Background: Ameloblastin plays a key role in the complex biomineralization process that forms tooth enamel, the hardest tissue of the body.
Results: Ameloblastin self-associates into ribbon-like supramolecular structures via a short segment encoded by exon 5.
Conclusion: Ameloblastin self-association may be essential for correct structural organization and mineralization of the enamel in vivo.
Significance: The results provide molecular insight into biology of tooth enamel formation.
Keywords: Extracellular Matrix Proteins, Intrinsically Disordered Proteins, Protein Purification, Protein Self-assembly, Tooth, Ameloblastin, Amelogenin
Abstract
Tooth enamel, the hardest tissue in the body, is formed by the evolutionarily highly conserved biomineralization process that is controlled by extracellular matrix proteins. The intrinsically disordered matrix protein ameloblastin (AMBN) is the most abundant nonamelogenin protein of the developing enamel and a key element for correct enamel formation. AMBN was suggested to be a cell adhesion molecule that regulates proliferation and differentiation of ameloblasts. Nevertheless, detailed structural and functional studies on AMBN have been substantially limited by the paucity of the purified nondegraded protein. With this study, we have developed a procedure for production of a highly purified form of recombinant human AMBN in quantities that allowed its structural characterization. Using size exclusion chromatography, analytical ultracentrifugation, transmission electron, and atomic force microscopy techniques, we show that AMBN self-associates into ribbon-like supramolecular structures with average widths and thicknesses of 18 and 0.34 nm, respectively. The AMBN ribbons exhibited lengths ranging from tens to hundreds of nm. Deletion analysis and NMR spectroscopy revealed that an N-terminal segment encoded by exon 5 comprises two short independently structured regions and plays a key role in self-assembly of AMBN.
Introduction
Intrinsically disordered proteins (IDPs)2 lack stable tertiary or secondary structures and exist as populations of quickly interconverting conformations that resemble the denatured states of ordered proteins (1). Recent bioinformatic predictions suggest that over 50% of proteins of higher eukaryotes contain long disordered regions, and more than 1300 mammalian proteins have been shown to be fully disordered (2, 3). IDPs are involved in normal physiological processes of replication, growth, regulation, signaling, defense, and biomineralization, and their functional repertoire complements the functions of ordered proteins (1, 4, 5).
Vertebrate biomineralization is closely associated with evolutionarily related proteins that are in majority encoded by genes belonging to the secretory calcium-binding phosphoprotein gene cluster 4q13-q21 of the human genome (6, 7). Most of the proteins related to hydroxyapatite crystal formation, growth control, and orientation are, indeed, largely or fully intrinsically disordered (5). These proteins are involved not only in initiating biomineralization and in promoting mineral crystal formation, but also in inflammatory responses in bones and teeth (8). Extracellular matrix proteins, primarily including amelogenin, ameloblastin, enamelin, and amelotin, play a key role in the complex biomineralization process forming tooth enamel, the hardest tissue in the body (9, 10). It was suggested that these component proteins must have specific protein-protein self-interactions and co-interactions to assemble into an organized organic matrix. This matrix is then competent to undergo mineral replacement and to form a highly ordered three-dimensional structure consisting of long, thin hydroxyapatite crystallites (9, 11–13). Two proteinases, the matrix metalloproteinase 20 (MMP-20, enamelysin) and kallikrein 4, were described to be secreted into the enamel matrix throughout amelogenesis and appear to be important for processing of extracellular matrix proteins (14, 15). Degraded protein fragments are then continuously removed from the developing enamel, which finally becomes hard, fully mineralized and mature (11–13).
Amelogenin (AMEL) is the most abundant protein of the developing enamel extracellular matrix and plays an essential role in the structural organization and mineralization of enamel (16, 17). AMEL is a globally unfolded IDP in solution with some regions containing extended β-sheets and polyproline type II helices (18, 19). Using cryoelectron microscopy, it has recently been demonstrated that AMEL undergoes stepwise hierarchical self-assembly, where oligomers of amelogenin stabilize mineral prenucleation clusters and guide their arrangement into linear chains that organize as parallel arrays (20).
Ameloblastin (AMBN; amelin or sheathlin) is the most abundant nonamelogenin enamel matrix protein; it is well conserved among species and is constantly present throughout evolution and ontogenesis of mineralized tissues (21–26). Bioinformatic analysis and CD spectroscopy studies demonstrate that AMBN is an IDP that consists of a basic N-terminal domain, having the tendency to adopt a helical conformation, and an acidic C-terminal domain, primarily responsible for the structural disorder of the entire AMBN molecule (27, 28). AMBN is processed by matrix proteinases during enamel formation, and the N- and C-terminal cleavage products of AMBN were found to exhibit a different localization in the enamel matrix (26, 29–31). The relatively stable N-terminal fragments accumulate in the sheath space between enamel rods, throughout the thickness of the enamel matrix, whereas the acidic C-terminal cleavage products appear to be short-lived and are observed only in the more immature regions of the matrix in the vicinity of the secreting ameloblasts (26, 29, 32). Studies involving mutant mice that expressed a truncated AMBN variant lacking the segment encoded by exons 5 and 6 (AMBNΔ5–6) revealed that AMBN plays a key role in correct enamel formation (33, 34). It was suggested that AMBN is a cell adhesion molecule that regulates proliferation and differentiation of ameloblasts, and several different mechanisms of AMBN interaction with cells were reported (33–37).
Although the structural and self-assembly properties of AMEL have been intensively explored (38), the experimental data on structural features of AMBN have been limited by the unavailability of sufficient amounts of the nondegraded protein. Therefore, we developed a procedure for production of highly purified full-length recombinant human AMBN in quantities allowing its structural characterization. We demonstrate here that AMBN self-associates into ribbon-like supramolecular structures via a short segment that is encoded on the exon 5 of the AMBN gene.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The Escherichia coli K12 strain XL1-Blue (Stratagene, La Jolla, CA) was used throughout this work for DNA manipulation and was grown in LB medium supplemented with 150 μg/ml ampicillin or 60 μg/ml kanamycin. The BL21(λDE3) E. coli strain (Novagen, Madison, WI) was used for expression of recombinant proteins and was grown at 30 °C in LB medium containing 150 μg/ml ampicillin or 60 μg/ml kanamycin.
Plasmid Constructs
To construct the plasmid for expression of recombinant human AMBN (lacking the N-terminal secretion signal, i.e., amino acid residues 1–26; numbering according to GenBankTM accession number BC106931), the gene for AMBN was amplified from cDNA (Image clone 40033703; Source BioScience), and the PCR product was cloned as an NheI-XhoI fragment into the multiple cloning site (MCS) of a pET28b-derived vector, pET28b-dH-Trx-TEV-MCS-TEV-sH. Thereby, the gene for AMBN was fused in frame to a sequence encoding a double His purification tag (6 × poly-His-loop-6 × poly-His) (39), thioredoxin (Trx), and a cleavable tobacco etch virus (TEV)-specific peptide at the 5′-end and the TEV peptide and 6 × poly-His purification tag at the 3′-end, respectively (see Fig. 1A). The resulting construct pET28b-Trx-AMBN was used for expression of the Trx-AMBN fusion protein in E. coli cells. The pET28b-Trx-AMBN-Nterm and pET28b-Trx-AMBN-Cterm plasmids, used for the expression of Trx fusion proteins harboring the N-terminal domain (encompassing residues 1–196, residue numbering refers to the AMBN sequence without the N-terminal secretion signal; Trx-AMBN-Nterm) and the C-terminal domain (encompassing residues 197–421; Trx-AMBN-Cterm) of AMBN, were constructed similarly.
FIGURE 1.
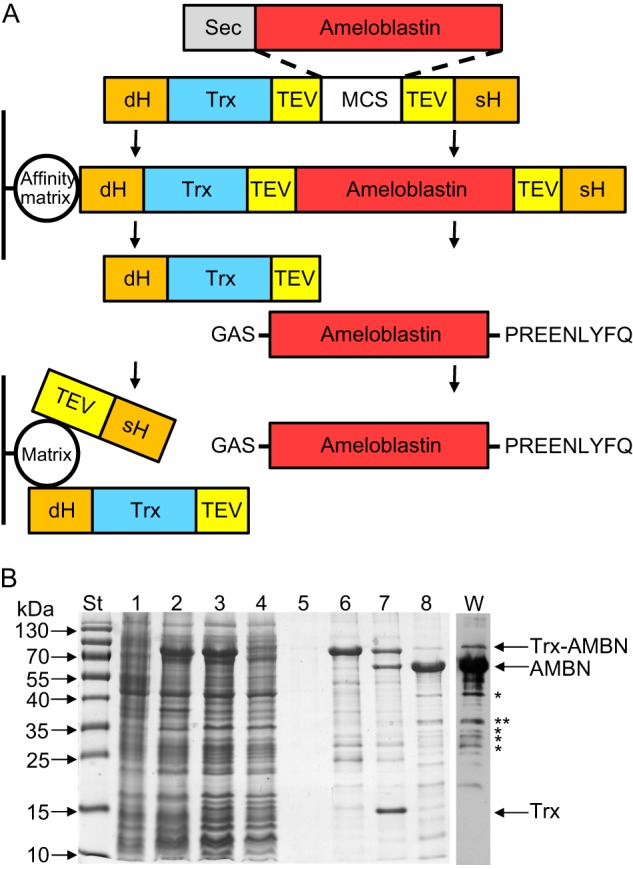
Production and analysis of recombinant human ameloblastin. A, scheme of the production and purification of AMBN. The fusion protein used for AMBN purification consists of AMBN (lacking the N-terminal secretion signal, Sec), which was genetically fused at the N terminus to a double His purification tag (dH), Trx, and a cleavable TEV specific peptide and at the C terminus to the TEV peptide and a single His purification tag (sH), respectively. After expression in E. coli BL21(λDE3) cells, the fusion protein is captured on an Ni-NTA affinity matrix, whereas the contaminating cell extract proteins are washed away from the column. After elution and dialysis, the fusion protein is processed in solution with TEV protease, and the excised AMBN is separated from Trx and purification tags by reversed phase chromatography on a PLRP-S column. The resulting purified recombinant AMBN harbors three extra residues (GAS) at the N terminus and a short nonapeptide (PREENLYFQ) at the C terminus. B, the samples collected throughout the AMBN production process were separated by SDS-PAGE (12.5%) and stained with Coomassie Blue. Lane St, molecular mass standards; lane 1, crude extract from uninduced cells; lane 2, crude extract from cells induced for production of the Trx-AMBN fusion protein; lane 3, clarified crude extract from induced cells; lane 4, Ni-NTA column flowthrough; lane 5, Ni-NTA column wash; lane 6, fraction of eluted Trx-AMBN fusion protein from Ni-NTA column; lane 7, products of Trx-AMBN fusion cleavage by TEV protease; lane 8, AMBN after reversed phase chromatography on a PLRP-S column. Lane W, purified AMBN (lane 8) was separated by SDS-PAGE (12.5%), transferred to a nitrocellulose membrane, and immunodetected by a polyclonal anti-AMBN rabbit serum. *, bands analyzed by MS.
The pET28b-Trx-AMBNΔX-Y plasmids, where X and Y stand for the numbers of the first and of the last amino acid residues of the AMBN segment deleted in a given construct (residue numbering refers to the AMBN sequence without the N-terminal secretion signal), were prepared by PCR mutagenesis and subsequent cloning of the NheI-XhoI-cleaved PCR fragments into the MCS of pET28b-dH-Trx-TEV-MCS (derived from the pET28b-dH-Trx-TEV-MCS-TEV-sH plasmid by deletion of a sequence encoding the TEV-sH peptide from the 3′-end of the MCS).
To construct the plasmid for expression of the fusion protein S36–72-AMBN-Cterm, consisting of the AMBN segment comprising residues 36–72 (segment 36–72) and the C-terminal domain of AMBN, a DNA fragment encoding the C-terminal domain of AMBN (encompassing residues 197–421) was amplified from cDNA, and the PCR product was cloned as an NheI-XhoI fragment into the MCS of pET11c-dHis (a modified pET11c vector (Novagen, Madison, WI) harboring a sequence encoding a double His purification tag). The resulting construct pET28b-dHis-AMBN-Cterm was digested with NheI and ligated in frame with a PCR NheI-NheI-digested fragment encoding segment 36–72. To construct the plasmid for expression of recombinant human amelogenin without the N-terminal secretion signal (i.e., lacking residues 1–16; numbering according to GenBankTM accession No. BC074951) in E. coli, the gene for amelogenin was amplified from cDNA (Image clone 30915277; Source BioScience), and the PCR product was cloned as an NheI-BamHI fragment into pET11c vector, yielding pET11c-AMEL.
Standard Techniques
Determination of protein concentration, SDS-PAGE, and Western blots were performed according to standard protocols (40). To obtain an anti-AMBN polyclonal serum, the highly purified recombinant human ameloblastin was separated on an analytical SDS-PAGE, and the full-length form of AMBN of an apparent size of ∼65 kDa was excised from 10 lanes of the gel (∼200 μg of AMBN). Polyacrylamide gel with the protein was mechanically disintegrated, mixed with aluminum hydroxide adjuvant (0.2% solution), and used for immunization. One rabbit was injected subcutaneously three times (200 μg of AMBN per dose) at 2-week intervals and was bled 2 weeks after the last injection. The peptide encompassing residues 36–72 of AMBN (peptide 36–72) was synthesized using an ABI 433A solid phase synthesizer (Applied Biosystems, Foster City, CA), and the purified deprotected peptide was dissolved in acetonitrile/H2O and dried.
Protein Production and Purification
All recombinant proteins were produced in E. coli BL21(λDE3) cells transformed with the appropriate plasmid. Exponential 500-ml cultures grown with shaking at 30 °C in MDO medium (yeast extract, 20 g/liter; glycerol, 20 g/liter; KH2PO4, 1 g/liter; K2HPO4, 3 g/liter; NH4Cl, 2 g/liter; Na2SO4, 0.5 g/liter; and thiamine hydrochloride, 0.01 g/liter) supplemented with 150 μg/ml of ampicillin or 60 μg/ml of kanamycin were induced at A600 = 0.5 with 1 mm isopropyl β-d-thiogalactopyranoside and grown for additional 4 h.
For Trx-AMBN, Trx-AMBN-Nterm, Trx-AMBN-Cterm, or mutant Trx-AMBNΔX-Y protein purification, the cells were harvested by centrifugation; washed twice with 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA; resuspended in 50 mm Tris-HCl (pH 7.4), 600 mm NaCl (TN buffer) containing 8 m urea and 5 mm imidazole; and disrupted by sonication at 4 °C. The homogenates were cleared at 20,000 × g for 30 min and loaded on an Ni-NTA-agarose column (Qiagen) equilibrated with TN buffer containing 8 m urea and 80 mm imidazole. The column was washed with TN containing 6 m urea and 80 mm imidazole. The fusion protein was eluted with TN containing 4 m urea and 600 mm imidazole and dialyzed against TN containing 5 mm EDTA and 1 mm DTT overnight at 4 °C. Processing by recombinant TEV protease (kind gift of L. Bumba, Institute of Microbiology, Academy of Sciences of the Czech Republic), was performed for 24 h at 4 °C. The cleaved protein mixture was dialyzed against 50 mm triethylamine, 5% aqueous acetonitrile (pH 8.0) overnight at 4 °C and loaded onto preparative PLRP-S column (Agilent Technologies; 8-μm particle size, 300-Å pore size, 7.5 mm × 30 mm). The excised free AMBN was eluted with a shallow gradient of 50 mm triethylamine, 95% aqueous acetonitrile (pH 8.0) as a major chromatographic peak at 38% of acetonitrile. Finally, AMBN was concentrated using a centrifugal evaporator; dialyzed against 10 mm Tris-HCl (pH 7.4), 40 mm NaCl; and stored at −20 °C. The S36–72-AMBN-Cterm fusion protein was purified by the same procedure with omission of the cleavage step. Amelogenin was purified to homogeneity from crude E. coli cell extracts by ammonium sulfate precipitation and reversed phase chromatography as previously described (38).
Mass Spectrometry Analyzes
Bands of interest were excised from SDS-polyacrylamide gels stained by Coomassie Brilliant Blue R-250 and cut into small cubes (approximately 1 mm3), and after complete distaining, washing, and partial drying in a SpeedVac concentrator, the gel pieces were reconstituted with cleavage buffer containing 25 mm 4-ethylmorpholine acetate, 10% acetonitrile, and sequencing grade trypsin (50 ng/μl; Promega) or Lys-C endoprotease (100 ng/μl; Roche Applied Science). In the third experiment, the protein cleavage was carried out with CNBr (20 μg/μl; Sigma-Aldrich) dissolved in 70% trifluoroacetic acid (v/v). All digestion mixtures were incubated for 16 h at 37 °C. The resulting peptides were extracted with 40% MeCN, 0.1% trifluoroacetic acid; desalted on a microtrap with OligoR3 resin (Applied Biosystems, Foster City, CA); and directly eluted onto a MALDI target with a solution of α-cyano-4-hydroxy-cinnamic acid (5 mg/ml) in 50% MeCN, 0.1% trifluoroacetic acid. The positive MALDI-TOF mass spectra and MS/MS LIFT spectra were acquired on a MALDI-TOF/TOF mass spectrometer ultraFLEX III (Bruker Daltonics, Bremen, Germany) equipped with a nitrogen laser (337 nm). The spectra were calibrated externally using the monoisotopic [M+H]+ ion of peptide standards PepMix II (Bruker Daltonics) and interpreted manually.
For mass determination of the full-length ameloblastin protein, the sample of purified AMBN was desalted using POROS R2/20 microtrap (Applied Biosystems) and measured in linear mode, using a saturated solution of sinapic acid in aqueous 50% acetonitrile, 0.1% trifluoroacetic acid as MALDI matrix. Spectra were externally calibrated by employing singly and doubly charged ions of BSA resulting in mass accuracy with an error of less than 0.1%.
High Resolution Size Exclusion Chromatography
Analytical size exclusion chromatography was performed using an Akta Prime FPLC system (GE Healthcare) and a Superdex 200 GL 10/300 column (GE Healthcare) pre-equilibrated with 50 mm Tris-HCl (pH 8.0), 150 mm NaCl and calibrated with standard molecular mass marker proteins (LMW calibration kit; GE Healthcare) at a flow rate of 0.4 ml/min.
Analytical Ultracentrifugation
Sedimentation velocity experiments were performed using a ProteomeLab XL-I Beckman Coulter analytical ultracentrifuge equipped with an AN50Ti rotor. Samples were dialyzed against buffer containing 20 mm Tris-HCl (pH 7.4), 200 mm NaCl prior to analysis. Buffer density, viscosity, and partial specific volume of all proteins were estimated using the program SEDNTERP 1.09. Data analysis was performed with the SEDFIT package (41). Experiments were conducted at several different loading concentrations at 20 °C and with rotor speeds of 25,000 rpm (AMBN, AMEL) or 48,000 rpm (AMBN-Cterm, AMBNΔ36–72), respectively. Scans were recorded at 280 nm in 1-min intervals with 30-μm spatial resolution. Sedimentation velocity data were analyzed using a sedimentation coefficient distribution model, c(s). Peaks were integrated to determine the weight-averaged sedimentation coefficients sw.
Circular Dichroism Spectroscopy
CD measurements were performed on a Jasco-815 spectropolarimeter equipped with the Peltier type temperature control system PTC-423S/L. CD spectra were recorded in 5 mm Tris-HCl (pH 7.4), 20 mm NaCl at an AMBN concentration of 62.5 μg/ml and an AMBN-derived peptide 36–72 concentration of 200 μg/ml, respectively. CD spectra were collected at temperatures of 5, 20, 37, 50, 75, and 90 °C, respectively, for wavelengths from 195 to 300 nm, at a standard instrument sensitivity and a scanning speed of 10 nm/min, a response time of 16 s and two spectra accumulations. Reversibility of the temperature dependence was checked by measurement of the samples chilled back to 20 °C.
NMR Spectroscopy and Structure Calculations
NMR experiments were acquired from 200-μl samples (3-mm NMR tube) of 5.8 mm peptide (residues 36–72 of AMBN) in a 25 mm sodium phosphate, 100 mm sodium chloride buffer (pH 6.0), containing 5% D2O, 95% H2O. The NMR spectra were collected at 25 °C on a 600-MHz Bruker Avance spectrometer equipped with a triple-resonance (15N/13C/1H) cryoprobe. A set of two-dimensional experiments was recorded to determine sequence-specific resonance assignments for the peptide. In particular, 15N/1H and 13C/1H heteronuclear single quantum coherence, heteronuclear multiple-bond correlation, and total correlation spectroscopy with 60-ms mixing time and NOESY, which were acquired with an NOE mixing time of 250 ms. The family of converged structures for the synthetic peptide was calculated using Cyana 2.1 (42). The combined automated NOE assignment and structure determination protocol was used to automatically assign the NOE cross-peaks identified in two-dimensional NOESY spectra and to produce preliminary structures. Subsequently, five cycles of simulated annealing combined with redundant dihedral angle constraints (43) were used to produce the sets of converged structures with no significant restraint violations (distance and van der Waals violations, <0.2 Å). Analysis of the obtained family of structures was carried out using the programs MOLMOL and PyMOL (45).
Transmission Electron Microscopy
Purified proteins were diluted prior to analysis with 10 mm Tris-HCl (pH 8.0) and 150 mm NaCl to the same protein concentration of 10 μg/ml. 5-μl drops of protein solutions were applied onto glow discharge-activated carbon-coated grids (46) and adsorbed for 30 s. The excess of solution was blotted with filter paper, and the grids were immediately negatively stained with 2% uranyl acetate in double-distilled H2O for 30 s. The grids were blotted with filter paper and air-dried. The samples were examined in Philips CM100 electron microscope at 80 kV and magnification of 64,000×. Digital images were recorded using MegaViewII slow scan camera at a magnification of 64,000×, giving a pixel size of ∼1 nm. The recorded images were processed in AnalySis3.2 software suite using embedded modules (Shading correction and Optimize 16-bit image for 8-bit display). No other image manipulation was used. Visualized objects were analyzed to determine an average width of ribbon-like structures of AMBN (n = 167), AMBN-Nterm (n = 115), and S36–72-AMBN-Cterm (n = 112), respectively, and an average diameter of AMEL spherical particles (n = 70).
Atomic Force Microscopy
Aliquots of the proteins were diluted in two steps in 4 mm PBS (pH 8.0). After the first dilution (47 μg/ml), the sample was centrifuged to eliminate aggregates originating from the freezing and melting process (6,700 × g for 10 min). Upon a second dilution, the concentration of the protein was adjusted to ∼0.5 μg/ml. The solution was kept on wet ice for 30 min and vortexed before 15-μl droplets were placed on freshly cleaved mica for 2 min. The mica was washed by distilled H2O and dried with nitrogen. It should be noted that the ribbon-like structures of AMBN formed and were already present in purified protein samples at concentrations ranging from 1 to 2 mg/ml (see results of analytical ultracentrifugation). These preformed ribbons then did not disassemble upon subsequent >1,000-fold dilution of the protein stocks, indicating a high affinity of interaction of ameloblastin subunits within the ribbon structure.
Atomic force microscopy (AFM) analysis was performed using an AFM Integra Probe Nanolaboratory (NT-MDT, Moscow, Russia). Analysis of dry samples was performed in semicontact mode with a 100 × 100-μm closed-loop scanner (scanning by sample). Samples were analyzed by high accuracy noncontact composite (HA_NC) ETALON silicon tip cantilevers (NT-MDT) at a typical resonant frequency of 280 kHz, using a tip radius of 10 nm and a force constant of 11.5 n/m in air and at ambient temperature and humidity. A scan rate of 0.5–1 Hz was used for the best resolution. The data were always collected at least from three different samples, with two different tips and in a minimum of 10 different positions on each sample.
Images were analyzed by Scanning Probe Image Processor software (Image Metrology A/S, Hørsholm, Denmark). Raw images were corrected for tilting of the sample stage and were zero-leveled based on the dominant height value in the distribution histogram. A built-in grain analysis module was used for analysis of height (protein monomers and ribbons, Zmax parameter), length (contour length of ribbons), or depth (defects in protein monolayers, Zmin) of objects, using the threshold method (at least 100 objects were analyzed for each parameter). The obtained values are presented as histograms.
RESULTS
Expression, Purification, and Analysis of Recombinant Ameloblastin
AMBN is an unstable protein that is rapidly degraded in the developing enamel (32). To isolate a highly purified form of nondegraded AMBN for structural and functional studies, the protein was produced in E. coli cells without its N-terminal secretion signal. Instead, a double His purification tag and the Trx moiety allowing high level expression and stability of the chimeric protein were fused to the N terminus of AMBN, whereas a single His purification tag was added to its C-terminal end (Fig. 1A). To enable separation of AMBN from Trx and purification tags, sequences recognized by the highly specific TEV protease were introduced at the N and C termini of AMBN, respectively (Fig. 1A).
The Trx-AMBN fusion protein was purified by metalloaffinity chromatography and processed with TEV protease, and the excised AMBN was further purified by reversed phase chromatography, as documented in Fig. 1. The final purified AMBN had three extra residues remaining at the N terminus (GAS) and a short nonapeptide (PREENLYFQ) added at the C terminus, thus consisting of 433 amino acid residues. Despite a calculated molecular mass of 46.7 kDa, the excised AMBN migrated on a 12.5% SDS-polyacrylamide gel as a protein with an apparent size of 65 kDa (Fig. 1B) that was unambiguously identified as ameloblastin by peptide mass fingerprint mapping (82% coverage of the AMBN sequence; supplemental Table S1). Moreover, MALDI-TOF spectrum of the purified undigested AMBN sample displayed a major peak at m/z = 46,840 (data not shown), confirming the presence of full-length AMBN, lacking Trx and purification tags.
In parallel with expected full-length AMBN, some lower molecular mass protein species were observed on the SDS-polyacrylamide gel (Fig. 1B). These were confirmed by Western blot analysis with an anti-AMBN rabbit polyclonal antibody to correspond to proteolytic fragments of ameloblastin that were likely generated by bacterial proteases already during AMBN production in E. coli cells (Fig. 1B). The most abundant cleavage products (indicated in Fig. 1B by asterisks) were isolated from the SDS-polyacrylamide gel and confirmed to be AMBN fragments by mass spectrometry (data not shown). It is noteworthy that one of the isolated AMBN fragments (indicated in Fig. 1B by two asterisks) resulted from cleavage between residues Arg-196 and Leu-197. Processing of the same Arg-Leu bond of porcine AMBN was previously shown to be catalyzed both in vitro and in vivo by MMP-20 (30, 31). Taken together, the used procedures allowed to prepare ∼2.5 mg of highly purified full-length AMBN per liter of bacterial culture.
Ameloblastin Forms Supramolecular Structures
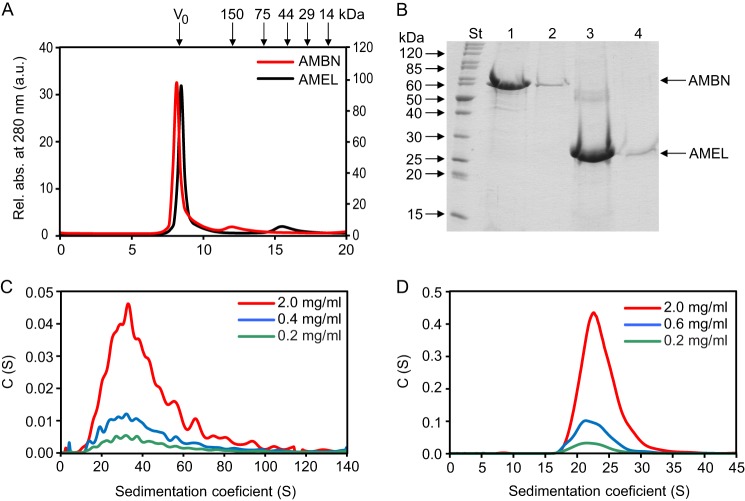
To examine whether ameloblastin, like amelogenin, can self-assemble into polymeric structures, the purified recombinant human AMBN protein was analyzed by high resolution size exclusion chromatography and by analytical ultracentrifugation. On a Superdex 200 column, precalibrated with globular protein standards, the recombinant ameloblastin eluted in the void (excluded) volume of the column (the exclusion limit of globular proteins for the column is ∼1.3 × 103 kDa) in a peak that contained 95% of the loaded protein (Fig. 2A). A minor amount of AMBN then eluted as an included peak of protein of an estimated size of 187 kDa. Identical chromatographic profiles were observed for different concentrations of AMBN at 0.1, 0.5, and 1.0 mg/ml, respectively (data not shown). As a control, the recombinant human AMEL (theoretical molecular mass, 20.2 kDa), known to self-assemble to multimeric structures, was analyzed on the same Superdex 200 column and was found to exhibit an elution profile similar to that of AMBN (Fig. 2A). A major fraction (88%) of purified AMEL eluted in the void volume, and a minor fraction of AMEL eluted as an included peak with an estimated size of 44 kDa (Fig. 2A). The identity of AMBN or AMEL in the excluded and included peaks was confirmed by SDS-PAGE (Fig. 2B).
FIGURE 2.
Ameloblastin forms supramolecular structures, similarly as amelogenin. A, the purified full-length AMBN and AMEL proteins were analyzed by size exclusion chromatography on a Superdex 200 10/30 column. The absorbances at 280 nm of eluted proteins are indicated for AMBN (red line, left y axis) or AMEL (black line, right y axis). The molecular masses of the globular protein standards are indicated above the chromatograms in kDa. V0, void volume. B, SDS-PAGE analysis of the AMBN and AMEL peak fractions collected during size exclusion chromatography. Lane St, molecular mass standards; lane 1, AMBN excluded peak; lane 2, AMBN included peak; lane 3, AMEL excluded peak; lane 4, AMEL included peak. The samples were analyzed on a 12.5% SDS-polyacrylamide gel and stained with Coomassie Blue. C and D, sedimentation velocity measurements were performed to analyze the high molecular mass structures of AMBN (C) and AMEL (D). Experiments were performed with different concentrations of AMBN or AMEL in 20 mm Tris-HCl (pH 7.4), 200 mm NaCl, at 20 °C and 25,000 rpm. Protein peaks were detected by using the absorption optical system at 280 nm. Rel. Abs., relative absorption.
To further characterize the multimerization state of AMBN, sedimentation velocity measurements were performed. The analysis of continuous sedimentation coefficient distributions c(s) revealed that AMBN formed a heterogeneous population of protein supramolecular assemblies exhibiting molecular masses ranging from ∼500 to ∼5000 kDa (Fig. 2C). The peak distribution was negligibly dependent on AMBN concentration within the range of 0.2–2.0 mg/ml (Fig. 2C). Under the same conditions, AMEL exhibited narrower peaks in sedimentation velocity measurements than AMBN, suggesting a lower heterogeneity of the supramolecular self-assemblies of AMEL that exhibited an average molecular mass of ∼1000 kDa (Fig. 2D). All of these results suggested that AMBN self-associated in the absence of other cellular components into a heterogeneous population of supramolecular structures.
A Segment Encompassing Residues 36–72 Encoded by Exon 5 Is Essential for the Formation of Ameloblastin Supramolecular Structures
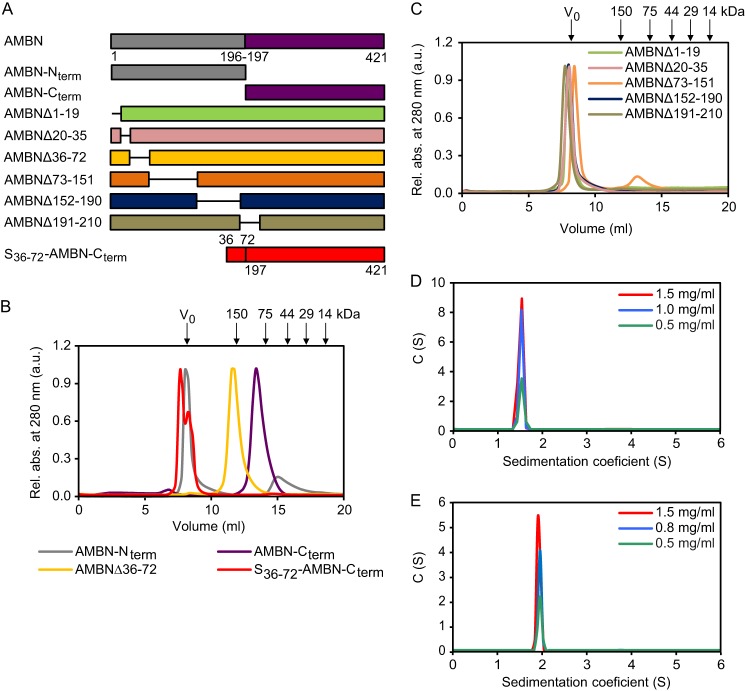
To localize the segment that accounts for self-assembly of AMBN to supramolecular structures, the N-terminal (AMBN-Nterm; theoretical molecular mass, 22.8 kDa) and C-terminal (AMBN-Cterm; theoretical molecular mass, 25.3 kDa) domains of AMBN were constructed (Fig. 3A), purified, and examined by size exclusion chromatography. As shown in Fig. 3B, the major portion of AMBN-Nterm eluted in the void volume peak on the Superdex 200 column, whereas AMBN-Cterm eluted as a single peak with an estimated size of 116 kDa. This suggested that the region essential for self-association of AMBN into supramolecular structures was located in the N-terminal domain of the protein. Therefore, truncated AMBN-Nterm variants with deletions corresponding to sequences encoded at the end of exon 2 and by entire exon 3 (AMBNΔ1–19), exon 4 (AMBNΔ20–35), exon 5 (AMBNΔ36–72), exon 6 (AMBNΔ73–151), exons 7–9 (AMBNΔ152–190), or exon 10 (AMBNΔ191–210) were prepared (Fig. 3A), and the respective purified proteins were subjected to size exclusion chromatography analysis. As summarized in Fig. 3B, only the AMBNΔ36–72 variant (theoretical molecular mass, 41.1 kDa) was retarded on Superdex 200 resin and eluted as a single peak with an apparent molecular mass corresponding to 233 kDa. All other truncated AMBN variants eluted in the void volume of the column (Fig. 3C), suggesting that their ability to form large supramolecular self-assemblies was preserved. When the AMBN segment containing residues 36–72 (segment 36–72) was genetically fused to AMBN-Cterm (S36–72-AMBN-Cterm; Fig. 3A), the C-terminal domain of AMBN acquired the ability to form supramolecular structures and eluted in the void volume of the column, as the entire AMBN molecule (Fig. 3B). All of these results demonstrate that segment 36–72 of AMBN, encoded by exon 5, is essential for self-assembly of ameloblastin and formation of its supramolecular structures.
FIGURE 3.
Residues 36–72 are essential for self-association of ameloblastin. A, schematic representation of truncated AMBN variants. The portions deleted in AMBN are indicated by the solid lines separating the colored bars. The numbers following the symbol Δ indicate the first and the last residue numbers of the respective portion deleted in the given protein construct. S36–72-AMBN-Cterm represents a fusion between segment 36–72 of AMBN and AMBN-Cterm. B and C, the purified AMBN mutant variants were analyzed by size exclusion chromatography on a Superdex 200 10/30 column. The molecular masses of the protein standards are indicated above chromatograms in kDa. V0, void volume. D and E, sedimentation velocity measurements were used to characterize the C-terminal domain of AMBN (D) and the AMBNΔ36–72 mutant variant (E). Experiments were performed with different concentrations of AMBN-Cterm or AMBNΔ36–72 in 20 mm Tris-HCl (pH 7.4), 200 mm NaCl, at 20 °C and 48,000 rpm. Protein peaks were detected by using the absorption optical system at 280 nm. Rel. abs., relative absorption.
It has to be noted that the estimation of molecular masses of nonglobular intrinsically disordered proteins from size exclusion chromatograms may be rather misleading when chromatographic columns are calibrated with globular protein standards (47). Therefore, sedimentation velocity measurements were performed with the AMBN-Cterm domain and the AMBNΔ36–72 variant. The analysis of continuous sedimentation coefficient distributions c(s) confirmed that AMBN-Cterm was unable to self-assemble into multimeric structures. At three different protein concentrations, a single peak of the protein was observed, exhibiting an average sw value of 1.5, which corresponded to a molecular mass of ∼28 kDa (Fig. 3D). Similarly, the deletion variant AMBNΔ36–72 (theoretical molecular mass, 41.1 kDa) was observed as a single peak with an average sw value of 1.9, corresponding to a molecular mass of ∼41 kDa (Fig. 3E). Thus, the molecular masses of AMBN-Cterm and AMBNΔ36–72 determined by analytical ultracentrifugation fitted almost exactly the calculated masses derived from the respective protein sequences, showing that both proteins were monomeric. These results hence suggest that segment 36–72 is crucial for self-association of ameloblastin to supramolecular structures and show that the AMBNΔ36–72 variant devoid of this segment was unable to form AMBN multimers.
The AMBN Segment Encoded by Exon 5 Consists of Two Independently Structured Regions
Recent bioinformatic analysis indicated and CD spectroscopy then demonstrated that the full-length AMBN is an intrinsically disordered protein (27, 28). As shown in supplemental Fig. S1, indeed, no well defined elements of regular secondary structure were observed in the CD spectra of the synthetic peptide encompassing residues 36–72 (peptide 36–72) of AMBN. In addition, thermal denaturation CD experiments with peptide 36–72 showed profiles characteristic for IDPs (supplemental Fig. S1). Therefore, the conformational behavior of the peptide fragment presumably accounting for AMBN self-assembly has been further probed using NMR spectroscopy. Preliminary analysis of NOESY spectra based on the abundant NOE contacts between backbone amide protons around the tryptophan residues Trp-50 and Trp-63 identified these regions as adopting a defined conformation. The subsequently obtained essentially complete 1H resonance assignments for the given peptide fragment then allowed for automated assignment of the NOEs identified in two-dimensional NOESY and for structural calculation using Cyana (42, 43). The numbers of distance constraints and structural statistics for the obtained structures are given in supplemental Table S2. A set of 20 satisfactorily converged structures obtained from 100 random starting conformations then revealed a relatively structured region in the central part of the peptide fragment between Phe-46 and Pro-66 residues, as shown in Fig. 4A. The first conformationally distinct region was found between residues Phe-46 and Pro-57, with a part of the backbone adopting a nearly regular α-helical conformation between residues Ser-48 and Gly-53 (Fig. 4B). The second region of the peptide with a defined conformation was found between residues Ser-59 and Pro-66 (Fig. 4C). Interestingly, both regions are structured around a tryptophan residue that is followed by a methionine. No stabilizing medium or long range NOE contacts were found in the N- and C-terminal regions that, hence, remain unstructured (Fig. 4).
FIGURE 4.
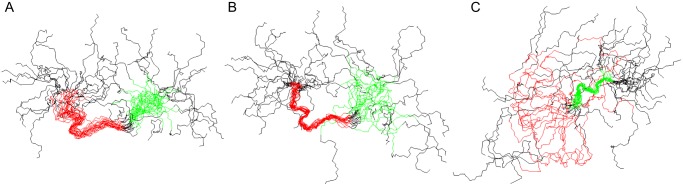
The AMBN-derived peptide involved in ameloblastin self-association consists of two regions with a relatively well defined conformation. Structures are represented by best fit superpositions of the peptide backbone for 20 converged structures with the region including residues Phe-46 to Pro-57 highlighted in red and the region including residues Ser-60 to Pro-66 highlighted in green. A, superposition over the region from Phe-46 to Pro-66. B, superposition over the first structured region (Phe-46 to Pro-57), which includes a short α-helix between residues Ser-48 and Gly-53. C, superposition over the second relatively structured region (Ser-59 to Pro-66). Both N and C termini are disordered.
Visualization of AMBN Supramolecular Structures Using Electron and Atomic Force Microscopy
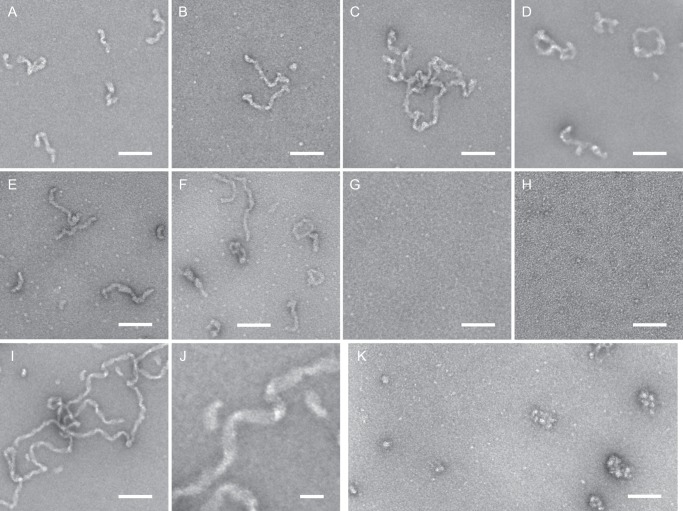
To extend the structural analysis, transmission electron microscopy (TEM) and AFM were used to visualize the polymeric structures formed by AMBN. Electron microscopy revealed that the AMBN high molecular mass self-assemblies were flat ribbon-like supramolecular structures with an average width of 18 ± 4 nm and with a variable size, ranging from tens to hundreds of nanometers in length (Fig. 5, A–D). No major differences in supramolecular structures of AMBN were observed at pH levels ranging from 6.0 to 8.0 and NaCl concentrations of 0 and 200 mm, respectively (supplemental Fig. S2). AMBN assemblies adopted various shapes, most of the structures being twisted and curled (Fig. 5A) and other forming pairs (Fig. 5B) or bundles (Fig. 5C). Occasionally the ends of a ribbon structure were found to be linked together and formed a loop-like morphology (Fig. 5D). Similar structures were visible when AMBN-Nterm was characterized by TEM, exhibiting an average width of 16 ± 4 nm (Fig. 5, E and F). In contrast, no polymeric structures were observed with AMBN-Cterm (Fig. 5G), indicating that the C-terminal domain of AMBN was not involved in self-association of the protein. Similarly, in agreement with the results of analytical ultracentrifugation showing that the AMBNΔ36–72 variant remained monomeric, no supramolecular structures were observed for AMBNΔ36–72 by TEM (Fig. 5H). However, when the segment encompassing residues 36–72 of AMBN was fused to AMBN-Cterm, the C-terminal domain gained the capacity to form flat ribbon-like supramolecular structures of an average width of 15 ± 2 nm (Fig. 5, I and J), which were rather similar to those formed by the full-length AMBN molecule. This clearly shows that the segment encompassing residues 36–72 confers the self-assembly capacity of AMBN. Examination of the control protein amelogenin by TEM revealed the previously described spherical supramolecular assemblies (20, 48, 49), exhibiting an average diameter of 21 ± 3 nm and aggregates of different sizes (Fig. 5K).
FIGURE 5.
Transmission electron micrographs of ameloblastin and its mutant variants. Purified AMBN (A–D), AMBN-Nterm (E and F), AMBN-Cterm (G), AMBNΔ36–72 (H), S36–72-AMBN-Cterm (I and J), and AMEL (K) were applied onto glow discharge-activated carbon coated grids and negatively stained with 2% uranyl acetate. Samples were examined at a magnification of 64,000×. J shows a 2.84-fold digitally magnified detail of the ribbon-like structure of the S36–72-AMBN-Cterm protein filament presented in I. Scale bars, 100 nm (A–I and K), or 25 nm (J).
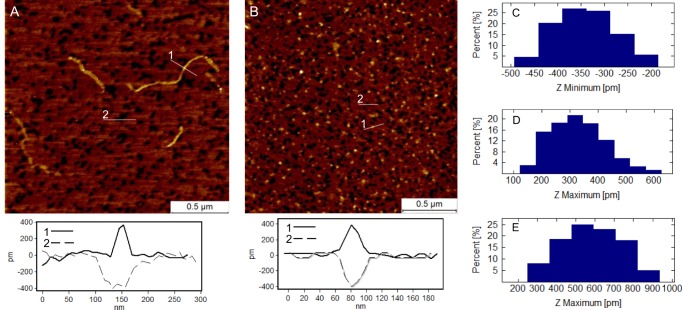
As further shown in Fig. 6, AFM analysis of AMBN and its AMBNΔ36–72 mutant variant performed on the atomically flat surface of mica corroborated the results obtained by analytical ultracentrifugation and TEM. Both AMBN and AMBNΔ36–72 exhibited the ability to self-assemble into organized protein monolayers on the surface of mica (Fig. 6, A and B). The thickness of the monolayers 0.43 ± 0.06 nm (AMBN) and 0.36 ± 0.07 nm (AMBNΔ36–72; Fig. 6C) was in the range comparable to the height of AMBNΔ36–72 monomers or of the AMBN ribbons observed on the top of the monolayer (see the cross-sectional profiles in Fig. 6). On the other hand, the C-terminal domain of AMBN (AMBN-Cterm) adsorbed in the form of isolated objects (average Zmax = 0.57 ± 0.16 nm; Fig. 6E), which were randomly distributed on the surface of mica, without exhibiting any signs of oriented self-assembly or any monolayer formation capacity (data not shown). Although not studied in detail, based on the determined thickness of AMBN and AMBNΔ36–72 deposits and the extremely low surface roughness of the monolayer, we assume that the proteins organized into a monolayer in a highly oriented manner. The building of the monolayer was observed already after short exposure (≈1 min) of the protein to mica. Besides the monolayer, the AMBN molecules were present also as self-assembled ribbon structures (Fig. 6A) observed exclusively on the top of the protein monolayer. The ribbons (average thickness Zmax = 0.34 ± 0.15 nm) were from several tens of nanometers to micrometers long, curved and occasionally crossed. In contrast, AMBNΔ36–72 was observed only as a monomeric protein with an average diameter Zmax = 0.32 ± 0.11 nm (Fig. 6, B and D). Again, single isolated monomers were observed on the top surface of the protein monolayer, and no ribbons of AMBNΔ36–72 were detected.
FIGURE 6.
Atomic force microscopy analysis of ameloblastin and its mutant variants. A, AMBN. A typical cross-section profile of a self-assembled ribbon (1) and a defect in protein monolayer (2) is shown on graph bellow the image. B, AMBNΔ36–72 mutant. A typical cross-section profile of a protein monomer (1) and a defect in protein monolayer (2) is shown on graph bellow the image. C, histogram of thickness of AMBNΔ36–72 monolayer measured at defects. D, height histogram of AMBNΔ36–72 monomers. E, height histogram of the C-terminal domain of AMBN.
DISCUSSION
AMBN belongs to the family of intrinsically disordered proteins that are involved in the highly organized process of biomineralization. Here we observed that AMBN self-associates into ribbon-like supramolecular structures and that this self-assembly process is abolished by the deletion of the 37 residues long segment 36–72 located in the N-terminal portion of the molecule.
In the developing enamel, AMBN is rapidly processed into lower molecular mass forms (32). The unstable character of AMBN was also observed in this study, when the protein was produced as recombinant protein in E. coli, in that degradation fragments of AMBN were present in producing bacteria and co-purified to some extent with the full-length molecule. The stability of AMBN was indeed increased when the N-terminal end of the molecule was protected from degradation by fusion to a stable protein, such as thioredoxin, or at least to a short purification tag, like the 6 × poly-His tag. In contrast, no further stabilization of AMBN was observed, when the C terminus of ameloblastin was protected from degradation by fusion to a short peptide or a proteinaceous partner (data not shown). Moreover, recombinant AMBN was cleaved by an E. coli protease between residues Arg-196 and Leu-197, separating the protein molecule into the N- and C-terminal domains. The cleavage of a corresponding Arg-Leu peptide bond was earlier shown to occur in porcine AMBN both in vitro and in vivo and was due to initial proteolytic processing by the MMP-20 protease (30, 31). This indicates that the peptide bond Arg-Leu found in the central region of AMBN is highly accessible to proteolytic processing and that it is cleaved not only by the natural MMP-20 protease present in the developing enamel but also by some irrelevant E. coli protease.
The recombinant ameloblastin, for which the identity and sequence-deduced size of 46.7 kDa was unambiguously confirmed here by mass spectrometry, repeatedly migrated aberrantly on SDS-polyacrylamide gels, where it exhibited a higher apparent molecular mass of ∼65 kDa. This is, indeed, a typical feature of disordered proteins that is explained by lower interaction of SDS with IDPs because of their unusual amino acid composition (2). It was also demonstrated that the estimation of molecular masses of nonglobular intrinsically disordered proteins by size exclusion chromatography is inaccurate, when calibrated with normal globular proteins (47). Indeed, we observed here that AMBN-Cterm runs on the size exclusion chromatography as a single peak with an estimated size of 116 kDa, corresponding to a pentameric form of the C-terminal domain and the AMBNΔ36–72 mutant variant eluted as a single peak with an estimated size of 233 kDa, suggesting a pentameric/hexameric form of the protein. However, using analytical ultracentrifugation, both AMBN-Cterm and AMBNΔ36–72 were clearly shown to remain monomeric (28 and 41 kDa, respectively). Similarly, the included peak of the entire AMBN with an estimated size of 187 kDa was most likely corresponding to a monomer than to a tetrameric form of the protein. Similar atypical behavior was, indeed, previously described for amelogenin in electrophoretic and chromatographic separations (50, 51), as also confirmed here.
Transmission electron, as well as atomic force microscopy confirmed the ability of AMBN to self-assemble into ribbon-like supramolecular structures and the key role of the sequence encompassing residues 36–72 in this activity. Surprisingly, the thickness of the ribbons was determined by AFM to be much lower than previously found for other self-assembled fibrous IDPs (52). The determined thickness of AMBN ribbons was comparable to the thickness of β-sheet structures or to that reported for ssDNA (53–55).
Another interesting feature of AMBN appears to be its capacity to form organized AMBN monolayers on the surface of mica. Although the biological relevance of this observation remains unclear, it is plausible to speculate that the AMBN monomers organized into surface monolayers because of electrostatic interactions. Indeed, freshly cleaved mica is an atomically flat hydrophilic surface of hexagonal lattice symmetry that exhibits a periodic distribution of negative charges (56). It has previously been shown that certain model peptides, such as those forming the cross-β-sheet ribbon structures in solution, can undergo reorganization on mica surface during water evaporation. This then results in formation of monomolecular highly ordered films because of alignment of these ribbons on mica surface (55). We hypothesize that a similar process could have been taking place also during AMBN monolayer formation on mica surface. Indeed, the observed thickness of the monolayer was comparable to the thickness of the AMBN ribbons and isolated ribbons were observed as attached in a second layer on the top of an AMBN monolayer. A protein monolayer could shield the negative charge of mica and allow the ribbon on its surface to maintain its native form. Although the ribbon formation is suppressed in the AMBNΔ36–72 mutant protein, the ordered monolayer formation was still preserved, which means that oriented interaction with surface was realized through the extended portion of the protein molecule, surpassing the deletion. The loss of the self-assembly capacity in the case of the C-terminal domain highlights the crucial role of the N-terminal domain in the self-assembly capacity of AMBN. Surprisingly, the measured thickness of the C-terminal domain was significantly higher than that of AMBN and AMBNΔ36–72. This indicates that the C-terminal domain underwent a structural rearrangement upon separation from the rest of the protein molecule and/or adopted an uneven orientation on mica surface.
Our results suggest that the AMBN self-association into ribbon-like supramolecular structures is realized by the segment encompassing residues 36–72, which serves as an association template for AMBN molecule self-assembly (Fig. 7). Indeed, the deletion of segment 36–72 completely abolished the capacity of AMBN to multimerize, and vice versa, the C-terminal domain of AMBN acquired the ability to form supramolecular structures, when fused to segment 36–72. As modeled in Fig. 7, it appears that AMBN self-assembly could be accomplished by the unfolded AMBN molecules both in a parallel (Fig. 7A), as well as in an antiparallel (Fig. 7B) monomer orientation. In both cases, segment 36–72 would likely be properly exposed and recognizable by the partner AMBN molecules. The requirement for an unfolded and extended character of the AMBN polypeptide chain would then be in good agreement with the observed extremely low thickness of the AMBN ribbon-like structures (0.34 ± 0.15 nm) observed by AFM.
FIGURE 7.
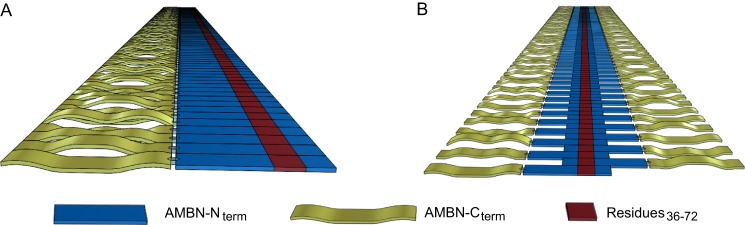
Schematic view of the possible arrangements of ameloblastin monomers within supramolecular structures. AMBN monomers with parallel (A) or antiparallel (B) orientation form supramolecular structures through interactions in the N-terminal segment comprising residues 36–72. The C-terminal domain of AMBN is apparently not critical for the self-assembly, and because of its highly charged character, it is likely to be important for protein solubility.
The self-assembly process was repeatedly described for amelogenin, the most abundant protein of the developing enamel extracellular matrix, which appears to be essential for the structural organization and mineralization of enamel (16, 17, 20). Moreover, the nonamelogenin enamel matrix protein tuftelin was also shown to self-assemble through its C-terminal region (57). It is worth noting that upon isolation of ameloblastin from newly formed porcine secretory enamel, even its N-terminal cleavage products of 13 kDa (Met-32 to Gln-130), 15 kDa (Val-1 to Gln-130), or 17 kDa (Val-1 to Arg-170), all appeared to be eluted from size exclusion chromatography columns in the void volume peak (58). Therefore, it has been suggested that the AMBN fragments formed large aggregates (58). Because all the above mentioned proteolytic fragments of AMBN harbored segment 36–72, it is plausible to assume that the previously observed aggregates could represent specific supramolecular structures of ameloblastin, similar to those described in this work. On the other hand, it should be noted that no interaction of ameloblastin molecules was observed in the yeast two-hybrid system (59).
Specific self-association of proteins into dimers or higher order oligomers is a common property in all biological systems, conferring new functions on the proteins that cannot be accomplished by isolated protein monomers (60–62). Thus, the here-described self-association of AMBN into supramolecular structures may also confer some new functions on the AMBN molecule that are characteristic for other self-assembling extracellular matrix proteins. These may comprise structural scaffolding and support, complexing with each other to increase structure stabilization, and/or multivalent binding to cellular receptors, which ensures high binding strength and regulates intercellular communication. Indeed, it was demonstrated that AMBN binds to the cell surface by several different mechanisms (35–37) and regulates proliferation and differentiation of ameloblasts (35).
It has previously been observed that ameloblastin isolated from porcine enamel harbors several post-translational modifications, including hydroxylation of Pro-11 and Pro-324, phosphorylation of Ser-17, and O-linked glycosylation of Ser-86 and Thr-361, respectively (63, 64). Because ameloblastin produced in a prokaryotic expression system is not post-translationally modified, the here-reported results would suggest that the described or as yet unidentified post-translational modifications of AMBN are not essential for the capacity to self-associate into supramolecular structures. It cannot, however, be excluded at present that the post-translational modifications might have some impact on the rate of the self-assembly process or on the final size and structure of the observed ameloblastin ribbons.
It has been previously suggested that AMBN serves as a nucleator of crystallization, because it is expressed at mineralization initiation sites within enamel (32, 44). Based on this observation and our results, it is plausible to hypothesize that intact AMBN may, upon secretion from ameloblasts, self-associate into macromolecular assemblies and interact with cellular receptor(s) and amelogenin and/or other enamel matrix proteins, so as to participate in formation of a highly organized supramolecular structure. This may then serve as a scaffold for initial crystal formation and growth. When the correct formation of the scaffold is disrupted because of a defect in AMBN self-assembly, this may translate into incorrect structural organization and mineralization of the enamel. Indeed, it has earlier been demonstrated in a mutant mouse model that AMBN plays an indispensable role in enamel formation (33, 34). The used mice produced an AMBN protein variant missing the sequence encoded by exons 5 and 6 (AMBNΔ5–6), thus encompassing residues 36–151. In the AMBNΔ5–6 mutant mouse, in fact, the dental epithelium differentiated into enamel-secreting ameloblasts, but the cells were detached from the enamel matrix surface, lost cell polarity, resumed proliferation, and formed multiple cell layers, producing abnormal, unstructured and calcified matrix (33, 34). In agreement with this phenotype, the N-terminal portion encoded by exon 5 was shown in this study to be crucial for AMBN self-association. Thus, the loss of AMBN self-association might explain at the molecular level the observed severe enamel hypoplasia in the AMBNΔ5–6 mutant mouse, similarly to what was observed in mutant mice expressing amelogenin variants that were unable to form AMEL self-assembled structures (17).
Acknowledgments
Helpful discussions with D. Kavan, L. Bumba, J. Prochazka, and P. Lyngstadaas; pilot NMR experiments by J. Chmelik; molecular dynamics simulations by J. Vymetal; the gift of the TEV protease by L. Bumba; and schematic drawings of AMBN supramolecular structures by J. Cisar are gratefully acknowledged. We thank H. Kubinova, S. Charvatova, I. Marsikova, and H. Dlouha for excellent technical help.
This work was supported by Grant P302/10/0427 from the Czech Science Foundation (to R. O. and J. V.), Czech National Cost Project OC10053 (to J. M.), Internal Grant Agency of Jan Evangelista Purkyne University (to J. M.), and Academy of Sciences of the Czech Republic Grant RVO61388971.

This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- IDP
- intrinsically disordered protein
- AMBN
- ameloblastin
- MMP
- matrix metalloproteinase
- AMEL
- amelogenin
- MCS
- multiple cloning site
- Trx
- thioredoxin
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid
- AFM
- atomic force microscopy
- TEM
- transmission electron microscopy.
REFERENCES
- 1. Uversky V. N., Oldfield C. J., Dunker A. K. (2008) Intrinsically disordered proteins in human diseases. Introducing the D2 concept. Annu. Rev. Biophys. 37, 215–246 [DOI] [PubMed] [Google Scholar]
- 2. Tompa P. (2002) Intrinsically unstructured proteins. Trends Biochem. Sci. 27, 527–533 [DOI] [PubMed] [Google Scholar]
- 3. Xie H., Vucetic S., Iakoucheva L. M., Oldfield C. J., Dunker A. K., Uversky V. N., Obradovic Z. (2007) Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome. Res. 6, 1882–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 5. Wojtas M., Dobryszycki P., Ożyhar A. (2012) Intrinsically disordered proteins in biomineralization, in Advanced Topics in Biomineralization (Seto J., ed) pp. 3–32, In Tech Europe, University Campus STeP Ri, Rijeka, Croatia [Google Scholar]
- 6. Kawasaki K., Suzuki T., Weiss K. M. (2004) Genetic basis for the evolution of vertebrate mineralized tissue. Proc. Natl. Acad. Sci. U.S.A. 101, 11356–11361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kawasaki K., Weiss K. M. (2003) Mineralized tissue and vertebrate evolution. The secretory calcium-binding phosphoprotein gene cluster. Proc. Natl. Acad. Sci. U.S.A. 100, 4060–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kalmar L., Homola D., Varga G., Tompa P. (2012) Structural disorder in proteins brings order to crystal growth in biomineralization. Bone 51, 528–534 [DOI] [PubMed] [Google Scholar]
- 9. Bartlett J. D., Ganss B., Goldberg M., Moradian-Oldak J., Paine M. L., Snead M. L., Wen X., White S. N., Zhou Y. L. (2006) 3. Protein-protein interactions of the developing enamel matrix. Curr. Top. Dev. Biol. 74, 57–115 [DOI] [PubMed] [Google Scholar]
- 10. Fincham A. G., Moradian-Oldak J., Simmer J. P. (1999) The structural biology of the developing dental enamel matrix. J. Struct. Biol. 126, 270–299 [DOI] [PubMed] [Google Scholar]
- 11. Moradian-Oldak J., Simmer J. P., Sarte P. E., Zeichner-David M., Fincham A. G. (1994) Specific cleavage of a recombinant murine amelogenin at the carboxy-terminal region by a proteinase fraction isolated from developing bovine tooth enamel. Arch. Oral Biol. 39, 647–656 [DOI] [PubMed] [Google Scholar]
- 12. Ryu O. H., Fincham A. G., Hu C. C., Zhang C., Qian Q., Bartlett J. D., Simmer J. P. (1999) Characterization of recombinant pig enamelysin activity and cleavage of recombinant pig and mouse amelogenins. J. Dent. Res. 78, 743–750 [DOI] [PubMed] [Google Scholar]
- 13. Shapiro J. L., Wen X., Okamoto C. T., Wang H. J., Lyngstadaas S. P., Goldberg M., Snead M. L., Paine M. L. (2007) Cellular uptake of amelogenin, and its localization to CD63, and Lamp1-positive vesicles. Cell Mol. Life Sci. 64, 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bartlett J. D., Simmer J. P., Xue J., Margolis H. C., Moreno E. C. (1996) Molecular cloning and mRNA tissue distribution of a novel matrix metalloproteinase isolated from porcine enamel organ. Gene 183, 123–128 [DOI] [PubMed] [Google Scholar]
- 15. Simmer J. P., Fukae M., Tanabe T., Yamakoshi Y., Uchida T., Xue J., Margolis H. C., Shimizu M., DeHart B. C., Hu C. C., Bartlett J. D. (1998) Purification, characterization, and cloning of enamel matrix serine proteinase 1. J. Dent. Res. 77, 377–386 [DOI] [PubMed] [Google Scholar]
- 16. Gibson C. W., Yuan Z. A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J. T., Decker S., Piddington R., Harrison G., Kulkarni A. B. (2001) Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 276, 31871–31875 [DOI] [PubMed] [Google Scholar]
- 17. Paine M. L., Zhu D. H., Luo W., Bringas P., Jr., Goldberg M., White S. N., Lei Y. P., Sarikaya M., Fong H. K., Snead M. L. (2000) Enamel biomineralization defects result from alterations to amelogenin self-assembly. J. Struct. Biol. 132, 191–200 [DOI] [PubMed] [Google Scholar]
- 18. Delak K., Harcup C., Lakshminarayanan R., Sun Z., Fan Y., Moradian-Oldak J., Evans J. S. (2009) The tooth enamel protein, porcine amelogenin, is an intrinsically disordered protein with an extended molecular configuration in the monomeric form. Biochemistry 48, 2272–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lakshminarayanan R., Fan D., Du C., Moradian-Oldak J. (2007) The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys. J. 93, 3664–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang P. A., Conway J. F., Margolis H. C., Simmer J. P., Beniash E. (2011) Hierarchical self-assembly of amelogenin and the regulation of biomineralization at the nanoscale. Proc. Natl. Acad. Sci. U.S.A. 108, 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cerný R., Slaby I., Hammarström L., Wurtz T. (1996) A novel gene expressed in rat ameloblasts codes for proteins with cell binding domains. J. Bone Miner. Res. 11, 883–891 [DOI] [PubMed] [Google Scholar]
- 22. Fong C. D., Hammarström L., Lundmark C., Wurtz T., Slaby I. (1996) Expression patterns of RNAs for amelin and amelogenin in developing rat molars and incisors. Adv. Dent. Res. 10, 195–200 [DOI] [PubMed] [Google Scholar]
- 23. Krebsbach P. H., Lee S. K., Matsuki Y., Kozak C. A., Yamada K. M., Yamada Y. (1996) Full-length sequence, localization, and chromosomal mapping of ameloblastin. A novel tooth-specific gene. J. Biol. Chem. 271, 4431–4435 [DOI] [PubMed] [Google Scholar]
- 24. Hu C. C., Fukae M., Uchida T., Qian Q., Zhang C. H., Ryu O. H., Tanabe T., Yamakoshi Y., Murakami C., Dohi N., Shimizu M., Simmer J. P. (1997) Sheathlin. Cloning, cDNA/polypeptide sequences, and immunolocalization of porcine enamel sheath proteins. J. Dent. Res. 76, 648–657 [DOI] [PubMed] [Google Scholar]
- 25. Simmer J. P., Hu J. C. (2002) Expression, structure, and function of enamel proteinases. Connect. Tissue Res. 43, 441–449 [DOI] [PubMed] [Google Scholar]
- 26. Uchida T., Murakami C., Dohi N., Wakida K., Satoda T., Takahashi O. (1997) Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies. J. Histochem. Cytochem. 45, 1329–1340 [DOI] [PubMed] [Google Scholar]
- 27. Vymetal J., Slabý I., Spahr A., Vondrásek J., Lyngstadaas S. P. (2008) Bioinformatic analysis and molecular modelling of human ameloblastin suggest a two-domain intrinsically unstructured calcium-binding protein. Eur. J. Oral Sci. 116, 124–134 [DOI] [PubMed] [Google Scholar]
- 28. Wald T., Bednárová L., Osička R., Pachl P., Sulc M., Lyngstadaas S. P., Slaby I., Vondrášek J. (2011) Biophysical characterization of recombinant human ameloblastin. Eur. J. Oral Sci. 119, 261–269 [DOI] [PubMed] [Google Scholar]
- 29. Murakami C., Dohi N., Fukae M., Tanabe T., Yamakoshi Y., Wakida K., Satoda T., Takahashi O., Shimizu M., Ryu O. H., Simmer J. P., Uchida T. (1997) Immunochemical and immunohistochemical study of the 27- and 29-kDa calcium-binding proteins and related proteins in the porcine tooth germ. Histochem. Cell Biol. 107, 485–494 [DOI] [PubMed] [Google Scholar]
- 30. Iwata T., Yamakoshi Y., Hu J. C., Ishikawa I., Bartlett J. D., Krebsbach P. H., Simmer J. P. (2007) Processing of ameloblastin by MMP-20. J. Dent. Res. 86, 153–157 [DOI] [PubMed] [Google Scholar]
- 31. Chun Y. H., Yamakoshi Y., Yamakoshi F., Fukae M., Hu J. C., Bartlett J. D., Simmer J. P. (2010) Cleavage site specificity of MMP-20 for secretory-stage ameloblastin. J. Dent. Res. 89, 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nanci A., Zalzal S., Lavoie P., Kunikata M., Chen W., Krebsbach P. H., Yamada Y., Hammarström L., Simmer J. P., Fincham A. G., Snead M. L., Smith C. E. (1998) Comparative immunochemical analyses of the developmental expression and distribution of ameloblastin and amelogenin in rat incisors. J. Histochem. Cytochem. 46, 911–934 [DOI] [PubMed] [Google Scholar]
- 33. Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., Krebsbach P. H., Nanci A., Kulkarni A. B., Yamada Y. (2004) Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 167, 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wazen R. M., Moffatt P., Zalzal S. F., Yamada Y., Nanci A. (2009) A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 28, 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sonoda A., Iwamoto T., Nakamura T., Fukumoto E., Yoshizaki K., Yamada A., Arakaki M., Harada H., Nonaka K., Nakamura S., Yamada Y., Fukumoto S. (2009) Critical role of heparin binding domains of ameloblastin for dental epithelium cell adhesion and ameloblastoma proliferation. J. Biol. Chem. 284, 27176–27184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beyeler M., Schild C., Lutz R., Chiquet M., Trueb B. (2010) Identification of a fibronectin interaction site in the extracellular matrix protein ameloblastin. Exp. Cell Res. 316, 1202–1212 [DOI] [PubMed] [Google Scholar]
- 37. Iizuka S., Kudo Y., Yoshida M., Tsunematsu T., Yoshiko Y., Uchida T., Ogawa I., Miyauchi M., Takata T. (2011) Ameloblastin regulates osteogenic differentiation by inhibiting Src kinase via cross talk between integrin beta1 and CD63. Mol. Cell Biol. 31, 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simmer J. P., Lau E. C., Hu C. C., Aoba T., Lacey M., Nelson D., Zeichner-David M., Snead M. L., Slavkin H. C., Fincham A. G. (1994) Isolation and characterization of a mouse amelogenin expressed in Escherichia coli. Calcif. Tissue Int. 54, 312–319 [DOI] [PubMed] [Google Scholar]
- 39. Khan F., He M., Taussig M. J. (2006) Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on Ni-nitrilotriacetic acid surfaces. Anal. Chem. 78, 3072–3079 [DOI] [PubMed] [Google Scholar]
- 40. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor, Cold Spring Harbor, NY [Google Scholar]
- 41. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 43. Güntert P., Wüthrich K. (1991) Improved efficiency of protein structure calculations from NMR data using the program DIANA with redundant dihedral angle constraints. J. Biomol. NMR 1, 447–456 [DOI] [PubMed] [Google Scholar]
- 44. Dhamija S., Krebsbach P. H. (2001) Role of Cbfa1 in ameloblastin gene transcription. J. Biol. Chem. 276, 35159–35164 [DOI] [PubMed] [Google Scholar]
- 45. Koradi R., Billeter M., Wuthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- 46. Benada O., Pokorný V. (1990) Modification of the Polaron sputter-coater unit for glow-discharge activation of carbon support films. J. Electron Microsc. Tech. 16, 235–239 [DOI] [PubMed] [Google Scholar]
- 47. Dawson R., Müller L., Dehner A., Klein C., Kessler H., Buchner J. (2003) The N-terminal domain of p53 is natively unfolded. J. Mol. Biol. 332, 1131–1141 [DOI] [PubMed] [Google Scholar]
- 48. Fincham A. G., Moradian-Oldak J., Diekwisch T. G., Lyaruu D. M., Wright J. T., Bringas P., Jr., Slavkin H. C. (1995) Evidence for amelogenin “nanospheres” as functional components of secretory-stage enamel matrix. J. Struct. Biol. 115, 50–59 [DOI] [PubMed] [Google Scholar]
- 49. Du C., Falini G., Fermani S., Abbott C., Moradian-Oldak J. (2005) Supramolecular assembly of amelogenin nanospheres into birefringent microribbons. Science 307, 1450–1454 [DOI] [PubMed] [Google Scholar]
- 50. Strawich E., Poon P. H., Renugopalakrishnan V., Glimcher M. J. (1985) Relative molecular mass determination of a major, highest relative molecular mass extracellular amelogenin of developing bovine enamel. FEBS Lett. 184, 188–192 [DOI] [PubMed] [Google Scholar]
- 51. Fincham A. G., Moradian-Oldak J., Simmer J. P., Sarte P., Lau E. C., Diekwisch T., Slavkin H. C. (1994) Self-assembly of a recombinant amelogenin protein generates supramolecular structures. J. Struct. Biol. 112, 103–109 [DOI] [PubMed] [Google Scholar]
- 52. Benseny-Cases N., Klementieva O., Malý J., Cladera J. (2012) Granular nonfibrillar aggregates and toxicity in Alzheimer's disease. Curr. Alzheimer Res. 9, 962–971 [DOI] [PubMed] [Google Scholar]
- 53. Klinov D., Dwir B., Kapon E., Borovok N., Molotsky T., Kotlyar A. (2007) High-resolution atomic force microscopy of duplex and triplex DNA molecules. Nanotechnology 18, 225102 [Google Scholar]
- 54. Makin O. S., Sikorski P., Serpell L. C. (2006) Diffraction to study protein and peptide assemblies. Curr. Opin. Chem. Biol. 10, 417–422 [DOI] [PubMed] [Google Scholar]
- 55. Whitehouse C., Fang J., Aggeli A., Bell M., Brydson R., Fishwick C. W., Henderson J. R., Knobler C. M., Owens R. W., Thomson N. H., Smith D. A., Boden N. (2005) Adsorption and self-assembly of peptides on mica substrates. Angew. Chem. Int. Ed. Engl. 44, 1965–1968 [DOI] [PubMed] [Google Scholar]
- 56. Hansma H. G. (2010) Possible origin of life between mica sheets. J. Theor. Biol. 266, 175–188 [DOI] [PubMed] [Google Scholar]
- 57. Paine M. L., Deutsch D., Snead M. L. (1996) Carboxyl-region of tuftelin mediates self-assembly. Connect. Tissue Res. 35, 157–161 [DOI] [PubMed] [Google Scholar]
- 58. Fukae M., Kanazashi M., Nagano T., Tanabe T., Oida S., Gomi K. (2006) Porcine sheath proteins show periodontal ligament regeneration activity. Eur. J. Oral. Sci. 114, 212–218 [DOI] [PubMed] [Google Scholar]
- 59. Paine M. L., Krebsbach P. H., Chen L. S., Paine C. T., Yamada Y., Deutsch D., Snead M. L. (1998) Protein-to-protein interactions. Criteria defining the assembly of the enamel organic matrix. J. Dent. Res. 77, 496–502 [DOI] [PubMed] [Google Scholar]
- 60. Ali M. H., Imperiali B. (2005) Protein oligomerization. How and why. Bioorg. Med. Chem. 13, 5013–5020 [DOI] [PubMed] [Google Scholar]
- 61. Namba K. (2001) Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells 6, 1–12 [DOI] [PubMed] [Google Scholar]
- 62. Matthews J. M., Sunde M. (2012) Dimers, oligomers, everywhere. Adv. Exp. Med. Biol. 747, 1–18 [DOI] [PubMed] [Google Scholar]
- 63. Kobayashi K., Yamakoshi Y., Hu J. C., Gomi K., Arai T., Fukae M., Krebsbach P. H., Simmer J. P. (2007) Splicing determines the glycosylation state of ameloblastin. J. Dent. Res. 86, 962–967 [DOI] [PubMed] [Google Scholar]
- 64. Yamakoshi Y., Tanabe T., Oida S., Hu C. C., Simmer J. P., Fukae M. (2001) Calcium binding of enamel proteins and their derivatives with emphasis on the calcium-binding domain of porcine sheathlin. Arch. Oral Biol. 46, 1005–1014 [DOI] [PubMed] [Google Scholar]