Background: Changes in gene expression mediate adaption to hypoxia.
Results: MMP1 was induced during prolonged hypoxia, and this induction was blocked after depletion of CREB and/or NF-κB.
Conclusion: CREB and NF-κB mediate the up-regulation of MMP1 during prolonged hypoxia.
Significance: CREB- and NF-κB-mediated gene transcription may play a major role in the cellular response to prolonged hypoxia.
Keywords: CREB, Hypoxia, Hypoxia-inducible Factor (HIF), Matrix Metalloproteinase (MMP), NF-κB Transcription Factor
Abstract
Responses to low levels of oxygen (hypoxia) are essential to maintain homeostasis. During the hypoxic response, gene expression is altered by various transcription factors. The transcription factor, hypoxia-inducible factor (HIF), plays a central role in the hypoxic response. The α subunit of HIF, which is actively degraded during normoxia, becomes stabilized during hypoxia, which leads to HIF activation. A microarray analysis of HeLa cells showed that expression of matrix metalloproteinase 1 (MMP1) was markedly induced during prolonged hypoxia. CREB and NF-κB binding sites were identified in the MMP1 promoter region between 1945 and 1896 nucleotides upstream of the transcription start site. Assays with luciferase reporters demonstrated that HIF activity was induced during the early phase of hypoxia, whereas CREB and NF-κB were activated during the later (prolonged) phase. Depletion of CREB and/or NF-κB reduced MMP1 induction during prolonged hypoxia both at the mRNA and protein levels. A chromatin immunoprecipitation assay demonstrated binding of CREB and NF-κB to the MMP1 promoter. Finally, cell migration and invasion on a collagen matrix and pulmonary metastasis in nude mice were inhibited after depletion of CREB and NF-κB in MDA-MB-231 cells. Taken together, these results suggest that the cooperative action of CREB and NF-κB plays an important role to induce MMP1 expression during prolonged hypoxia and regulates cell migration and invasion in cancer cells.
Introduction
Organisms utilize oxygen to efficiently produce energy. Homeostasis is maintained during exposure to low levels of oxygen (hypoxia) by the hypoxic response. The transcription factor hypoxia-inducible factor (HIF)2 plays a key role in the hypoxic response (1). HIF consists of two subunits, HIF-α and -β. Whereas the β subunit is constitutively expressed, the α subunit is mainly expressed during hypoxia, and it is actively degraded during normoxia by the ubiquitin-proteasome system. The von Hippel-Lindau (pVHL) ubiquitin ligase complex ubiquitinates HIF-α when the proline residues of the α subunit are hydroxylated (2). This prolyl-hydroxylation of HIF-α is mediated by prolyl-hydroxylases (PHD, also called HPH (HIF-prolyl hydroxylase) or EGLN (egg-laying abnormal nine)), which belong to the 2-oxoglutarate-dependent dioxygenase family and require co-factors such as 2-oxoglutarate, Fe2+, ascorbic acid, and oxygen for their enzymatic activity (3). PHD activity is attenuated during hypoxia, leading to a reduction in HIF-α ubiquitination and stabilization of the protein. Consequently, a heterodimer of HIF-α and -β is able to form, which induces the expression of a number of hypoxia-regulated genes.
Regulation of gene expression during hypoxia is also mediated by other transcription factors, such as CREB and NF-κB. NF-κB is activated during acute hypoxia in macrophages and pulmonary artery smooth muscle cells (4, 5). In macrophages the activation of NF-κB during hypoxia precedes the stabilization of HIF-1α; NF-κB (p50/p65) binds the promoter region of HIF-1α and up-regulates HIF-1α mRNA. In contrast, HIF-1α appears to regulate NF-κB expression in neutrophils (6). These results suggest there is a regulatory loop between the two factors. CREB, which mediates a cAMP-dependent gene expression, is activated when it is phosphorylated by kinases, such as protein kinase A or Ca2+/calmodulin-dependent kinase (7). However, precisely when CREB and NF-κB is activated during hypoxia still remains unknown.
Pre-mRNA processing factor 19 (PRP19) is a component of the spliceosome. We previously demonstrated that the PHD enzyme, PHD3, interacts with PRP19 and inhibits cell death during hypoxia (8). To further examine the role of PRP19 during hypoxia, we performed microarray analysis of PRP19-depleted cells that had been cultured in prolonged hypoxic conditions. The matrix metalloproteinase 1 (MMP1) gene was highly up-regulated in these cells, whereas there was little change in the level of MMP1 protein. Nevertheless, MMP1 was clearly induced independently of PRP19 after prolonged hypoxic treatment. Because it remains largely unknown how gene expression is regulated during prolonged hypoxia, we studied how the expression of MMP1 is up-regulated during prolonged hypoxic treatment.
MMP1 is a member of the matrix metalloproteinase family and requires zinc for its activity (9). MMP1 is subgrouped with MMP8 and MMP13, and all three proteins have collagenase activity. Through its enzymatic activity, MMP1 plays roles in various processes including tumor invasion, blood vessel remodeling, and wound healing (9). Previous reports indicate that NF-κB, p38, and JNK pathways are involved in MMP1 expression by cytokine stimulation in fibroblast or chondrocytes (10, 11). It was also reported that induction of MMP1 is mediated by HIF-1 during hypoxia in fibroblasts (12, 13) or by HIF-2 in chondrocytes upon IL-1β stimulation (14). Although HIF is a key regulator of the hypoxic response, it remains unclear if MMP1 is induced by these factors during prolonged hypoxia or if any other factor(s) is involved in MMP1 induction. In this study it is reported that MMP1 is specifically induced during prolonged hypoxia in multiple cancer cells and that this process is cooperatively regulated by CREB and NF-κB.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa cells and the breast tumor cell lines MCF7 and MDA-MB-231 were cultured in Dulbecco's modified Eagle's medium (high glucose) (Wako, Japan) containing 10% fetal bovine serum and antibiotics. Human umbilical vein endothelial cells (HUVECs) were cultured in complete endothelial cell growth medium (211K-500) (Cell Applications inc., San Diego, CA).
Hypoxia Treatment
Cells (HeLa, MCF7, MDA-MB231, or HUVEC) were treated under hypoxic conditions (1% or 5% O2, 5% CO2, and the rest were balanced with N2) in a hypoxia workstation (Hirasawa Works, Tokyo, Japan). An oxygen sensor was used to ensure the oxygen concentration inside the work station, which was maintained at 1 or 5% throughout the experiment (MC-8G-S, Iijima Electrics, Gamagori, Japan).
siRNA Treatment
Initially, three kinds of siRNA for each gene were tested, and the most effective ones were selected. HeLa or MDA-MB-231 cells were transfected with negative control siRNA (#45-2001, Invitrogen), HIF-1α siRNA (HSS #179231 from Invitrogen), HIF-2α siRNA (HSS #103261 from Invitrogen), Rel A (NF-κB) siRNA (SASI Hs01 00171091 from Sigma), CREB siRNA (SASI Hs01 00116988 from Sigma), and MMP1 siRNA (HSS #106611 from Invitrogen) using LipofectamineTM RNAiMAX transfection reagent according to manufacturer's instructions (Invitrogen). The day after transfection, cells were plated onto collagen-coated dishes or plates and subjected to the scratch and invasion assays, respectively. For the luciferase assay, cells were transfected with luciferase vectors the day after siRNA transfection and were cultured in normoxic or hypoxic conditions for 24 or 48 h. A portion of the cells were collected, and total RNA or protein was extracted. For Western blotting and quantitative real-time PCR (qPCR), cells were cultured in normoxic or hypoxic conditions for up to 48 h, and protein or RNA was extracted, respectively. The efficiency of gene silencing was determined by qPCR or Western blotting.
Reagents and Antibodies
The following antibodies were used: anti-β-actin antibody (Sigma); anti-HIF-1α antibody (Novus, Littleton, CO); anti-HIF-2α antibody (Novus); anti-NF-κB p65 antibody (Cell Signaling Technology, Danvers, MA); anti-CREB antibody (Cell Signaling Technology); anti-phosphoSer-133-CREB antibody (Cell Signaling Technology); anti-IκBα antibody (Cell Signaling Technology). For the chromatin immunoprecipitation (ChIP) assay, control rabbit IgG (Millipore Upstate, Temecula, CA), anti-CREB antibody (Millipore Upstate), and anti-NF-κB p65 antibody (Santa Cruz, CA) were used. An anti-MMP1 antibody was generated by immunizing rabbits with a mixture of peptides corresponding to amino acids 131–144 and 375–388 of human MMP1. Its specificity to recognize MMP1 was characterized by Western blotting (Fig. 3, D and E). Staurosporine was purchased from Wako Pure Chemical (Osaka, Japan).
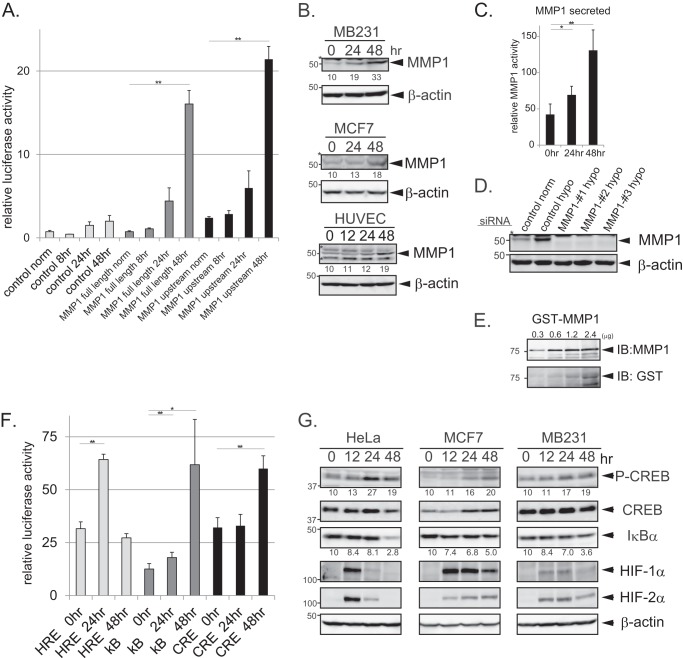
FIGURE 3.
Increased activity of NF-κB and CREB during prolonged hypoxia coincides with the induction of MMP1. A, shown is enhancement of MMP1 promoter activity during prolonged hypoxia. The luciferase activities of the full-length and upstream MMP1 promoter constructs were measured in HeLa cells during hypoxia (n = 3). The ratio between the firefly luciferase activity (MMP1 promoter) and the control Renilla luciferase activity (pRL-null internal control) is shown (A and F). Error bars indicate S.D., and significance was statistically analyzed with a t test (**, p < 0.02). B, shown is induction of MMP1 protein expression during prolonged hypoxia. MDA-MB-231, MCF7, and HUVECs were cultured in 1% O2 for the indicated times. MMP1 expression was detected by Western blotting. The numbers below the bands represent the relative intensities quantified using densitometric analyses. Asterisks indicate nonspecific bands detected by the antibody. C, shown is increased MMP1 activity in the cell culture medium during hypoxia. An ELISA assay was performed on cell culture medium collected from MDA-MB-231 cells incubated with hypoxia for different times to measure MMP1 activity (n = 3). The graph shows relative MMP1 activity. Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02). D, shown is decreased expression of MMP1 by MMP1 siRNA. MDA-MB-231 cells transfected with control or three kinds of MMP1 siRNA were treated with hypoxic condition for 24 h, and cell lysate was subjected to Western blotting. The antibody detected the expression of MMP1, which was induced by hypoxia and depleted by the three siRNAs. The asterisk indicates nonspecific bands detected by the antibody. E, shown is detection of GST-MMP1. GST-MMP1 (75 kDa) purified from Escherichia coli was subjected to Western blotting (IB). Different amounts of the protein were loaded to the lanes as indicated in the figure. The blot was detected with anti-MMP1 antibody (upper panel) and anti-GST antibody (lower panel). F, increased luciferase activity of HIF, NF-κB (kB), and CREB target sequences during prolonged hypoxia is shown. The luciferase activities of the typical target sequences of HIF, CREB, and NF-κB were monitored in HeLa cells during hypoxia (n = 3). Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02). G, shown is activation of CREB and NF-κB signaling pathways during hypoxia. Phosphorylation of CREB and the protein expression levels of CREB and IκBα were monitored at the indicated time points by Western blotting after hypoxic treatment in HeLa, MCF7, and MDA-MB-231 cells. The numbers below the bands represent the relative intensities quantified using densitometric analyses. Immunoblots of HIF-1α, HIF-2α, and β-actin are shown in the bottom panels.
Western Blotting
Cells (HeLa, MCF7, MDA-MB231, or HUVEC) were harvested on ice and lysed in lysis buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1 μg/ml PMSF, and 2 μg/ml leupeptin). Total cell lysates were subjected to SDS-PAGE (50 μg of total lysate/lane) and were transferred onto a nitrocellulose membrane (PALL, East Hills, NY).
qPCR
Total RNA was isolated from cultured cells (HeLa, MCF7, MDA-MB231, or HUVEC) using an RNeasy® kit (Qiagen, Valencia, CA). First strand cDNA synthesis was performed with 2 μg of total RNA using the PrimeScriptTM II kit (Takara Bio). Synthesized cDNA was used for PCR analysis using SsoFastTM EvaGreen® Supermix (Bio-Rad) with a Chromo4TM real-time PCR detecting system (Bio-Rad). Relative expression levels were calculated using the ΔCt method and normalized against β-actin or cyclophilin internal control. Primer sequences are listed in the supplemental table.
Luciferase Assay
HeLa or MCF7 cells were plated on 24-well culture plates. The MMP1 promoter luciferase vector and its deletion mutants HRE-luc, κB-luc, or CRE-luc (500 ng) were transiently transfected with the Renilla control pRL-null vector (160 ng) using the FuGENE® transfection reagent (Promega, Madison, WI). Cells were cultured in normoxia or hypoxia, and each well was lysed in 100 μl of passive lysis buffer (Promega) 2 days after transfection. The luciferase assay was performed using 30 μl of cell lysate with the Dual-Luciferase® Reporter Assay (Promega). Luciferase activity was measured using a Microplate reader Centro LB960 (Berthold, Bad Wildbad, Germany). The relative luciferase activity was calculated by dividing the firefly luciferase activity with that of the Renilla control.
ChIP Assay
HeLa cells were cultured in normoxia or hypoxia (1% O2) for 24 h, and cross-linking was performed using 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by the addition of 150 mm glycine. Cells were lysed and sonicated 5 times to obtain 200–800-bp chromatin fragments. The protein-chromatin complex was immunoprecipitated by incubating the cell extract with 20 μg of antibodies overnight and then with 30 μl of protein G magnetic beads (Cell Signaling Technology) for 2 h at 4 °C. After eight washes, cross-links of the protein/chromatin complex were resolved, and chromatin was purified using phenol-chloroform extraction followed by ethanol precipitation. DNA was prepared and used as a template in PCR analyses using the specific primer sets listed in the supplemental table.
MMP1 ELISA
MMP1 activity in the cell culture medium was measured using the SensoLyte® MMP1 assay kit (Anaspec, Fremont, CA). Briefly, cell culture medium of MDA-MB-231 cells containing the secreted form of MMP1 is incubated with a substrate that fluoresces when cleaved by MMP1. The fluorescence was measured by ARVO-MX plate reader (PerkinElmer Life Sciences).
Pulmonary Metastasis Studies
MDA-MB-231 cells were transfected with siRNA (control or CREB and NF-κB). One day after the transfection, cells were suspended in PBS at 5 × 106 cells/ml, and 100 μl of cell suspension was injected to the tail vein of BALB/cAJcl-nu/nu mice (4-week-old female, Japan CLEA, Inc.). Mice were sacrificed after 6 weeks of injection, and the lungs were collected and fixed with Bouin's fixative. Metastatic nodules on the lung surface were counted under the dissection microscope. All the animal experiments have been performed in accordance with a protocol approved by the Tokyo Medical and Dental University Animal Care Committee.
Cell Migration Assay
MDA-MB-231 cells were transfected with control, CREB, NF-κB, or MMP1 siRNAs. One day later cells were re-plated on collagen-coated dishes (coated with collagen type I, Asahi Glass Co. Ltd., Tokyo, Japan) and grown until fully confluent. The cell layer was scratched using a 1-ml pipette tip, and cells were cultured in normoxic or hypoxic conditions for up to 24 h in the presence of mitomycin C (5 μg/ml). Cells were imaged 0, 12, and 24 h after the scratch wound was introduced, and the width of gap was measured. Results presented are the means of three independent experiments.
Invasion Assay
A cell invasion assay was performed using a CytoSelectTM 96-well Cell Invasion assay kit (Cell Biolabs, San Diego, CA). MDA-MB-231 cells were transfected with siRNA, and 1 day later cells were detached and 2 × 105 cells were placed in the upper chamber of the assay plate that was covered with a layer of collagen. The assay plate was cultured in hypoxic conditions for 48 h, and cells that migrated toward the lower chamber were collected and quantified using CyQUANT® GR fluorescent dye. Each experiment was performed at least three times, and the representative data is shown.
RESULTS
Up-regulation of MMP1 during Hypoxia
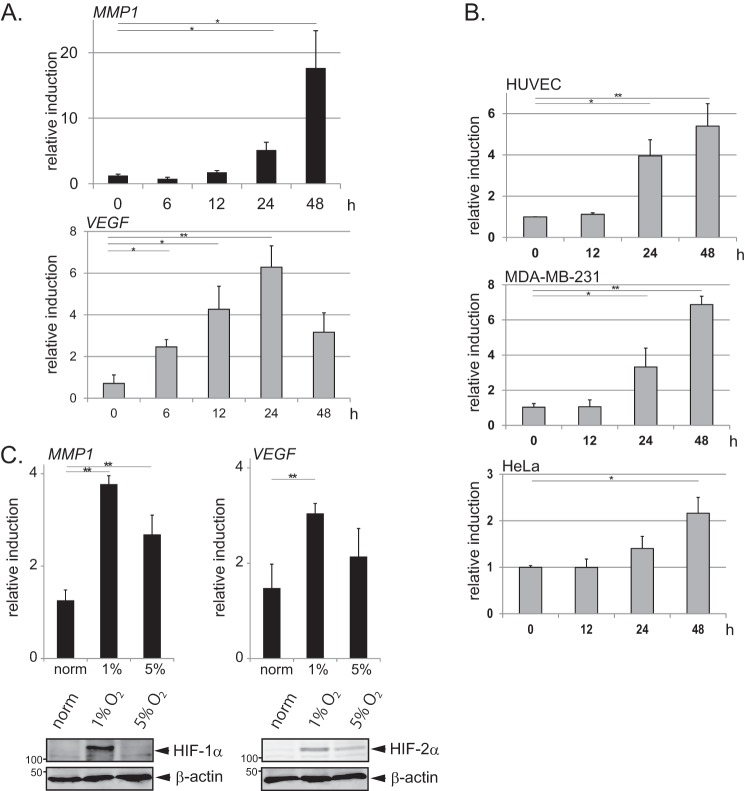
MMP1 was initially identified in a microarray analysis as a gene induced in PRP19-depleted cells after 24 h of hypoxic treatment. Although the level of MMP1 mRNA was found to be increased in PRP19-depleted cells, the level of MMP1 protein was unaffected (data not shown). Alternatively, it is identified that MMP1 was markedly induced after prolonged hypoxia, and the level of MMP1 induction was greater after 48 h of hypoxia than after 24 h in MCF7 cells (Fig. 1A). Thus, MMP1 induction during prolonged hypoxia is further characterized. In contrast to MMP1, the induction of VEGF, a typical HIF target gene, was highest after 24 h of hypoxia, after which the level of induction fell (Fig. 1A). Induction of MMP1 during prolonged hypoxia was detected in several cell types, including HUVECs, the breast cancer cell line MDA-MB-231, and HeLa cells (Fig. 1B). These results suggest that MMP1 is a hypoxia-inducible gene that is mainly up-regulated during the prolonged phase of hypoxia. The induction of MMP1 was observed both in 5% O2 (mild hypoxia) and in 1% O2 (Fig. 1C). In contrast, whereas VEGF was clearly induced in 1% O2, it was not significantly induced in 5% O2. The induction of VEGF was similar to the expression of HIF-1α and HIF-2α, which were greatly increased in 1% O2 and moderately increased in 5% O2 (Fig. 1C).
FIGURE 1.
MMP1 gene expression is induced during prolonged hypoxia. A, shown is induction of MMP1 and VEGF during hypoxia. MCF7 cells were cultured in 1% O2 for the indicated times, and expression of MMP1 and VEGF was determined by qPCR analyses (n = 3). Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02) (A–C). B, shown is induction of MMP1 in different cell lines during hypoxia. HUVEC, MDA-MB-231, and HeLa cells were cultured in 1% O2 for the indicated times, and expression of MMP1 was determined by qPCR analyses (n = 3). C, shown is the effect of mild hypoxia on MMP1 and VEGF expression. MCF7 cells were cultured in 1 or 5% O2 for 24 h. Expression of MMP1 and VEGF was determined by qPCR analyses (n = 4). HIF-1α and HIF-2α were detected by Western blotting.
Characterization of the MMP1 Promoter Region Required for Hypoxic Induction
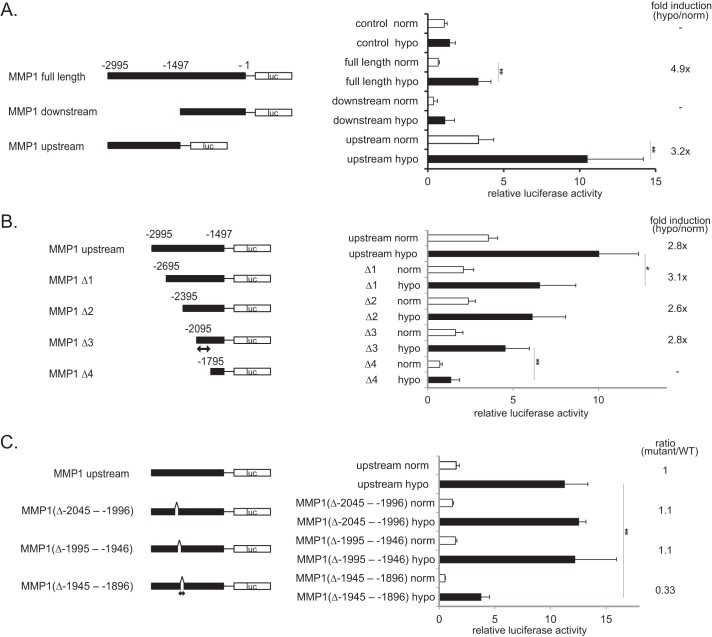
The promoter of MMP1 was analyzed to determine the region that is responsible for the induction of MMP1 during prolonged hypoxia. The full-length promoter region of the MMP1 gene, defined as the 2995-bp upstream of the transcription start site, was cloned into the pGL3 luciferase vector (pGL3-MMP1 promoter (−2995 to −1 bp), referred to as “full-length”) (Fig. 2A). The luciferase vector containing the hypoxia response element (HRE) efficiently induced luciferase activity after hypoxic treatment for 24 h (Fig. 3F). Under the same conditions, the full-length vector induced luciferase activity in a hypoxia-dependent manner (Fig. 2A). The two halves of the full-length promoter were cloned; the upstream half (−2995 to −1497 bp) and the downstream half (−1496 to −1 bp). Only the upstream region increased luciferase activity in response to hypoxia (Fig. 2A). These results suggest that the upstream region of the MMP1 promoter contains the sequence responsible for the induction of MMP1 expression in response to hypoxia. Next, we generated a series of deletion mutants of the upstream construct that lacked 300, 600, 900, or 1200 bp starting from the first nucleotide of the promoter (position −2995), which were named Δ1, Δ2, Δ3, and Δ4, respectively (Fig. 2B). The Δ1, Δ2, and Δ3 mutants were activated during hypoxia, whereas the Δ4 mutant was not (Fig. 2B). These results indicate that the 300 bp region from −2095 to −1796 bp in the MMP1 promoter is responsible for the hypoxic induction. A database search detected conserved binding sequences for CREB, NF-κB, and C/EBPβ within this region (15, 16). To further narrow down the region required for promoter activity, three deletion mutants of the full-length construct were generated that each lacked 50 bp. These mutants were named Δ −2045 to −1996, Δ −1995 to −1946, and Δ −1945 to −1896 (the numbers indicate the deleted regions). The induction of MMP1 during hypoxia was significantly reduced with the Δ −1945 to −1896 mutant (Fig. 2C). The consensus binding sequences for CREB, NF-κB, and C/EBPβ locate in this region.
FIGURE 2.
Identification of the region of the MMP1 promoter required for MMP1 induction during prolonged hypoxia. A, shown is the requirement of −2995 to −1497 bp for the hypoxic activation of the MMP1 promoter. The region of the MMP1 promoter required for induction of the MMP1 during hypoxia (24 h) was characterized by performing a luciferase assay in HeLa cells using the deletion constructs indicated (n = 6). The ratio between the firefly luciferase activity (MMP1 promoter) and the Renilla luciferase activity (pRL-null internal control) is shown (A–C). Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02) (A–C). -Fold induction upon hypoxic treatment (hypo/norm) is shown. Promoter activity below the activity of control (basal promoter) was considered to be background and is indicated as a dash (A and B). B, shown is the requirement of −2095 to −1796 bp for the hypoxic activation of the MMP1 promoter. The activities of a series of MMP1 promoter deletion constructs were monitored using a luciferase assay in HeLa cells after 24 h of hypoxia (n = 6). The position of the sequence relevant for the promoter activation is marked with a double-headed arrow. C, shown is the requirement of −1945 to −1896 bp for the hypoxic activation of the MMP1 promoter. A series of deletion mutants of the upstream construct was generated. Their activities were monitored using a luciferase assay in HeLa cells after 48 h of hypoxia (n = 3). The position of the sequence relevant for the promoter activation is marked with a double-headed arrow. Relative activity of mutant promoters to the wild type promoter (mutant/wild type) is shown as ratio.
Activation of MMP1 and CREB- and NF-κB-dependent Promoters during Prolonged Hypoxia
Next, activity of the MMP1 promoter constructs at different time points during hypoxic treatment was compared in HeLa cells. The full-length construct and the upstream construct were activated in a time-dependent manner during hypoxia; they were moderately activated (around 5-fold) after 24 h of hypoxia and were activated further (up to 20-fold) after 48 h of hypoxia (Fig. 3A). MMP1 protein expression was also found to be up-regulated during prolonged hypoxia in MDA-MB-231 and MCF7 cells as well as in HUVECs (Fig. 3B). Similarly, MMP1 secreted in the culture medium, which was measured by its activity, increased during hypoxia, which corresponds to the increase of MMP1 in cell lysate (Fig. 3C). Specificity of the MMP1 antibody was assured by the depletion of MMP1 band with three independent siRNAs against MMP1 and its ability to detect purified GST-MMP1 in Western blotting (Fig. 3, D and E). Cancerous MCF7 and MDA-MB-231 cells grew efficiently in high glucose medium compared with non-cancerous HUVECs (supplemental Fig. 1), yet the induction profile of MMP1 was comparable. Of note, overall expression level of MMP1 in MDA-MB-231 cells was higher than that in MCF7 cells. To determine the activities of CREB, NF-κB, and HIF, luciferase assays using vectors driven by HRE (HRE-luc), κB-response element (κB-luc), and CRE (CRE-luc) were performed at different time points during hypoxic treatment. Whereas the activity of HRE-luc peaked at 24 h and began to decrease at 48 h, the activities of κB-luc and CRE-luc were greater after 48 h of hypoxia than after 24 h (Fig. 3F and supplemental Fig. 2). These results suggest that HRE, κB-response element, and CRE are maximally active at different time points during hypoxia. Based on this result, prolonged hypoxia is defined in this report as being more than 24 h of hypoxic treatment, after which the activity of HIF-1 started to decrease.
The activation profiles of κB-luc or CRE-luc were similar to that of the MMP1 promoter (Fig. 3, A and F, and supplemental Fig. 2). Next, the CREB, NF-κB, and HIF signaling pathways were examined by Western blotting. CREB activation was examined by monitoring the phosphorylation of Ser-133 using a phospho-specific antibody. CREB phosphorylation was induced after 24 h of hypoxic treatment in HeLa, MCF7, and MDA-MB-231 cells (Fig. 3G). To evaluate NF-κB activity, the expression of its inhibitory subunit IκBα was monitored. The level of IκBα protein decreased after hypoxic treatment and was lowest after 48 h of hypoxia, suggesting that NF-κB activity had increased (Fig. 3G). In contrast, the protein expression of HIF-1α and HIF-2α in most cases peaked at earlier time points and decreased during prolonged hypoxia (Fig. 3G), which is similar to the pattern of HRE luciferase activity observed (Fig. 3F).
Inhibition of the CREB and NF-κB Signaling Pathways Suppresses MMP1 Expression during Hypoxia
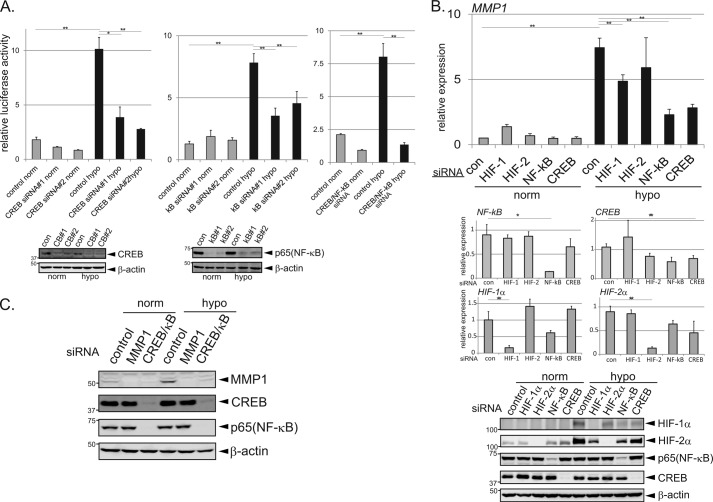
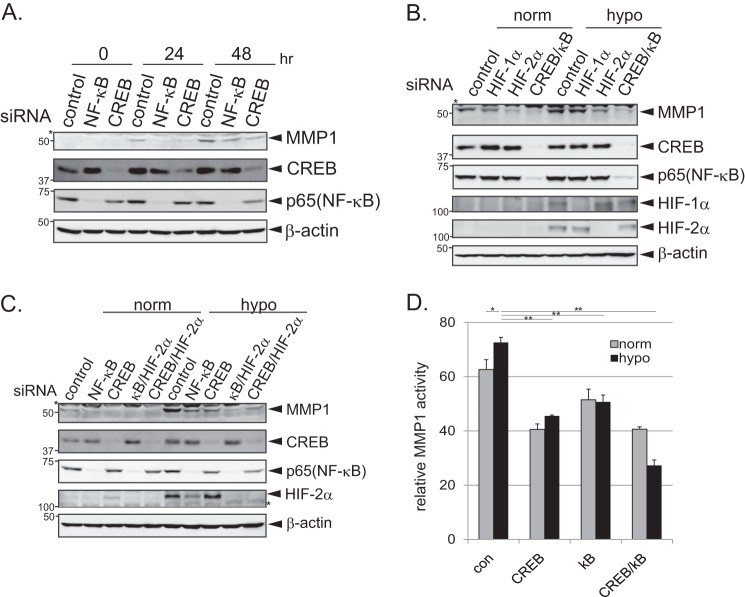
Because the timing of MMP1 promoter activation was synchronized with CREB and NF-κB activation and consensus binding sequences for CREB and NF-κB were found in the MMP1 promoter, it was examined whether CREB and/or NF-κB are involved in up-regulating MMP1 during prolonged hypoxia. Western blotting confirmed that the protein levels of CREB and NF-κB were effectively depleted after transfection of siRNAs targeting these molecules (Fig. 4A). Massive cell death was observed when C/EBPβ-depleted cells were cultured in hypoxic conditions, and so the effects of C/EBPβ could not be studied. The activation of the MMP1 promoter during hypoxia was significantly inhibited after depletion of CREB and NF-κB in HeLa cells (Fig. 4A, left and middle). When CREB or NF-κB was depleted separately, luciferase activity was reduced by ∼50%; however, simultaneous depletion of CREB and NF-κB reduced luciferase activity by up to 80% (Fig. 4A, right). These results suggest that CREB and NF-κB cooperatively activate the MMP1 promoter during prolonged hypoxia. It is possible that the 20% activity remaining after depletion of CREB and NF-κB is due to the activity of another transcription factor(s) (e.g. C/EBPβ). Of note, depletion of CREB moderately reduced the basal activity of MMP1 luciferase reporter (Fig. 4A, left), which implicates the involvement of CREB in the basal expression of MMP1. The effect of CREB and NF-κB depletion on MMP1 gene expression was examined by qPCR in HeLa cells. Depletion of HIF-1α and HIF-2α was also performed to examine the previously reported involvement of the HIF pathway in MMP1 expression (14). Although expression of MMP1 was enhanced in control cells after 48 h of hypoxia, this increase was significantly inhibited after depletion of CREB or NF-κB (Fig. 4B, top). Depletion of HIF-1α or HIF-2α also inhibited MMP1 induction, although to a lesser extent than CREB or NF-κB depletion (Fig. 4B, top). The efficient depletion of each molecule after siRNA treatment was confirmed by qPCR and Western blotting (Fig. 4B, middle four panels and bottom). Taken together, these results suggest that the induction of MMP1 during prolonged hypoxia mainly depends on the CREB and NF-κB signaling pathways but is also affected by HIF-1α and HIF-2α. MMP1 protein expression after depletion of CREB and NF-κB was examined by Western blotting in MDA-MB-231 cells (Fig. 4C). Simultaneous depletion of CREB and NF-κB efficiently decreased MMP1 protein levels during both normoxia and hypoxia; however, the effect was less pronounced than that observed after transfection of MMP1-targeting siRNA (Fig. 4C). These results indicate that whereas the CREB and NF-κB signaling pathways play a certain role in the regulation of basal MMP1 expression during normoxia, they play a major role in up-regulating MMP1 expression during prolonged hypoxia.
FIGURE 4.
CREB and NF-κB regulate the induction of MMP1 during prolonged hypoxia. A, mediation of MMP1 promoter activation during prolonged hypoxia by CREB and NF-κB is shown. MMP1 promoter activity was monitored after depletion of CREB and/or NF-κB (left, CREB: middle, NF-κB; right, CREB and NF-κB) in HeLa cells (n = 3). Two siRNAs targeted against each gene were used, and the depletion efficiency was monitored by Western blotting (bottom panels). Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02). B, induction of MMP1 during prolonged hypoxia is mainly dependent on CREB and NF-κB. HeLa cells treated with control, HIF-1α, HIF-2α, NF-κB, or CREB siRNAs were cultured in normoxic or hypoxic conditions for 48 h. Expression of MMP1 was monitored by qPCR analyses (n = 3). Middle graphs confirm the inhibition of gene expression after siRNA transfection. Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02). Depletion of each protein by the corresponding siRNAs was confirmed by Western blotting (bottom). C, shown is inhibition of MMP1 induction after depletion of CREB and NF-κB. Western blotting was used to monitor MMP1 protein expression in MDA-MB-231 cells depleted of MMP1 or CREB and NF-κB and grown in normoxic or hypoxic conditions for 24 h.
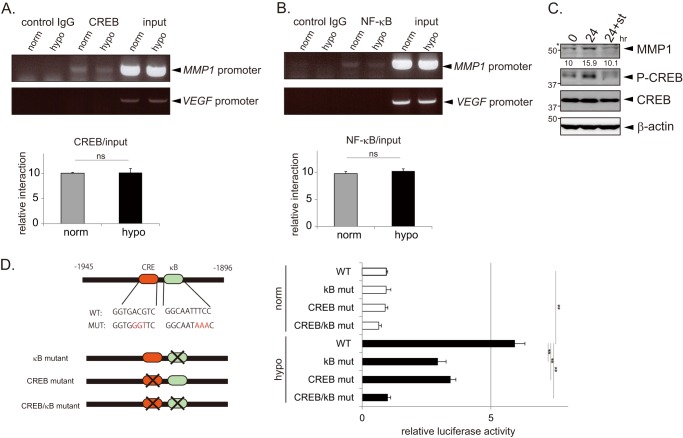
Recruitment of CREB and NF-κB to the MMP1 Promoter
To determine if CREB and NF-κB interact with the promoter region of MMP1, ChIP assays were performed using lysates prepared from HeLa cells grown under normoxic or hypoxic conditions. The MMP1 promoter region (−2095 to −1796 bp) was amplified from chromatin immunoprecipitated with the CREB and NF-κB antibodies (Fig. 5, A and B, top). Both transcription factors bound to the MMP1 promoter, and the binding efficiency between normoxia and hypoxia was found not to be significantly different. Because hypoxic-treated samples grew slower and had less cell numbers, the signals obtained in the ChIP PCR were normalized against the input and shown in the graph (Fig. 5, A and B, bottom). Inhibition of CREB phosphorylation by staurosporine treatment attenuated the hypoxic induction of MMP1 (Fig. 5C). These results suggest that the hypoxic induction of MMP1 may depend on activation of CREB and NF-κB, which is mediated by post-translational modification(s) or recruitment of co-factors to the promoter region through protein-protein interaction and is not due to enhanced binding of them to the promoter. MMP1 promoter luciferase constructs were generated that had mutations in the NF-κB -binding site, the CREB binding site, or in both sites. Hypoxic-dependent activation of the MMP1 promoter was reduced in all mutants, and the largest reduction was observed with the CREB/NF-κB double mutant (Fig. 5D).
FIGURE 5.
Binding of CREB and NF-κB to the MMP1 promoter. A and B, CREB and NF-κB bind the MMP1 promoter between −2095 and −1796 bp. A ChIP assay was performed using lysates prepared from HeLa cells cultured in normoxic or hypoxic conditions for 24 h using an anti-CREB (A) or an anti-NF-κB (B) antibody. The promoter region was specifically amplified from the immunoprecipitates of the CREB and NF-κB antibodies and was not amplified from the immunoprecipitate of the control IgG. VEGF promoter was monitored as a negative control. The ratio of ChIP/input was calculated, and an average of three experiments is shown in the graph (n = 3). Error bars indicate S.D., and significance was statistically analyzed with a t test (ns, significant difference was not found). C, hypoxic induction of MMP1 is inhibited by staurosporine. MDA-MB-231 cells were incubated under hypoxic condition for 24 h in the presence or absence of staurosporine (st, 10 nm). Expression of MMP1 and CREB and status of CREB phosphorylation was monitored by Western blotting. The numbers below the bands represent the relative intensities quantified using densitometric analyses. The asterisk indicates a nonspecific band detected by the antibody. D, the binding sites of CREB and NF-κB (kB) on the MMP1 promoter are important for the hypoxic activation of the MMP1 promoter. The activities of MMP1 promoter luciferase constructs containing point mutations in the NF-κB or CREB binding sites (shown in the left figure) were compared with that of the wild type (WT) full-length vector using the luciferase assay in HeLa cells (n = 3). Error bars indicate S.D., and significance was statistically analyzed with a t test (**, p < 0.02).
Increased Expression and Activity of MMP1 during Hypoxia
Because the hypoxic induction of MMP1 gene expression appears to be regulated by CREB, NF-κB, and HIF-1/2, the role of these molecules in the hypoxic induction of MMP1 protein expression was examined in MDA-MB-231 cells. MMP1 protein levels increased during hypoxia, and this was partially blocked after depletion of NF-κB or CREB (Fig. 6A). This inhibition was observed after 24 and 48 h of hypoxic treatment. However, single knockdown of NF-κB or CREB did not completely block MMP1 expression, suggesting that another factor(s) also contributes to the induction of MMP1. Depletion of CREB/NF-κB or HIF-2α efficiently blocked MMP1 expression in MDA-MB-231 cells, whereas depletion of HIF-1α blocked it moderately (Fig. 6B). Moreover, simultaneous depletion of NF-κB/HIF-2α or CREB/HIF-2α further enhanced the degree of MMP1 inhibition, indicating that CREB, NF-κB, and HIF-2 function in parallel to regulate MMP1 expression during prolonged hypoxia (Fig. 6C). Next, the secreted form of MMP1 was monitored in the cell culture medium derived from CREB- and/or NF-κB-depleted cells. A moderate but significant increase in MMP1 activity in the culture medium was observed in MDA-MB-231 cells grown in hypoxic conditions (Fig. 6D). MMP1 activity was diminished after depletion of NF-κB and/or CREB, further indicating that the hypoxic induction of MMP1 is dependent on these transcription factors (Fig. 6D).
FIGURE 6.
CREB and NF-κB signaling pathways play an important role in the induction of MMP1 expression during prolonged hypoxia. A, shown is inhibition of MMP1 induction after depletion of NF-κB or CREB. MDA-MB-231 cells were transfected with control, NF-κB, or CREB siRNA and were cultured in hypoxic conditions for up to 48 h. Cell lysates were subjected to Western blotting. The asterisk indicates a nonspecific band detected by the antibody. B, shown is the effect of HIF-1α, HIF-2α, and CREB/NF-κB depletion on MMP1 induction. MDA-MB-231 cells transfected with HIF-1α, HIF-2α, or CREB/NF-κB siRNAs were cultured in normoxic or hypoxic conditions for 24 h, and cell lysates were subjected to Western blotting. The asterisk indicates a nonspecific band detected by the antibody. C, shown is the effect of NF-κB/HIF-2α and CREB/HIF-2α siRNA on MMP1 induction. MDA-MB-231 cells transfected with corresponding siRNAs were cultured in normoxic or hypoxic conditions for 24 h, and cell lysates were subjected to Western blotting. Asterisks indicate nonspecific bands detected by the antibodies. D, shown is decreased MMP1 activity in cell culture medium after depletion of CREB and/or NF-κB (kB). An ELISA assay was performed on cell culture medium collected from MDA-MB-231 cells transfected with CREB and/or NF-κB siRNA and treated with normoxia or hypoxia (24 h) to measure MMP1 activity (n = 3). The graph shows relative collagenase activity. Error bars indicate S.D. Significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02).
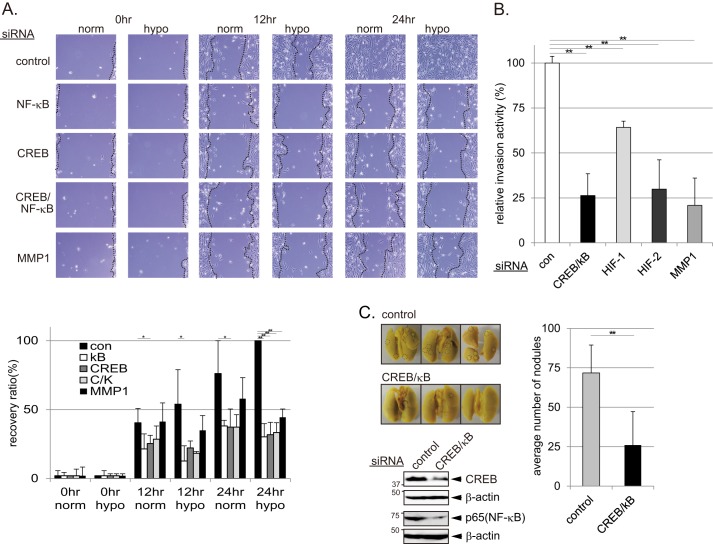
Inhibition of the CREB and NF-κB Signaling Pathways Reduces Cell Migration, Invasion, and Metastasis
Next, cell migration and invasion assays were performed to characterize the biological significance of the hypoxic induction of MMP1. MDA-MB-231 cells transfected with control, NF-κB, CREB, CREB/NF-κB, or MMP1 siRNAs were plated on collagen-coated dishes, and a single scratch was introduced. Depletion of CREB and/or NF-κB reduced the ability of cells to migrate into the gap throughout the entire duration of the experiment (24 h) (Fig. 7A). Depletion of MMP1 also reduced the cell migration, yet the effect was moderate compared with CREB and/or NF-κB depletion (Fig. 7A). Reduced migratory activity of CREB/NF-κB-depleted cells was partly recovered when cultured in conditioned medium derived from MDA-MB-231 cells treated with hypoxia but not by that derived from MMP1 siRNA cells (supplemental Fig. 3). Significant reduction of migratory activity was only found on the collagen-coated dishes but not on fibronectin or poly-l-lysine surfaces (supplemental Fig. 4). Furthermore, the invasive activity of MDA-MB-231 cells was monitored using the transwell migration assay with collagen-coated wells. Control cells actively passed through the collagen layer and moved to the lower well. This movement was inhibited after depletion of CREB and NF-κB or HIF-2α to a level comparable to that observed after MMP1 depletion (Fig. 7B). Similar inhibition was also observed in MDA-MB-231 cells stably expressing shRNA of CREB, NF-κB, or MMP1 (supplemental Fig. 5). These results indicate that MMP1 is one of the critical factors to control cell migration and invasion on a collagen layer during prolonged hypoxia and whose expression is regulated by CREB/NF-κB and HIF-2. Finally, the requirement of CREB and NF-κB for the lung metastasis was examined in the mouse pulmonary metastasis model using MDA-MB-231 cells. Whereas control cells formed an average of 72 metastatic nodules in the lung, CREB- and NF-κB-depleted cells formed significantly reduced numbers of nodules compared with the control cells (Fig. 7C). This result further indicates the importance of CREB and NF-κB, which induces MMP1, for tumor metastasis under pathophysiological conditions.
FIGURE 7.
Inhibition of CREB and NF-κB impairs cell migration, invasion, and metastasis. A, cell migration on collagen-coated dishes decreased after depletion of MMP1, CREB, and/or NF-κB (kB). MDA-MB-231 cells transfected with siRNAs (as indicated in the figure) were plated on collagen-coated dishes and cultured in normoxic or hypoxic conditions for the indicated times. Representative images for each time point are shown. Recovery ratio (%) during the time course is shown in the graph (A, bottom) (n = 3). Error bars indicate S.D., and significance was statistically analyzed with a t test (*, p < 0.05; **, p < 0.02). B, cell invasion decreased after depletion of CREB and NF-κB. MDA-MB-231 cells transfected with siRNAs (as indicated in the figure) were subjected to the transwell migration assay. Cells were cultured in hypoxic conditions for 48 h, and the average number of cells that passed through the collagen-coated membrane was determined (n = 3). Error bars indicate S.D., and significance was statistically analyzed with a t test (**, p < 0.02). C, lung metastasis was decreased by depletion of CREB and NF-κB. Control or CREB/NF-κB depleted MDA-MB-231 cells were injected to the tail vein of athymic nu/nu mice. Representative images of the lungs and the graph of the average number of metastatic nodules formed on the lung surface are shown (n = 7). Error bars indicate S.D., and significance was statistically analyzed with a t test (**, p < 0.02). Inhibition of CREB and NF-κB in MDA-MB-231 cells was examined by Western blotting.
DISCUSSION
In the present study, it has been demonstrated that CREB and NF-κB are activated during prolonged hypoxia and cooperatively induce MMP1 expression. Binding sites for CREB and NF-κB were located adjacently in the MMP1 promoter, and these sites are important for the induction of MMP1 after prolonged hypoxia. Although the hypoxic activation of HIF is mainly mediated by stabilization of its α subunit, the molecular mechanism underlying the hypoxic activation of CREB and NF-κB is not well understood. NF-κB is activated after the IKK-dependent phosphorylation and degradation of IκBα (17). It has been shown that hypoxia activates NF-κB signaling and that this is decreased in IKK-β−/− mice (5), indicating that the hypoxic activation of NF-κB occurs through a canonical pathway mediated by the IKK complex (IKK-β, IKK-α, and NF-κB essential modulator (NEMO)) (17). IKK-β is a PHD substrate (mainly PHD1), and prolyl-hydroxylation of IKK-β negatively regulates its kinase activity (18). PHD-dependent prolyl-hydroxylation may regulate IKK-β activity and thereby NF-κB signaling during hypoxia. Because NF-κB signaling was found to be mainly activated during prolonged hypoxia in MCF7, MDA-MB-231, and HeLa cells, it is possible that PHD activity is maximally inhibited during this phase. However, it has been shown that PHD2 and PHD3 become reactivated during prolonged hypoxia, leading to the down-regulation of HIF-1α (19). This is thought to be due in part to the up-regulation of PHD2 and PHD3 during prolonged hypoxia. In contrast, PHD1 is not up-regulated during hypoxia (20); therefore, it is possible that the PHD1 activity remains inhibited during prolonged hypoxia leading to activation of IKK-β and thereby NF-κB signaling.
One of the hallmarks of CREB activation is phosphorylation of Ser-133, and this is significantly increased during prolonged hypoxia. This phosphorylation is mediated by several kinases, including protein kinase A, protein kinase C, Akt, Ca2+/calmodulin-dependent kinase, and MAPKAP (MAPK-activated protein) kinase 2 (7). Some of these kinases, including Akt, Ca2+/calmodulin-dependent kinase, and MAPKAP kinase are activated in hypoxic/ischemic conditions, and may, therefore, contribute to the hypoxic activation of CREB (4, 21, 22). However, it is also reported that CREB activation in PC12 cells is mediated by none of these kinases (23), and the kinase responsible for regulating CREB activity during prolonged hypoxia still remains unclear. In addition to the phosphorylation-dependent activation of CREB, the stability of CREB is also regulated during hypoxia. CREB is reported to be ubiquitinated and degraded during the acute phase of hypoxia (24). However, it has been reported that SUMOylation of CREB increases during prolonged hypoxia, causing CREB to be stabilized and activated, and thereby leading to the induction of various genes such as amphiregulin (25, 26). Thus, CREB activation during prolonged hypoxia is mediated by at least two mechanisms, phosphorylation and stabilization.
There are several examples of CREB and NF-κB acting cooperatively. For example, simultaneous activation of CREB and NF-κB occurs in endothelial cells and astrocytes by phospholipase activation and β2-adrenergic receptor signaling (27, 28). Moreover, protein kinase A expression is induced by NF-κB, leading to activation of CREB and cooperation between these molecules (29). Although HIF signaling was induced/activated during acute hypoxia, CREB and NF-κB signaling pathways were maximally activated during prolonged hypoxia (Fig. 3, F and G). When CREB or NF-κB were activated separately, the activity of the MMP1 promoter increased by ∼3-fold, and this was increased to 6-fold when CREB and NF-κB were activated together (Fig. 5D). Additionally, inhibition of CREB phosphorylation by staurosporine decreased the hypoxic induction of MMP1 (Fig. 5C). Importantly, possible roles of other kinases and their substrates besides the CREB pathway cannot be excluded in this experiment, as staurosporine inhibits multiple kinases. Yet, the synchronized activation of CREB and NF-κB might be an important regulatory mechanism to enhance gene expression during prolonged hypoxia.
Generally, when cells are exposed to hypoxic conditions, HIF-1α is immediately stabilized and induces the expression of various hypoxia-responsive genes (1). It is also reported that the expression of HIF-1 target genes is maintained during prolonged hypoxia (30). However, because HIF-1α expression tends to decrease during prolonged hypoxia (30), expression of some HIF-1 target genes may similarly decrease, such as VEGF observed in this study (Fig. 1A). Therefore, maintenance of the gene expression during prolonged hypoxia may be one of the important roles for prolonged hypoxic signaling, which involves CREB and NF-κB (Fig. 8). Additionally, the prolonged hypoxic signaling induces a specific set of genes including MMP1, which may regulate changes in cell shape, migration, and invasion that are often observed during prolonged hypoxia (31). Thus, it is likely that the combinatory effect of genes regulated by CREB and NF-κB as well as another transcription factor(s) determines the migratory and invasive activity of cancer cells under such condition. The time window during which HIF-1 activity decreases and the activities of CREB and NF-κB increase may signify the shift from acute to prolonged hypoxic responses. Changes in the activities of signaling pathways would be responsible for alterations in global gene expression at different stages of hypoxia.
FIGURE 8.
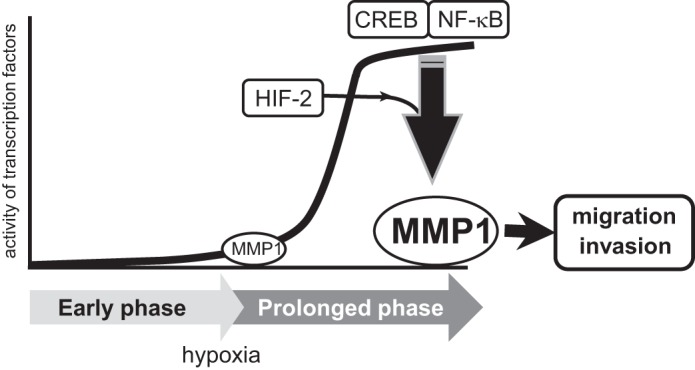
Schematic diagram of the study. Activity of CREB and NF-κB is increased during prolonged phase of hypoxia. CREB and NF-κB together with HIF-2 induces the expression of MMP1under such condition. Induction of MMP1 is involved in enhanced migration and invasion of cancer cells.
Finally, this study demonstrates that MMP1 expression is up-regulated during prolonged hypoxia. Because MMP1 possesses type I collagenase activity, its expression is linked to the invasion and metastasis of cancer cells (9). NF-κB is reported to also induce several MMPs upon cytokine activation, including MMP9, MMP13, and MT1-MMP (10, 32, 33). Similarly, CREB induces MMP2, MMP9, and MMP13 (34, 35). Therefore, synchronized activation of CREB and NF-κB during prolonged hypoxia might control the timing of metastasis, which is mediated by the orchestration of MMP1 and other MMPs (9). Because induction of MMP1 was observed in both cancerous MCF7/MB231 cells and non-cancerous HUVECs, which have a different metabolic status, hypoxic induction of MMP1 would be involved in various physiological processes besides tumor invasion and metastasis. In articular cartilage cells, IL-1β increases the level of HIF-2α and induces MMP1, leading to the destruction of osteoarthritic cartilage (14). This is consistent with our results showing that HIF-2 is an important regulator of MMP1 during prolonged hypoxia, although HIF-1 also appears to be involved in this process moderately. HIF-1 also regulates MMP1 expression in different cell lines, such as mesenchymal stem cells and lung adenocarcinoma cells (36, 37). Thus, the requirement of HIF-1 or HIF-2 to induce MMP1 might also depend on cell types. Similarly, induction profile of MMP1 may also be different between the cell types depending on the activity of CREB and NF-κB. It has been reported that the activity of HIF-2 is higher than that of HIF-1 during prolonged hypoxia in certain cell lines (38, 39). Therefore, CREB, NF-κB, and HIF-2 might constitute a network that regulates gene expression during prolonged hypoxia (Fig. 8). Further study is needed to identify the target genes regulated by the interaction of these transcription factors during prolonged hypoxia.
Acknowledgments
I am grateful to Drs. Ikuo Morita and Olga Safronova for the HRE-luc vector and Dr. Toshio Kitamura for the retrovirus packaging PLAT-A cell. I thank Dr. Hiroshi Nishina for valuable advice during the course of study. I also thank Dr. Ei-nan Boku for technical assistance and members of the Nakayama Laboratory for helpful discussions.
This study was supported by a Grant-in-Aid for Scientific Research C (Japan Society for the Promotion of Science).

This article contains a supplemental table and Figs. 1–5.
- HIF
- hypoxia-inducible factor
- HUVEC
- human umbilical vein endothelial cells
- PRP
- pre-mRNA processing factor
- MMP
- matrix metalloproteinase
- PHD
- prolyl-hydroxylase domain containing
- HRE
- hypoxia response element
- CRE
- cAMP-responsive element
- CREB
- CRE-binding protein
- norm
- normoxia
- hypo
- hypoxia
- qPCR
- quantitative PCR
- IKK
- IκB kinase.
REFERENCES
- 1. Wenger R. H., Stiehl D. P., Camenisch G. (2005) Integration of oxygen signaling at the consensus HRE. Sci. STKE 2005, re12. [DOI] [PubMed] [Google Scholar]
- 2. Semenza G. L. (2003) Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 [DOI] [PubMed] [Google Scholar]
- 3. Kaelin W. G., Jr., Ratcliffe P. J. (2008) Oxygen sensing by metazoans. The central role of the HIF hydroxylase pathway. Mol Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 4. Belaiba R. S., Bonello S., Zähringer C., Schmidt S., Hess J., Kietzmann T., Görlach A. (2007) Hypoxia up-regulates hypoxia-inducible factor-1α transcription by involving phosphatidylinositol 3-kinase and nuclear factor κB in pulmonary artery smooth muscle cells. Mol. Biol. Cell 18, 4691–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rius J., Guma M., Schachtrup C., Akassoglou K., Zinkernagel A. S., Nizet V., Johnson R. S., Haddad G. G., Karin M. (2008) NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature 453, 807–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walmsley S. R., Print C., Farahi N., Peyssonnaux C., Johnson R. S., Cramer T., Sobolewski A., Condliffe A. M., Cowburn A. S., Johnson N., Chilvers E. R. (2005) Hypoxia-induced neutrophil survival is mediated by HIF-1α-dependent NF-κB activity. J. Exp. Med. 201, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johannessen M., Delghandi M. P., Moens U. (2004) What turns CREB on? Cell. Signal. 16, 1211–1227 [DOI] [PubMed] [Google Scholar]
- 8. Sato M., Sakota M., Nakayama K. (2010) Human PRP19 interacts with prolyl-hydroxylase PHD3 and inhibits cell death in hypoxia. Exp. Cell Res. 316, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 9. Kessenbrock K., Plaks V., Werb Z. (2010) Matrix metalloproteinases. Regulators of the tumor microenvironment. Cell 141, 52–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mengshol J. A., Vincenti M. P., Coon C. I., Barchowsky A., Brinckerhoff C. E. (2000) Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor κB. Differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum. 43, 801–811 [DOI] [PubMed] [Google Scholar]
- 11. Vincenti M. P., Coon C. I., Brinckerhoff C. E. (1998) Nuclear factor κB/p50 activates an element in the distal matrix metalloproteinase 1 promoter in interleukin-1β-stimulated synovial fibroblasts. Arthritis Rheum. 41, 1987–1994 [DOI] [PubMed] [Google Scholar]
- 12. Ahn J. K., Koh E. M., Cha H. S., Lee Y. S., Kim J., Bae E. K., Ahn K. S. (2008) Role of hypoxia-inducible factor-1α in hypoxia-induced expressions of IL-8, MMP-1, and MMP-3 in rheumatoid fibroblast-like synoviocytes. Rheumatology 47, 834–839 [DOI] [PubMed] [Google Scholar]
- 13. Yamanaka M., Ishikawa O. (2000) Hypoxic conditions decrease the mRNA expression of proα1(I) and (III) collagens and increase matrix metalloproteinases-1 of dermal fibroblasts in three-dimensional cultures. J. Dermatol. Sci. 24, 99–104 [DOI] [PubMed] [Google Scholar]
- 14. Yang S., Kim J., Ryu J. H., Oh H., Chun C. H., Kim B. J., Min B. H., Chun J. S. (2010) Hypoxia-inducible factor-2α is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 16, 687–693 [DOI] [PubMed] [Google Scholar]
- 15. Heinemeyer T., Wingender E., Reuter I., Hermjakob H., Kel A. E., Kel O. V., Ignatieva E. V., Ananko E. A., Podkolodnaya O. A., Kolpakov F. A., Podkolodny N. L., Kolchanov N. A. (1998) Databases on transcriptional regulation. TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 26, 362–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akiyama Y. (1995) TFSEARCH, Searching transcription factor binding sites, RWCP, Japan [Google Scholar]
- 17. Oeckinghaus A., Hayden M. S., Ghosh S. (2011) Cross-talk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 18. Cummins E. P., Berra E., Comerford K. M., Ginouves A., Fitzgerald K. T., Seeballuck F., Godson C., Nielsen J. E., Moynagh P., Pouyssegur J., Taylor C. T. (2006) Prolyl hydroxylase-1 negatively regulates IκB kinase-β, giving insight into hypoxia-induced NFκB activity. Proc. Natl. Acad. Sci. U.S.A. 103, 18154–18159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ginouvès A., Ilc K., Macías N., Pouysségur J., Berra E. (2008) PHDs overactivation during chronic hypoxia “desensitizes” HIFα and protects cells from necrosis. Proc. Natl. Acad. Sci. U.S.A. 105, 4745–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. (2004) Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465 [DOI] [PubMed] [Google Scholar]
- 21. Kayyali U. S., Pennella C. M., Trujillo C., Villa O., Gaestel M., Hassoun P. M. (2002) Cytoskeletal changes in hypoxic pulmonary endothelial cells are dependent on MAPK-activated protein kinase MK2. J. Biol. Chem. 277, 42596–42602 [DOI] [PubMed] [Google Scholar]
- 22. Mabuchi T., Kitagawa K., Kuwabara K., Takasawa K., Ohtsuki T., Xia Z., Storm D., Yanagihara T., Hori M., Matsumoto M. (2001) Phosphorylation of cAMP response element-binding protein in hippocampal neurons as a protective response after exposure to glutamate in vitro and ischemia in vivo. J. Neurosci. 21, 9204–9213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beitner-Johnson D., Millhorn D. E. (1998) Hypoxia induces phosphorylation of the cyclic AMP response element-binding protein by a novel signaling mechanism. J. Biol. Chem. 273, 19834–19839 [DOI] [PubMed] [Google Scholar]
- 24. Taylor C. T., Furuta G. T., Synnestvedt K., Colgan S. P. (2000) Phosphorylation-dependent targeting of cAMP response element binding protein to the ubiquitin/proteasome pathway in hypoxia. Proc. Natl. Acad. Sci. U.S.A. 97, 12091–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Comerford K. M., Leonard M. O., Karhausen J., Carey R., Colgan S. P., Taylor C. T. (2003) Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc. Natl. Acad. Sci. U.S.A. 100, 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Reilly S. M., Leonard M. O., Kieran N., Comerford K. M., Cummins E., Pouliot M., Lee S. B., Taylor C. T. (2006) Hypoxia induces epithelial amphiregulin gene expression in a CREB-dependent manner. Am. J. Physiol. Cell Physiol. 290, C592–C600 [DOI] [PubMed] [Google Scholar]
- 27. Hadad N., Tuval L., Elgazar-Carmom V., Levy R. (2011) Endothelial ICAM-1 protein induction is regulated by cytosolic phospholipase A2α via both NF-κB and CREB transcription factors. J. Immunol. 186, 1816–1827 [DOI] [PubMed] [Google Scholar]
- 28. Spooren A., Kooijman R., Lintermans B., Van Craenenbroeck K., Vermeulen L., Haegeman G., Gerlo S. (2010) Cooperation of NFκB and CREB to induce synergistic IL-6 expression in astrocytes. Cell. Signal. 22, 871–881 [DOI] [PubMed] [Google Scholar]
- 29. Kaltschmidt B., Ndiaye D., Korte M., Pothion S., Arbibe L., Prüllage M., Pfeiffer J., Lindecke A., Staiger V., Israël A., Kaltschmidt C., Mémet S. (2006) NF-κB regulates spatial memory formation and synaptic plasticity through protein kinase A/CREB signaling. Mol. Cell. Biol. 26, 2936–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stiehl D. P., Wirthner R., Köditz J., Spielmann P., Camenisch G., Wenger R. H. (2006) Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 281, 23482–23491 [DOI] [PubMed] [Google Scholar]
- 31. Koh M. Y., Lemos R., Jr., Liu X., Powis G. (2011) The hypoxia-associated factor switches cells from HIF-1α- to HIF-2α-dependent signaling promoting stem cell characteristics, aggressive tumor growth, and invasion. Cancer Res. 71, 4015–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han Y. P., Tuan T. L., Wu H., Hughes M., Garner W. L. (2001) TNF-α stimulates activation of pro-MMP2 in human skin through NF-(κ)B mediated induction of MT1-MMP. J. Cell Sci. 114, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yokoo T., Kitamura M. (1996) Dual regulation of IL-1 β-mediated matrix metalloproteinase-9 expression in mesangial cells by NF-κB and AP-1. Am. J. Physiol. 270, F123–F130 [DOI] [PubMed] [Google Scholar]
- 34. Park J. K., Park S. H., So K., Bae I. H., Yoo Y. D., Um H. D. (2010) ICAM-3 enhances the migratory and invasive potential of human non-small cell lung cancer cells by inducing MMP-2 and MMP-9 via Akt and CREB. Int. J. Oncol. 36, 181–192 [PubMed] [Google Scholar]
- 35. Bui C., Barter M. J., Scott J. L., Xu Y., Galler M., Reynard L. N., Rowan A. D., Young D. A. (2012) cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB J. 26, 3000–3011 [DOI] [PubMed] [Google Scholar]
- 36. Lin J. L., Wang M. J., Lee D., Liang C. C., Lin S. (2008) Hypoxia-inducible factor-1α regulates matrix metalloproteinase-1 activity in human bone marrow-derived mesenchymal stem cells. FEBS Lett. 582, 2615–2619 [DOI] [PubMed] [Google Scholar]
- 37. Shyu K. G., Hsu F. L., Wang M. J., Wang B. W., Lin S. (2007) Hypoxia-inducible factor 1α regulates lung adenocarcinoma cell invasion. Exp. Cell Res. 313, 1181–1191 [DOI] [PubMed] [Google Scholar]
- 38. Holmquist-Mengelbier L., Fredlund E., Löfstedt T., Noguera R., Navarro S., Nilsson H., Pietras A., Vallon-Christersson J., Borg A., Gradin K., Poellinger L., Påhlman S. (2006) Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma. HIF-2α promotes an aggressive phenotype. Cancer Cell 10, 413–423 [DOI] [PubMed] [Google Scholar]
- 39. Koh M. Y., Powis G. (2012) Passing the baton. The HIF switch. Trends Biochem. Sci 37, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]