbiology, Polysaccharide, Protein Structure, X-ray Crystallography, SGNH Hydrolase, Acetylation, Alginate, Exopolysaccharide Biosynthesis
Abstract
The exopolysaccharide alginate, produced by mucoid Pseudomonas aeruginosa in the lungs of cystic fibrosis patients, undergoes two different chemical modifications as it is synthesized that alter the properties of the polymer and hence the biofilm. One modification, acetylation, causes the cells in the biofilm to adhere better to lung epithelium, form microcolonies, and resist the effects of the host immune system and/or antibiotics. Alginate biosynthesis requires 12 proteins encoded by the algD operon, including AlgX, and although this protein is essential for polymer production, its exact role is unknown. In this study, we present the X-ray crystal structure of AlgX at 2.15 Å resolution. The structure reveals that AlgX is a two-domain protein, with an N-terminal domain with structural homology to members of the SGNH hydrolase superfamily and a C-terminal carbohydrate-binding module. A number of residues in the carbohydrate-binding module form a substrate recognition “pinch point” that we propose aids in alginate binding and orientation. Although the topology of the N-terminal domain deviates from canonical SGNH hydrolases, the residues that constitute the Ser-His-Asp catalytic triad characteristic of this family are structurally conserved. In vivo studies reveal that site-specific mutation of these residues results in non-acetylated alginate. This catalytic triad is also required for acetylesterase activity in vitro. Our data suggest that not only does AlgX protect the polymer as it passages through the periplasm but that it also plays a role in alginate acetylation. Our results provide the first structural insight for a wide group of closely related bacterial polysaccharide acetyltransferases.
Introduction
Exopolysaccharides have numerous and varied functions and play a key role in bacterial survival as major components of biofilm matrices (1). Biofilms are of great medical importance because, once formed, they are extremely difficult to eradicate. Bacteria within the matrix are less prone to dehydration and have increased resistance to antibiotics and the host immune system (2, 3). The architecture and properties of the matrix are determined by the components present, which can include not only exopolysaccharides but also extracellular DNA and protein (2, 4, 5). The matrix composition is species-dependent and can change in response to different external pressures (2, 6). The properties or characteristics of the biofilm matrix can also be altered by the embedded bacteria using a variety of modifications, which can either alter the exopolysaccharides directly or alter the proteins involved in the formation, maintenance, and dispersion of the biofilm (5, 7–12). For example, in Escherichia coli, Staphylococcus aureus, and Staphylococcus epidermidis, biofilm formation requires the partial de-N-acetylation of the exopolysaccharide poly-β-1,6-N-acetyl-d-glucosamine (5, 7, 9). In the Gram-negative bacterium E. coli, this modification is a prerequisite for the export of poly-β-1,6-N-acetyl-d-glucosamine through the outer membrane (9), whereas, in Gram-positive Staphylococcus species, partial deacetylation is necessary for retention of the polymer on the cell surface (5). In contrast, although acetylation of the cellulose that forms Pseudomonas fluorescens biofilms does not affect biofilm attachment to solid surfaces or biofilm initiation at the air-liquid interface, it does produce biofilms that are thicker and almost 10-fold stronger than its non-acetylated counterpart (12). In addition, the rate of increase in biofilm mass was observed to be higher when cellulose was acetylated, and more cells were retained within the matrix (12). Dispersion of cells from a Pseudomonas putida biofilm in nutrient-limiting conditions also requires modifications, but in this case to the associated proteins. A starvation-led decrease in intracellular bis-(3′,5′)-cyclic dimeric guanosine monophosphate levels triggers a regulatory cascade that ultimately leads to cleavage of the protein LapA by the cysteine protease LapG. LapA is associated with both the outer membrane of the cell and the exopolysaccharide biofilm components and therefore links the cells to the biofilm matrix. LapA cleavage dissociates bacterial cells from the exopolysaccharide in the biofilm, thus enabling cellular dispersion to occur (8). Given the importance of such modifications, research into the mechanisms by which bacteria regulate and perform these modifications is required if we are to fully understand the function and purpose of specific biofilms.
Pseudomonas aeruginosa is an opportunistic pathogen that infects the lungs of cystic fibrosis patients and is responsible for much of the morbidity and mortality associated with this disease (13–16). The conditions in the cystic fibrosis lung cause the bacterium to produce the exopolysaccharide alginate, a key component of P. aeruginosa biofilms in this environment. Current understanding of alginate biosynthesis, polymerization, and export indicates the involvement of 13 proteins, 12 of which are encoded by genes located on the algD operon (17, 18). Two of the genes encoded in the operon, algA and algD, are involved in alginate precursor synthesis. The other 10 proteins are hypothesized to form a complex through which alginate is biosynthesized and secreted (19). Alginate is initially produced as a negatively charged polymer of β-1,4-linked d-mannuronate. Postpolymerization, alginate undergoes two types of modification as it is exported through the periplasm. The C5-epimerase AlgG converts selected mannuronate residues to l-guluronate (20, 21), which increases the viscosity of the polymer and hence of the biofilm (22), whereas the concerted actions of AlgI, AlgJ, and AlgF are responsible for the selective O-acetylation of mannuronate residues (23–25). Acetylation of the hydroxyl groups can occur at the C2 and/or C3 positions and alters biofilm architecture, augments bacterial resistance to antibiotics and opsonic phagocytosis, enhances microcolony formation, and improves biofilm adhesion (10, 26, 27). Currently, it is not known whether epimerization occurs prior to or after acetylation, but varying amounts of both modifications are known to occur on the same exopolysaccharide strand (28).
The periplasmic protein AlgX, encoded by the algD operon, has been proposed to have a number of roles in the alginate biosynthetic machinery. Previous work has demonstrated that AlgX protects alginate from degradation as the polymer traverses the periplasm (29). AlgX has also been shown to have at least two interaction partners in the periplasm. It interacts with AlgK, another essential component of the biosynthetic machinery and a protein product that is also encoded by the algD operon (30), as well as the periplasmic protease MucD (30, 31), a protein that primarily acts as a negative regulator of alginate production. Although the AlgX-MucD interaction is intriguing from a regulatory/feedback possibility, the functional basis for the interaction has not yet been determined (30–32), and interestingly, ΔmucD mutants actually have increased alginate production when compared with the wild-type strain (32, 33). Despite the work that has been carried out on AlgX, its exact role has yet to be defined. Bioinformatics analyses suggest that AlgX is a two-domain protein, with an N-terminal region that is predicted to share structural homology with members of the SGNH hydrolase superfamily (34). This region of the protein also shares ∼69% similarity and 30% identity with AlgJ, the putative alginate acetyltransferase (24), and has therefore been proposed to have a similar function (19). The C-terminal domain has no apparent sequence homology to any known protein (34). To determine the role of AlgX in alginate biosynthesis, we have determined its structure to 2.15 Å resolution. The structure reveals that the protein has bilobal architecture with an N-terminal SGNH hydrolase-like domain and a C-terminal carbohydrate-binding module (CBM).5 Although the topology and some features of the N-terminal domain are different from those found in a canonical SGNH hydrolase domain, and no enzymatic activity has previously been ascribed to AlgX, this domain contains the Ser-His-Asp triad that is typically found in members of this enzyme superfamily. Site-specific mutation in vivo of the Ser-His-Asp triad and other key residues in the putative active site results in either non-acetylated or acetylation-reduced alginate polymer, respectively. In vitro analysis demonstrates that the protein exhibits acetylesterase activity and that the Ser-His-Asp triad is required for this activity. Taken together, our data suggest that AlgX plays two roles in alginate biosynthesis; not only does the protein protect the polymer as it passages through the periplasm (29), but it also plays a role in its acetylation, an activity that is controlled by the Ser-His-Asp catalytic triad. In addition, the structure of AlgX provides insight into the function of AlgJ and other polysaccharide acetyltransferases involved in the acetylation of biofilm polymers and other bacterial polysaccharides, such as cellulose and peptidoglycan, respectively.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Media
The bacterial strains and plasmids used in this study are listed in supplemental Table S1. For expression of native or selenomethionine-incorporated protein, E. coli BL21-CodonPlus® (DE3)-RP or B834 Met− cells transformed with pPLHJTWX were grown in rich medium (32 g of tryptone, 20 g of yeast extract, and 5 g of NaCl) at 37 °C or in M9 medium according to the protocol of Lee et al. (35), respectively. DNA manipulations were performed in E. coli DH5α. E. coli DH5α and P. aeruginosa strains were grown in either LB medium or LPIA medium consisting of a 1:1 mixture of l-agar and Pseudomonas isolation agar at 37 °C, unless otherwise stated. Where appropriate, antibiotics were added to media at the following concentrations: ampicillin at 100 μg ml−1, carbenicillin at 200 μg ml−1, chloramphenicol at 30 μg ml−1, gentamicin at 15 μg ml−1 for E. coli and 100 μg ml−1 for P. aeruginosa, and kanamycin at 40 μg ml−1 for E. coli.
DNA Manipulations
The Gateway® recombinase-based multisite cloning system (Invitrogen) was used as described by the manufacturer. PCR amplification was carried out using the high fidelity DNA polymerase, PfuTurbo® (Stratagene), and the primers were designed to contain appropriate attB sequences for recombinational cloning into the relevant Gateway vector (supplemental Table S1). P. aeruginosa PAO1 genomic sequences for primer design were obtained from the Pseudomonas Genome Database (36). Restriction enzymes were from New England Biolabs. Plasmid DNA was extracted from E. coli using the Qiaprep Spin Miniprep kit (Qiagen). DNA fragments for cloning were purified from agarose gels using the QIAEX II gel extraction system (Qiagen). Sequences of cloned DNA were determined commercially (ACGT, Inc., Wheeling, IL). Plasmids carrying an origin of transfer (oriT) were transferred from E. coli to P. aeruginosa by triparental mating as described (37), using the conjugal helper plasmid pRK2013, and selection for P. aeruginosa was on LPIA with appropriate antibiotics.
The AlgX expression plasmid pPLHJTWX (34) was used to generate point mutants of Asp-174, His-176, and Ser-269 using the QuikChange site-directed mutagenesis Lightning kit (Agilent Technologies). In each case, the native residue was substituted with alanine. Each construct was verified by DNA sequencing (ACGT DNA Technologies Corp., Toronto, Canada).
Protein Expression and Purification of AlgX
The expression and purification of recombinant C-terminally His6-tagged, codon-optimized AlgX (AlgX-His6) for structural studies was performed as described previously (34) with minor modifications. In brief, bacterial cell pellets were thawed and resuspended in Buffer A (500 mm NaCl, 50 mm Tris-HCl, pH 7.8, 2% (v/v) glycerol, 2 mm dithiothreitol) containing 1 mg ml−1 lysozyme. This suspension was incubated at 4 °C for 30 min prior to homogenization with an Avestin Eumulsiflex C3 homogenizer (three passages at 15,000 p.s.i.). Unlysed cells were removed from the lysate by centrifugation (5,000 × g for 15 min at 4 °C), and the resultant supernatant was applied to a 5-ml Ni2+-nitrilotriacetic acid Superflow affinity column (Qiagen) pre-equilibrated with Buffer A. The column was washed extensively with Buffer A to remove contaminating proteins that did not bind to the affinity resin. This wash step was continued until the absorbance at 280 nm returned to zero. Bound protein was eluted from the column using a 0–150 mm imidazole (in Buffer A) gradient. The fractions containing AlgX were pooled and concentrated by Amicon ultrafiltration using a 30,000-Da molecular weight cut-off concentrator (Millipore). The protein was further purified and buffer-exchanged into Buffer B (50 mm Tris-HCl, pH 7.8, 2% (v/v) glycerol, 2 mm dithiothreitol) by size exclusion chromatography using a HiLoad 16/60 Supradex 200 prep-grade gel filtration column (GE Healthcare).
Protein used to determine the crystal structure was expressed as a selenomethionine (SeMet)-incorporated version of AlgX according to the protocol of Lee et al. (35) and purified as described above. Wild-type and the single site-directed mutants used for in vitro enzymatic analysis were expressed in rich medium at 18 °C and purified as described above with the modification that no dithiothreitol was added to the buffers used. Prior to assessing the folding of the wild-type and mutant variants using CD spectroscopy, the protein was dialyzed into Buffer C (20 mm HEPES, pH 7.8). Experiments were performed with 6 μm protein in Buffer C at room temperature on a Jasco J-810 spectrometer (Jasco Spectroscopic Co.) and repeated in triplicate.
Crystallization and Structure Determination of AlgX
SeMet-incorporated AlgX was concentrated to ∼8–10 mg ml−1 by Amicon ultrafiltration (30,000 Da molecular mass cut-off) prior to crystallization. Diffraction quality crystals of SeMet-incorporated AlgX were grown by optimizing the conditions found previously for the wild-type sulfur-containing protein (100 mm sodium citrate, pH 5.5, 20% (w/v) PEG 3000) (34). Crystals were grown with 13–16% (w/v) PEG 3000 at temperatures of 18–20 °C with a pH between 5 and 5.3 with successive rounds of seeding as outlined previously (34). Crystals were soaked for 2–3 min in cryoprotectant, crystallization buffer supplemented with 25% (v/v) glycerol, prior to vitrification in liquid nitrogen (34).
In an attempt to determine an AlgX-alginate complex, the native sulfur-containing protein was co-crystallized in the presence of synthetic tetramannuronate (38). This tetramer has an acetylated ethylamine linker from the synthesis still attached at the reducing end. The tetramer was added to the protein solution (8 mg ml−1) such that the tetramer had a final concentration of 1 mm. Hanging drop vapor diffusion crystallization trials were subsequently set up using 2 μl of the protein/substrate mixture and 2 μl of mother liquor. Crystals from the co-crystallization experiments were prepared for data collection as outlined by Weadge et al. (34) with no synthetic substrate present during the cryoprotection and flash-freezing processes.
All data were collected at Station X29 of the National Synchrotron Light Source. A 0.16-mm collimator was used to collect a total of 360 images of 1° Δϕ oscillations on an ADSC Quantum-315 detector with an exposure time of 0.5 s/image. SeMet and native data were integrated, reduced, and scaled using d*TREK and HKL2000, respectively (Table 1). To determine the phases of the SeMet data, the positions of all 10 expected selenium atoms (5 seleniums/molecule) were located using HKL2MAP (39), and density-modified phases were calculated using SOLVE/RESOLVE (40). The resulting electron density map was of sufficient quality to allow automatic model building using PHENIX (41), followed by manual model building with COOT (42). Refinement was carried out using PHENIX.REFINE (41), and progress was assessed by monitoring the reduction (and convergence) of the Rwork and Rfree. When Rwork reached 25%, waters and ligands were added, and TLS groups as determined by the TLSMD Web server (43, 44) were applied. The native structure was solved by molecular replacement with PHASER (45) from the PHENIX package using molecule A of the SeMet-SAD structure as a search model. Refinement of the native structure was as described for the SeMet model. The refinement statistics are given in Table 1. DSSP was used to assign secondary structure elements of AlgX (46, 47). All figures relating to the structure of AlgX were created in PyMOL (PyMOL Molecular Graphics System, version 1.2r3pre, Schrödinger, LLC). Surface representations showing electrostatics and residue conservation include all residue side chains, including those not present in the PDB deposition that could not be modeled due to lack of suitable density.
TABLE 1.
Summary of data collection and refinement statistics
| Native | SeMet-SAD | |
|---|---|---|
| Data collection | ||
| Beamline | NSLS X29 | NSLS X29 |
| Wavelength (Å) | 1.075 | 0.979 |
| Space group | P21212 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 123.7, 82.4, 92.7 | 46.4, 121.4, 92.1 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 95.2, 90.0 |
| Total no. of reflections | 506,683 | 437,204 |
| No. of unique reflections | 52,314 | 68,397 |
| Resolution (Å) | 50.0–2.15 (2.23–2.15)a | 43.22–2.00 (2.07–2.02) |
| Average I/σI | 44.7 (4.0) | 23.0 (2.1) |
| Completeness (%) | 98.82 (89.26) | 90.7 (64.5) |
| Redundancy | 9.7 (9.8) | 7.1 (6.2) |
| Rmerge (%)b | 8.1 (64.0) | 8.3 (68.1) |
| Refinement | ||
| Rwork/Rfree (%)c | 17.60/23.10 | |
| No. of atoms | ||
| Protein | 6,504 | |
| Water | 372 | |
| Average B-factor (Å2) | ||
| Protein | 50.10 | |
| Water | 50.20 | |
| RMSDs | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (degrees) | 1.040 | |
| Ramachandran plotd | ||
| Total favored (%) | 97.0 | |
| Total allowed (%) | 100 | |
| Coordinate error (Å)e | 0.24 | |
| PDB code | 4KNC | |
a Values in parentheses correspond to highest resolution shell
b Rmerge = ΣΣ|I(k) − 〈I〉|/ΣI(k), where I(k) and 〈I〉 represent the intensity values of the individual measurements and the corresponding mean values. The summation is over all unique measurements.
c Rwork = Σ‖Fo| − k|Fc‖/|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Rfree is the sum extended over a subset of reflections (5%) excluded from all stages of the refinement.
d As calculated using MOLPROBITY (95).
e Maximum likelihood-based coordinate error as determined by PHENIX (41).
Transformation of P. aeruginosa
Competent cells were prepared by growing a culture of PAO1 to A600 of 0.200. The flask was chilled on ice for 10 min. A cold centrifuge tube was filled with the culture and centrifuged at 7,649 × g for 10 min at 5 °C. The supernatant was discarded, and the pellet was resuspended in 10 ml of ice-cold 150 mm MgCl2 and centrifuged at 7,649 × g for 10 min. The pellet was again resuspended in 10 ml of cold 150 mm MgCl2 and incubated on ice for 60 min. The sample was then centrifuged at 7,649 × g for 10 min at 5 °C, and the pellet was resuspended in 1.0 ml of cold 150 mm MgCl2, on ice. To increase competency ∼10-fold, the cell suspension was stored on ice overnight. Competent cells were stored for ∼2 months by adding 70 μl of DMSO (7% final concentration) and freezing aliquots at −80 °C. For transformations, plasmid DNA (1–5 μl) was added to 100 μl of competent cells and incubated on ice for 60 min, followed by a 60-s heat shock at 42 °C. The sample was transferred to 2 ml of LB broth and allowed to shake with aeration at 37 °C for 60 min for expression of selective markers. Aliquots (50–100 μl) were spread onto selective agar and incubated at 37 °C overnight.
Construction of PAO1 algX::GmR
A non-polar algX::GmR mutant of PAO1 was constructed using the Gateway cloning system (Invitrogen). Three entry clones were created, and inserts were sequenced for verification. The first was ∼1 kb of the 5′ region of algX (amplified with primers attB4–5′X-F and attB1r-5′X-R (supplemental Table S1)) in pDONR-P4-P1R. The second was the gentamicin resistance cassette (GmR) from pUCGM (amplified with primers attB1-Gm-F and attB2-Gm-R (supplemental Table S1)) in pDONR-221. Finally, ∼1 kb of the 3′ region of algX (amplified with primers attB2r-3′X-F and attB3–3′X-R (supplemental Table S1)) was cloned into pDONR-P2R-P3. The three cloned fragments were assembled in the correct order in the multisite destination and gene replacement vector, pEX183-R4R3, constructed for this study by amplification of ∼2.3 kb of the multisite destination vector pDEST-R4R3 (primers NsiI-M13F and Nsi-M13R (supplemental Table S1)), containing attR4-ccdB-CmR-attR3, and cloning it into the NsiI site of pEX100T. Competent ccdB-resistant E. coli cells (Invitrogen) were transformed with pEX183-R4R3 containing the cloned fragments, and cells were screened for chloramphenicol resistance. One clone (pEX205-algX::GmR) was transferred to the chromosome of PAO1 by triparental mating with selection on LPIA agar containing carbenicillin to obtain single crossover recombinants, which were plated again on carbenicillin. Double recombinants were selected for on LPIA-sucrose (5%) agar, and GmRCbS phenotypes were verified.
Replacement of Chromosomal algX with algX* Mutant Alleles
Chromosomal algX was replaced with algX* site-directed mutants using the Gateway cloning system (Invitrogen). An entry vector was created with a 1.4-kb amplicon of algX, including the Shine-Delgarno sequence and stop codon (amplified with primers attB1-SDalgX-F and attB2-SDalgX-R (supplemental Table S1)), in pDONR221. A positive clone was subjected to site-directed mutagenesis using the QuikChange Lightning kit (Agilent) and mutagenic primers (supplemental Table S1). Residues chosen for mutagenesis were selected in order to mutate the wild-type amino acid to alanine, using the codon GCC or GCG. All clones were sequenced. The algX wild-type and mutant algX* clones were transferred to the destination vector pEXApGW, which is derived from the gene replacement vector pEX100T (48). The pEXApGW-algX clones were transferred to the chromosome of PAO1 algX::GmR by triparental mating, with selection on LPIA agar containing carbenicillin. Double recombinants were selected on LPIA-sucrose agar, and GmSCbS phenotypes were confirmed.
Activation of Alginate Production in PAO1 Strains
The generalized transducing phage F116L (49) was used to infect PDO351 (50), a mucA::GmR strain, by incubating 10 μl of F116L lysate (at various dilutions) with 0.2 ml of PDO351 (mid-log growth in LB broth) in the presence of 10 mm MgSO4 and 5 mm CaCl2. The samples were incubated at room temperature for 10 min, and then molten top agar at 50 °C (3 ml of LB broth with 0.7% noble agar) was added to each tube, mixed, and immediately poured onto the surface of an l-agar plate. Plates were incubated for 24 h at 37 °C. LB broth (3 ml) was added to the plate showing the best confluent plaques, the top agar was broken up, and the sample was centrifuged for 10 min at 7,649 × g. The supernatant was passed through a 0.45-μm filter, and 0.1 ml of the filtrate was mixed with 0.2 ml of P. aeruginosa PAO1 algX* mutants (grown to mid-log in LB broth). The samples were incubated at ambient temperature for 15 min and then placed at 37 °C for 1 h, shaking at 225 rpm. The samples were plated on LPIA + gentamicin and incubated at 37 °C overnight.
Construction of an algX+ Expression and Complementation Plasmid
A pBAD-araC controlled expression Gateway plasmid, pBAD-DEST49 (Invitrogen) permits arabinose-inducible expression and is compatible with cloning the algX+ fragment in pDONR199-algX via the LR reaction. To allow pBAD-DEST49 to replicate in P. aeruginosa and complement chromosomal mutations in trans, the mini-stabilization fragment (mSF) in pSS124 was amplified by PCR (using primers NdeI-pUC19MCS-F and NdeI-pUC19MCS-R (supplemental Table S1)) and initially cloned blunt-ended with T4 DNA ligase (New England Biolabs) into the SmaI site of pLG339; this was detected by insertional inactivation of its KanR gene and called pLG369-mSF. From there, the mSF was removed by NdeI digestion and ligated into the single NdeI site of pBAD-DEST49. The ligation mix was transformed directly into competent PAO1 cells to select for mSF+ clones, and one was named pBAD506-SF. The algX+ fragment in pDONR199-algX was transferred to pBAD506-SF in an LR reaction, and one clone, pBAD510-algX+, was transferred to the mucA23 mutant PDO300 algX::GmR.
Enzyme Assay
All enzyme assays were performed at least in triplicate, in a 96-well microtiter plate, using a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA). Standard reactions contained 3.0 mm 3-carboxyumbelliferyl acetate, dissolved in DMSO, and 40 μg of AlgX in 100 μl of 50 mm sodium HEPES buffer (pH 8.0) at 25 °C. The final DMSO concentration did not exceed 10% (v/v). Reactions were initiated by the addition of substrate and were monitored in real time for 10 min, using an excitation of 386 nm and an emission of 447 nm as described previously (51). Background hydrolysis rates, in the absence of enzyme, were also monitored and subtracted from the enzyme-catalyzed reaction. A calibration curve for 7-hydroxycourmarin-3-carboxylic acid, the fluorescent hydrolysis product of 3-carboxyumbelliferyl acetate, was obtained under the reaction conditions and used to calculate the reaction rate. The protein concentration of each variant was determined using the Pierce® BCA protein assay kit (Thermo Scientific, Rockford, IL).
Sequence Alignments, Residue Conservation, and Homology Searches
Homologues of AlgX were identified using BLAST, and sequence alignments were carried out using TCOFFEE (52, 53). Conservation of residues was determined by ConSurf (54, 55). Sequence identity and similarity scores were determined using FASTA SSEARCH (56). The SCOP Superfamily database was searched to identify fold families similar to the N-terminal domain of AlgX (57). The CAZYmes Analysis Toolkit was used to carry out a sequence similarity-based annotation of the AlgX C-terminal domain, to determine whether the AlgX CBM aligned with any of the families currently in the Carbohydrate-active Enzymes (CAZy) database (58).
Alginate Purification and Concentration
P. aeruginosa PAO1 mucA::GmR algX+ or its mutant strains were grown overnight at 37 °C in modified alginate-producing medium (7.5 mm NaH2PO4, 16.8 mm K2HPO4, 10 mm MgSO4, 100 mm monosodium glutamate, 100 mm d-gluconate) medium supplemented with gentamicin. The cells were removed by centrifugation at 10,000 × g for 1 h. The concentration of alginate in the supernatants was determined using a modification of the carbazole assay of Knutson and Jeanes (59). Briefly, 30 μl of purified alginate was layered on top of 1 ml of ice-cold borate-sulfuric acid reagent (100 mm H3BO4 in concentrated H2SO4). The samples were vortexed for 4 s and placed on ice. An equal volume (30 μl) of carbazole solution (0.1% (w/v) in ethanol) was added. The mixture was heated to 55 °C for 30 min and then cooled on ice. The alginate concentration was determined spectrophotometrically at 530 nm with Macrocystis pyrifera alginate (Sigma) as a standard.
Acetylation Assay
A chemical method described by Hestrin (60) was used to measure alginate acetylation. Briefly, 250 μl of an alginate solution was incubated with 250 μl of alkaline hydroxylamine (35 mm NH2OH, 75 mm NaOH) for 10 min at 25 °C. The reaction mixture was acidified with 250 μl of 1.0 m perchloric acid, followed by the addition of 250 μl of 70 mm ferric perchlorate (10 ml of 0.5 m perchloric acid, 0.248 g of iron perchlorate). The concentration of acetyl groups was determined spectrophotometrically at 500 nm from a standard curve, with ethyl acetate as the substrate (10 mm acetyls = 192 μl of ethyl acetate in 100 ml of H2O).
NMR Analysis of Alginate
Alginate for characterization by 1H NMR spectroscopy was harvested from cultures grown in LB medium at 28 °C for 72 h. The cultures were diluted 2-fold with 145 mm NaCl. The cells were pelleted by centrifugation, and the supernatant was retained. The supernatant was mixed with an equal volume of isopropyl alcohol to precipitate alginate, which was harvested by centrifugation. The alginate was dissolved in 145 mm NaCl and precipitated and harvested as before. The procedure was repeated two more times, dissolving the alginate in 1 m NaCl and then again in 145 mm NaCl. The final solution of alginate was incubated with 10 mg liter−1 Pronase for 1 h prior to precipitation. The purified alginate was dissolved in H2O and dialyzed against several changes of H2O, in 6,000–8,000 molecular weight cut-off dialysis tubing.
The alginate for 1H NMR analysis was prepared by adjusting 5–10-mg samples to pH 3.5 and heating to reflux for 1 h. The samples were cooled, and the pH was adjusted to 5.5. Each sample was lyophilized and then dissolved in 0.57 ml of D2O containing 2 mm EDTA and 10% DMSO-d6. Spectra were acquired at 80 °C using a pulse sequence to suppress the water peak. Peak assignments for mannuronate and guluronate residues were based on the published values (61, 62).
Alginate Epimerization Assay
The relative extent of epimerization was measured using the G-lyase assay as previously described by Chitnis and Ohman (63) and Douthit et al. (64). This assay measures the relative number of guluronate (G) residues/g of alginate by assaying the abundance of cleaved, unsaturated residues present after treatment with a guluronate-specific lyase. Briefly, alginate (65 μg) in 150 μl of water was deacetylated with 65 μl of 1 m NaOH for 15 min at 65 °C and then neutralized with 50 μl of 1 m HCl and 100 μl of lyase buffer (50 mm Tris-HCl, pH 8.0, 10 mm MgCl2·6H2O, 5.0 mm CaCl2). Alginate G-lyase (10 μl) from Klebsiella aerogenes was added to the alginate, and the samples were incubated for 1 h at 25 °C. To assay for unsaturated residues, periodic acid (250 μl 0.2 m) was added, and the samples were incubated at 25 °C for 40 min. After the addition of sodium arsenite (100 μl of 2% in 0.5 n HCl) for 1 min at 25 °C, thiobarbiturate (1 ml of 0.6% solution, pH 2.0) was added, and the reactions were incubated at 65 °C for 30 min. The samples were cooled for 1 h at room temperature and centrifuged for 5 min to remove precipitate. Guluronate residue abundances were measured spectrophotometrically at 535 nm, and the results were compared with the value obtained with PAO1 mucA::GmR algX+ alginate.
Antibody Generation and Western Blot Analysis
AlgX was expressed and purified as described above and was used to generate antiserum from rabbits using a 70-day standard protocol (Cedarlane). Prior to use, the polyclonal antisera were purified as described by Salamitou et al. (65).
PAO1 mucA::GmR algX* strains were grown to A600 of ∼1.0 in LB-no salt medium. Aliquots of 1.5 ml were removed, and the cells were harvested. The cells were resuspended in 2× Laemmli buffer (∼100 μl, depending upon exact OD) and heated to 100 °C for 5 min. Equal quantities of cell lysate were loaded and separated on 4% stacking, 10% resolving polyacrylamide gels prior to being transferred to polyvinylidene difluoride (PVDF). The proteins of interest were detected using rabbit polyclonal antibodies to AlgX (1:1000) or PilF (1:1000) (66). The secondary goat anti-rabbit antibody conjugated to alkaline phosphatase was used as per the manufacturer's instructions, and the blots were developed with 5-bromo-4-chloro-3′-indolyphosphate p-toluidine and nitro blue tetrazolium chloride (BioShop).
RESULTS
AlgX Is a Two-domain Protein
To probe the function and role of AlgX in alginate biosynthesis, we have determined its structure to 2.15 Å resolution. SeMet-incorporated protein was expressed, purified, and crystallized, and the structure was determined using the single-wavelength anomalous dispersion (SAD) method. The SAD model, in conjunction with the molecular replacement technique, was subsequently used to determine the structure of the native (sulfur-containing) protein that had been crystallized in the presence of a synthetic tetramer of polymannuronate (34, 38) (Fig. 1). The presence of the alginate polymer probably explains the differences in unit cell dimensions and space group observed between the native and SeMet crystals (Table 1). Native AlgX crystallized in space group P21212, with two molecules (molecules A and B) in the asymmetric unit. Extensive rounds of model building and refinement yielded models with good geometry and an Rwork and Rfree of 17.6 and 23.1%, respectively (Table 1). Analysis using size exclusion chromatography and dynamic light scattering (data not shown) indicates that AlgX is a monomer in solution, suggesting that the dimerization observed in the crystal is not biologically relevant. The synthetic tetramer was not visible in the final electron density maps, although a region of positive density was observed in the active site of molecule A that could be best fitted with citrate. Although positive density was located in the active site of molecule B, we were unable to model this density satisfactorily.
FIGURE 1.
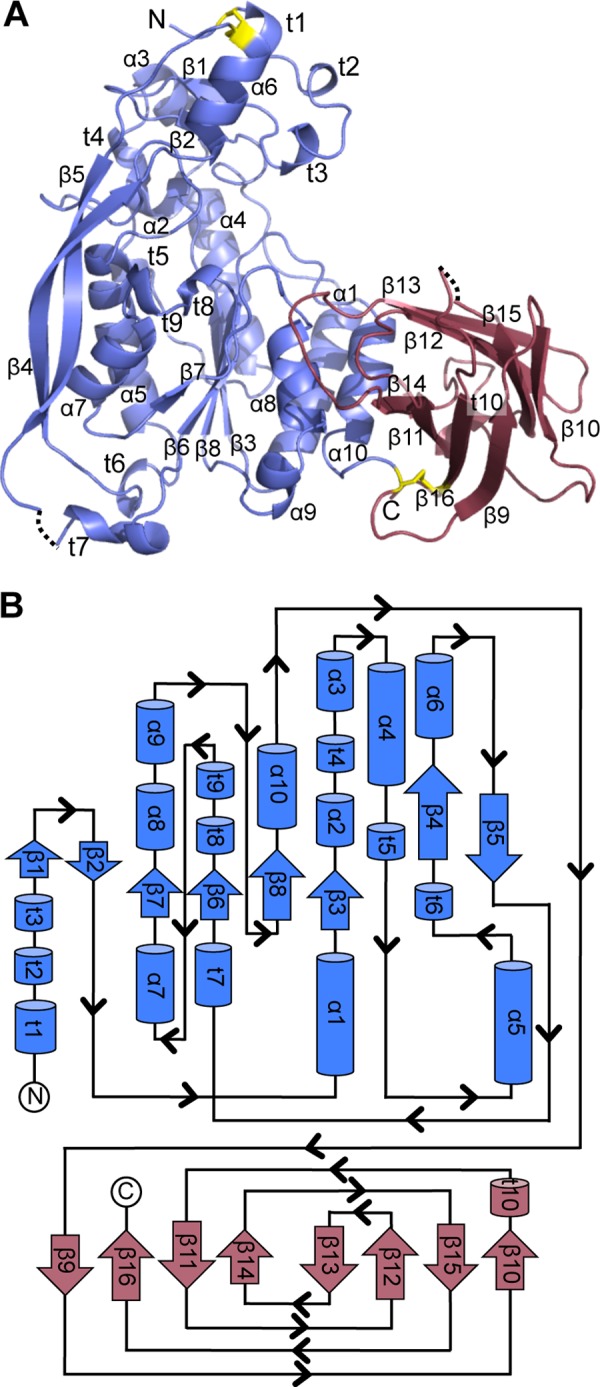
Overall structure of AlgX. A, schematic representation of AlgX with secondary structure elements labeled as follows: α-helix (α), β-strand (β), 310 helix (t). The N-terminal α/β/α and C-terminal CBM domains are represented in blue and red, respectively. The two disulfide bonds present in the structure are colored in yellow. The dashed lines represent residues that cannot be built or modeled due to poor quality electron density. N and C, N and C termini of the protein. B, topology representation of AlgX with the secondary structural elements labeled and colored as in A.
The structure of AlgX revealed a bilobal architecture, composed of an N-terminal hydrolase domain with α/β/α topology followed by a C-terminal CBM domain with a β-sandwich jelly roll fold (Fig. 1). In molecule A, residues 247–258 and residue 447 were unable to be modeled due to the quality of the electron density in this region. Similarly, residues 41, 250, 251, and 447 of molecule B were unable to be modeled. The root mean square deviation (RMSD) between the two molecules in the asymmetric unit is 0.67 Å over 409 equivalent Cα positions. Because molecule B is the most complete model, this model has been used in the structural analysis presented below. Two disulfide bonds are present in the structure. The first, between Cys-44 and Cys-229, positions two long β-strands, β4 and β5, alongside the N-terminal domain. The second, between Cys-347 and Cys-460, appears to fix the relative orientation of the two domains with respect to each other (Fig. 1A).
Examination of the surface potential of AlgX reveals that the surface has areas of both positive and negative electrostatic potential. However, there are two distinct regions of electropositive charge, one in each domain, that lie on the same face of the protein and overlap with regions of high sequence conservation, including critical residues identified for binding and/or catalysis in both the N-terminal and C-terminal domains (Fig. 2). Because alginate is negatively charged (67), it is anticipated that the polymer may associate with AlgX via these positively charged, highly conserved regions. Structural homology searches revealed no proteins in the FATCAT or DALI databases that resemble the entire AlgX protein. Structural homologues to each of the domains, separately, were identified, which makes the combination of the two domains found in AlgX unique among structures solved to date.
FIGURE 2.
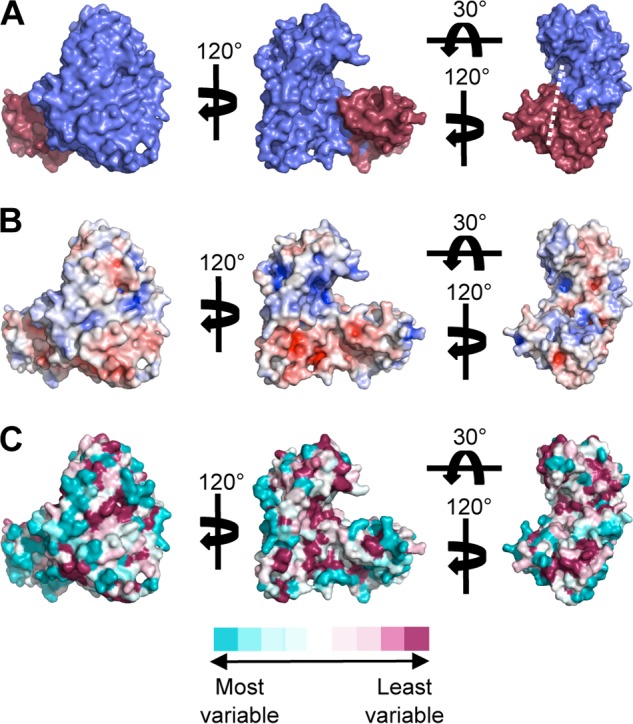
Surface representations of AlgX. A, surface representation of AlgX depicting the N-terminal α/β/α domain in blue and the C-terminal CBM domain in red. The white line indicates the proposed path of alginate across AlgX, between the two domains. When compared with the representations in B and C, it can be seen that the path crosses conserved, mainly electropositive residues. B, electrostatic surface representation of AlgX. The electrostatic potential, calculated using APBS (91–93), is contoured from −7 to 7 kT/e and colored on a gradient from negative (red) to positive (blue). C, representation of the surface residue conservation in eight pseudomonads and A. vinelandii as calculated using ConSurf and colored on a gradient from high (dark pink) to low (cyan) sequence conservation (54, 55).
C-terminal Domain Is a Carbohydrate-binding Module
The C-terminal CBM domain of AlgX (residues 348–474) adopts a β-sandwich jelly roll fold, with each β-sheet consisting of four antiparallel β-strands (Figs. 1B and 3A). This fold is the most common CBM fold found to date (68–70), and a structural homology search of the Protein Data bank using FATCAT (71) revealed that this domain of AlgX is similar to other known and putative CBMs, with the highest homology being to (i) the C-terminal galactose-binding-like domain of alkaline serine protease, KP-43, from Bacillus sp. KSM-KP43 (PDB code 1WMD; RMSD 3.06 Å over 99 equivalent Cα positions), (ii) CBM29-2 of the non-catalytic protein 1 from Piromyces equi (PDB code 1GWL; RMSD 3.14 Å over 97 equivalent Cα positions), and (iii) the C-terminal β-sandwich domain of Thermatoga maritima β-fructosidase (PDB code 1UYP; RMSD 3.13 Å over 92 equivalent Cα positions). Sequence alignments of P. aeruginosa AlgX with homologues from other Pseudomonas species (Pseudomonas syringae, P. fluorescens, P. putida, Pseudomonas brassicacaerum, P. mendocina, P. alkylphenolia, P. entomophila) and the alginate-producing bacterium Azotobacter vinelandii reveal that conservation across the CBM is high, with 18% identity and 57% similarity across all nine species (Fig. 2, B and C). Because alginate is a linear polysaccharide, we predict that this domain will be a Type B CBM. Characteristic features of this type of CBM include the presence of aromatic amino acids and conserved polar residues located in a groove on the surface of the protein that form a distinctive “pinch point,” through which the polysaccharide chain passes (69, 70). The aromatic residues have been shown to stack with the rings of the sugar, thereby stabilizing the polysaccharide-protein complex, and are important in determining specificity for particular polysaccharides. The polar residues form direct hydrogen bonds with the polymer chain to increase the stability of the complex (69, 70). The pinch point of the AlgX CBM is formed by several residues, primarily a surface-exposed tryptophan, Trp-400 (Fig. 3, B and C), which is 100% conserved across the aligned Pseudomonas spp. and A. vinelandii (data not shown). Within the pinch point, there is a surface-exposed polar residue, Thr-398, which is highly conserved among alginate-producing species (Fig. 3, B and C). There are also two positive residues that may aid in binding alginate, Arg-406 and Arg-364, which are located beside Trp-400 and on the opposite side of the groove, respectively (Fig. 3C). Arg-406 is conserved in charge, with all aligned alginate-producing species containing either an arginine or a histidine at this position (Fig. 3, B and C). Arg-364 is less conserved, but in all aligned species other than A. vinelandii, a basic or polar residue is found at this position (Fig. 3, B and C). Two lysine residues, Lys-396 and Lys-410, are positioned just outside the pinch point (Fig. 3, B and C). It is postulated that these positively charged residues could be important for directing the polymer along the face of the CBM toward the N-terminal hydrolase domain. Lys-410 is conserved in all aligned species except A. vinelandii, and Lys-396 is conserved in charge except in A. vinelandii and P. mendocina. Superposition of the CBM domain with P. equi CBM29-2 in complex with mannohexaose supports the hypothesis that this domain of AlgX may bind alginate because the superposed mannohexaose passes through the AlgX pinch point and along the conserved path without the necessity for structural rearrangements (Fig. 3, B and C).
FIGURE 3.

C-terminal domain of AlgX is a carbohydrate-binding module. Shown are schematic representations of the CBM, in red, showing the two antiparallel β-sheets organized into a β-jelly roll fold (A) and the superposition of the CBM (red) and P. equi CBM29-2 (PDB code 1GWK; gray) (B) in complex with mannohexaose. The Trp-400, Thr-398, Arg-364, and Arg-406 residues that comprise the substrate recognition pinch point are shown in yellow. Lys-396 and Lys-410, located outside the pinch point and postulated to play a role in directing the alginate polymer across the protein, are colored orange. C, surface representation of the CBM of AlgX showing the location of the superposed mannohexaose from P. equi CBM29-2. The oligosaccharide passes through the pinch point on the surface of the CBM.
N-terminal Domain Has an SGNH Hydrolase-like Core
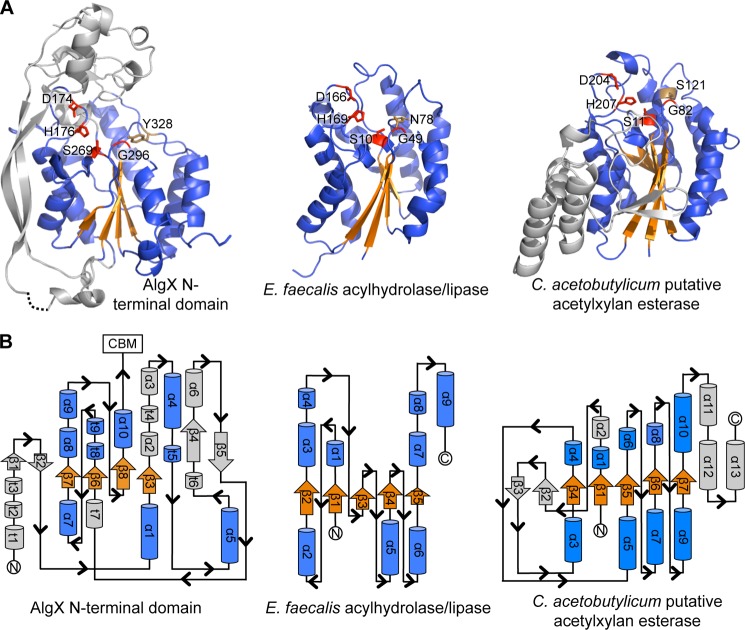
The N-terminal hydrolase domain of AlgX (residues 1–347) has α/β/α topology. Structural homology searches using FATCAT (71) revealed that the core of the domain shares features with proteins from the SGNH (GDSL) hydrolase superfamily (Fig. 4A), including (i) Enterococcus faecalis acyl hydrolase/lipase (PDB code 1YZF; RMSD 3.11 Å over 127 equivalent Cα positions), (ii) an isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae (72) (PDB ID 3MIL, RMSD 3.05 Å over 100 equivalent Cα positions), and (iii) platelet-activating factor acetylhydrolase Ib γ subunit (73) (PDB ID 1ES9; RMSD 3.15 Å over 88 equivalent Cα positions). Structurally, the core of the N-terminal AlgX domain has a number of differences relative to the canonical SGNH hydrolase fold, as exemplified by the acyl hydrolase/lipase from E. faecalis (PDB code 1YZF) (Fig. 4). Typically, SGNH hydrolases contain a five-stranded parallel β-sheet, surrounded by seven α-helices. Based on secondary structure assignment (46, 47), AlgX has the seven core α-helices seen in members of the SGNH fold family, helices α2, α5, α6, α8, α9, α10, and α11, but contains only the first four strands of the five-stranded parallel β-sheet. The fifth strand is replaced by a one-residue β-bridge (Trp-153). Although AlgX structurally resembles SGNH hydrolases, examination of the topology reveals that the arrangement of the secondary structural elements found in the N-terminal domain is different from that of members of the SGNH hydrolase superfamily (Fig. 4B). In particular, the four core β-strands of AlgX are in a different order when compared with the arrangement (from the N to the C terminus) of canonical SGNH hydrolases (Fig. 4B). In addition to the variation in the topology of the α/β/α domain, AlgX also contains a number of insertions not found in a canonical SGNH hydrolase fold. These include (i) two long, antiparallel β-strands on the side of the core hydrolase domain, strands β4 and β5, and (ii) a “cap” over the central part of the core domain formed by two short antiparallel β-strands, β1 and β2, and a series of short α-helices, helices α1, α3, α4, and α7. The structure also contains 10 310 helices, labeled t1–t10 (Fig. 4B).
FIGURE 4.
The N-terminal domain of AlgX has an SGNH-like hydrolase fold. A, the N-terminal SGNH hydrolase-like domain of AlgX and two representative members of the SGNH hydrolase superfamily: E. faecalis acylhydrolase/lipase (PDB code 1YZF) and the putative acetylxylan esterase from Clostridium acetobutylicum (PDB code 1ZMB). In all three proteins, the Ser-His-Asp triad and signature Gly residues are structurally conserved and shown in red in stick representations. The conserved SGNH Asn residue and the equivalent Tyr in AlgX are shown in brown in stick representations. The parallel β-sheet and surrounding α-helices of the core SGNH hydrolase domains and equivalent features in AlgX are colored in orange and blue, respectively. The secondary structural elements of AlgX and the C. acetobutylicum protein that are not part of the core SGNH hydrolase fold are colored in gray. B, topology of the N-terminal domain of AlgX and its comparison with the typical topologies found in the SGNH hydrolases, E. faecalis acylhydrolase/lipase and C. acetobutylicum putative acetylxylan esterase. The topology diagrams are colored as in A.
Typically, SGNH hydrolases contain a Ser-His-Asp/Glu catalytic triad, a conserved glycine, and a conserved asparagine residue located in the core central β-sheet (74). The Ser-Gly-Asn-His residues, for which this family is named, are found in four conserved blocks, blocks I, II, III, and V, respectively (Table 2). In block I, the serine residue is part of the GDSL consensus sequence (letters in boldface type are expected to be 100% conserved) and serves not only as the nucleophile but also as a proton donor in the oxyanion hole. The glycine and asparagine residues in blocks II (NXSXXGXT) and III (GXND), respectively, serve as proton donors in the oxyanion hole, whereas block V (DXXHP) contains a highly conserved histidine that acts as the catalytic base, deprotonating the hydroxyl group of the serine to make it more nucleophilic. Block V also typically contains an aspartate or glutamate three residues upstream of the histidine, which is the third member of the catalytic triad and is positioned to form a salt bridge with the imidazolium ring of the conserved histidine. Although not found in any of the predicted blocks, the SGNH-associated catalytic triad and the key Gly residue are structurally conserved in AlgX (Table 2). Superposition of Asp-174, His-176, Ser-269, and Gly-296 residues in AlgX with the catalytic triad and the Gly residue of the oxyanion hole in SGNH superfamily member E. faecalis acylhydrolase/lipase was performed (Fig. 5A). The superposition (RMSD of 0.60 Å across all equivalent Cα) confirms the presence and orientation of a Ser-His-Asp triad in AlgX, composed of residues Ser-269, His-176, and Asp-174, respectively (Fig. 5, A and B). The spatial positioning of the catalytic center is similar to that of serine esterases (75) and serine proteases (76). The superposition also indicates that Gly-296 is positioned and oriented correctly to participate in oxyanion hole formation in AlgX. Residues Tyr-328 (AlgX) and Asn-78 (E. faecalis acylhydrolase/lipase) were not included in the superposition, but the alignment suggests that they occupy the same relative positions in the respective proteins (Fig. 5A). Thus, it appears that the conserved asparagine found in block III has been replaced by Tyr-328 in AlgX. AlgX is not the only protein with an SGNH or SGNH-like hydrolase fold that lacks the conserved asparagine because the putative acetylxylan esterase from C. acetyobutylicum has a Ser residue in this position (PDB 1ZMB) (Fig. 4A). In addition, whereas the conserved aspartic acid in SGNH hydrolases is typically found three residues upstream of the catalytic histidine, in AlgX, Asp-174 and His-176 are only two residues apart. This is not unprecedented because the N-terminal SGNH hydrolase domain of a cellulase from Cellvibrio japonicus (CtCel5C-CE2, PDB ID 2WAA) has the same Asp-X-His arrangement (77). Like AlgX, CtCel5C-CE2 is also a two-domain protein with an N-terminal cellulase domain and C-terminal CBM with non-catalytic cellulose-binding function.
TABLE 2.
SGNH hydrolase conserved blocks
The conserved active site SGNH hydrolase residues in E. faecalis acylhydrolase/lipase are found in the canonical blocks. The blocks in AlgX are not conserved, and the active site residues are found in a different order in the protein. Residues in boldface type are 100% conserved in SGNH hydrolases. Underlined residues are those found in the canonical block motifs or the equivalent in AlgX.
| Classic SGNH hydrolase block | Classic SGNH hydrolase motifs | E. faecalis motifs | AlgX motifs | AlgX signature residues |
|---|---|---|---|---|
| I | GDSL | GDSI | GTSN | Ser-269 |
| II | NXXSXXGXT | NAGMPGDT | NAVSGGGF | Gly-296 |
| III | GXND | GAND | THYD | Tyr-328 |
| V | DXXHP | DGLHF | DHHW | Asp-174, His-176 |
FIGURE 5.
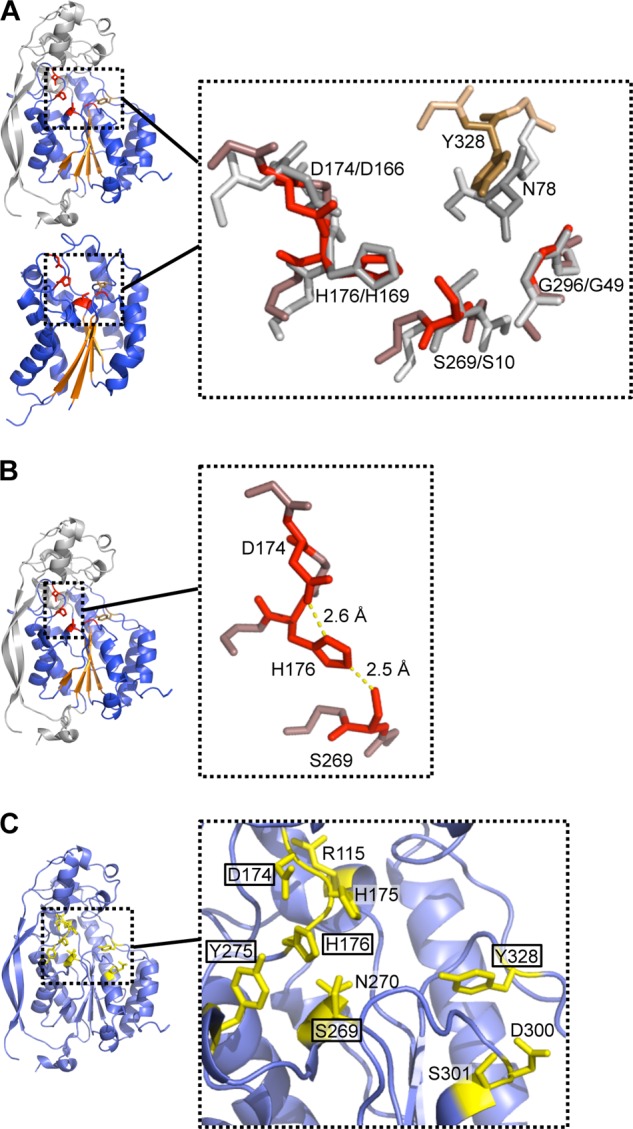
Architecture of the active site. A, superposition of the Asp-His-Ser triad and conserved Gly of AlgX (shown in red) and E. faecalis acylhydrolase/lipase (in gray). Asn and Tyr residues of E. faecalis acylhydrolase/lipase and AlgX, not used in the superposition, are shown in dark gray and brown, respectively. Main chain atoms surrounding the residues of interest are colored in pale gray and in light red/brown for the E. faecalis acylhydrolase/lipase and AlgX structures, respectively. B, the bonds formed between the catalytic triad residues are shown in yellow. C, 10 residues of AlgX were independently mutated on the chromosome to alanine to probe their role in O-acetylation of alginate. These residues, depicted in stick representations and colored in yellow, were selected for mutation based upon their location within the putative active site. The residues found to be important for the acetylation of alginate are boxed.
Sequence alignments of the N-terminal domain of P. aeruginosa PAO1 AlgX with seven other Pseudomonas spp. (P. syringae, P. fluorescens, P. putida, P. brassicacaerum, P. mendocina, P. alkylphenolia, and P. entomophila) and A. vinelandii reveal that the residues in this domain share 33% identity and 65% similarity (Fig. 2C). The catalytic triad and Gly residues as well as the tyrosine that replaces the asparagine are 100% conserved across these species. The AlgX N-terminal domain also has 30% sequence identity and ∼69% sequence similarity to AlgJ, which has previously been shown to play a role in the O-acetylation of the alginate polymer (24). The SGNH-associated residues identified in AlgX are all conserved in P. aeruginosa AlgJ.
The Ser-His-Asp Triad Is Essential for Acetylation of Alginate
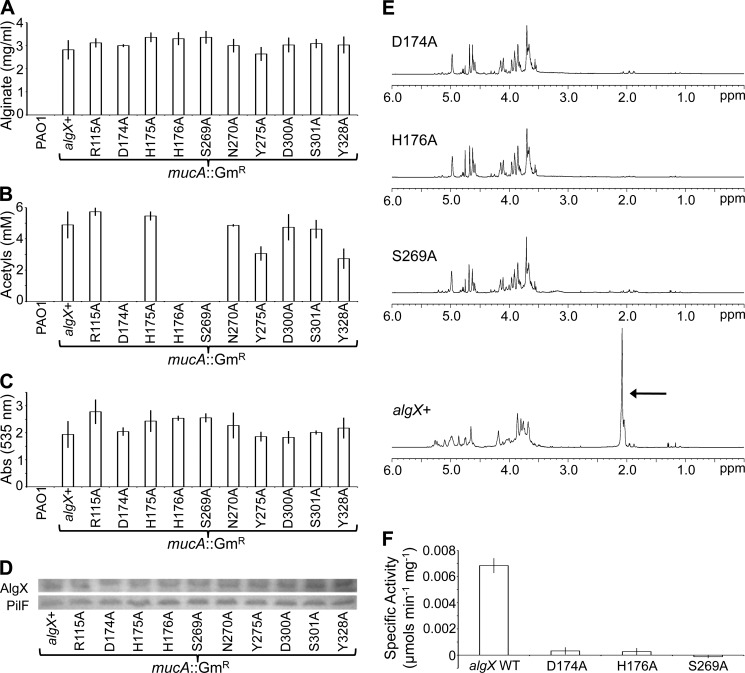
The presence of SGNH-associated residues in AlgX and its sequence similarity to the putative alginate acetyltransferase AlgJ (24, 29) suggested that AlgX may have a role in alginate acetylation. To probe the in vivo function of AlgX, a series of alanine chromosomal point mutant strains were constructed in P. aeruginosa PAO1 by replacing a marked chromosomal copy of algX with plasmid-borne algX mutant alleles through homologous recombination. Alginate production in these strains was then constitutively turned on by disruption of mucA. mucA encodes an anti-σ factor, and its disruption leads to activation of the algD operon and alginate production (33). In addition to substitutions in the Ser-His-Asp triad, mutations were also made in seven residues in close proximity to the putative active site that could play a role in alginate binding (Fig. 5C). These individual mutant AlgX strains were then tested for their ability to produce polymeric alginate. The level of AlgX expression was also assessed to ensure that any variations in alginate content were not due to reduced levels of protein production. All of the mutant strains produced alginate at a level equivalent to that of the P. aeruginosa PAO1 mucA::GmR algX+ strain and had comparable levels of AlgX expression (Fig. 6, A and D). Because all mutant strains expressed alginate, the polymer was further analyzed to determine whether its acetylation state had been altered. Using a spectrophotometric assay and NMR analysis, it was found that the alginate produced by each of the S269A, H176A, and D174A AlgX mutant strains was not acetylated (Fig. 6, B and E). When a plasmid expressing algX was transformed into these three mutants, the acetylation of alginate was restored (data not shown). Alginate acetylation was observed to be reduced by ∼50 and ∼40% for the Y328A and Y275A mutant strains, respectively (Fig. 6B). Mutation of residues Arg-115, His-175, Asn-270, Asp-300, and Ser-301 had no effect on either alginate production or its acetylation (Fig. 6, A and B). To determine if the lack of acetylation affected the second postpolymerization modification that alginate undergoes, epimerization, the alginate polymers produced were deacetylated and incubated with alginate G-lyase. This enzyme is only able to cleave the alginate polymer if mannuronate residues within the polymer have been epimerized to guluronate. Analysis of the hydrolyzed alginate revealed that the level of epimerization in the mutant strains was equivalent to that observed in the wild-type algX+ strain, regardless of whether the alginate was acetylated in vivo or not (Fig. 6C). This suggests that the lack of acetylation in the S269A, H176A, and D174A mutant strains does not affect epimerization of the polymer.
FIGURE 6.
Role of AlgX in alginate O-acetylation. Analysis of the PAO1 mucA::GmR mutant strains for alginate production (A), O-acetylation of the produced alginate (B), and epimerization (C). The absorbance at 535 nm is directly related to the guluronate content of alginate. D, algX was expressed at equivalent levels in all of the mutants, as indicated by Western blotting analysis. PilF, from the Type IV pilus system of P. aeruginosa, was used as a loading control. E, NMR spectra of the alginate produced by the wild-type algX+ strain and D174A, H176A, and S269A variants. The peak at ∼2.1 ppm in the algX+ spectrum indicates the presence of acetyl groups on the alginate. F, in vitro activity assay showing the specific activities of the recombinant AlgX protein and the D174A, H176A, and S269A protein variants.
Acetyltransferase activity has not yet been demonstrated in vitro. However, acetylesterase activity (Fig. 7, A and B) can be assayed using a pseudosubstrate, 3-carboxyumbelliferyl acetate. Using a fluorometric assay, the ability of the wild-type AlgX and D174A, H176A, and S269A AlgX mutant proteins to remove the acetate group from this substrate was assessed. The specific activity of wild-type AlgX was found to be low (0.0068 ± 0.00057 μmol min−1 mg−1) (Fig. 6F), although this was not unexpected because 3-carboxyumbelliferyl acetate has been substituted for the acetate donor that would be used in vivo, the identity of which is currently unknown. Furthermore, the inability to detect acetate release when AlgX was incubated with alginate and the low specific activity suggest that AlgX is not an acetylesterase because much higher in vitro activity would have been expected, as has been observed for other esterases assayed with similar, commercially available analogues (78). The acetylesterase activity of each of the three mutant proteins was found to be close to zero. The D174A and H176A mutants had specific activities of 0.00032 ± 0.00028 μmol min−1 mg−1 and 0.00027 ± 0.00027 μmol min−1 mg−1, respectively, whereas the activity was completely abolished in the S269A mutant (Fig. 6F). The catalytic activity of AlgX followed Michaelis-Menten kinetics (data not shown). CD spectroscopy indicates that the differences in catalytic activity observed are not due to structural changes in the protein variants and that the mutant proteins are folded (data not shown). These data suggest that each of these residues is required for in vitro removal of an acetate group from a donor. In addition, the results corroborate our in vivo data, which indicate that the Ser-His-Asp catalytic triad is required for acetylation of alginate and add weight to the suggestion that AlgX is an acetyltransferase.
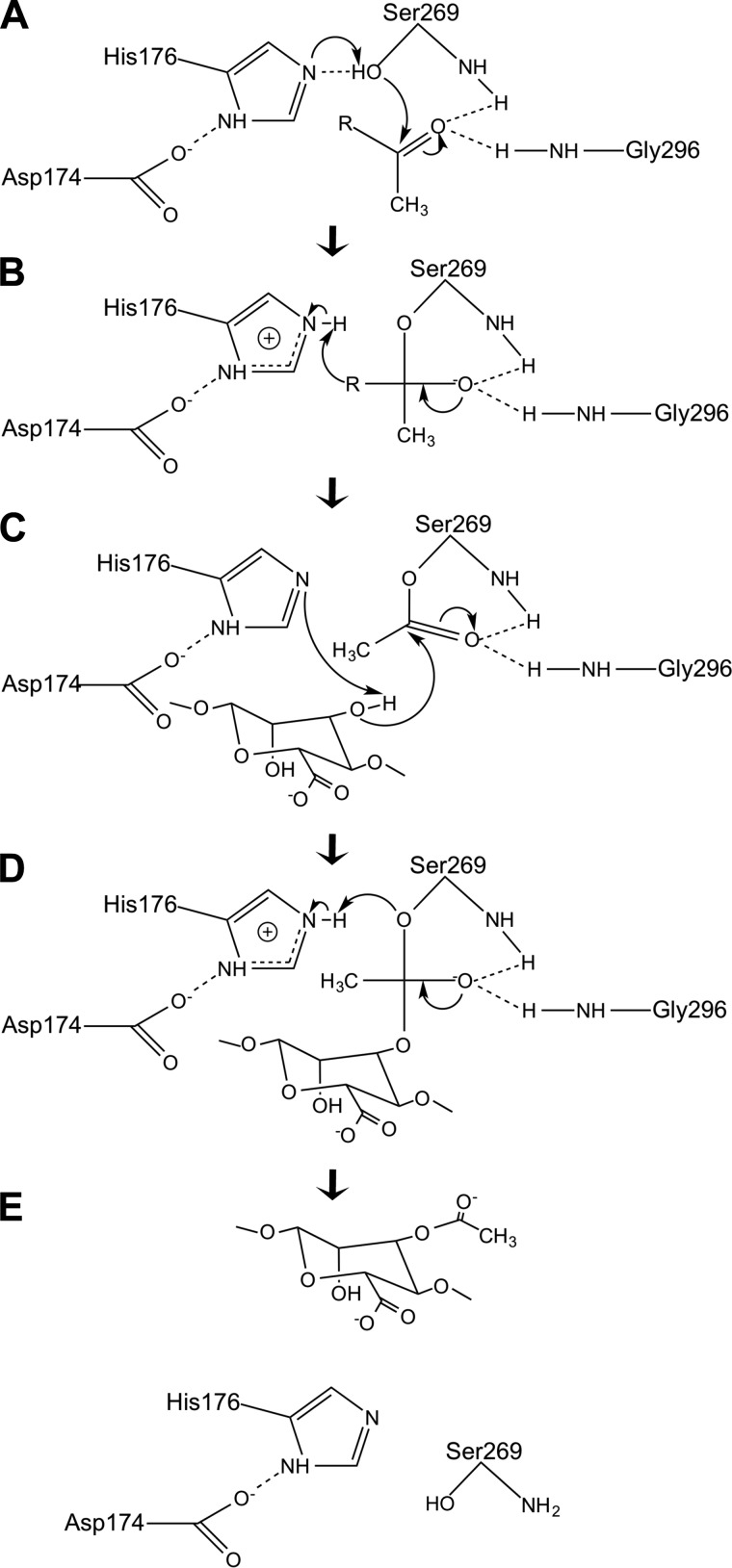
FIGURE 7.
Proposed mechanism of action of AlgX. The Ser-269 nucleophile attacks the carbonyl carbon of an ester-linked acetate group on the substrate acetyl donor (A), leading to the formation of a tetrahedral intermediate (B). His-176 abstracts the hydroxyl proton of a mannuronate residue, and the nucleophilic hydroxyl oxygen of the mannuronate residue attacks the carbonyl carbon, forming an acyl-enzyme intermediate (C). This may involve the C2 or C3 hydroxyl group on the mannuronate residue. The carbonyl double bond is reformed, releasing the acetyl group from the donor (D), and the residues revert to their resting state (E). During the reaction, the amide nitrogens of Ser-269 and Gly-296 stabilize the oxyanion of the tetrahedral intermediate. His-176 acts as a base, increasing the nucleophilic properties of Ser-269, and Asp-174 neutralizes the charge that develops on His-176 in the course of the reaction. R represents the, as yet unidentified, acetyl donor.
DISCUSSION
In this study, we have presented the crystal structure of AlgX and have demonstrated that this protein plays a role in the O-acetylation of alginate in vivo and that a Ser-His-Asp triad governs this activity. Our in vitro enzymatic analyses also support the hypothesis that AlgX is catalytically active and functions as an acetyltransferase. AlgX contains two domains, an N-terminal SGNH hydrolase-like domain and a C-terminal carbohydrate-binding module. This combination is not unexpected because CBMs are commonly appended to enzymatic domains, including SGNH hydrolases, where they function to bring the relevant polysaccharide into close proximity to the active site (68, 79). The CBM domain of AlgX does not share sequence homology with any of the 67 CBM families currently assigned in the CAZy database, but it does have structural homology with other CBMs. Superposition of the CBM of AlgX and a cellulose-binding domain-mannohexaose complex indicates that the binding region of the AlgX CBM comprises a pinch point containing a conserved tryptophan, a threonine, and two arginine residues and that the proposed path for polymer binding includes two conserved lysines (Fig. 3, B and C). Crystal contacts between Trp-400 and Leu-214 of neighboring molecules occur near the pinch point region of the CBM. Because CBMs are often appended to catalytic domains to enhance substrate binding, our efforts to co-crystallize the protein with the substrate may have been hindered by these occluding crystal contacts. However, the arrangement of polar, positively charged, and aromatic residues at the pinch point of the CBM is consistent with the hypothesis that this domain binds a single chain of alginate. Furthermore, the surface potential of AlgX is such that the negatively charged alginate chain could conceivably bind along a readily apparent path that joins the conserved residues of the pinch point in the CBM to a positively charged region on the surface of the N-terminal domain, which is in close proximity to the active site residues (Fig. 2).
The N-terminal domain of AlgX has an SGNH hydrolase-like fold with some additional structural features. Although the importance of these additional features is not known, it is believed that they are relevant for the function of AlgX as part of the alginate biosynthetic complex, as potential binding sites for the known interaction partners AlgK and MucD (30, 31) or other members of the acetylation machinery, AlgI/J/F. Members of the SGNH hydrolase superfamily are primarily esterases and lipases, although enzymes with transferase activity and dual activity enzymes have also been identified (74, 80–85). The canonical description of the SGNH hydrolase fold includes the presence of particular residues, Ser, Gly, Asn, His, and Asp/Glu, in conserved blocks (74). However, the superfamily now encompasses members with atypical or missing blocks, suggesting that sequence divergence has occurred within this superfamily, whereas the tertiary structure has been closely maintained. AlgX contains the Ser, His, Asp, and Gly residues orientated and in the same tertiary positions as the equivalent residues in canonical SGNH hydrolases. However, these residues in AlgX do not follow the primary sequence conservation of the consensus blocks. The increasing number of proteins with SGNH hydrolase-like tertiary architecture that lack some of the canonical residues suggests that the definition of the SGNH hydrolase superfamily may need to be broadened or that subgroups within the superfamily need to be defined based on activity, substrate preference, and structure of the protein.
AlgX residues Asp-174, His-176, and Ser-269 comprise the catalytic triad common to SGNH hydrolases, and we propose that these three residues mediate transfer of an acetyl group to alginate (Figs. 7 and 8). Ser-269 is believed to have dual functionality, with a direct role in the catalytic mechanism and also acting, with Gly-296, to form an oxyanion hole. The donated acetyl group sits in the oxyanion hole, and the Ser and Gly residues act to stabilize the negative charges on the acetyl oxyanion during the formation of the tetrahedral intermediates in the reaction (Fig. 7). In the majority of SGNH hydrolases, a conserved asparagine is also important for oxyanion hole stability, but AlgX has Tyr-328 located at this position instead. Examination of the structure suggests that it is perhaps more likely that Tyr-328 as well as Tyr-275, which when mutated to alanine reduce acetylation by 40 and 50%, respectively, are important for the correct positioning of the alginate polymer across the putative active site. These residues are located on either side of the catalytic triad, and aromatic residues frequently bind to sugar molecules via hydrophobic interactions (70). If Tyr-328 is involved in substrate binding and not oxyanion stabilization, this suggests that AlgX may use a chymotrypsin/trypsin-like serine protease mechanism in which the oxyanion hole is formed by just the serine and glycine residues (76, 86) (Fig. 7).
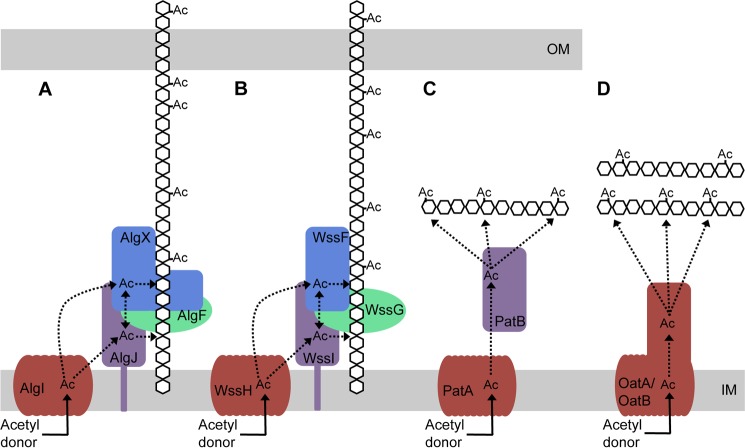
FIGURE 8.
Model for O-acetylation of alginate and other bacterial polysaccharides. A, AlgI receives an acetyl group from a cytoplasmic acetyl donor. From AlgI, the acetyl group is transferred to AlgJ and/or AlgX and then on to the alginate polymer. AlgJ and AlgX may be able to pass the acetyl group between themselves. The role of AlgF is currently unknown. B, the four proteins of the P. fluorescens Wss system acetylate cellulose and may act in a similar manner to the four Alg proteins. C, proposed mechanism of peptidoglycan acetylation in Gram-negative bacteria as presented by Moynihan and Clarke (81). This mechanism requires two proteins, an AlgI homologue, PatA, which transports the acetate across the cytoplasmic membrane to the periplasm, and an AlgJ homologue, PatB, which transfers the acetate to the sugar. D, in Gram-positive bacteria, the N-acetyl muramic acid residues of the peptidoglycan are O-acetylated by the two-domain protein, OatA (80, 94). The N-terminal domain of OatA is homologous to AlgI, and the C-terminal domain is homologous to AlgJ (80). More recently, a second protein, OatB, has been identified in Lactobacillus plantarum, which O-acetylates N-acetylglucosamine peptidoglycan residues (80). OatB also has two domains homologous to AlgI and AlgJ. Ac, acetate. Dotted lines, potential movement of the acetate after transport across the inner membrane. Proteins with similar predicted structure and function are colored the same.
AlgX is the fourth protein in the algD operon that has now been implicated in alginate acetylation as previous studies have suggested that AlgI, AlgJ, and AlgF are all essential for this process to occur (23–25, 87). AlgI and AlgJ are proposed to be a membrane-bound MBOAT family O-acetyltransferase and a periplasmic O-acetyltransferase, respectively (25, 88). The function of AlgF is currently not known. Franklin and Ohman (24) showed that AlgJ and AlgX share ∼69% similarity and 30% identity. It is predicted that AlgJ is also an SGNH hydrolase-like protein but differs from AlgX in that AlgJ does not have a CBM domain and is anchored to the inner membrane by a single N-terminal transmembrane helix (25). It is not apparent why four proteins are required for alginate acetylation and in particular why two periplasmic acetyltransferases would be needed. Homologues of AlgI and AlgJ have been identified in a number of different bacteria (88), including the peptidoglycan acetylation proteins PatA and PatB in Neisseria gonorrhoeae (81) and the bidomain proteins OatA and OatB in Gram-positive bacteria, each of which have domains resembling both AlgI and AlgJ (80). Homologues of AlgF and AlgX are less common, but the acetylation of cellulose during the formation of the P. fluorescens pellicle also requires four proteins that are located together in the wss operon (12, 89). WssGHI are homologous to AlgFIJ, with amino acid sequence identities of 24, 46, and 33%, respectively. WssF shares no sequence homology with any of the Alg proteins but is structurally predicted to belong to the SGNH hydrolase superfamily and has the three residues comprising the catalytic triad as well as the Gly residue required for oxyanion hole formation. WssF may therefore have a role similar to that of AlgX (12). Interestingly, like AlgX, examination of the Phyre2 model of WssF (90) reveals that the conserved Asn residue that is found in most SGNH hydrolases is missing and is replaced with a Thr, suggesting that the oxyanion hole of WssF is formed by the Ser and Gly residues only. This provides another example of an putative SGNH hydrolase that may have a role in acetylation and that uses the chymotrypsin/trypsin-like oxyanion stabilization mechanism.
Phenotypic characterization of site-specific mutants of both AlgX and AlgJ suggest that each protein plays a role in the acetylation of the nascent alginate polymer (88). AlgX and AlgJ are predicted to be structurally similar to PatB and the SGNH hydrolase domains of OatA and OatB (80, 81). Although PatB acetyltransferase activity has been demonstrated in vivo (81), like us, the authors of this study were unable to develop an in vitro assay to demonstrate full transferase activity. Instead, comparable with our use of 3-carboxyumbelliferyl acetate, Moynihan and Clarke (81) used p-nitrophenyl acetate to characterize the first part of the reaction in vitro and, comparable with our results, found that PatB has only weak esterase activity. OatA and OatB acetyltransferase activity has also been shown in vivo (80). The functional equivalency and similarity of the predicted structures of PatB, OatA, and OatB reinforce our hypothesis that AlgX and AlgJ are also acetyltransferases. The resemblance of alginate acetylation to the OatA/B and PatA-PatB systems supports the theory that the integral inner membrane protein AlgI receives an acetyl group from a cytoplasmic acetyl donor. This is then passed to AlgJ and/or AlgX. These proteins may pass the acetyl group between themselves or acetylate the nascent polymer. The role of AlgF has not been elucidated (Fig. 8) but is known through mutational analysis to be an important part of the acetylation process (23, 25). The alginate model can also be used to illustrate a potential method of cellulose acetylation by WssFGHI, in which the acetyl group is received from an acetyl donor and transported to the periplasm by WssH. WssI and/or WssF receive the acetyl group, and one or both of the proteins transfer it to the growing cellulose chain (Fig. 8). Like AlgF, no role has been attributed to WssG in this model.
We have described the structure of AlgX, a two-domain protein that we propose binds to alginate via its C-terminal CBM and is then able to add acetyl groups to the nascent polymer through the catalytic triad in the N-terminal SGNH hydrolase-like domain. This is the first structure of a probable polysaccharide acetyltransferase with SGNH hydrolase-like architecture, which could serve as a prototype for other proteins that have been shown to be involved in the acetylation of peptidoglycan (12, 80, 81). The hypothesis that AlgX is an acetyltransferase and the acetylation model presented provide avenues for future studies aimed at better understanding this key polysaccharide modification.
Acknowledgments
We thank Yura Lobsanov, Francis Wolfram, Dustin J. Little, Jason Koo, John C. C. Whitney, and G. David Smith for helpful discussions and Patrick Yip for technical assistance. National Synchrotron Light Source beamline X29 is supported by the United States Department of Energy Office of Biological and Environmental Research and the National Institutes of Health National Center for Research Resources.
This work was supported, in whole or in part, by Canadian Institutes of Health Research (CIHR) Grant 13337 (to P. L. H.). This work was also supported by National Institutes of Health, NIAID Grant AI-19146 (to D. E. O.), National Institutes of Health, NIGMS Grant GM081419 (to P. A. T.), Veterans Administration Medical Research Grant IO1BX000477 (to D. E. O.), and Netherlands Organization for Scientific Research Grant NWO-vidi (to J. D. C. C.).

This article contains supplemental Table S1.
The atomic coordinates and structure factors (code 4KNC) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CBM
- carbohydrate-binding module
- SeMet
- selenomethionine
- mSF
- mini-stabilization fragment
- SAD
- single-wavelength anomalous dispersion
- RMSD
- root mean square deviation
- PDB
- Protein Data Bank.
REFERENCES
- 1. Cuthbertson L., Mainprize I. L., Naismith J. H., Whitfield C. (2009) Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol. Mol. Biol. Rev. 73, 155–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flemming H. C., Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 3. Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010) Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35, 322–332 [DOI] [PubMed] [Google Scholar]
- 4. Ghafoor A., Hay I. D., Rehm B. H. (2011) Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77, 5238–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vuong C., Kocianova S., Voyich J. M., Yao Y., Fischer E. R., DeLeo F. R., Otto M. (2004) A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279, 54881–54886 [DOI] [PubMed] [Google Scholar]
- 6. Das T., Sharma P. K., Busscher H. J., van der Mei H. C., Krom B. P. (2010) Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl. Environ. Microbiol. 76, 3405–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerca N., Jefferson K. K., Maira-Litrán T., Pier D. B., Kelly-Quintos C., Goldmann D. A., Azeredo J., Pier G. B. (2007) Molecular basis for preferential protective efficacy of antibodies directed to the poorly acetylated form of staphylococcal poly-N-acetyl-β-(1–6)-glucosamine. Infect. Immun. 75, 3406–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gjermansen M., Nilsson M., Yang L., Tolker-Nielsen T. (2010) Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Genetic elements and molecular mechanisms. Mol. Microbiol. 75, 815–826 [DOI] [PubMed] [Google Scholar]
- 9. Itoh Y., Rice J. D., Goller C., Pannuri A., Taylor J., Meisner J., Beveridge T. J., Preston J. F., 3rd, Romeo T. (2008) Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 190, 3670–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nivens D. E., Ohman D. E., Williams J., Franklin M. J. (2001) Role of alginate and its O-acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183, 1047–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sauer K., Cullen M. C., Rickard A. H., Zeef L. A., Davies D. G., Gilbert P. (2004) Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186, 7312–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spiers A. J., Bohannon J., Gehrig S. M., Rainey P. B. (2003) Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50, 15–27 [DOI] [PubMed] [Google Scholar]
- 13. Hentzer M., Teitzel G. M., Balzer G. J., Heydorn A., Molin S., Givskov M., Parsek M. R. (2001) Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Z., Kosorok M. R., Farrell P. M., Laxova A., West S. E., Green C. G., Collins J., Rock M. J., Splaingard M. L. (2005) Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA 293, 581–588 [DOI] [PubMed] [Google Scholar]
- 15. Linker A., Jones R. S. (1966) A new polysaccharide resembling alginic acid isolated from pseudomonads. J. Biol. Chem. 241, 3845–3851 [PubMed] [Google Scholar]
- 16. Govan J. R., Deretic V. (1996) Microbial pathogenesis in cystic fibrosis. Mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chitnis C. E., Ohman D. E. (1993) Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8, 583–593 [DOI] [PubMed] [Google Scholar]
- 18. Darzins A., Wang S. K., Vanags R. I., Chakrabarty A. M. (1985) Clustering of mutations affecting alginic acid biosynthesis in mucoid Pseudomonas aeruginosa. J. Bacteriol. 164, 516–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franklin M. J., Nivens D. E., Weadge J. T., Howell P. L. (2011) Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Franklin M. J., Chitnis C. E., Gacesa P., Sonesson A., White D. C., Ohman D. E. (1994) Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176, 1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain S., Franklin M. J., Ertesvåg H., Valla S., Ohman D. E. (2003) The dual roles of AlgG in C-5-epimerization and secretion of alginate polymers in Pseudomonas aeruginosa. Mol. Microbiol. 47, 1123–1133 [DOI] [PubMed] [Google Scholar]
- 22. Gacesa P. (1988) Alginates. Carbohydr. Polym. 8, 161–182 [Google Scholar]
- 23. Franklin M. J., Ohman D. E. (1993) Identification of algF in the alginate biosynthetic gene cluster of Pseudomonas aeruginosa which is required for alginate acetylation. J. Bacteriol. 175, 5057–5065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Franklin M. J., Ohman D. E. (1996) Identification of algI and algJ in the Pseudomonas aeruginosa alginate biosynthetic gene cluster which are required for alginate O-acetylation. J. Bacteriol. 178, 2186–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Franklin M. J., Ohman D. E. (2002) Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O-acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184, 3000–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pier G. B., Coleman F., Grout M., Franklin M., Ohman D. E. (2001) Role of alginate O-acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69, 1895–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tielen P., Strathmann M., Jaeger K.-E., Flemming H.-C., Wingender J. (2005) Alginate acetylation influences initial surface colonization by mucoid Pseudomonas aeruginosa. Microbiol. Res. 160, 165–176 [DOI] [PubMed] [Google Scholar]
- 28. Evans L. R., Linker A. (1973) Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 116, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robles-Price A., Wong T. Y., Sletta H., Valla S., Schiller N. L. (2004) AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186, 7369–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hay I. D., Schmidt O., Filitcheva J., Rehm B. H. (2012) Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl. Microbiol. Biotechnol. 93, 215–227 [DOI] [PubMed] [Google Scholar]
- 31. Gutsche J., Remminghorst U., Rehm B. H. (2006) Biochemical analysis of alginate biosynthesis protein AlgX from Pseudomonas aeruginosa. Purification of an AlgX-MucD (AlgY) protein complex. Biochimie 88, 245–251 [DOI] [PubMed] [Google Scholar]
- 32. Damron F. H., Yu H. D. (2011) Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J. Bacteriol. 193, 286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wood L. F., Ohman D. E. (2006) Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J. Bacteriol. 188, 3134–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weadge J. T., Yip P. P., Robinson H., Arnett K., Tipton P. A., Howell P. L. (2010) Expression, purification, crystallization and preliminary x-ray analysis of Pseudomonas aeruginosa AlgX. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 588–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee J. E., Cornell K. A., Riscoe M. K., Howell P. L. (2001) Structure of E. coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases. Structure 9, 941–953 [DOI] [PubMed] [Google Scholar]
- 36. Winsor G. L., Van Rossum T., Lo R., Khaira B., Whiteside M. D., Hancock R. E., Brinkman F. S. (2009) Pseudomonas Genome Database. Facilitating user-friendly, comprehensive comparisons of microbial genomes. Nucleic Acids Res. 37, D483–D488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suh S. J., Silo-Suh L. A., Ohman D. E. (2004) Development of tools for the genetic manipulation of Pseudomonas aeruginosa. J. Microbiol. Methods 58, 203–212 [DOI] [PubMed] [Google Scholar]
- 38. Codée J. D., van den Bos L. J., de Jong A. R., Dinkelaar J., Lodder G., Overkleeft H. S., van der Marel G. A. (2009) The stereodirecting effect of the glycosyl C5-carboxylate ester. Stereoselective synthesis of β-mannuronic acid alginates. J. Org. Chem. 74, 38–47 [DOI] [PubMed] [Google Scholar]
- 39. Pape T., Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 40. Terwilliger T. C., Berendzen J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 43. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 44. Painter J., Merritt E. A. (2006) TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 45. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joosten R. P., te Beek T. A., Krieger E., Hekkelman M. L., Hooft R. W., Schneider R., Sander C., Vriend G. (2011) A series of PDB related databases for everyday needs. Nucleic Acids Res. 39, D411–D419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure. Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 48. Choi K. H., Schweizer H. P. (2005) An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holloway B. W. (1969) Genetics of Pseudomonas. Bacteriol. Rev. 33, 419–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wood L. F., Leech A. J., Ohman D. E. (2006) Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa. Roles of σ (AlgT) and the AlgW and Prc proteases. Mol. Microbiol. 62, 412–426 [DOI] [PubMed] [Google Scholar]
- 51. Chibba A., Poloczek J., Little D. J., Howell P. L., Nitz M. (2012) Synthesis and evaluation of inhibitors of E. coli PgaB, a polysaccharide de-N-acetylase involved in biofilm formation. Org. Biomol. Chem. 10, 7103–7107 [DOI] [PubMed] [Google Scholar]
- 52. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 53. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee. A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 54. Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. (2005) ConSurf 2005. The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pearson W. R. (1996) Effective protein sequence comparison. Methods Enzymol. 266, 227–258 [DOI] [PubMed] [Google Scholar]
- 57. Wilson D., Pethica R., Zhou Y., Talbot C., Vogel C., Madera M., Chothia C., Gough J. (2009) SUPERFAMILY. Sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 37, D380–D386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Park B. H., Karpinets T. V., Syed M. H., Leuze M. R., Uberbacher E. C. (2010) CAZymes Analysis Toolkit (CAT). Web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20, 1574–1584 [DOI] [PubMed] [Google Scholar]
- 59. Knutson C. A., Jeanes A. (1968) A new modification of the carbazole analysis. Application to heteropolysaccharides. Anal. Biochem. 24, 470–481 [DOI] [PubMed] [Google Scholar]
- 60. Hestrin S. (1949) The reaction of acetylcholine and other carboxylic acid derivatives with hydroxylamine, and its analytical application. J. Biol. Chem. 180, 249–261 [PubMed] [Google Scholar]
- 61. Grasdalen H. (1979) A P.M.R. study of the composition and sequence of uronate residues in alginate. Carbohydr. Res. 68, 23–31 [Google Scholar]
- 62. Grasdalen H. (1983) High-field, 1H n.m.r. spectroscopy of alginate: sequential structure and linkage conformations. Carbohydr. Res. 118, 255–260 [Google Scholar]
- 63. Chitnis C. E., Ohman D. E. (1990) Cloning of Pseudomonas aeruginosa algG, which controls alginate structure. J. Bacteriol. 172, 2894–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Douthit S. A., Dlakic M., Ohman D. E., Franklin M. J. (2005) Epimerase active domain of Pseudomonas aeruginosa AlgG, a protein that contains a right-handed β-helix. J. Bacteriol. 187, 4573–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salamitou S., Lemaire M., Fujino T., Ohayon H., Gounon P., Béguin P., Aubert J. P. (1994) Subcellular localization of Clostridium thermocellum ORF3p, a protein carrying a receptor for the docking sequence borne by the catalytic components of the cellulosome. J. Bacteriol. 176, 2828–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koo J., Tammam S., Ku S. Y., Sampaleanu L. M., Burrows L. L., Howell P. L. (2008) PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa Type IV pilus secretin. J. Bacteriol. 190, 6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Høiby N., Krogh Johansen H., Moser C., Song Z., Ciofu O., Kharazmi A. (2001) Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microbes Infect. 3, 23–35 [DOI] [PubMed] [Google Scholar]
- 68. Hashimoto H. (2006) Recent structural studies of carbohydrate-binding modules. Cell. Mol. Life Sci. 63, 2954–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guillén D., Sánchez S., Rodríguez-Sanoja R. (2010) Carbohydrate-binding domains. Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 85, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 70. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ye Y., Godzik A. (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19, ii246–ii255 [DOI] [PubMed] [Google Scholar]
- 72. Ma J., Lu Q., Yuan Y., Ge H., Li K., Zhao W., Gao Y., Niu L., Teng M. (2011) Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity. Proteins 79, 662–668 [DOI] [PubMed] [Google Scholar]
- 73. McMullen T. W., Li J., Sheffield P. J., Aoki J., Martin T. W., Arai H., Inoue K., Derewenda Z. S. (2000) The functional implications of the dimerization of the catalytic subunits of the mammalian brain platelet-activating factor acetylhydrolase (Ib). Protein Eng. 13, 865–871 [DOI] [PubMed] [Google Scholar]
- 74. Akoh C. C., Lee G. C., Liaw Y. C., Huang T. H., Shaw J. F. (2004) GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552 [DOI] [PubMed] [Google Scholar]
- 75. Pfeffer J. M., Weadge J. T., Clarke A. J. (2013) Mechanism of action of Neisseria gonorrhoeae O-acetylpeptidoglycan esterase, an SGNH serine esterase. J. Biol. Chem. 288, 2605–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hedstrom L. (2002) Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 77. Montanier C., Money V. A., Pires V. M., Flint J. E., Pinheiro B. A., Goyal A., Prates J. A., Izumi A., Stålbrand H., Morland C., Cartmell A., Kolenova K., Topakas E., Dodson E. J., Bolam D. N., Davies G. J., Fontes C. M., Gilbert H. J. (2009) The active site of a carbohydrate esterase displays divergent catalytic and noncatalytic binding functions. PLoS Biol. 7, e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Weadge J. T., Clarke A. J. (2006) Identification and characterization of O-acetylpeptidoglycan esterase. A novel enzyme discovered in Neisseria gonorrhoeae. Biochemistry 45, 839–851 [DOI] [PubMed] [Google Scholar]
- 79. Bolam D. N., Ciruela A., McQueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. (1998) Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 331, 775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Bernard E., Rolain T., Courtin P., Guillot A., Langella P., Hols P., Chapot-Chartier M. P. (2011) Characterization of O-acetylation of N-acetylglucosamine. A novel structural variation of bacterial peptidoglycan. J. Biol. Chem. 286, 23950–23958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Moynihan P. J., Clarke A. J. (2010) O-Acetylation of peptidoglycan in Gram-negative bacteria. Identification and characterization of peptidoglycan O-acetyltransferase in Neisseria gonorrhoeae. J. Biol. Chem. 285, 13264–13273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pickens L. B., Sawaya M. R., Rasool H., Pashkov I., Yeates T. O., Tang Y. (2011) Structural and biochemical characterization of the salicylyl-acyltranferase SsfX3 from a tetracycline biosynthetic pathway. J. Biol. Chem. 286, 41539–41551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reina J. J., Guerrero C., Heredia A. (2007) Isolation, characterization, and localization of AgaSGNH cDNA. A new SGNH-motif plant hydrolase specific to Agave americana L. leaf epidermis. J. Exp. Bot. 58, 2717–2731 [DOI] [PubMed] [Google Scholar]
- 84. Robertson D. L., Hilton S., Wong K. R., Koepke A., Buckley J. T. (1994) Influence of active site and tyrosine modification on the secretion and activity of the Aeromonas hydrophila lipase/acyltransferase. J. Biol. Chem. 269, 2146–2150 [PubMed] [Google Scholar]
- 85. Teutschbein J., Gross W., Nimtz M., Milkowski C., Hause B., Strack D. (2010) Identification and localization of a lipase-like acyltransferase in phenylpropanoid metabolism of tomato (Solanum lycopersicum). J. Biol. Chem. 285, 38374–38381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Di Cera E. (2009) Serine proteases. IUBMB Life 61, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shinabarger D., May T. B., Boyd A., Ghosh M., Chakrabarty A. M. (1993) Nucleotide sequence and expression of the Pseudomonas aeruginosa algF gene controlling acetylation of alginate. Mol. Microbiol. 9, 1027–1035 [DOI] [PubMed] [Google Scholar]
- 88. Franklin M. J., Douthit S. A., McClure M. A. (2004) Evidence that the algI/algJ gene cassette, required for O-acetylation of Pseudomonas aeruginosa alginate, evolved by lateral gene transfer. J. Bacteriol. 186, 4759–4773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Winsor G. L., Lam D. K., Fleming L., Lo R., Whiteside M. D., Yu N. Y., Hancock R. E., Brinkman F. S. (2011) Pseudomonas Genome Database. Improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39, D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 91. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems. Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR. Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) PDB2PQR. An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Bera A., Herbert S., Jakob A., Vollmer W., Götz F. (2005) Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787 [DOI] [PubMed] [Google Scholar]
- 95. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]