Abstract
Development of high-throughput sequencing-based methods has enabled us to examine nuclear architecture at unprecedented resolution, allowing further examination of the function of long-range chromosomal interactions. Here, we review methods used to investigate novel long-range chromosomal interactions and genome-wide organization of chromatin. We further discuss transcriptional activation and silencing in relation to organization and positioning of gene loci and regulation of chromatin organization through protein complexes and noncoding RNAs.
Keywords: Chromatin, Chromatin Regulation, Chromatin Structure, Nuclear Organization, Nuclear Structure, Chromosome Conformation Capture
Introduction
The nucleus is a three-dimensional structure that is often simplified to a one-dimensional linear structure, which overlooks the importance of nuclear organization in biological functions. Early microscopy studies on nuclear architecture focused on general organizing principles such as chromosome positioning, which led to the hypothesis that each chromosome remains within its own territory during interphase (1, 2). However, it is now recognized that chromosome territories frequently intermingle and coincide with genome function and stability (3, 4). The advent of chromosome conformation capture (3C)2-based methods has transformed our understanding of long-range chromosomal interactions, revealing through sequencing analysis a highly complex landscape consisting of specific localization patterns that correlate with transcriptional activation, repression, translocation, and other biological events. Together with traditional imaging-based methods (5), these newer technologies are starting to uncover more general principles of chromosomal organization among a variety of cell types (6–8).
In this minireview, we will first examine recent technologies used to define long-range interactions. We will then discuss the functional implications of these long-range interactions and the different factors involved in organizing long-range interactions in the nucleus. Our focus will be mainly on chromatin conformation. Many other excellent reviews focusing on global nuclear organization relating to chromosome positioning (9) and nuclear bodies (10) are available for interested readers.
Studying Long-range Chromosomal Interactions with FISH- and 3C-based Methods
Nuclear organization was initially explored using imaging analysis such as FISH-based methods (11). Multicolor FISH provided direct evidence for chromosome territories (5). FISH was also used to correlate transcriptional activity with position of genes within a chromosome territory in several studies. Actively transcribed genes at the HoxB locus were more likely seen looped out of its chromosome territory (12). Positioning of genes at the periphery of a chromosome territory reflected a repressed transcriptional state (13). Genes located far from one other can often aggregate and colocalize when activated (14). Interestingly, FISH studies have also characterized in trans long-range interactions as rare events, occurring in 5–15% of cells (15–20). However, FISH requires fixation of cells, leading to two interpretations for these data (21). It may be that long-range interactions are so transient that, at any given time, we can observe only a small percentage of these interactions or that long-range interactions are stable but present in only a subset of cells. The latter observation is supported when transactivation, measured based on accumulation of cytoplasmic mRNA in interchromosomal interactions, occurs only in a small percentage of cells (22). Although requiring further study, both confirm that interactions are heterogeneous within cells.
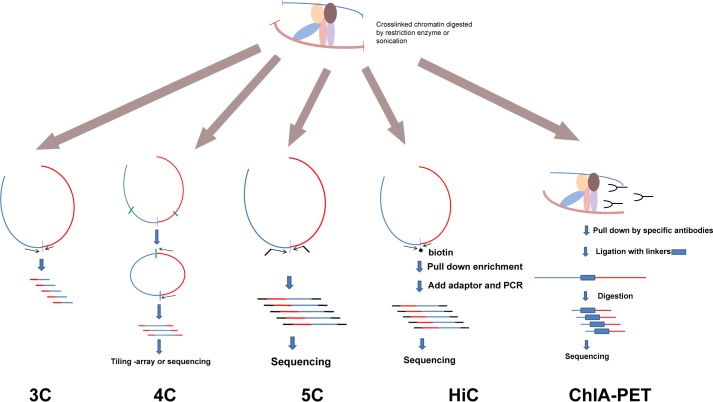
Recently, new approaches have been developed based mostly on 3C technology, introduced by Dekker et al. (23) to quantify interactions between distant DNA regions (Fig. 1). In the original protocol, cells are fixed with formaldehyde, and chromatin is digested with either “six cutter” restriction enzymes such as HindIII, BamHI, and EcoRI or “four cutter” enzymes such as DpnII, MboI, and Csp6I. Then, to ensure that only DNA fragments within the same DNA-protein complex will ligate to each other, the fragments are diluted to favor in cis ligation. Ligation products are identified by PCR using primers targeting sequences flanking the restriction enzyme cutting sites. Usually, various primer combinations targeting every restriction site in a specific region are used to construct a matrix of ligation efficiency, revealing interaction frequencies within that region. The traditional 3C approach captures “one-to-one” interactions, where the gene of interest is probed for probable interacting partner loci; however, in many cases, there is no prior indication of where these potential interacting sites are.
FIGURE 1.
Schematic representation of 3C-based methods. There are many methods derived from the original 3C design. Here, we present a few popular methods. In brief, cells are cross-linked, and chromatin is digested by restriction enzymes or sonicated. The structures of protein complexes containing DNA are preserved. These complexes are then diluted to a very low concentration, and ligation reactions are performed. Different amplification strategies are used to measure the relative cross-linking efficiency between loci. 3C is used to detect one specific interaction. 4C detects all possible interacting regions of one given locus. 5C and HiC provide “many-to-many” interacting efficiencies in a large genomic region or the whole genome. ChIP-PET includes immunoprecipitation to specifically examine the long-range interactions associated with a specific protein.
Building on the 3C procedure, chromosome conformation capture-on-chip (4C) defines a bait region to discover novel interacting regions (15, 16). A variety of 4C strategies have been developed, but generally, the fixation and digestion steps are similar to 3C analysis, where circular DNA loops are also formed by ligation (Fig. 1). Inverse PCR primers designed to amplify all unknown sequences are ligated to the bait region. The amplification product is then subjected to microarray or next-generation sequencing. In addition to using enzyme digestion, sonication can also be adapted to 4C to avoid systematic bias of enzyme cutting (24, 25).
To look at all contact frequencies between any two points within a single large genomic region, chromosome conformation capture carbon copy (5C) was designed to generate a matrix of interaction frequencies (7, 26) using oligonucleotide pairs matched to every ligation site between interacting 3C fragments. After amplification, readout of the junctions using microarray or next-generation sequencing can generate a three-dimensional organization map of a large genomic region at high resolution. 5C is powerful because it can theoretically measure the interaction efficiency between any two digestion sites; however, a very large number of oligonucleotides are needed to evaluate the conformation of a whole chromosome or an entire genome, so the cost of synthesizing such a large amount of primers prevents this technology from being applied to genome-wide studies.
A similar but more powerful method, HiC can generate an “all-to-all” genome-wide interaction frequency matrix (6, 7). The fixation and digestion steps are similar to those in the basic 3C protocol, but after digestion, the restriction ends are filled in with biotin-labeled nucleotides, which after blunt end ligation can be pulled down for high-throughput sequencing. This removes the need to design specific oligonucleotide pairs (7) and increases the resolution to ∼1 Mb based on 10 million pair-end reads (7). However, increasing HiC resolution is difficult because a 10-fold increase in resolution requires a 100-fold increase in sequence depth (27). Because HiC is only able to resolve on the Mb level, correlation with specific genes or epigenetic marks still remains unrealistic. Only in organisms with smaller genomes such as yeast can a kb level resolution be reached (28). Nevertheless, HiC remains a powerful tool for revealing chromosome territories and genome compartmentalization.
To specifically screen “point-to-point” interactions, another strategy called ChIA-PET was developed combining ChIP with 3C to discover loops bound by particular proteins (29). In the initial study, Fullwood et al. reported thousands of intrachromosomal contacts between estrogen receptor α-binding sites. Recently, this technique was extended to delineate looping activity between CCCTC-binding factor (CTCF) and RNA polymerase II (30, 31). ChIA-PET is different from most other 3C-based protocols (Fig. 1). First, sonication is used instead of enzyme digestion, which rules out the digestion inefficiency at different restriction enzyme cutting sites. Second, an immunoprecipitation step is added after ligation. The interactions detected by ChIA-PET can be validated by traditional 3C and 4C (29), suggesting that ChIA-PET is robust and reproducible.
These 3C-based techniques are powerful but limited because 3C-based methods can capture only the average chromatin contact frequency in all cells. Whether these contact frequencies are significant will require validation in individual cells through FISH, which remains the only way to detect organization at the single cell level so far.
Long-range Interactions and Transcription
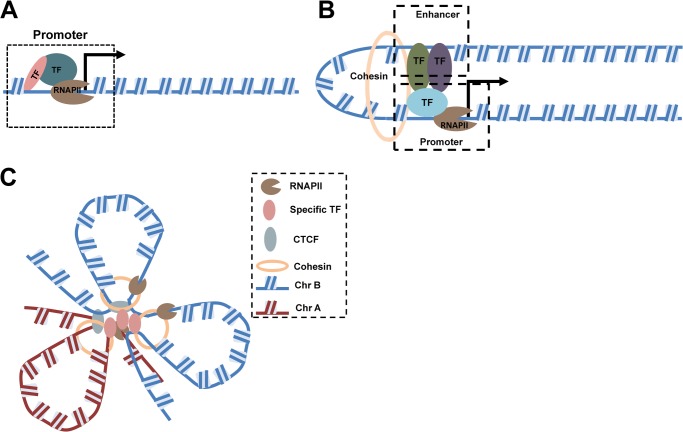
The classic explanation of transcription imagines a one-dimensional process in which RNA polymerase slides through a region to be transcribed (Fig. 2, A and B), but transcriptional mechanisms in vivo are likely very different. Early immunolabeling experiments of nascent RNA transcripts identified uneven distribution of transcripts in nuclei. These discrete foci were found to be sensitive to transcriptional inhibition (32). Even at saturation, the number of foci observed in such labeling studies ranged from hundreds to several thousand per nucleus (14, 33, 34), but there were far fewer foci than active transcription units observed (33, 35). Thus, regions in which several transcription units shared the same foci came to be called “transcription factories” (36).
FIGURE 2.
Transcriptional architecture: from a one-dimensional linear model to a three-dimensional transcription factory model. A, the simplest model of transcription organization considers chromatin structure as one-dimensional and posits that the transcriptional machinery acts mainly on promoters or enhancers immediately upstream of a transcription start site. TF, transcription factor. B, the looping model usually describes interaction between a promoter and a distant upstream enhancer. The cohesin complex is considered essential to maintain the looping structure. C, a transcription factory consists of immobilized and enriched RNAPII foci. DNA loci in cis and in trans are recruited to the factory by specific transcription factors as well as by CTCF and the cohesin complex. Different chromosomes are represented as Chr A and Chr B.
Immunogold and immunofluorescent detection of RNA polymerase II (RNAPII) coupled with nascent RNA revealed that many RNAPII foci and nascent RNA foci overlap (37). However, not all RNAPII sites are positive for nascent RNA labeling, indicating that not all RNAPII-enriched foci are equally active. RNAPII phosphorylated at Ser-2 and Ser-5 label active transcription factories, whereas RNAPII foci phosphorylated only at Ser-5 are transcription poised sites (13, 34). Recent studies showed that when transcription initiation is inhibited by heat shock, RNAPII remains at the transcription foci 30 min after active genes have moved away (38), suggesting that transcription factories are not self-assembled RNAPII proteins brought together by transcription but are relatively stable subnuclear compartments.
A key feature of the transcription factory model is that disparate transcription units can share the same factory at a given frequency, a concept initially referred to as “aggregation neighboring active genes” (39). Subsequent studies revealed that colocalization of “neighboring genes” up to 40 Mb away can share the same factories at higher-than-expected frequencies, as confirmed by both RNA FISH/RNAPII Ser-5 colocalization and 3C assays (14). Colocalization of active loci is not limited to genes on the same chromosome but can be expanded to the whole genome, although trans colocalization occurs with reduced frequency (40). Studies comparing transcription of α-globin (Hba) and β-globin (Hbb) “super genes,” constitutively expressed genes in all erythroid cells from nascent RNA FISH analysis (14), with other active genes suggested that although super genes constantly occupy transcription factories, other active genes transiently move into these factories only when transcription is needed. Supporting this “burst” model, kinetic analysis of immediate-early genes such as Fos and Myc indicated that induction as short as 5 min is sufficient for Fos relocation from outside of transcription factories into the constitutively transcribed Igh locus (41). Although the concept of transcription factories has become popular in recent years, debate continues on whether transcription is the direct and rapidly acting force that drives loci to transcription factories. One study showed that inhibition of transcription elongation for 5 h does not perturb colocalization (42). The interpretation may be that although correlated with transcription, colocalization is independent of transcriptional bursts. Other examples argue that colocalization of active genes may be mediated through nuclear speckles enriched in splicing factor SC35 (43). However, this may be an experimental artifact because active RNAPII redistributes with nuclear speckles in protocols utilizing chromatin fixation (44).
If active alleles “gather” to share basic transcriptional machinery, one would expect that active alleles in the same foci would behave similarly, but evidence appears to the contrary (40). Specialized transcription factories seem to be the organizers of coregulated genes (45). As shown by an immuno-RNA FISH tracking experiment, a minichromosome containing active genes introduced into cells localized with endogenous genes within a subset of transcription factories depending on the promoters of the active genes, suggesting that coregulated genes share the same factory (46). Schoenfelder et al. (40) provided more direct evidence of in vivo clustering of active genes; when a human HBB allele was introduced into a mouse genome, the integrated HBB preferentially localized with the endogenous mouse Hbb locus compared with the mouse Hba locus. When an enhanced 4C assay was applied to screen loci occupied by active RNAPII and interacting with Hbb or Hba, hundreds of overlapping loci were observed that contained both in cis and in trans transcription partners. These colocalization events were regulated by Klf1, which, together with RNAPII, formed these specialized transcription factories. Similarly, NFκB factories transcribing microRNAs were observed during TNFα induction (47).
In summary, a functionally organized nucleus consists of specialized transcription factories containing active RNAPII, specific transcription factors, and various genes active or poised for transcription. Organization of the nucleus' intra- and interchromosomal interactions ensures compartmentalization of nonrandom transcription. These conformations may be tissue-specific and dynamically regulated in particular biological events.
Nuclear Lamins and Silenced Chromosome Regions
Nuclear lamins are type V intermediate filament proteins that are located between the inner nuclear membrane and peripheral heterochromatin (48). They are involved in various functions, including regulation of nuclear envelope shape (49), formation of the mitotic spindle (50), DNA replication (51), and transcription (52). More recently, lamins were shown to associate with transcriptionally inactive chromatin in humans and Drosophila (53, 54). These large lamin-interacting domains (LADs) span >1 Mb and are mostly heterochromatic. The location of these LADs depends partly on their interaction with lamins, although definitive evidence is still lacking. For example, in lamin B null (LMNB−/−) mouse embryonic fibroblasts, despite having lower gene density, chromosome 18 is positioned away from the nuclear periphery (55, 56). Apart from LADs, centromeres and telomeres also interact with lamins. Centromeres are usually anchored near the nuclear periphery by B-type lamins (57, 58). Similarly, the distribution and stability of telomeres are influenced by lamins as well (59).
Although the nuclear lamina associates with largely repressed regions, whether it causes transcriptional silencing remains an unanswered question. Transcriptional activity at the nuclear lamina has been shown to be similar to that within internal regions of the nucleus (52). Moreover, when chromosome regions relocate to the nuclear periphery through tethering to the inner nuclear membrane, only a fraction of the genes show reduced expression (60). Additionally, genes can relocate to other organizing structures such as pericentromeric heterochromatin loci and be silenced (61, 62). Further investigation will clarify what role nuclear lamins play in transcriptional silencing.
Potential Role of Long-range Interactions in Chromosomal Translocation
Long-range interactions may be involved in creating chromosomal translocations. As a hallmark of cancer, specific chromosome regions frequently become translocated (63) through DNA single- or double-strand breaks (DSBs) (64, 65). In many cases, this results in fusion proteins that confer selective advantage to cells harboring the translocations, contributing to cell transformation. Particular chromosomal translocations seem to recur due to the nonrandom compartmentalization of the genome. Interestingly, translocations are often associated with active transcription (66), which may correlate certain translocation patterns with transcription factory colocalization (67). As mentioned previously, although Myc and Igh are located on different chromosomes, they share the same transcription factory, with 25% of Myc alleles colocalizing with the Igh locus after Fos induction. This preferential colocalization may lead to more frequent chromosomal translocation as seen in Burkitt lymphoma (41). Recently, two studies assessing genome-wide translocation compared translocation sites using 4C- or HiC-based sequencing (68, 69). To experimentally manipulate translocation frequency, both studies applied a knock-in or integrated I-SceI locus, which can be cut when restriction enzyme is introduced into the cells to cause artificial DSBs. Using high-throughput genome-wide translocation sequencing or translocation-capture sequencing, numerous translocations were identified after introducing DSBs at various loci. When comparing translocation frequency with spatial proximity, the studies reached complementary conclusions. Zhang et al. (69) utilized HiC to investigate translocation frequencies of various integrated I-SceI loci and found that both in cis and in trans translocation correlated with spatial proximity. Using 4C on a more defined system, Hakim et al. (68) found that whereas Aid−/− (Aid encodes an enzyme that create DNA breaks) translocation correlated with nuclear contact frequency, Aid-dependent translocations, which corresponded to recurrent translocations, did not correlate with nuclear contacts. Instead, it was DNA damage itself detected through AID activity in specific chromatin regions that correlated with recurrent translocations (68). Therefore, both studies concluded that although most certain types of translocation are associated with contact frequency, some are not.
Regulatory Mechanisms in Nuclear Organization
As discussed, the nucleus is highly organized and exhibits nonrandom conformations. Many powerful tools such as 3C-based methods and FISH have helped model nuclear organization in detail; however, there is still no efficient way to systematically identify the proteins that organize these interactions. Nonetheless, we discuss selected examples of protein and RNA factors that have been shown to influence chromatin conformation and may contribute to the organization of nuclear architecture.
Control of the β-Globin Locus by Transcription Factors EKLF, GATA-1, and FOG-1
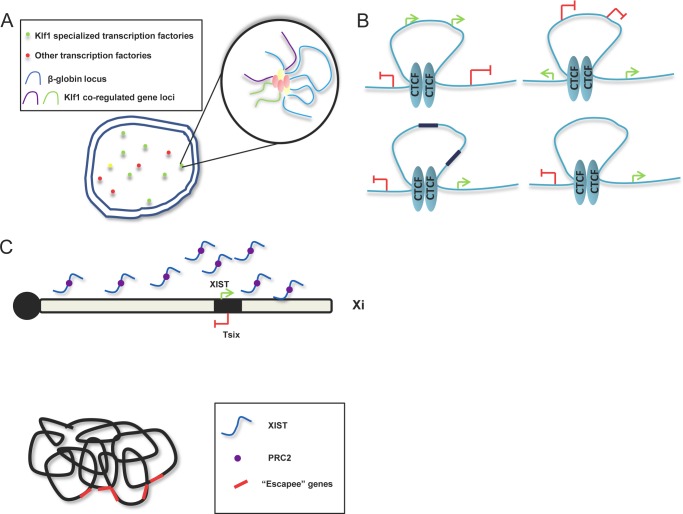
The β-globin locus is one of the most extensively studied examples of long-range interaction. The locus control region (LCR), located 25 kb from the closest gene, regulates a set of genes encoding variants of the β-chain of hemoglobin as far as 80 kb away. As determined by 3C analysis, LCR loops out genes to interact with the γ-globin loci in fetal cells and switches to regulating the β-globin loci in adult cells (70). LCR contains binding sites for transcription factors EKLF (Klf1) and GATA-1, both essential regulators of β-globin expression (71). EKLF knock-out mice show severely reduced β-globin expression and loss of long-range interactions between LCR and β-globin (72). Induction of GATA-1 fused with the estrogen receptor in a GATA-1−/− background promotes long-range interactions and GATA-1 occupancy of LCR/β-globin loci. Interaction of its cofactor FOG-1 with GATA-1 is also essential to mediate looping (73). More recently, enhanced 4C analysis of genome-wide long-range interactions involving β-globin loci showed that many of these loci colocalize with EKLF staining and contain EKLF-regulated genes (Fig. 3A) (40). These results validate the role of EKLF in organizing specialized transcription, including β-globin and its in cis and in trans interacting partners GATA-1 and FOG-1.
FIGURE 3.
Regulatory mechanisms governing nuclear architecture in various systems. A, β-globin loci in erythroid cells are enriched in Klf1-specialized transcription factories. Other Klf1-coregulated gene loci are dynamically colocalized with the β-globin locus and cotranscribed in specialized transcription factories. B, four categories of CTCF-mediated looping functions. Green arrows indicate genes with active epigenetic marks. Red brackets represent genes with repressive histone modification. Black boxes represent active enhancer elements. C, XIST-mediated XCI. The XIST lncRNA is expressed on Xi and spreads to cover the full chromosome (upper panel). The PRC2 complex is recruited by XIST and also loaded onto Xi. Xi is compacted and forms unique higher order chromosome structure (lower panel), with inactivated genes located inside, and active genes (“escapees” represented by red boxes) located on the outer surface.
CTCF Regulates Genome Organization in Pluripotent Stem Cells
Pluripotent stem cells are distinct from somatic cells in transcriptional and epigenetic status. Recent studies using 3C and 4C methods have shown that pluripotency factors Nanog and Klf4 have distinct interaction networks that maintain stem cell identity (74–76). Additionally, Dixon et al. (6) systematically explored genome-wide organization by HiC and provided significant insights into genome structure in human and mouse pluripotent stem cells. Using an optimized HiC method and computational algorithms, a high-resolution interaction map generated in stem cells and differentiated progenitors established the concept of the “topological domain,” defined as a region bound by narrow segments where chromatin interactions end. By determining long-range interacting partners at each specific domain and demarcating transition regions, this work provided a simple but elegant way of modeling genome organization.
HiC mapping of embryonic stem (ES) cells identified CTCF to be strongly enriched at topological boundary regions. As the only insulator characterized in vertebrates, CTCF is unique in its ability to block enhancers and demarcate euchromatin/heterochromatin boundaries (77). However, genome-wide occupancy profiling indicated that CTCF binds many more sites than anticipated, suggesting roles other than as an insulator (78, 79). On the local level, CTCF binding reportedly participates in long-range interactions such as colocalization of gene loci Igf2/H19 and Wsb1/Nf1, organization of the β-globin loci, and pairing of X chromosomes during X inactivation (19, 80, 81). Direct evidence that CTCF regulates long-range interactions on a genome-wide level was not available until Handoko et al. (30) characterized CTCF-mediated functional chromatin interactomes. The authors applied ChIA-PET through pulldown of CTCF in biological and technical replicates and deep sequenced for in cis and in trans long-range interactions (30). Altogether, 3306 CTCF-binding sites with 1480 intrachromosomal interactions and 336 interchromosomal interactions were identified and validated by 4C and FISH assays. These interactions were CTCF-dependent, demonstrated by CTCF knockdown cells analyzed by 3C and FISH. Most importantly, the authors defined four categories of looping with corresponding biological function based on assessment of epigenetic marks inside and outside the loop (Fig. 3B). The first category featured active epigenetic marks inside the loop and repressive marks outside, whereas the second described the opposite. The third category consisted of enhancer promoter loops, and the fourth considered loops acting as barriers between active and repressive regions. Defining these types of looping showed CTCF with multiple roles regulating higher order chromatin structure and linked chromatin loops with local transcription.
We are beginning to understand how CTCF functions in organizing chromatin structure, but several questions remain. First, CTCF has >68,000 binding sites in mouse ES cells, but only 3000 of them are identified to participate in looping (30). These 3000 sites cannot be distinguished from other CTCF sites, suggesting that cofactors may be involved. Another explanation may be that binding interactions are transient or occur only in a subpopulation of cells, thus requiring deeper sequencing depth to capture those interactions. Furthermore, although CTCF is ubiquitously expressed, no comprehensive studies have looked at whether transient depletion of CTCF has a significant influence on transcription, thus leaving the full biological impact of looping elusive for now.
Long Noncoding RNAs in X Chromosome Inactivation
Perhaps the best example of long noncoding RNAs (lncRNAs) mediating higher order chromatin structure is in X chromosome inactivation (XCI). XIST was the first lncRNA identified to be essential for XCI, which was later followed by the discovery of other lncRNA players such as RepA, Tsix, and Jpx (82).
XCI proceeds stepwise through development. Before differentiation of ES cells, the lncRNA Tsix is expressed biallelically and prevents binding of the RepA-PRC2 protein complex to the Xist promoter (83). When differentiation initiates, Tsix expression becomes limited to only one randomly selected allele (83). This leads to RepA-PRC2 complex loading onto the Xist locus on the to-be inactivated X chromosome (Xi) and activates Xist expression. Xist transcripts recruit PRC2 as transcription proceeds (84), while at the same time, transcription factor YY1 binds to the “nucleation center” of Xi but not to the activated X chromosome (85). Finally, the PRC2-XIST complex is loaded onto the YY1 protein and begins the spread of heterochromatic regions from the nucleation center to neighboring regions, eventually “coating” PRC2 and XIST over the surface of the entire Xi (Fig. 3C) (86). Although XIST clearly functions in almost every step of XCI, XIST depletion after XCI has little effect on chromosome conformation (87).
The central role of lncRNAs in XCI may not be unique but rather an example of their universal role as mediators of higher order chromatin structure because lncRNAs appear to be inherently suitable to mediate these types of interactions. lncRNAs can tether to transcription units to serve as allele-specific tags, as their binding sites are more selective than those of DNA-binding proteins (88), or serve as adaptors and interact with multiple proteins (89, 90). For example, a recent study showed HOTTIP lncRNA as an essential mediator of the WDR5-MLL complex in regulating long-range interactions and gene expression at the HoxA cluster (90). lncRNAs are more likely to be retained in the nucleus, making them essential regulators of nuclear architecture (91). Further characterization of lncRNA function through knock-out studies is needed to conclude which specific lncRNAs are involved in nuclear organization because some knock-out studies have shown that some lncRNAs are dispensable for nuclear organization (92–94).
How Dynamic Are the Long-range Interactions? Insights from Mobility Studies
To date, most chromatin interaction studies use fixed cells, which can only reflect interaction efficiency at a population average. FISH experiments face limitations as well, where well defined long-range interactions are seen only in a small population of cells (92–94). To fully understand chromatin dynamics, chromatin motility studies provide us with a glimpse. One study in yeast revealed that chromatin undergoes Brownian diffusion within a confined nuclear subregion (95), which accords with the concept of chromosome territories (96). In mammalian cells, it is estimated that the radius of rapid motion is ∼40 nm and requires ATP in an active mode of Brownian diffusion, not a passive one (97). Long-range motion, occurring usually at the micrometer level, is also much rarer than short-range motion (98) as concluded by 4C and HiC studies showing that most interactions occur within domains at the Mb level. Thus, these interchromosomal interactions and their overall biological significance still remain to be determined.
Conclusion
The significance of higher order chromatin structure and long-range chromosomal interactions has opened up new fields of research in molecular biology. A combination of FISH- and 3C-based technology has helped probe nuclear architecture at unprecedented resolution, and instead of imagining chromatin as a randomly compacted structure, we can now treat chromatin as a dynamic structure that loops within and between compartments in the nucleus. Many distal elements control specialized cellular functions such as transcription, gene silencing, and translocation through long-range interactions. Although the details of how features of nuclear architecture regulate these functions are still largely unknown, studies on insulators, transcription factors, the polycomb complex, and noncoding RNAs provide some preliminary insight into the regulation of biological function by these chromatin structure components.
Footnotes
- 3C
- chromosome conformation capture
- 4C
- chromosome conformation capture-on-chip
- 5C
- chromosome conformation capture carbon copy
- ChIA-PET
- chromatin interaction analysis with paired-end tag sequencing
- CTCF
- CCCTC-binding factor
- RNAPII
- RNA polymerase II
- LAD
- lamin-interacting domain
- DSB
- double-strand break
- EKLF
- erythroid Krüppel-like factor
- LCR
- locus control region
- ES
- embryonic stem
- lncRNA
- long noncoding RNA
- XCI
- X chromosome inactivation
- Xi
- to-be inactivated X chromosome.
REFERENCES
- 1. Cremer T., Cremer C. (2006) Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur. J. Histochem. 50, 223–272 [PubMed] [Google Scholar]
- 2. Hübner M. R., Spector D. L. (2010) Chromatin dynamics. Annu. Rev. Biophys. 39, 471–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Branco M. R., Pombo A. (2006) Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4, e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bian Q., Belmont A. S. (2012) Revisiting higher-order and large-scale chromatin organization. Curr. Opin. Cell Biol. 24, 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cremer T., Cremer M. (2010) Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N. L., Meisig J., Sedat J., Gribnau J., Barillot E., Blüthgen N., Dekker J., Heard E. (2012) Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferrai C., de Castro I. J., Lavitas L., Chotalia M., Pombo A. (2010) Gene positioning. Cold Spring Harb. Perspect. Biol. 2, a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spector D. L., Lamond A. I. (2011) Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cremer C., Cremer T., Gray J. W. (1982) Induction of chromosome damage by ultraviolet light and caffeine: correlation of cytogenetic evaluation and flow karyotype. Cytometry 2, 287–290 [DOI] [PubMed] [Google Scholar]
- 12. Chambeyron S., Bickmore W. A. (2004) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferrai C., Xie S. Q., Luraghi P., Munari D., Ramirez F., Branco M. R., Pombo A., Crippa M. P. (2010) Poised transcription factories prime silent uPA gene prior to activation. PLoS Biol. 8, e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Osborne C. S., Chakalova L., Brown K. E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J. A., Lopes S., Reik W., Fraser P. (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36, 1065–1071 [DOI] [PubMed] [Google Scholar]
- 15. Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 16. Zhao Z., Tavoosidana G., Sjölinder M., Göndör A., Mariano P., Wang S., Kanduri C., Lezcano M., Sandhu K. S., Singh U., Pant V., Tiwari V., Kurukuti S., Ohlsson R. (2006) Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38, 1341–1347 [DOI] [PubMed] [Google Scholar]
- 17. Spilianakis C. G., Lalioti M. D., Town T., Lee G. R., Flavell R. A. (2005) Interchromosomal associations between alternatively expressed loci. Nature 435, 637–645 [DOI] [PubMed] [Google Scholar]
- 18. Lomvardas S., Barnea G., Pisapia D. J., Mendelsohn M., Kirkland J., Axel R. (2006) Interchromosomal interactions and olfactory receptor choice. Cell 126, 403–413 [DOI] [PubMed] [Google Scholar]
- 19. Ling J. Q., Li T., Hu J. F., Vu T. H., Chen H. L., Qiu X. W., Cherry A. M., Hoffman A. R. (2006) CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 [DOI] [PubMed] [Google Scholar]
- 20. Sandhu K. S., Shi C., Sjölinder M., Zhao Z., Göndör A., Liu L., Tiwari V. K., Guibert S., Emilsson L., Imreh M. P., Ohlsson R. (2009) Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 23, 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams A., Spilianakis C. G., Flavell R. A. (2010) Interchromosomal association and gene regulation in trans. Trends Genet. 26, 188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noordermeer D., de Wit E., Klous P., van de Werken H., Simonis M., Lopez-Jones M., Eussen B., de Klein A., Singer R. H., de Laat W. (2011) Variegated gene expression caused by cell-specific long-range DNA interactions. Nat. Cell Biol. 13, 944–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Capturing chromosome conformation. Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 24. Gao F., Wei Z., Lu W., Wang K. (2013) Comparative analysis of 4C-Seq data generated from enzyme-based and sonication-based methods. BMC Genomics 14, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao F., Wei Z., An W., Wang K., Lu W. (2013) The interactomes of POU5F1 and SOX2 enhancers in human embryonic stem cells. Sci. Rep. 3, 1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dostie J., Richmond T. A., Arnaout R. A., Selzer R. R., Lee W. L., Honan T. A., Rubio E. D., Krumm A., Lamb J., Nusbaum C., Green R. D., Dekker J. (2006) Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Wit E., de Laat W. (2012) A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., Lee C., Shendure J., Fields S., Blau C. A., Noble W. S. (2010) A three-dimensional model of the yeast genome. Nature 465, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., Mohamed Y. B., Orlov Y. L., Velkov S., Ho A., Mei P. H., Chew E. G., Huang P. Y., Welboren W. J., Han Y., Ooi H. S., Ariyaratne P. N., Vega V. B., Luo Y., Tan P. Y., Choy P. Y., Wansa K. D., Zhao B., Lim K. S., Leow S. C., Yow J. S., Joseph R., Li H., Desai K. V., Thomsen J. S., Lee Y. K., Karuturi R. K., Herve T., Bourque G., Stunnenberg H. G., Ruan X., Cacheux-Rataboul V., Sung W. K., Liu E. T., Wei C. L., Cheung E., Ruan Y. (2009) An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Handoko L., Xu H., Li G., Ngan C. Y., Chew E., Schnapp M., Lee C. W., Ye C., Ping J. L., Mulawadi F., Wong E., Sheng J., Zhang Y., Poh T., Chan C. S., Kunarso G., Shahab A., Bourque G., Cacheux-Rataboul V., Sung W. K., Ruan Y., Wei C. L. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., Wang P., Poh H. M., Goh Y., Lim J., Zhang J., Sim H. S., Peh S. Q., Mulawadi F. H., Ong C. T., Orlov Y. L., Hong S., Zhang Z., Landt S., Raha D., Euskirchen G., Wei C. L., Ge W., Wang H., Davis C., Fisher-Aylor K. I., Mortazavi A., Gerstein M., Gingeras T., Wold B., Sun Y., Fullwood M. J., Cheung E., Liu E., Sung W. K., Snyder M., Ruan Y. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wansink D. G., Schul W., van der Kraan I., van Steensel B., van Driel R., de Jong L. (1993) Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 122, 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackson D. A., Hassan A. B., Errington R. J., Cook P. R. (1993) Visualization of focal sites of transcription within human nuclei. EMBO J. 12, 1059–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pombo A., Jackson D. A., Hollinshead M., Wang Z., Roeder R. G., Cook P. R. (1999) Regional specialization in human nuclei: visualization of discrete sites of transcription by RNA polymerase III. EMBO J. 18, 2241–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackson D. A., Iborra F. J., Manders E. M., Cook P. R. (1998) Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol. Biol. Cell 9, 1523–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iborra F. J., Pombo A., McManus J., Jackson D. A., Cook P. R. (1996) The topology of transcription by immobilized polymerases. Exp. Cell Res. 229, 167–173 [DOI] [PubMed] [Google Scholar]
- 37. Iborra F. J., Pombo A., Jackson D. A., Cook P. R. (1996) Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J. Cell Sci. 109, 1427–1436 [DOI] [PubMed] [Google Scholar]
- 38. Mitchell J. A., Fraser P. (2008) Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22, 20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cook P. R. (2002) Predicting three-dimensional genome structure from transcriptional activity. Nat. Genet. 32, 347–352 [DOI] [PubMed] [Google Scholar]
- 40. Schoenfelder S., Sexton T., Chakalova L., Cope N. F., Horton A., Andrews S., Kurukuti S., Mitchell J. A., Umlauf D., Dimitrova D. S., Eskiw C. H., Luo Y., Wei C. L., Ruan Y., Bieker J. J., Fraser P. (2010) Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42, 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Osborne C. S., Chakalova L., Mitchell J. A., Horton A., Wood A. L., Bolland D. J., Corcoran A. E., Fraser P. (2007) Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 5, e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palstra R. J., Simonis M., Klous P., Brasset E., Eijkelkamp B., de Laat W. (2008) Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS ONE 3, e1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown J. M., Green J., das Neves R. P., Wallace H. A., Smith A. J., Hughes J., Gray N., Taylor S., Wood W. G., Higgs D. R., Iborra F. J., Buckle V. J. (2008) Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 182, 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guillot P. V., Xie S. Q., Hollinshead M., Pombo A. (2004) Fixation-induced redistribution of hyperphosphorylated RNA polymerase II in the nucleus of human cells. Exp. Cell Res. 295, 460–468 [DOI] [PubMed] [Google Scholar]
- 45. Chakalova L., Fraser P. (2010) Organization of transcription. Cold Spring Harb. Perspect. Biol. 2, a000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu M., Cook P. R. (2008) Similar active genes cluster in specialized transcription factories. J. Cell Biol. 181, 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Papantonis A., Kohro T., Baboo S., Larkin J. D., Deng B., Short P., Tsutsumi S., Taylor S., Kanki Y., Kobayashi M., Li G., Poh H. M., Ruan X., Aburatani H., Ruan Y., Kodama T., Wada Y., Cook P. R. (2012) TNFα signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J. 31, 4404–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dechat T., Adam S. A., Taimen P., Shimi T., Goldman R. D. (2010) Nuclear lamins. Cold Spring Harb. Perspect. Biol. 2, a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goldman R. D., Grin B., Mendez M. G., Kuczmarski E. R. (2008) Intermediate filaments: versatile building blocks of cell structure. Curr. Opin. Cell Biol. 20, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tsai M. Y., Wang S., Heidinger J. M., Shumaker D. K., Adam S. A., Goldman R. D., Zheng Y. (2006) A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science 311, 1887–1893 [DOI] [PubMed] [Google Scholar]
- 51. Shumaker D. K., Solimando L., Sengupta K., Shimi T., Adam S. A., Grunwald A., Strelkov S. V., Aebi U., Cardoso M. C., Goldman R. D. (2008) The highly conserved nuclear lamin Ig-fold binds to PCNA: its role in DNA replication. J. Cell Biol. 181, 269–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kumaran R. I., Spector D. L. (2008) A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 180, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. (2006) Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014 [DOI] [PubMed] [Google Scholar]
- 54. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 55. Malhas A., Lee C. F., Sanders R., Saunders N. J., Vaux D. J. (2007) Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 176, 593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meaburn K. J., Cabuy E., Bonne G., Levy N., Morris G. E., Novelli G., Kill I. R., Bridger J. M. (2007) Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell 6, 139–153 [DOI] [PubMed] [Google Scholar]
- 57. Shimi T., Pfleghaar K., Kojima S., Pack C. G., Solovei I., Goldman A. E., Adam S. A., Shumaker D. K., Kinjo M., Cremer T., Goldman R. D. (2008) The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 22, 3409–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Solovei I., Schermelleh L., Düring K., Engelhardt A., Stein S., Cremer C., Cremer T. (2004) Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma 112, 410–423 [DOI] [PubMed] [Google Scholar]
- 59. Gonzalez-Suarez I., Redwood A. B., Gonzalo S. (2009) Loss of A-type lamins and genomic instability. Cell Cycle 8, 3860–3865 [DOI] [PubMed] [Google Scholar]
- 60. Finlan L. E., Sproul D., Thomson I., Boyle S., Kerr E., Perry P., Ylstra B., Chubb J. R., Bickmore W. A. (2008) Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 4, e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brown K. E., Baxter J., Graf D., Merkenschlager M., Fisher A. G. (1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell 3, 207–217 [DOI] [PubMed] [Google Scholar]
- 62. Hewitt S. L., Yin B., Ji Y., Chaumeil J., Marszalek K., Tenthorey J., Salvagiotto G., Steinel N., Ramsey L. B., Ghysdael J., Farrar M. A., Sleckman B. P., Schatz D. G., Busslinger M., Bassing C. H., Skok J. A. (2009) RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat. Immunol. 10, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y., Gostissa M., Hildebrand D. G., Becker M. S., Boboila C., Chiarle R., Lewis S., Alt F. W. (2010) The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv. Immunol. 106, 93–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nussenzweig A., Nussenzweig M. C. (2010) Origin of chromosomal translocations in lymphoid cancer. Cell 141, 27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tsai A. G., Lu H., Raghavan S. C., Muschen M., Hsieh C. L., Lieber M. R. (2008) Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell 135, 1130–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Klein I. A., Resch W., Jankovic M., Oliveira T., Yamane A., Nakahashi H., Di Virgilio M., Bothmer A., Nussenzweig A., Robbiani D. F., Casellas R., Nussenzweig M. C. (2011) Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Radford I. R. (2002) Transcription-based model for the induction of interchromosomal exchange events by ionizing irradiation in mammalian cell lines that undergo necrosis. Int. J. Radiat. Biol. 78, 1081–1093 [DOI] [PubMed] [Google Scholar]
- 68. Hakim O., Resch W., Yamane A., Klein I., Kieffer-Kwon K. R., Jankovic M., Oliveira T., Bothmer A., Voss T. C., Ansarah-Sobrinho C., Mathe E., Liang G., Cobell J., Nakahashi H., Robbiani D. F., Nussenzweig A., Hager G. L., Nussenzweig M. C., Casellas R. (2012) DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 484, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y., McCord R. P., Ho Y. J., Lajoie B. R., Hildebrand D. G., Simon A. C., Becker M. S., Alt F. W., Dekker J. (2012) Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Palstra R. J., Tolhuis B., Splinter E., Nijmeijer R., Grosveld F., de Laat W. (2003) The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 35, 190–194 [DOI] [PubMed] [Google Scholar]
- 71. Stamatoyannopoulos J. A., Goodwin A., Joyce T., Lowrey C. H. (1995) NF-E2 and GATA binding motifs are required for the formation of DNase I hypersensitive site 4 of the human β-globin locus control region. EMBO J. 14, 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Drissen R., Palstra R. J., Gillemans N., Splinter E., Grosveld F., Philipsen S., de Laat W. (2004) The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 18, 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vakoc C. R., Letting D. L., Gheldof N., Sawado T., Bender M. A., Groudine M., Weiss M. J., Dekker J., Blobel G. A. (2005) Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell 17, 453–462 [DOI] [PubMed] [Google Scholar]
- 74. Apostolou E., Ferrari F., Walsh R. M., Bar-Nur O., Stadtfeld M., Cheloufi S., Stuart H. T., Polo J. M., Ohsumi T. K., Borowsky M. L., Kharchenko P. V., Park P. J., Hochedlinger K. (2013) Genome-wide chromatin interactions of the Nanog locus in pluripotency, differentiation, and reprogramming. Cell Stem Cell 12, 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wei Z., Gao F., Kim S., Yang H., Lyu J., An W., Wang K., Lu W. (2013) Klf4 organizes long-range chromosomal interactions with the Oct4 locus in reprogramming and pluripotency. Cell Stem Cell 10.1016/j.stem.2013.05.010 [DOI] [PubMed] [Google Scholar]
- 76. Zhang H., Jiao W., Sun L., Fan J., Chen M., Wang H., Xu X., Shen A., Li T., Niu B., Ge S., Li W., Cui J., Wang G., Sun J., Fan X., Hu X., Mrsny, Randall J., Hoffman, Andrew R., Hu J.-F. (2013) Intrachromosomal looping is required for activation of endogenous pluripotency genes during reprogramming. Cell Stem Cell 10.1016/j.stem.2013.05.012 [DOI] [PubMed] [Google Scholar]
- 77. Phillips J. E., Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 79. Herold M., Bartkuhn M., Renkawitz R. (2012) CTCF: insights into insulator function during development. Development 139, 1045–1057 [DOI] [PubMed] [Google Scholar]
- 80. Donohoe M. E., Silva S. S., Pinter S. F., Xu N., Lee J. T. (2009) The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature 460, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Splinter E., Heath H., Kooren J., Palstra R. J., Klous P., Grosveld F., Galjart N., de Laat W. (2006) CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes Dev. 20, 2349–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lee J. T. (2010) The X as model for RNA's niche in epigenomic regulation. Cold Spring Harb. Perspect. Biol. 2, a003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Lee J. T., Davidow L. S., Warshawsky D. (1999) Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21, 400–404 [DOI] [PubMed] [Google Scholar]
- 84. Wutz A., Rasmussen T. P., Jaenisch R. (2002) Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167–174 [DOI] [PubMed] [Google Scholar]
- 85. Jeon Y., Lee J. T. (2011) YY1 tethers Xist RNA to the inactive X nucleation center. Cell 146, 119–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lee J. T. (2011) Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12, 815–826 [DOI] [PubMed] [Google Scholar]
- 87. Splinter E., de Wit E., Nora E. P., Klous P., van de Werken H. J., Zhu Y., Kaaij L. J., van Ijcken W., Gribnau J., Heard E., de Laat W. (2011) The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25, 1371–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mancini-Dinardo D., Steele S. J., Levorse J. M., Ingram R. S., Tilghman S. M. (2006) Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 20, 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rinn J. L., Kertesz M., Wang J. K., Squazzo S. L., Xu X., Brugmann S. A., Goodnough L. H., Helms J. A., Farnham P. J., Segal E., Chang H. Y. (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang K. C., Yang Y. W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B. R., Protacio A., Flynn R. A., Gupta R. A., Wysocka J., Lei M., Dekker J., Helms J. A., Chang H. Y. (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Djebali S., Davis C. A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G. K., Khatun J., Williams B. A., Zaleski C., Rozowsky J., Röder M., Kokocinski F., Abdelhamid R. F., Alioto T., Antoshechkin I., Baer M. T., Bar N. S., Batut P., Bell K., Bell I., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Falconnet E., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M. J., Gao H., Gonzalez D., Gordon A., Gunawardena H., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Luo O. J., Park E., Persaud K., Preall J. B., Ribeca P., Risk B., Robyr D., Sammeth M., Schaffer L., See L. H., Shahab A., Skancke J., Suzuki A. M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Ruan X., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T., Reymond A., Antonarakis S. E., Hannon G., Giddings M. C., Ruan Y., Wold B., Carninci P., Guigo R., Gingeras T. R. (2012) Landscape of transcription in human cells. Nature 489, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nakagawa S., Naganuma T., Shioi G., Hirose T. (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol. 193, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nakagawa S., Ip J. Y., Shioi G., Tripathi V., Zong X., Hirose T., Prasanth K. V. (2012) Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang B., Arun G., Mao Y. S., Lazar Z., Hung G., Bhattacharjee G., Xiao X., Booth C. J., Wu J., Zhang C., Spector D. L. (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Marshall W. F., Straight A., Marko J. F., Swedlow J., Dernburg A., Belmont A., Murray A. W., Agard D. A., Sedat J. W. (1997) Interphase chromosomes undergo constrained diffusional motion in living cells. Curr. Biol. 7, 930–939 [DOI] [PubMed] [Google Scholar]
- 96. Cremer T., Cremer M., Dietzel S., Müller S., Solovei I., Fakan S. (2006) Chromosome territories–a functional nuclear landscape. Curr. Opin. Cell Biol. 18, 307–316 [DOI] [PubMed] [Google Scholar]
- 97. Levi V., Ruan Q., Plutz M., Belmont A. S., Gratton E. (2005) Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys. J. 89, 4275–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Soutoglou E., Misteli T. (2007) Mobility and immobility of chromatin in transcription and genome stability. Curr. Opin. Genet. Dev. 17, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]