Background: Nrf2 activation is regulated by ERK.
Results: IQGAP1 interacts with Nrf2 and mediates Nrf2 activation by ERK.
Conclusion: Activation of Nrf2 by kinases is mediated by scaffold protein.
Significance: IQGAP1-mediated MEK-ERK activation of Nrf2 may represent a means of regulating the response of Nrf2 to oxidative stress.
Keywords: Antioxidants, ERK, Gene Silencing, Nrf2, Scaffold Proteins, IQGAP1, MEK, PEITC
Abstract
Nrf2 plays a critical role in the regulation of cellular oxidative stress. MEK-ERK activation has been shown to be one of the major pathways resulting in the activation of Nrf2 and induction of Nrf2 downstream targets, including phase II detoxifying/antioxidant genes in response to oxidative stress and xenobiotics. In this study, IQGAP1 (IQ motif-containing GTPase-activating protein 1), a new Nrf2 interaction partner that we have published previously, was found to modulate MEK-ERK-mediated Nrf2 activation and induction of phase II detoxifying/antioxidant genes. Nrf2 binds directly to the IQ domain (amino acids 699–905) of IQGAP1. Knockdown of IQGAP1 significantly attenuated phenethyl isothiocyanate- or MEK-mediated activation of the MEK-ERK-Nrf2 pathway. Knockdown of IQGAP1 also attenuated MEK-mediated increased stability of Nrf2, which in turn was associated with a decrease in the nuclear translocation of Nrf2 and a decrease in the expression of phase II detoxifying/antioxidant genes. In the aggregate, these results suggest that IQGAP1 may play an important role in the MEK-ERK-Nrf2 signaling pathway.
Introduction
The Nrf2 (NF-E2-related factor 2) signaling pathway, which is activated in response to different extracellular stimuli (1–4), modulates the oxidative stress responses in many cell types (5, 6). Nrf2, a transcription factor belongs to the CNC (Cap“n”Collar) family, has been shown to regulate the induction of phase II/antioxidant genes, including GST, NQO1 (NAD(P)H:quinone oxidoreductase 1), UDP-glucuronosyltransferase (UGT),3 γ-glutamylcysteine synthetase, and HO-1 (heme oxygenase-1) (7, 8). In unstimulated conditions, Nrf2 levels are generally low because Nrf2 is sequestrated by Keap1 (Kelch-like ECH-associated protein 1) and is targeted by ubiquitinase for ubiquitination. The ubiquitinated Nrf2 proteins are then degraded by proteasomes (9). However, in response to oxidative stress, Nrf2 dissociates from Keap1 and translocates into nucleus, where Nrf2 binds to a cis-acting element called the antioxidant response element (ARE)/electrophile response element (5′-(A/G)TGACNNNGC(A/G)-3′) at 5′-flanking promoter regions of the phase II/antioxidant genes (10).
Nrf2 activation has been shown to be regulated by different kinases, including MAPKs (11, 12) and PKC (13). MAPK cascades are activated in response to a variety of extracellular stimuli. The most widely studied MAPK pathway is the ERK cascade. ERK has been shown to activate Nrf2 and is required for activation of Nrf2 induced by different stimuli, including dietary compounds such as phenethyl isothiocyanate (PEITC) (1) or inducers of oxidative stress. In vitro kinase assays showed that PEITC can induce the phosphorylation of ERK, which in turn phosphorylates and activates Nrf2 (1).
We have recently identified IQGAP1 (IQ motif-containing GTPase-activating protein 1) as a novel binding partner of Nrf2 (14). Like other IQGAP family proteins such as IQGAP2 and IQGAP3, IQGAP1 shares conserved protein structure, including calmodulin-binding IQ motifs and the GTPase-activating protein homology domain (15). IQGAP1 was first identified as a Rac1- and Cdc42-binding protein coupled with actin. It is widely accepted that IQGAP1 inhibits its GTPase activity by recruiting Cdc42 and Rac1 (16, 17). IQGAP protein interacts with numerous cellular signaling molecules and coordinates various cellular events in actin and tubulin cytoskeletons through four major domains (18): the calponin homology domain for actin binding (19); the WW domain for ERK2 binding (20); IQ motifs for calmodulin (21), myosin light chain (22), and S100B (23) binding; the GTPase-activating protein-related domain for Cdc42 and Rac1 binding (16, 24); and the distal C-terminal domain for E-cadherin and β-catenin (17) and CLIP-170 (17, 25) binding.
Previous reports indicate that IQGAP1 acts as a scaffold protein and modulates MEK-ERK activity (26). In contrast, our preliminary results showed that IQGAP1 binds Nrf2 (14). Taking these results together, we hypothesize that IQGAP1 acts as a scaffold for Nrf2 and MEK-ERK machinery and modulates Nrf2 activity in response to the MAPK activator PEITC. We present evidence that IQGAP1 and Nrf2 associate both in vitro and in cells. The knockdown of IQGAP1 by shRNA attenuates PEITC- or MEK-induced activation of the MEK-ERK pathway, interaction of ERK and Nrf2, Nrf2 nuclear translocation, and phase II gene expression. These data suggest that IQGAP1 plays an important role in MEK-ERK-Nrf2 signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK-293 cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air in minimum essential medium supplemented with 10% heat-inactivated FBS and 50 units/ml penicillin/streptomycin mixture (Invitrogen). Cells were grown to 60–80% confluence before experiments.
Plasmids
IQGAP1 siRNA-DNA duplex was inserted into the pRNAT-CMV3.2/neo/cGFP vector (GenScript, Piscataway, NJ). The sequence for pRNAT-siIQ was 5′-GATCCTTCTACGGTCAATAGCTTCATTTGATATCCGATGAAGCTATTGACCGTAGAACGC-3′. The duplex was synthesized and purified by HPLC (Integrated DNA Technologies, San Diego, CA). Synthesized DNA was annealed to form double strands, cut with BamHI and XhoI restriction enzyme sites, and ligated into the pRNAT-CMV3.2/neo/cGFP vector. IQGAP1 cDNA was purchased from the I.M.A.G.E Clone Consortium (Invitrogen) and amplified by PCR with specifically designed primers containing appropriate restriction enzyme sites. The amplified IQGAP1 gene was cut with XhoI/ApaI restriction enzymes and then subcloned into the pDsRed-Mono-C1 vector (Clontech, Mountain View, CA). Myc-tagged constructs of human IQGAP1, IQGAP1-N, IQGAP1-N1, IQGAP1-N2, IQGAP1ΔCHD, IQGAP1ΔIQ, and IQGAP1ΔWW were described previously (26). Constitutively active MEK1 (pcDNA3-DNEE-MEK1) was a kind gift from Dr. Rony Seger (Weizmann Institute of Science) (35); HA-ERK (pcDNA3-HA-ERK) and GFP-Nrf2 plasmids were routinely used in our laboratory and were described previously (1).
Transient Transfection
Cells (0.5 × 106/well) were seeded in 6-well plates the day before transfection. The cells were then transfected using GeneJuice (EMD Chemicals) or FuGENE 6 (Roche) according to the manufacturers' instructions. For each well, plasmids were mixed with transfection reagents at a 1:6 (μg:μl) ratio and then added into 100 μl of Opti-MEM I. Various amounts of plasmids were also applied for certain experiments as indicated in the figure legends. pcDNA3.1 was added to make the total amount of plasmids equal. The transfection mixtures were mixed vigorously and incubated at room temperature for 15 min. The cells were then incubated with the transfection mixtures for 20–24 h in a 37 °C incubator. The cells were subjected to further treatments.
Reporter Gene Activity Assays
The ARE TATA-Inr luciferase reporter plasmid pARE-luc was obtained from Dr. William Fahl (University of Wisconsin, Madison, WI). Cells (1 × 105/well) were seeded in 24-well plates the day before transfection. Cells transfected with pARE-luc were washed once with PBS, scraped, and lysed in 200 μl of reporter lysis buffer (Promega) on ice for 10 min. After centrifugation at 12,000 rpm for 5 min at 4 °C, 10 μl of lysate was mixed with luciferase substrate (Promega), and ARE-luciferase activity was measured using a Sirius luminometer (Berthold Detection Systems, Huntsville, AL).
Western Blot Analysis
Cells were plated in 6-well culture dishes at ∼4.0 × 105 cells/well for 16 h prior to plasmid transfection. After transfection, cells were scraped and lysed with radioimmune precipitation assay lysis buffer (150 mm NaCl, 0.5% Triton X-100, 50 mm Tris-HCl (pH 7.4), 25 mm NaF, 20 mm EGTA, 1 mm DTT, 1 mm Na3VO4, and one protease inhibitor mixture tablet) for 30 min on ice, followed by centrifugation at 14,800 × g for 15 min. The protein concentration of the supernatant was measured using the BCA reagent. Protein (20 μg) was loaded on 4–15% CriterionTM Tris-HCl gel (Bio-Rad) and transferred onto PVDF membrane in Tris/glycine buffer (pH 8.4) containing 20% methanol. The membrane was then blocked in 5% fat-free dry milk in phosphate-buffered saline with 0.1% Tween 20 for 1 h and probed with primary antibodies and horseradish peroxidase-conjugated secondary antibody by standard Western blotting procedures. The proteins were visualized with the Femto signal chemiluminescent substrate (Thermo Fisher Scientific) with an image analyzer (Bio-Rad). Primary antibodies against IQGAP1 (H-109), Nrf2 (C-20), HO-1 (C-20), actin (C-11), lamin B (C-20), c-Myc (9E10), HA (Y-11), and ubiquitin (P4D1) and secondary antibodies were purchased from Santa Cruz Biotechnology. Anti-ERK, anti-phospho-ERK, anti-MEK, and anti-phospho-MEK antibodies were purchased from Cell Signaling. Anti-phospho-Ser/Thr-Pro antibody (05-368) was purchased from Millipore. Anti-GFP antibody (ab920) was purchased from Abcam.
Immunoprecipitation
Cells were plated on 100-mm dishes and cultured until >60–70% confluent. Next, the cells were treated, harvested, and lysed with M-PER buffer (Pierce) for the cytosolic extract. Cytosolic samples (400 μg) were subjected to immunoprecipitation assay using different antibodies. In this experiment, Dynabeads Protein G (Invitrogen) were used for immunoprecipitation. The protocol provided by the manufacturer was followed. Briefly, antibodies (10 μg) were incubated with Dynabeads Protein G for 10 min to make an antibody-bead complex. This complex was then incubated with cytosolic lysates for 10 min at room temperature. After the immunoprecipitation complexes were washed, they were resuspended with 200 μl of 2× SDS sample buffer, and 20-μl aliquots were subjected to Western blotting using different antibodies.
In Vitro Binding Assays
Full-length IQGAP1, truncated IQ fragments, and deletion mutants were produced with the TnT quick coupled transcription/translation system (Promega). The proteins were then incubated with His-tagged Nrf2 protein in 500 μl of buffer for 1 h at 4 °C. Anti-C-Myc antibody and Dynabeads Protein G were then added and incubated overnight. Alternatively, nickel-nitrilotriacetic acid (Ni-NTA) beads were incubated to pull down IQGAP1-Nrf2 complexes. After centrifugation, samples were washed and resolved by SDS-PAGE.
Quantitative RT-PCR
RNA extraction and quantitative RT-PCR analysis were performed following the SYBR Green quantitative PCR protocol. The primer sequences used were as follows: HO-1, 5′-AGAGGGAATTCTCTTGGCTGGCTT-3′ (forward) and 5′-ATGCCATAGGCTCCTTCCTCCTTT-3′ (reverse); UGT, 5′-ATGACCCGTGCCTTTATCACCCAT-3′ (forward) and 5′-ATGACCCGTGCCTTTATCACCCAT-3′ (reverse); and NQO1, 5′-AAGGATGGAAGAAACGCCTGGAGA-3′ (forward) and 5′-GGCCCACAGAAAGGCCAAATTTCT-3′ (reverse). The results were normalized to β-actin (forward, 5′-GCACAGAGCCTCGCCTT-3′; and reverse, 5′-GTTGTCGACGACGAGCG-3′). The relative expression levels of samples were calculated using the 2−ΔΔCt method.
RESULTS
IQGAP1 Binds Nrf2 in HEK-293 Cells, and PEITC Further Stimulates the Interaction
Previously, we identified that IQGAP1 and Nrf2 interact in vitro (14). To ascertain whether IQGAP1 and Nrf2 interact in a normal cellular environment, immunoprecipitation was performed using anti-IQGAP1 antibody. We observed that endogenous Nrf2 bound to IQGAP1 in HEK-293 cells (Fig. 1A). Little Nrf2 was detected in samples immunoprecipitated with nonimmune rabbit serum, confirming that IQGAP1 and Nrf2 specifically interact in HEK-293 cells. Upon stimulation with PEITC for 20 min, the interaction between IQGAP1 and Nrf2 increased and was attenuated in IQGAP1-knockdown cells (Fig. 1B).
FIGURE 1.
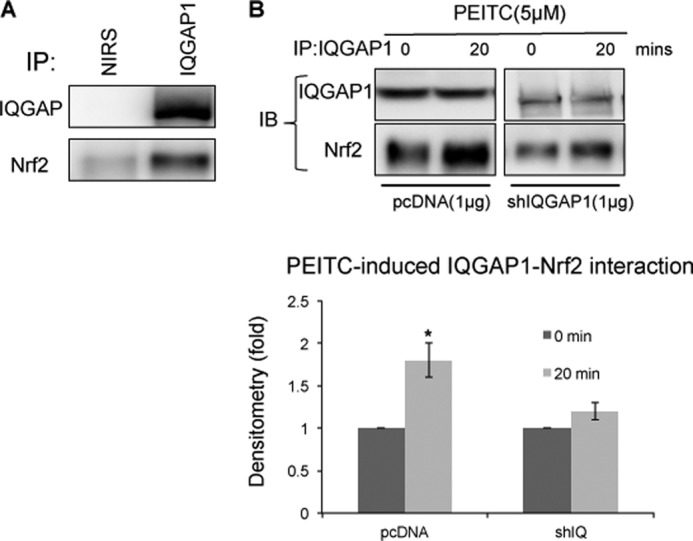
IQGAP1 and Nrf2 interact in a normal cellular environment. A, HEK-293 cell lysate was immunoprecipitated (IP) with anti-IQGAP1 antibody and nonimmune rabbit serum (NIRS). B, HEK-293 cells were transfected with pcDNA3.1 and IQGAP1 shRNA (shIQGAP1, shIQ) for 24 h and then treated with PEITC for 20 min, and cell lysates were immunoprecipitated with anti-IQGAP1 antibody. IQGAP1-Nrf2 interaction 20 min after PEITC treatment is expressed relative to interaction immediately after treatment and represents the mean ± S.D. (n = 3). *, p < 0.05. IB, immunoblot.
IQGAP1 Mediates PEITC-induced Expression of Phase II/Antioxidant Genes
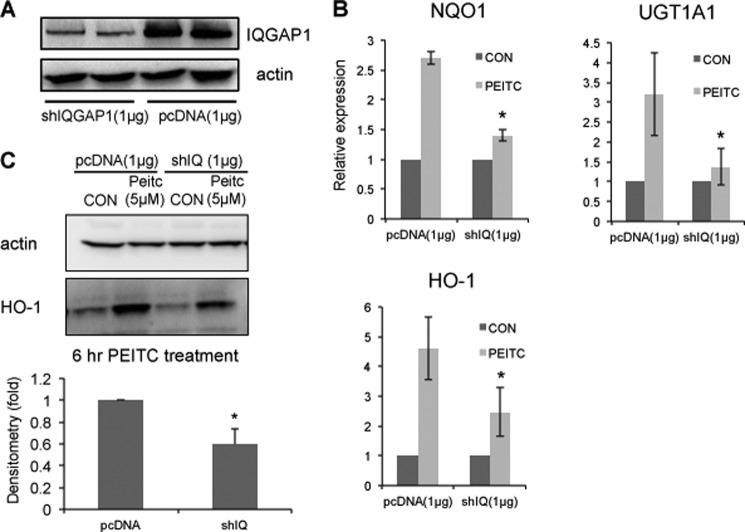
Transfection of HEK-293 cells with IQGAP1 shRNA in FuGENE 6 transfection reagent resulted in >50% knockdown of IQGAP1 protein (Fig. 2A). PEITC induced activation of Nrf2 and expression of phase II/antioxidant genes, including HO-1, NQO1, and UGT mRNAs. The phase II/antioxidant mRNAs were induced 2–5-fold by PEITC in HEK-293 cells but were attenuated in IQGAP1-knockdown HEK-293 cells (Fig. 2B). Specifically, Western blotting of HO-1 revealed a similar result. Expression of HO-1 protein was induced ∼3.5-fold by PEITC in HEK-293 cells but attenuated in IQGAP1-knockdown HEK-293 cells (Fig. 2C). Taking these results together, IQGAP1 is needed for PEITC-induced expression of phase II/antioxidant genes.
FIGURE 2.
IQGAP1 mediates PEITC-induced expression of phase II/antioxidant genes. A, HEK-293 cells were transfected with pcDNA3.1 and IQGAP1 shRNA (shIQGAP1) for 24 h. The down-regulation of IQGAP1 protein was measured. Data are representative of four independent experiments. B, HEK-293 cells transfected with IQGAP1 shRNA (shIQ) or pcDNA3.1 were treated with PEITC for 6 h. mRNA expression of NQO1, UGT1A1, and HO-1 was determined by quantitative PCR. Data represent the mean ± S.D. (n = 3). *, p < 0.05. CON, control. C, HEK-293 cells transfected with IQGAP1 shRNA or pcDNA3.1 were treated with PEITC for 6 h. Protein expression of HO-1 was determined by Western blotting. PEITC-induced HO-1 protein expression in IQGAP1-knockdown cells is expressed relative to wild-type cells and represents the mean ± S.D. (n = 3). *, p < 0.05.
IQGAP1 Mediates PEITC-induced Nuclear Accumulation of Nrf2
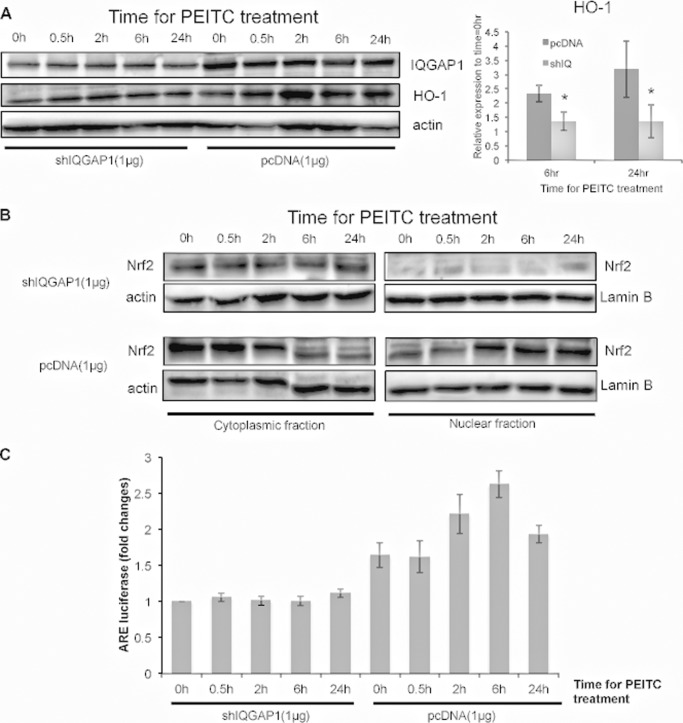
PEITC time-dependently induced HO-1 protein. However, the induction of HO-1 protein was substantially attenuated in IQGAP1-knockdown HEK-293 cells (Fig. 3A). To determine whether IQGAP1 modulates nuclear accumulation of Nrf2 induced by PEITC, the amount of Nrf2 in the nucleus after PEITC treatment was determined in IQGAP1-knockdown HEK-293 cells. Maximal Nrf2 nuclear translocation occurred in HEK-293 cells from 2 to 24 h after PEITC treatment, but the amount of nuclear Nrf2 was attenuated in IQGAP1-knockdown HEK-293 cells during this period (Fig. 3B). PEITC-induced ARE activity was also attenuated in IQGAP1-knockdown cells (Fig. 3C). These results indicate that IQGAP1 mediates PEITC-induced nuclear translocation of Nrf2.
FIGURE 3.
IQGAP1 mediates PEITC-induced nuclear accumulation of Nrf2. HEK-293 cells transfected with pcDNA3.1 or IQGAP1 shRNA (shIQGAP1, shIQ) were treated with PEITC. Cytoplasmic and nuclear fractions were collected 0, 0.5, 2, 6, and 24 h after treatment. A, protein expression of HO-1 in the cytoplasm was determined. Data are representative of three independent experiments. B, nuclear accumulation of Nrf2 was determined. Data are representative of three independent experiments. C, HEK-293 cells were transfected with pcDNA3.1 or IQGAP1 shRNA and ARE plasmids for 24 h. ARE induction was determined using luciferase assays. ARE activities induced by PEITC are expressed relative to IQGAP1 shRNA-transfected cells at 0 h after PEITC treatment and represent the mean ± S.D. (n = 2).
Nrf2 Binds to the IQ Domain of IQGAP1
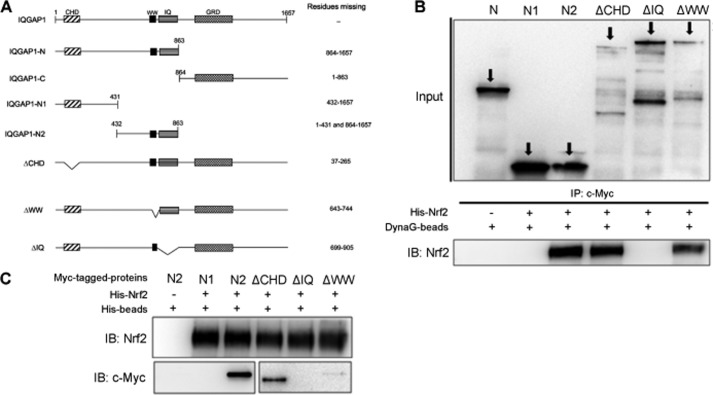
We next aimed to determine which IQGAP1 domain Nrf2 binds to using the IQGAP1 constructs (Fig. 4A) provided by one of us (D. B. S.) Myc-tagged full-length IQGAP1, truncated IQGAP1, and deletion mutant proteins were generated following to the of TnT quick coupled transcription/translation system protocol and were mixed with purified His-tagged Nrf2. The mixtures were then subjected to immunoprecipitation using anti-Myc antibody and pulled down with Dynabeads Protein G. We showed that the truncated N-terminal half (amino acid residues 1–863) of IQGAP1 bound to Nrf2, and further examination revealed the specific binding of Nrf2 to the IQ domain of IQGAP1 (Fig. 4B). Similarly, immunoprecipitation using Ni-NTA beads to pull down His-tagged Nrf2 together with the various truncated IQGAP1 proteins confirmed that Nrf2 specifically bound to the IQ domain (amino acid residues 699–905) of IQGAP1 (Fig. 4C).
FIGURE 4.
Nrf2 binds to the IQ domain of IQGAP1. A, schematic diagram of the IQGAP1 constructs. CHD, calponin homology domain; GRD, GTPase-activating protein-related domain. B, full-length IQGAP1 proteins, truncated IQGAP1 proteins, and deletion mutants were generated according to the protocol of the TnT® quick coupled transcription/translation system. Dynabeads Protein G (DynaG-beads) and anti-c-Myc antibody were used to pull down proteins that formed complexes with Myc-tagged IQGAP1 proteins. Samples were resolved by SDS-PAGE and probed with anti-Nrf2 antibody. IP, immunoprecipitate; IB, immunoblot. C, Ni-NTA beads were used to pull down proteins that formed complexes with His-tagged Nrf2. Samples were resolved by SDS-PAGE and probed with anti-c-Myc antibody.
IQGAP1 Mediates PEITC-induced MEK-ERK Activation and MEK-ERK-induced ARE Expression
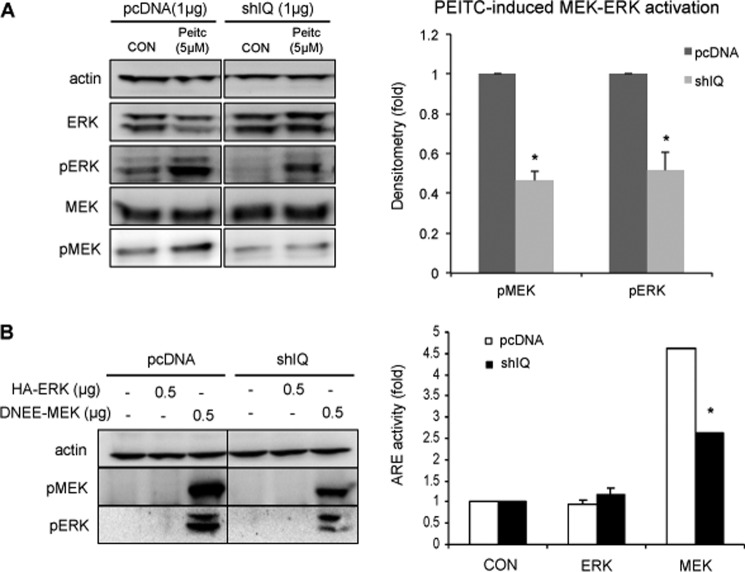
We have shown previously that activation of the MEK-ERK pathway is required for PEITC-induced phase II/antioxidant gene expression (27). PEITC-induced MEK-ERK activation was dependent on IQGAP1, with an increase in phosphorylation of MEK and ERK 30 min after PEITC treatment of HEK-293 cells, but was attenuated in IQGAP1-knockdown HEK-293 cells (Fig. 5A). Activation of MEK-ERK concomitantly induced ARE-luciferase by ∼4.5-fold but was attenuated in IQGAP1-knockdown HEK-293 cells (Fig. 5B), indicating that activation of the MEK-ERK pathway is required for Nrf2 activation and that IQGAP1 mediates this MEK-ERK-Nrf2 pathway.
FIGURE 5.
IQGAP1 mediates PEITC-induced MEK-ERK activation and MEK-ERK-induced ARE expression. A, HEK-293 cells transfected with pcDNA3.1 or IQGAP1 shRNA (shIQ) were treated with PEITC for 10 min. MEK and ERK phosphorylation was probed using anti-phospho-MEK (pMEK) and anti-phospho-ERK (pERK) antibodies. PEITC-induced MEK and ERK phosphorylation in IQGAP1-knockdown cells is expressed relative to wild-type cells and represents the mean ± S.D. (n = 3). *, p < 0.05. CON, control. B, HEK-293 cells were transfected with pcDNA3.1, ARE, HA-ERK, and DNEE-MEK plasmids for 24 h. MEK and ERK phosphorylation was probed using anti-phospho-MEK and anti-phospho-ERK antibodies. ARE induction was determined using luciferase assays. ARE activities induced by ectopic expression of HA-ERK or DNEE-MEK are expressed relative to the control and represent the mean ± S.D. (n = 2). *, p < 0.05.
IQGAP1 Mediates MEK-ERK Activation and ERK-Nrf2 Interaction
The possible functional significance of IQGAP1 in MEK-ERK-Nrf2 interaction was investigated. Ectopic expression of MEK and ERK significantly activated the MEK-ERK pathway, which was attenuated when IQGAP1 was knocked down (Fig. 6A). Next, the interaction between ERK and Nrf2 was examined. Activation of MEK was required to increase the interaction between ERK and Nrf2, and knockdown of IQGAP1 attenuated the interaction between ERK and Nrf2 (Fig. 6A). In agreement with these results, Nrf2 phosphorylation by activation of the MEK-ERK pathway was also attenuated in the IQGAP1-knockdown cells (Fig. 6B).
FIGURE 6.
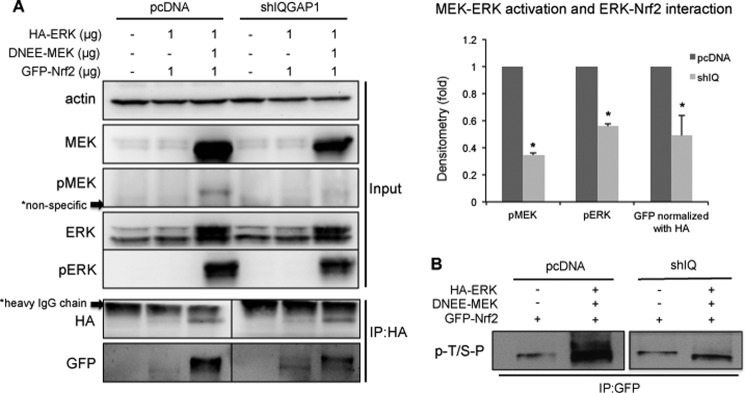
IQGAP1 mediates MEK-ERK activation and ERK-Nrf2 interaction. A, HEK-293 cells and IQGAP1-knockdown HEK-293 cells were transfected with pcDNA3.1, GFP-Nrf2, HA-ERK, and DNEE-MEK plasmids. Cell lysates were immunoprecipitated (IP) with anti-HA antibody, the samples were resolved by SDS-PAGE, and the GFP-Nrf2 protein pulled down was probed with GFP. MEK-ERK activation and ERK-Nrf2 interaction in IQGAP1-knockdown cells are expressed relative to wild-type cells and represent the mean ± S.D. (n = 3). *, p < 0.05. shIQGAP1 and shIQ, IQGAP1 shRNA; pMEK, phospho-MEK; pERK, phospho-ERK. B, HEK-293 cells and IQGAP1-knockdown HEK-293 cells were transfected with pcDNA3.1, GFP-Nrf2, HA-ERK, and DNEE-MEK plasmids. Cell lysates were immunoprecipitated with anti-GFP antibody to pull down the GFP-tagged Nrf2 proteins, and the phosphorylation level was determined by anti-phospho-(Ser/Thr)-Pro (p-T/S-P) antibody. Data are representative of two independent experiments.
IQGAP1 Mediates MEK-ERK-induced Stabilization of Nrf2 by Interfering with Ubiquitination of Nrf2
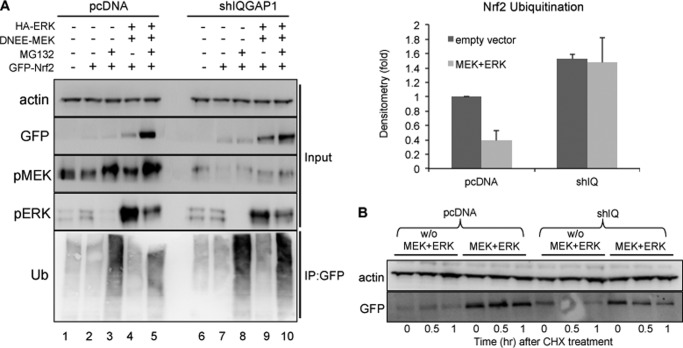
MEK-ERK activation stabilized the ectopically expressed GFP-Nrf2 protein (Fig. 7A, lanes 3 and 5), and IQGAP1 appeared to attenuate the stabilization (lanes 8 and 10). Cells were treated with MG132 to stabilize the ubiquitinated Nrf2 for detection. Our results showed that activation of MEK-ERK decreased ubiquitination of Nrf2, suggesting interference with the Nrf2-Keap1 interaction. In contrast, the decreased ubiquitination of Nrf2 was not observed in IQGAP1-knockdown cells. In fact, there was a higher basal level of ubiquitination of Nrf2 in IQGAP1-knockdown cells (Fig. 7A), indicating that IQGAP1 plays a role in MEK-ERK stabilization of basal Nrf2 levels. Stabilization of Nrf2 by MEK-ERK activation was IQGAP1-dependent. After cycloheximide treatment, the level of GFP-Nrf2 in HEK-293 cells was compared with that in IQGAP1-knockdown cells. MEK-ERK activation stabilized Nrf2 protein (Fig. 7B, lanes 4–6), but Nrf2 protein degraded with a half-life of ∼30 min in IQGAP1-knockdown cells (lanes 10–12).
FIGURE 7.
IQGAP1 mediates MEK-ERK-induced stabilization of Nrf2 by interfering with ubiquitination of Nrf2. A, HEK-293 cells were transfected as indicated, and the GFP-Nrf2 proteins were immunoprecipitated (IP) with anti-GFP antibody. Ubiquitination of Nrf2 was probed using anti-ubiquitin (Ub) antibody. Nrf2 ubiquitination in IQGAP1-knockdown cells is expressed relative to wild-type cells and represents the mean ± S.D. (n = 2). shIQGAP1 and shIQ, IQGAP1 shRNA; pMEK, phospho-MEK; pERK, phospho-ERK. B, HEK-293 cells were transfected as indicated for 24 h and treated with cycloheximide (CHX) for 0, 0.5 and 1 h. The cells were lysed, and GFP-Nrf2 expression was blotted. Data are representative of two independent experiments.
DISCUSSION
Previously, we isolated and identified several possible Nrf2 signaling partner proteins using the One-STrEP-tag purification system (14). Among the newly identified Nrf2 partners, the scaffolding protein IQGAP1 was further investigated in Nrf2 signaling. Here, we found that IQGAP1 modulated PEITC-induced Nrf2 activation. In that regard, we postulated that IQGAP1 protein may serve as a scaffold for MEK-ERK-Nrf2 signaling, leading to activation and nuclear translocation of Nrf2 to induce phase II/antioxidant genes.
Nrf2 has been considered to be a key transcription factor for the regulation of ARE-mediated gene expression in response to oxidative and electrophilic insults. MAPK signaling has been shown to be activated by various Nrf2-inducing chemicals such as PEITC (1), butylated hydroxyanisole (28), and 3H-1,2-dithiole-3-thione (29). Interestingly, ERK2 activation has been shown to be necessary for induction of Nrf2-regulated genes induced by these chemicals. We have identified the MEK-ERK-Nrf2 sequential phosphorylation as the major pathway of Nrf2 activation,4 but the mechanism by which these proteins come together to form a functional module remains unclear.
Previously, using the One-STrEP-tag purification system, we observed that Nrf2 bound IQGAP1 (14). Co-immunoprecipitation of IQGAP1 and Nrf2 in HEK-293 cells under unstimulated or stimulated conditions (PEITC treatment) further confirmed their binding in a physiological environment. IQGAP1 binds to an array of proteins, including calmodulin, Cdc42, Rac1, actin, β-catenin, E-cadherin, and CLIP-170. Evidence has been provided recently that IQGAP1 acts a scaffold protein and modulates the Raf-MEK-ERK pathway. It has been shown that IQGAP1 binds Raf, MEK, and ERK directly and modulates their functions and activation (30). Changing the IQGAP1 concentration impairs the ability of EGF to activate MEK and ERK. Given the experimental evidence suggesting that IQGAP1 binds to Nrf2 and mediates the MEK-ERK signaling pathway and that MEK-ERK activates Nrf2, we hypothesized that IQGAP1 may be an unrecognized scaffold for the MEK-ERK-Nrf2 signaling pathway. The evidence presented here supports this hypothesis.
Decreasing the intracellular IQGAP1 concentration leads to marked impairment of inducible Nrf2 activity. The activation of Nrf2 is modulated by IQGAP1, supported by the data showing that PEITC-induced HO-1, UGT, and NQO1 mRNA expression was abrogated in IQGAP1-knockdown HEK-293 cells. The decrease in phase II/antioxidant genes in IQGAP1-knockdown cells is due to a decrease in nuclear accumulation of Nrf2. PEITC induced a maximal nuclear translocation of Nrf2 2 h after treatment, but the translocation was significantly attenuated in IQGAP1-knockdown cells.
Upon PEITC stimulation, increased binding of Nrf2 to IQGAP1 was observed, suggesting the functional significance of the binding between Nrf2 and IQGAP1. Our data show that under stimulated conditions (activation of MEK-ERK), Nrf2 binding to ERK increased, and this interaction was attenuated if IQGAP1 was knocked down. The decreased ERK-Nrf2 interaction in IQGAP1-knockdown HEK-293 cells could be due to decreased MEK-ERK signaling as observed by the decrease in phosphorylation of MEK and ERK in IQGAP1-knockdown cells. Overexpression of constitutively active MEK increased phosphorylation of MEK together with ERK, correlating with an increase in ERK-Nrf2 interaction. In contrast, overexpression of ERK alone had no effect on MEK-ERK activation, and ERK-Nrf2 interaction was not observed.
Another line of evidence also indicates that a functional module of MEK-ERK-Nrf2 could take place on IQGAP1. Other groups have shown that both MEK and ERK bind to the N terminus (amino acids 1–600) of IQGAP1 (20). Additional studies showed that MEK binds to the IQ domain and that ERK binds to WW domain of IQGAP1 (26). Interestingly, our data show that Nrf2 also bound directly to the IQ domain of IQGAP1, suggesting that IQGAP1 could bring Nrf2 and the MEK-ERK complex in close proximity and that they are regulated in the same fashion. Maximal activation of Nrf2 by PEITC was observed only when all interacting proteins (MEK, ERK, and Nrf2) were assembled on IQGAP1. The components have to be present in an appropriate stoichiometric ratio. It was reported that the concentrations of IQGAP1, MEK2, and ERK2 in MCF-7 cells were 0.8, 6.9, and 15.5 μm (26). Thus, changing the amount of IQGAP1 may result in a large change in the stoichiometry of IQGAP1 to MEK, ERK, and Nrf2. However, the stoichiometry required for optimal activation could vary from cell type to cell type and would need further study.
Here, we proposed a model of IQGAP1 as a scaffold in the MEK-ERK-Nrf2 pathway. When the concentration of IQGAP1 is too low, the functional complexes are unable to form. In IQGAP1-knockdown HEK-293 cells, suboptimal activation of MEK-ERK signaling leads to a decrease in the recruitment of Nrf2 to ERK, which correlates with increased ubiquitination of Nrf2 and decreased Nrf2 stabilization (half-life of 30 min). The decreased Nrf2 in the cytoplasm correlates with decreased nuclear accumulation of Nrf2 and ARE activity.
In summary, we have identified IQGAP1 as a binding partner of Nrf2 and a regulator of Nrf2 activity. We provided evidence that IQGAP1 participates in and modulates activation of Nrf2. Furthermore, our data suggest that IQGAP1 modulates Nrf2 activation through regulation of MEK-ERK signaling. Our study provides the novel hypothesis that IQGAP1 links the MAPK and Nrf2 signaling together. Our data also suggest that a change in intracellular IQGAP1 concentration could severely impair Nrf2 activation ability, thus diminishing cellular defense against oxidative damages or carcinogens. IQGAP1 has been shown to be involved in organizing reactive oxygen species-dependent VEGF signaling, and it promotes endothelial cell migration and proliferation (31), reactive oxygen species-dependent tyrosine phosphorylation of VE-cadherin and loss of cell-cell contacts (32), and hyperoxia-induced p47phox translocation and reactive oxygen species generation in lung endothelial cells (33). Knowing the interplay between IQGAP1, Nrf2, and oxidative stress will provide more insights into how IQGAP1 mediates our cellular defense against oxidative damages or carcinogens, helping us to explain disease mechanisms such as oxidative-stress induced carcinogenesis, including hepatotumorigenesis (34). In addition, the fact that IQGAP1 mediates PEITC-induced phase II gene expression implies that the chemopreventive effects of a wide range of compounds could also be IQGAP1-dependent. In fact, we also found that IQGAP1-knockdown cells have decreased phase II gene expression (NQO1) in response to other compounds,4 demonstrating the importance of IQGAP1-Nrf2 interplay in chemoprevention.
K. L. Cheung, J. H. Lee, L. Shu, J.-H. Kim, D. B. Sacks, and A.-N. T. Kong, unpublished data.
- UGT
- UDP-glucuronosyltransferase
- ARE
- antioxidant response element
- PEITC
- phenethyl isothiocyanate
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Xu C., Yuan X., Pan Z., Shen G., Kim J. H., Yu S., Khor T. O., Li W., Ma J., Kong A. N. (2006) Mechanism of action of isothiocyanates: the induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol. Cancer Ther. 5, 1918–1926 [DOI] [PubMed] [Google Scholar]
- 2. Higgins L. G., Kelleher M. O., Eggleston I. M., Itoh K., Yamamoto M., Hayes J. D. (2009) Transcription factor Nrf2 mediates an adaptive response to sulforaphane that protects fibroblasts in vitro against the cytotoxic effects of electrophiles, peroxides and redox-cycling agents. Toxicol. Appl. Pharmacol. 237, 267–280 [DOI] [PubMed] [Google Scholar]
- 3. Keum Y. S., Han Y. H., Liew C., Kim J. H., Xu C., Yuan X., Shakarjian M. P., Chong S., Kong A. N. (2006) Induction of heme oxygenase-1 (HO-1) and NAD(P)H:quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA), and its metabolite, tert-butylhydroquinone (tBHQ), in primary cultured human and rat hepatocytes. Pharm. Res. 23, 2586–2594 [DOI] [PubMed] [Google Scholar]
- 4. Wang X. J., Sun Z., Chen W., Li Y., Villeneuve N. F., Zhang D. D. (2008) Activation of Nrf2 by arsenite and monomethylarsonous acid is independent of Keap1-C151: enhanced Keap1-Cul3 interaction. Toxicol. Appl. Pharmacol. 230, 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreger H., Westphal K., Wilck N., Baumann G., Stangl V., Stangl K., Meiners S. (2010) Protection of vascular cells from oxidative stress by proteasome inhibition depends on Nrf2. Cardiovasc. Res. 85, 395–403 [DOI] [PubMed] [Google Scholar]
- 6. Chen P. C., Vargas M. R., Pani A. K., Smeyne R. J., Johnson D. A., Kan Y. W., Johnson J. A. (2009) Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: critical role for the astrocyte. Proc. Natl. Acad. Sci. U.S.A. 106, 2933–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wakabayashi N., Slocum S. L., Skoko J. J., Shin S., Kensler T. W. (2010) When NRF2 talks, who's listening? Antioxid. Redox Signal. 13, 1649–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hayes J. D., McMahon M., Chowdhry S., Dinkova-Kostova A. T. (2010) Cancer chemoprevention mechanisms mediated through the Keap1-Nrf2 pathway. Antioxid. Redox Signal. 13, 1713–1748 [DOI] [PubMed] [Google Scholar]
- 9. McMahon M., Itoh K., Yamamoto M., Hayes J. D. (2003) Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 278, 21592–21600 [DOI] [PubMed] [Google Scholar]
- 10. Kobayashi M., Yamamoto M. (2006) Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 46, 113–140 [DOI] [PubMed] [Google Scholar]
- 11. Shen G., Hebbar V., Nair S., Xu C., Li W., Lin W., Keum Y. S., Han J., Gallo M. A., Kong A. N. (2004) Regulation of Nrf2 transactivation domain activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J. Biol. Chem. 279, 23052–23060 [DOI] [PubMed] [Google Scholar]
- 12. Keum Y. S., Yu S., Chang P. P., Yuan X., Kim J. H., Xu C., Han J., Agarwal A., Kong A. N. (2006) Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 66, 8804–8813 [DOI] [PubMed] [Google Scholar]
- 13. Huang H. C., Nguyen T., Pickett C. B. (2000) Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. U.S.A. 97, 12475–12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J. H., Xu E. Y., Sacks D. B., Lee J., Shu L., Xia B., Kong A. N. (2013) Identification and functional studies of a new Nrf2 partner IQGAP1: a critical role in the stability and transactivation of Nrf2. Antioxid. Redox Signal. 19, 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weissbach L., Settleman J., Kalady M. F., Snijders A. J., Murthy A. E., Yan Y. X., Bernards A. (1994) Identification of a human RasGAP-related protein containing calmodulin-binding motifs. J. Biol. Chem. 269, 20517–20521 [PubMed] [Google Scholar]
- 16. Hart M. J., Callow M. G., Souza B., Polakis P. (1996) IQGAP1, a calmodulin-binding protein with a RasGAP-related domain, is a potential effector for Cdc42Hs. EMBO J. 15, 2997–3005 [PMC free article] [PubMed] [Google Scholar]
- 17. Kuroda S., Fukata M., Nakagawa M., Fujii K., Nakamura T., Ookubo T., Izawa I., Nagase T., Nomura N., Tani H., Shoji I., Matsuura Y., Yonehara S., Kaibuchi K. (1998) Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin-mediated cell-cell adhesion. Science 281, 832–835 [DOI] [PubMed] [Google Scholar]
- 18. Briggs M. W., Sacks D. B. (2003) IQGAP1 as signal integrator: Ca2+, calmodulin, Cdc42 and the cytoskeleton. FEBS Lett. 542, 7–11 [DOI] [PubMed] [Google Scholar]
- 19. Brill S., Li S., Lyman C. W., Church D. M., Wasmuth J. J., Weissbach L., Bernards A., Snijders A. J. (1996) The Ras GTPase-activating protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 16, 4869–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roy M., Li Z., Sacks D. B. (2004) IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. 279, 17329–17337 [DOI] [PubMed] [Google Scholar]
- 21. Li Z., Sacks D. B. (2003) Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J. Biol. Chem. 278, 4347–4352 [DOI] [PubMed] [Google Scholar]
- 22. Weissbach L., Bernards A., Herion D. W. (1998) Binding of myosin essential light chain to the cytoskeleton-associated protein IQGAP1. Biochem. Biophys. Res. Commun. 251, 269–276 [DOI] [PubMed] [Google Scholar]
- 23. Mbele G. O., Deloulme J. C., Gentil B. J., Delphin C., Ferro M., Garin J., Takahashi M., Baudier J. (2002) The zinc- and calcium-binding S100B interacts and co-localizes with IQGAP1 during dynamic rearrangement of cell membranes. J. Biol. Chem. 277, 49998–50007 [DOI] [PubMed] [Google Scholar]
- 24. McCallum S. J., Wu W. J., Cerione R. A. (1996) Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J. Biol. Chem. 271, 21732–21737 [DOI] [PubMed] [Google Scholar]
- 25. Fukata M., Watanabe T., Noritake J., Nakagawa M., Yamaga M., Kuroda S., Matsuura Y., Iwamatsu A., Perez F., Kaibuchi K. (2002) Rac1 and Cdc42 capture microtubules through IQGAP1 and CLIP-170. Cell 109, 873–885 [DOI] [PubMed] [Google Scholar]
- 26. Roy M., Li Z., Sacks D. B. (2005) IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol. Cell. Biol. 25, 7940–7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu C., Shen G., Yuan X., Kim J. H., Gopalkrishnan A., Keum Y. S., Nair S., Kong A. N. (2006) ERK and JNK signaling pathways are involved in the regulation of activator protein 1 and cell death elicited by three isothiocyanates in human prostate cancer PC-3 cells. Carcinogenesis 27, 437–445 [DOI] [PubMed] [Google Scholar]
- 28. Yuan X., Xu C., Pan Z., Keum Y. S., Kim J. H., Shen G., Yu S., Oo K. T., Ma J., Kong A. N. (2006) Butylated hydroxyanisole regulates ARE-mediated gene expression via Nrf2 coupled with ERK and JNK signaling pathway in HepG2 cells. Mol. Carcinog. 45, 841–850 [DOI] [PubMed] [Google Scholar]
- 29. Manandhar S., Cho J. M., Kim J. A., Kensler T. W., Kwak M. K. (2007) Induction of Nrf2-regulated genes by 3H-1,2-dithiole-3-thione through the ERK signaling pathway in murine keratinocytes. Eur. J. Pharmacol. 577, 17–27 [DOI] [PubMed] [Google Scholar]
- 30. Ren J. G., Li Z., Sacks D. B. (2007) IQGAP1 modulates activation of B-Raf. Proc. Natl. Acad. Sci. U.S.A. 104, 10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamaoka-Tojo M., Ushio-Fukai M., Hilenski L., Dikalov S. I., Chen Y. E., Tojo T., Fukai T., Fujimoto M., Patrushev N. A., Wang N., Kontos C. D., Bloom G. S., Alexander R. W. (2004) IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species-dependent endothelial migration and proliferation. Circ. Res. 95, 276–283 [DOI] [PubMed] [Google Scholar]
- 32. Yamaoka-Tojo M., Tojo T., Kim H. W., Hilenski L., Patrushev N. A., Zhang L., Fukai T., Ushio-Fukai M. (2006) IQGAP1 mediates VE-cadherin-based cell-cell contacts and VEGF signaling at adherence junctions linked to angiogenesis. Arterioscler. Thromb. Vasc. Biol. 26, 1991–1997 [DOI] [PubMed] [Google Scholar]
- 33. Usatyuk P. V., Gorshkova I. A., He D., Zhao Y., Kalari S. K., Garcia J. G., Natarajan V. (2009) Phospholipase D-mediated activation of IQGAP1 through Rac1 regulates hyperoxia-induced p47phox translocation and reactive oxygen species generation in lung endothelial cells. J. Biol. Chem. 284, 15339–15352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsubota A., Matsumoto K., Mogushi K., Nariai K., Namiki Y., Hoshina S., Hano H., Tanaka H., Saito H., Tada N. (2010) IQGAP1 and vimentin are key regulator genes in naturally occurring hepatotumorigenesis induced by oxidative stress. Carcinogenesis 31, 504–511 [DOI] [PubMed] [Google Scholar]
- 35. Jaaro H., Rubinfeld H., Hanoch T., Seger R. (1997) Nuclear translocation of mitogen-activated protein kinase kinase (MEK1) in response to mitogenic stimulation. Proc. Natl. Acad. Sci. U.S.A. 94, 3742–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]