Background: The molecular basis for sodium channel inhibition by spider venom peptides is poorly understood.
Results: Key toxin residues and structural features important for activity of huwentoxin-IV are identified.
Conclusion: Toxin activity involves a hydrophobic protrusion surrounded by a ring of basic residues.
Significance: New structure-function information may provide a foundation for the design of peptides with therapeutic potential.
Keywords: Electrophysiology, Ion Channels, Sodium Channels, Structural Biology, Toxins
Abstract
Voltage-gated sodium channels (VGSCs) are essential to the normal function of the vertebrate nervous system. Aberrant function of VGSCs underlies a variety of disorders, including epilepsy, arrhythmia, and pain. A large number of animal toxins target these ion channels and may have significant therapeutic potential. Most of these toxins, however, have not been characterized in detail. Here, by combining patch clamp electrophysiology and radioligand binding studies with peptide mutagenesis, NMR structure determination, and molecular modeling, we have revealed key molecular determinants of the interaction between the tarantula toxin huwentoxin-IV and two VGSC isoforms, Nav1.7 and Nav1.2. Nine huwentoxin-IV residues (F6A, P11A, D14A, L22A, S25A, W30A, K32A, Y33A, and I35A) were important for block of Nav1.7 and Nav1.2. Importantly, molecular dynamics simulations and NMR studies indicated that folding was normal for several key mutants, suggesting that these amino acids probably make specific interactions with sodium channel residues. Additionally, we identified several amino acids (F6A, K18A, R26A, and K27A) that are involved in isoform-specific VGSC interactions. Our structural and functional data were used to model the docking of huwentoxin-IV into the domain II voltage sensor of Nav1.7. The model predicts that a hydrophobic patch composed of Trp-30 and Phe-6, along with the basic Lys-32 residue, docks into a groove formed by the Nav1.7 S1-S2 and S3-S4 loops. These results provide new insight into the structural and molecular basis of sodium channel block by huwentoxin-IV and may provide a basis for the rational design of toxin-based peptides with improved VGSC potency and/or selectivity.
Introduction
Over 40,000 species of spiders have been described to date, making them one of the most successful species on the planet. Spiders owe this success to their ability to produce complex venoms for predation and defense (1). The major components of spider venoms that have been investigated are small, disulfide-rich peptides, with each venom containing as many as 1,000 peptides, the majority of which have not been characterized in any detail (2). The spider venom peptides that have been described to date are detailed in the ArachnoServer database (3, 4).
Many of the toxins listed in ArachnoServer target voltage-gated sodium channels (VGSCs).2 Because the majority of spiders prey primarily on invertebrates, it is likely that these toxins target VGSCs in the insect nervous system, providing a highly efficient mechanism for spiders to incapacitate their prey. However, given the structural similarity between many insect and vertebrate sodium channels, it is not surprising that many spider venom peptides also modulate the activity of vertebrate VGSCs (5–9). In vertebrates, there are nine VGSC isoforms, Nav1.1–Nav1.9, with each isoform exhibiting a unique pattern of tissue distribution. VGSCs are essential to the initiation and propagation of electrical signals in the vertebrate nervous system, and aberrant sodium channel function is thought to underlie a variety of medical disorders, including epilepsy (10), arrhythmia (11), myotonia (12), and pain (13). VGSC-blocking spider toxins, therefore, may have significant therapeutic potential, and their detailed characterization is clearly warranted (14).
Twelve families of spider venom peptides that target VGSCs (so-called “NaSpTx” families 1–12) have recently been defined (15), based on either documented VGSC activity or sequence similarity to confirmed VGSC-modulating peptides. Several of the best characterized peptides are from NaSpTx family 1. All peptides from this family were isolated from the venom of tarantulas (Theraphosidae), and they consist of 33–35 amino acid residues with three disulfide bridges that form an inhibitory cysteine knot (ICK) motif (14). Peptides from family 1 include huwentoxin-1 (μ/ω-TRTX-Hh1a) (16), hainantoxin-IV (μ-TRTX-Hhn1b) (9), phrixotoxin-3 (β-TRTX-Ps1a) (5), ceratotoxin-1 (β-TRTX-Cm1a) (5), and huwentoxin-4 (μ-TRTXHh2a, HwTX-IV) (8, 17). All of these peptides are inhibitors of vertebrate VGSCs, and their mechanism of action probably involves a specific interaction with the voltage sensor region(s), impeding its movement and inhibiting channel activation (5, 8). However, very little is known about the channel residues involved in these interactions. The only studies addressing this question are from Cummins and colleagues (8, 18, 19), who have identified several residues in the S3-S4 linker region of domain II that are crucial for huwentoxin-IV inhibition of Nav1.7. Even less is known about the toxin residues that interact with VGSCs. The only studies published on this subject suggest that charged residues (Lys-27, His-28, Arg-29, and Lys-32) are important for the activity of hainantoxin-IV on VGSCs in isolated rat dorsal root ganglion cells (15, 20, 21).
Given the poor understanding of the structure-activity relationships for NaSpTx family 1 toxins and the pharmacological and therapeutic importance of sodium channel blockers, we sought to characterize the molecular determinants of the interaction between the NaSpTx family 1 member, HwTX-IV, and the VGSC isoforms, Nav1.7 and Nav1.2, in more detail. We combined functional and binding studies with peptide-scanning mutagenesis, NMR structure determination, molecular dynamics simulations, and docking in order to better understand the structure-activity relationships for the HwTX-IV-VGSC interaction. Our results provide the most comprehensive evaluation of the structure-activity relationships (SAR) for a family 1 toxin described to date as well as the first systematic evaluation of toxin SAR against two closely related VGSCs. Our experimental findings, coupled with the predictions of our model, provide a structural framework for the interaction between HwTX-IV and Nav1.7/Nav1.2 and may provide a basis for rational design of toxin-based peptides with improved VGSC potency and/or selectivity with potential for the treatment of a variety of human disorders.
EXPERIMENTAL PROCEDURES
Peptide Production and Purification
Generation and Transfection of Expression Constructs
Wild-type huwentoxin-IV and the alanine-scanning variant clones were designed as His6-HSA-(G4S)4-HRV3C-huwentoxin-IV fusion proteins and cloned into an in-house, pcDNA-derived mammalian expression vector in frame with an IgG2a signal peptide to direct secretion of the fusion into the expression medium. In the absence of a method to C-terminally amidate the recombinant peptides, we chose to include the C-terminal Gly-Lys in the peptide coding region to substitute for the amidation. The gene sequences for wild-type huwentoxin-IV and the designed variants were generated using in-house gene assembly technology (US6521427B1). The amino acid sequences of the designed peptide variants were back-translated to DNA sequences using human high frequency codons. Standard molecular biology techniques were used to generate and identify a positive clone for each designed variant.
Plasmid DNA for transfection was generated in milligram quantities using Qiagen Gigapreps. HEK 293F cells were transfected at 2–3-liter scale using FreeStyle Max (Invitrogen) transfection reagent according to the manufacturer's specifications; grown in 2-liter shake flasks for 96 h at 37 °C, 8% CO2, and 120 rpm; and harvested by centrifugation. The cell pellets were discarded, and the supernatants were filtered for purification.
Purification of Huwentoxin-IV Amino Acid-scanning Library Variants
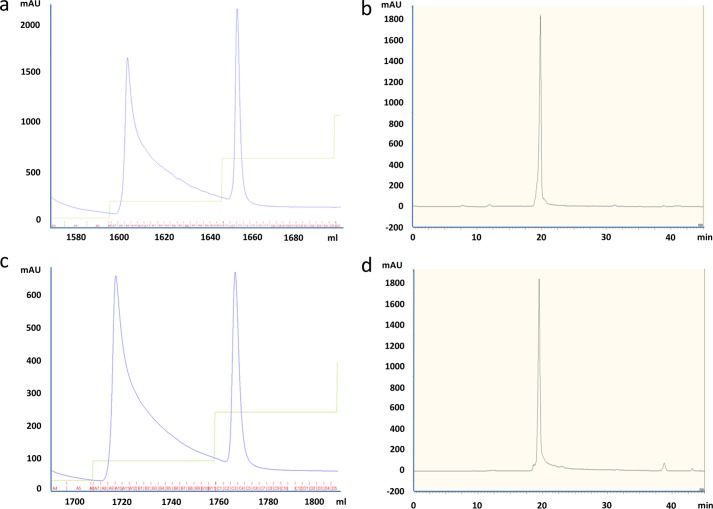
The secreted proteins were purified from the expression supernatants via immobilized metal affinity chromatography using 5-ml HisTrap HP columns (GE Healthcare). The chromatography method was run using an AKTA Explorer or FPLC, and protein was eluted from the column using a step gradient of imidazole. Peak fractions were pooled and concentrated using Amicon Ultra-15 centrifugal concentrators (Millipore), dialyzed against Dulbecco's phosphate-buffered saline, pH 7.2 (Invitrogen), and digested overnight with HRV3C (EMD; 1 unit/100 μg of protein). The cleaved fusion was purified by immobilized metal affinity chromatography using 5-ml HisTrap HP columns. The peptide was collected in the flow-through fraction. Pooled peptide was concentrated and polished via RP-HPLC using a C18(2) column (Phenomenex) (Fig. 1). The chromatography method was run on an Agilent 1100 HPLC system, and the bound peptide was eluted using a linear gradient of acetonitrile.
FIGURE 1.
Purification of parent and alanine-scanning variants of huwentoxin-IV. Shown are representative chromatograms for immobilized metal affinity chromatography purification of intact HSA fusions (a and c) and of RP-HPLC purification of free peptides (b and d) for WT huwentoxin-IV and S12A.
Each peak fraction was analyzed by RP-HPLC on an analytical C18(2) column (Phenomenex) using an acetonitrile linear gradient. Fractions with the same retention times were pooled and lyophilized. Lyophilized peptides were resuspended in HEPES-buffered saline, pH 7.4 (10 mm HEPES, 137 mm NaCl, 5.4 mm KCl, 5 mm glucose, 2 mm CaCl2, 1 mm MgCl2). In cases where multiple peaks were present in the RP-HPLC chromatogram, all fractions were assayed, and the one with the highest potency against Nav1.7 was the value reported. Without exception, the largest peak from the preparative HPLC corresponded to the most active fraction.
Absorbance was measured at 280 nm, and concentrations were calculated using each peptide's extinction coefficient. Final peptides were analyzed by electrospray ionization mass spectrometry. Retention of Gly-36 and Lys-37 varies; however, its presence (or absence) appears to have little or no impact on activity on VGSCs (data not shown).
Electrophysiology
In preliminary studies, synthetic HwTX-IV (Peptides International) exhibited similar potency regardless of whether activity was measured using manual or automated (QPatch HT, Sophion) whole-cell patch clamp techniques (pIC50 values for block of hNav1.7 were 7.9 ± 0.07 and 7.8 ± 0.03 in manual and automated patch-clamp, respectively). Given the excellent agreement between manual and automated patch clamp recordings, all subsequent studies were performed on the QPatch.
HEK293 cells stably expressing human Nav1.7 (Millipore) or human Nav1.2 (supplied by Dr. H. A. Hartmann, University of Maryland Biotechnology Institute) were cultured in DMEM/F-12 medium (1:1), supplemented with 10% fetal bovine serum, 400 μg/ml Geneticin, and 100 μm non-essential amino acids (all reagents from Invitrogen) at 37 °C and in 5% CO2. Cell preparation for QPatch assays was performed as described previously (22). For whole-cell patch clamp recording, the extracellular solution contained 137 mm NaCl, 5.4 mm KCl, 1 mm MgCl2, 2 mm CaCl2, 5 mm glucose, and 10 mm HEPES, pH 7.4, 315 mosm. The intracellular solution contained 135 mm CsF, 10 mm CsCl, 5 mm EGTA, 5 mm NaCl, and 10 mm HEPES, pH 7.3, 290 mosm. To elicit sodium currents, cells were first hyperpolarized from the holding potential to −120 mV for 2 s and then depolarized to 0 mV for 5 ms before returning to the holding potential (−75 mV for Nav1.7 and −65 mV for Nav1.2, which are the V½ values for steady-state inactivation for the respective channels; data not shown). This protocol was repeated once every 60 s during liquid applications (see below). Cells were otherwise held at −75 mV (Nav1.7) or −65 mV (Nav1.2) when the voltage protocol above was not executed. Upon establishment of the whole-cell recording configuration, a total of five applications of the extracellular solution (1× control buffer, 3× test compound/control, 1 μm tetrodotoxin (positive control), all containing 0.1% bovine serum albumin (BSA) except for the tetrodotoxin solution) were made on each cell. The voltage protocol was executed 10 times after each application. Currents were sampled at 25 kHz and filtered at 5 kHz with an 8-pole Bessle filter. The series resistance compensation level was set at 80%. All experiments were performed at room temperature (∼22 °C). pIC50 values were determined from logistic fits of concentration-response data using GraphPad Prism.
Radioligand Binding
HwTX-IV (Peptide Institute Inc., Osaka, Japan) was radioiodinated using previously described methods (23). The specific activity of the product was estimated to be 2,200 Ci/mmol. A cold iodination was performed using the same stoichiometry and methods but substituting NaI (Sigma) for Na125I.
HEK-293 cells stably expressing Nav1.7 or Nav1.2 were seeded at a density of 100,000 cells/well in 96-well poly-d-lysine-coated plates and were allowed to attach for 16–20 h at 37 °C. For saturation experiments, cells were incubated with increasing concentrations of 125I-HwTX-IV for 3 h, and replicate (at least triplicate) samples were averaged for each experimental point. At the end of the incubation, cells were washed twice with 100 μl of Dulbecco's phosphate-buffered saline plus 10% fetal bovine serum. 100 μl of liquid scintillation fluid (Microscint-40, PerkinElmer Life Sciences) was added, and radioactivity was counted using the TopCountTM microplate scintillation counter. Nonspecific binding was defined as binding in the presence of 1 μm synthetic HwTX-IV. Total binding was defined as binding in the presence of radioisotope alone. Specific binding was obtained by subtracting nonspecific binding from total binding. Data were fit to the equation, Beq = (BmaxL*)/(Kd + L*), where L* is the free 125I-HwTX-IV concentration, to determine Kd values. To determine kinetics of ligand association, cells were incubated with 125I-HwTX-IV, and specific binding was determined at varying time points thereafter. To determine the kinetics of dissociation, cells were incubated with 125I-HwTX-IV for 3 h before dissociation was initiated by washing twice with media. Specific binding was determined at varying time points after washing. kobs and koff were derived from monoexponential fits of the kinetic data. The association rate constant, kon, was determined from the equation, kobs = kon[B] + koff. Kd values were calculated from the equation, Kd = koff/kon. For competition binding experiments, total binding of 4 nm 125I-HwTX-IV was measured after 3 h in the absence or presence of increasing concentrations of either synthetic HwTX-IV or the recombinant peptides. Binding pIC50 values were derived from logistic fits of concentration-inhibition curves. The absolute inhibition constant (Ki) values were determined from the equation, Ki = binding IC50/(1 + (L*/Kd)).
NMR Structure Determination
All NMR experiments were performed using Bruker Avance 600-, 700-, or 950-MHz spectrometers. The peptides were dissolved in aqueous buffer containing 10% D2O. The buffer maintained a pH of 6.7 using 20 mm phosphate, 0.1 mm deuterated EDTA, and 0.002% NaN3. All spectra were collected at 298 K unless otherwise stated. Individual residue spin systems were assigned using total correlation spectroscopy (24) spectra using spin-lock (Malcolm Levitt) with mixing times of 75 ms. Sequential residue assignments were made from NOESY (25) experiments collected with a mixing time of 150 ms. In addition, 15N HSQC (26) experiments aided assignment, and cystine oxidation states were elucidated via 13C HSQC (26) spectra. Shifted sinebell squared weighting and zero filling was applied before Fourier transformation using NMRPipe (27) during data processing.
Interproton distance restraints were derived from through-space interactions observed in the NOESY spectra and automatically assigned by CYANA (28). In addition, peptides containing Trp-32 that showed significant (>0.2 ppm) ring current anisotropy on neighboring amino acids have aromatic side-chain restraints applied. The applications PREDITOR (29) and DANGLE (30) were used to predict φ and ψ angle ranges based on chemical shift data. Backbone ω angle restraints were set to 180º. Based on data derived from the NOESY and 13C HSQC experiments, disulfide bonds were fixed between Cys-2 and Cys-17, Cys-9 and Cys-24, and Cys-16 and Cys-31.
Homology models of the peptides were used as input (Cycle 1) to CYANA followed by six cycles of combined automated NOESY assignment and structure calculation. During each cycle, 1,000 conformers were calculated using a standard simulated annealing schedule with 10,000 torsion angle dynamics steps per conformer followed by 50,000 steps of energy minimization.
Molecular Dynamics
An NMR Structure of Native HwTX-IV (PDB code 1MB6) was used as the starting point to characterize the stability of HwTX-IV using molecular dynamics simulations. In addition to simulations of the native HwTX-IV, simulations were performed to determine the changes in peptide stability due to single alanine point mutations. For each molecular dynamics simulation, the HwTX-IV was solvated in explicit water (with a minimum of 12 Å of padding) and neutralized to 0.1 m NaCl. The protein was minimized and equilibrated for 50 ns, using NAMD version 2.8 (31). CHARMM version 22 CMAP (32) parameters were used for the simulations with a multiple time-stepping algorithm for evaluating electrostatics with bonded interactions computed every 1 fs, short range non-bonded interactions computed every 2 fs, and long range interactions computed every 4 fs. Long range electrostatic forces were evaluated using the particle mesh Ewald summation method with a grid spacing of less than 1 Å. Temperature was maintained at 300 K using Langevin dynamics, and a constant pressure of 1 atm was maintained using a Nóse-Hoover Langevin piston. Periodic boundary conditions were assumed, and non-bonded interactions were calculated using scaled 1–4 exclusion with shifting starting at 8 Å and a complete cut-off at 12 Å. Following simulation, the molecular dynamics trajectories were aligned based on the backbone α-carbon (CA) atoms and the root mean square deviation (RMSD) per residue calculated over the entire simulation relative to the initial NMR structure using VMD (33).
Homology Modeling of Nav1.7 and Docking of HwTX-IV
A homology model of Nav1.7 DII S1–S4 was built with NavAb (PDB code 3RVY) as a template using the Modeler component in Discovery Studio version 3.1 (Accelrys) (34). The model was then further refined to generate a resting state Nav1.7 structure using the crystal structure of Kv1.2 in the ground state as a guide (35). S4 was manually moved down into a resting state configuration, the S1-S2 and S3-S4 loops were regenerated, and the entire model was energy-minimized. Native HwTX-IV was manually docked into the Nav1.7 homology model based on the results of the alanine scan of HwTX-IV inhibition against Nav1.7 and on published Nav1.7 mutations that affect HwTX-IV binding (8, 18, 19). Following the manual docking, the entire Nav1.7 DII S1–S4 with docked HwTX-IV system was minimized, and an implicit membrane molecular dynamics simulation was performed using the CHARMm force field with Generalized Born Implicit Membrane (Discovery Studio (36)) to further refine the docked structure.
RESULTS
Alanine-scanning Mutagenesis of HwTX-IV
In order to understand the structural requirements for VGSC block by HwTX-IV, we generated a recombinant library of HwTX-IV alanine mutants, in which each non-cysteine position in the toxin was changed to alanine. Because we observed yields as high as hundreds of mg/liter for human serum albumin (HSA) fusions and because expression was poor for the peptide alone and in other fusion constructs, we produced the library variants initially as HRV3C-cleavable HSA fusion proteins. The sequences of the purified, cleaved variants are summarized in Table 1.
TABLE 1.
Sequences of the 30 recombinant alanine-scanning variants
Residues in parentheses indicate that the sample was partially lacking the terminal glycine in question, as measured by electrospray ionization-MS.
| Mutation | Sequence |
|---|---|
| WT | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCKYQI(G) |
| E1A | GPACLEIFKACNPSNDQCCKSSKLVCSRKTRWCKYQIG |
| L3A | PECAEIFKACNPSNDQCCKSSKLVCSRKTRWCKYQIG |
| E4A | GPECLAIFKACNPSNDQCCKSSKLVCSRKTRWCKYQIG |
| I5A | GPECLEAFKACNPSNDQCCKSSKLVCSRKTRWCKYQIG |
| F6A | GPECLEIAKACNPSNDQCCKSSKLVCSRKTRWCKYQIGK |
| K7A | GPECLEIFAACNPSNDQCCKSSKLVCSRKTRWCKYQIG |
| N10A | GPECLEIFKACAPSNDQCCKSSKLVCSRKTRWCKYQI(G) |
| P11A | GPECLEIFKACNASNDQCCKSSKLVCSRKTRWCKYQI(G) |
| S12A | GPECLEIFKACNPANDQCCKSSKLVCSRKTRWCKYQIG |
| N13A | GPECLEIFKACNPSADQCCKSSKLVCSRKTRWCKYQI(G) |
| D14A | GPECLEIFKACNPSNAQCCKSSKLVCSRKTRWCKYQI(G) |
| Q15A | GPECLEIFKACNPSNDACCKSSKLVCSRKTRWCKYQIG |
| K18A | GPECLEIFKACNPSNDQCCASSKLVCSRKTRWCKYQIGK |
| S19A | GPECLEIFKACNPSNDQCCKASKLVCSRKTRWCKYQIGK |
| S20A | GPECLEIFKACNPSNDQCCKSAKLVCSRKTRWCKYQIG |
| K21A | GPECLEIFKACNPSNDQCCKSSALVCSRKTRWCKYQI(G) |
| L22A | GPECLEIFKACNPSNDQCCKSSKAVCSRKTRWCKYQI(G) |
| V23A | GPECLEIFKACNPSNDQCCKSSKLACSRKTRWCKYQIGK |
| S25A | GPECLEIFKACNPSNDQCCKSSKLVCARKTRWCKYQI(G) |
| R26A | GPECLEIFKACNPSNDQCCKSSKLVCSAKTRWCKYQI(G) |
| K27A | GPECLEIFKACNPSNDQCCKSSKLVCSRATRWCKYQI(G) |
| T28A | GPECLEIFKACNPSNDQCCKSSKLVCSRKARWCKYQI(G) |
| R29A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTAWCKYQI(G) |
| W30A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRACKYQIG |
| K32A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCAYQIG |
| Y33A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCKAQIG |
| Q34A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCKYAI(G) |
| I35A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCKYQA(G) |
| Q36A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCKYQIA |
| K37A | GPECLEIFKACNPSNDQCCKSSKLVCSRKTRWCKYQIGA |
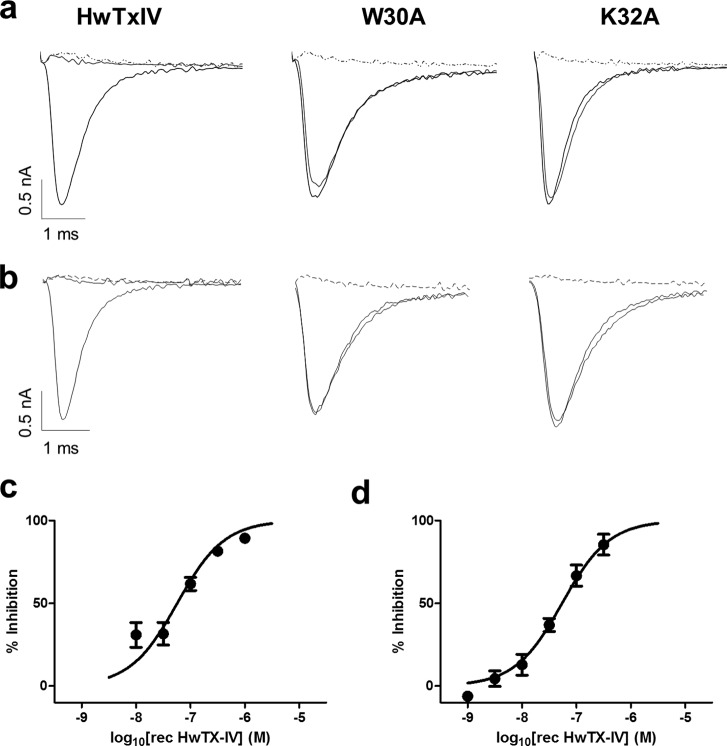
The activity of purified recombinant HwTX-IV and 30 mutants was measured against Nav1.7 and Nav1.2, and the results are summarized in Fig. 2 and Table 2. Recombinant HwTX-IV was less potent than synthetic HwTX-IV, probably mostly reflecting the importance of C-terminal amidation, because unamidated HwTX-IV has significantly lower potency (data not shown). pIC50 values for block of Nav1.7 were 7.8 ± 0.03 and 7.27 ± 0.1 for synthetic and recombinant HwTX-IV, respectively. A similar reduction of potency for recombinant material was also observed for block of Nav1.2 (pIC50 values were 8.0 ± 0.03 and 7.27 ± 0.04 for synthetic and recombinant HwTX-IV, respectively). Essential residues were defined as those resulting in at least a 10-fold loss of activity by the alanine substitution. For Nav1.7, nine mutant toxins were at least 10-fold less potent than parent recombinant HwTX-IV: F6A, P11A, D14A, L22A, S25A, W30A, K32A, Y33A, and I35A. In particular, W30A and K32A were completely devoid of activity at concentrations up to 1 μm. Of the remaining 21 mutants, 20 exhibited activity similar to that of parent recombinant HwTX-IV against Nav1.7, whereas one, E4A, showed marginally improved potency. Most of the mutants that substantially altered Nav1.7 activity also showed parallel changes in Nav1.2 activity. F6A, P11A, D14A, L22A, S25A, W30A, K32A, and Y33A were all at least 10-fold less active on Nav1.2 than the recombinant parent HwTX-IV, and E4A was again marginally more active. Interestingly, some positions were identified that differentially modified Nav1.7 and Nav1.2 potency. F6A, K18A, R26A, and K27A all resulted in a preferential loss of Nav1.2 activity, whereas I35A resulted in preferential loss of Nav1.7 activity.
FIGURE 2.
a and b, representative recordings of hNav1.7 (a) or hNav1.2 (b) currents in the absence or presence of 1 μm recombinant HwTX-IV (left), W30A (middle), or K32A (right). Note the complete block of current in the presence of HwTX-IV (left) but lack of effect of W30A (middle) or K32A (right). Tetrodotoxin (1 μm; dotted line) completely inhibited sodium currents in all cells. Concentration-response curves were generated for recombinant HwTX-IV against hNav1.7 (c) or hNav1.2 (d), using automated patch clamp (QPatch) techniques. pIC50 values were 7.26 ± 0.05 and 7.27 ± 0.04 for hNav1.7 and hNav1.2, respectively. Values shown are mean ± S.E.
TABLE 2.
Alanine-scanning mutagenesis of HwTx-IV
pIC50 and pKi values were determined for recombinant parent HwTx-IV (rec HwTx-IV) and 31 single amino acid mutants using Qpatch and 125I-HwTx-IV competition binding assays. For peptides that did not exhibit sufficient activity to determine a pIC50 or pKi, percentage inhibition at the top concentration tested (1 μm) is shown (percentage inhibition numbers are shown in boldface type and indicated with %). Data are the means ± S.E., n > 3.
| Alanine scan | QPatch |
125I-HwTx-IV binding |
||||||
|---|---|---|---|---|---|---|---|---|
| hNav1.7 |
hNav1.2 |
hNav1.7 |
hNav1.2 |
|||||
| pIC50 | Hill slope | pIC50 | Hill slope | pKi | Hill slope | pKi | Hill slope | |
| rec HwTx-IV | 7.3 ± 0.05 | 0.7 ± 0.10 | 7.3 ± 0.04 | 1.1 ± 0.10 | 6.5 ± 0.05 | 1.2 ± 0.20 | 6.7 ± 0.05 | 1.0 ± 0.10 |
| E1A | 7.7 ± 0.03 | 1.4 ± 0.10 | 7.6 ± 0.14 | 1.1 ± 0.40 | 6.7 ± 0.05 | 1.1 ± 0.20 | 7.0 ± 0.10 | 1.1 ± 0.20 |
| L3A | 6.6 ± 0.10 | 0.9 ± 0.30 | 6.9 ± 0.05 | 1.0 ± 0.10 | 6.0 ± 0.08 | 0.7 ± 0.10 | 7.0 ± 0.05 | 1.0 ± 0.10 |
| E4A | 7.9 ± 0.20 | 1.2 ± 0.40 | 7.5 ± 0.01 | 1.8 ± 0.10 | 7.3 ± 0.03 | 1.1 ± 0.10 | 6.7 ± 0.07 | 0.8 ± 0.10 |
| I5A | 7.1 ± 0.08 | 0.7 ± 0.10 | 6.7 ± 0.17 | 0.7 ± 0.20 | 5.9 ± 0.05 | 1.9 ± 0.60 | 6.1 ± 0.20 | 0.7 ± 0.30 |
| F6A | 6.3 ± 0.04 | 0.7 ± 0.07 | 5.1 ± 0.03 | 1.0 | 5.3 ± 0.4 | 1.0 ± 0.60 | 13.0 ± 6.0% | |
| K7A | 6.8 ± 0.08 | 0.7 ± 0.13 | 7.1 ± 0.20 | 1.0 ± 0.40 | 6.4 ± 0.03 | 1.2 ± 0.10 | 6.6 ± 0.10 | 0.8 ± 0.10 |
| N10A | 7.0 ± 0.14 | 0.8 ± 0.20 | 6.7 ± 0.05 | 1.0 ± 0.10 | 6.4 ± 0.04 | 1.1 ± 0.10 | 6.6 ± 0.10 | 0.7 ± 0.10 |
| P11A | 6.0 ± 0.10 | 0.8 ± 0.20 | 5.9 ± 0.10 | 0.8 ± 0.10 | 13.5 ± 4.8% | 21.1 ± 10.0% | ||
| S12A | 7.0 ± 0.13 | 0.8 ± 0.20 | 6.8 ± 0.10 | 0.9 ± 0.30 | 6.8 ± 0.06 | 1.0 ± 0.10 | 6.6 ± 0.04 | 1.1 ± 0.10 |
| N13A | 7.0 ± 0.10 | 0.7 ± 0.10 | 6.8 ± 0.04 | 1.3 ± 0.20 | 7.2 ± 0.07 | 0.8 ± 0.10 | 7.2 ± 0.10 | 0.8 ± 0.20 |
| D14A | 5.9 ± 0.01 | 1.6 ± 0.06 | 5.5 ± 0.08 | 1.0 | 11.7 ± 11.1% | 32.3 ± 4.3% | ||
| Q15A | 7.3 ± 0.13 | 0.6 ± 0.14 | 7.2 ± 0.10 | 1.1 ± 0.20 | 7.1 ± 0.05 | 1.0 ± 0.10 | 7.0 ± 0.05 | 0.8 ± 0.10 |
| K18A | 7.3 ± 0.04 | 0.7 ± 0.05 | 6.3 ± 0.06 | 1.5 ± 0.30 | 6.2 ± 0.07 | 1.1 ± 0.20 | 6.2 ± 0.05 | 1.9 ± 0.50 |
| S19A | 7.0 ± 0.05 | 0.7 ± 0.06 | 7.3 ± 0.10 | 0.8 ± 0.20 | 6.5 ± 0.04 | 0.8 ± 0.10 | 6.9 ± 0.03 | 1.0 ± 0.10 |
| S20A | 7.2 ± 0.08 | 0.7 ± 0.09 | 6.9 ± 0.06 | 0.9 ± 0.10 | 6.2 ± 0.04 | 1.4 ± 0.20 | 6.6 ± 0.05 | 0.8 ± 0.10 |
| K21A | 6.9 ± 0.06 | 1.0 ± 0.14 | 7.0 ± 0.10 | 1.1 ± 0.20 | 6.8 ± 0.05 | 1.1 ± 0.10 | 7.0 ± 0.04 | 1.2 ± 0.20 |
| L22A | 6.1 ± 0.01 | 1.0 ± 0.01 | 5.5 ± 0.2 | 1.0 | 5.7 ± 3.2% | 2.8 ± 2.2% | ||
| V23A | 6.9 ± 0.07 | 0.7 ± 0.09 | 7.2 ± 0.10 | 0.9 ± 0.20 | 6.5 ± 0.03 | 1 ± 0.10 | 6.4 ± 0.05 | 0.8 ± 0.10 |
| S25A | 5.1 ± 0.2 | 1.1 ± 0.40 | 5.8 ± 0.17 | 1.0 | 24.7 ± 7.6% | 24.5 ± 12.7% | ||
| R26A | 6.9 ± 0.04 | 0.8 ± 0.07 | 5.7 ± 0.30 | 0.9 ± 0.10 | 6.1 ± 0.04 | 1.5 ± 0.30 | 16.1 ± 2.2% | |
| K27A | 6.7 ± 0.03 | 0.7 ± 0.05 | 5.9 ± 0.10 | 1.0 ± 0.40 | 5.8 ± 0.07 | 1.9 ± 0.60 | 13.9 ± 6.5% | |
| T28A | 7.2 ± 0.13 | 1.1 ± 0.30 | 6.5 ± 0.20 | 0.7 ± 0.20 | 6.2 ± 0.05 | 1.4 ± 0.30 | 6.5 ± 0.04 | 1.2 ± 0.20 |
| R29A | 7.3 ± 0.04 | 0.9 ± 0.10 | 6.7 ± 0.03 | 0.9 ± 0.10 | 6.6 ± 0.1 | 1.0 ± 0.10 | 5.6 ± 0.10 | 0.7 ± 0.12 |
| W30A | 2.0 ± 6.0% | 14.0 ± 7.0% | 3.3 ± 10.8% | 3.3 ± 1.6% | ||||
| K32A | 5.0 ± 1.8% | 7.9 ± 3.8% | 2.8 ± 7.7% | 2.9 ± 6.2% | ||||
| Y33A | 6.1 ± 0.03 | 1.1 ± 0.10 | 6.0 ± 0.05 | 0.8 ± 0.10 | 11.9 ± 3.3% | 23.6 ± 6.3% | ||
| Q34A | 7.1 ± 0.03 | 0.9 ± 0.20 | 7.0 ± 0.10 | 0.8 ± 0.10 | 6.2 ± 0.04 | 1.3 ± 0.20 | 6.2 ± 0.04 | 1.3 ± 0.20 |
| I35A | 6.2 ± 0.06 | 0.9 ± 0.10 | 6.8 ± 0.10 | 0.5 ± 0.10 | 5.7 ± 0.20 | 1.7 ± 1.20 | 5.7 ± 0.20 | 1.1 ± 0.50 |
| G36A | 6.4 ± 0.20 | 0.7 ± 0.20 | 6.5 ± 0.10 | 1.0 ± 0.30 | 6.0 ± 0.10 | 1.1 ± 0.30 | 6.6 ± 0.10 | 1.0 ± 0.20 |
| K37A | 7.0 ± 0.10 | 0.5 ± 0.10 | 6.7 ± 0.05 | 1.2 ± 0.20 | 6.2 ± 0.04 | 1.2 ± 0.20 | 6.7 ± 0.10 | 1.2 ± 0.40 |
In order to determine whether changes in potency reflect changes in toxin affinity or efficacy, we developed a novel 125I-HwTX-IV binding assay for Nav1.7 and Nav1.2. Tyr-33 is the only tyrosine residue native to HwTX-IV that can be iodinated. Because alanine-scanning mutagenesis of HwTX-IV showed that this residue may be important for the interaction with Nav1.7 and Nav1.2, we first tested the activity of the iodinated peptide on Nav1.7 and Nav1.2. Synthetic cold iodinated HwTX-IV inhibited Nav1.7 and Nav1.2 currents with pIC50 values of 8.1 ± 0.07 and 8.4 ± 0.04, respectively. These values were similar to those determined for native, non-iodinated synthetic HwTX-IV (7.8 ± 0.03 and 8.0 ± 0.03 for Nav1.7 and Nav1.2, respectively).
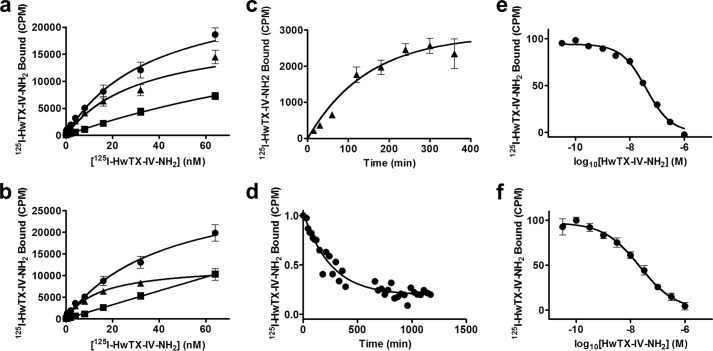
To characterize the binding of synthetic 125I-HwTX-IV to Nav1.7 and Nav1.2, HEK293 cells stably expressing these sodium channel isoforms were incubated with increasing concentrations of the radiolabeled toxin for 3 h in the absence or presence of 1 μm unlabeled synthetic HwTX-IV. Nonspecific binding increased linearly with increasing tracer concentrations for both channel types, whereas specific binding was saturable with pKd values of 7.5 ± 0.1 and 7.8 ± 0.06 (n = 4) for Nav1.7 and Nav1.2, respectively (Fig. 3, a and b). Bmax values were 19,122 ± 2,586 (n = 4) and 12,343 ± 778.2 (n = 4) for Nav1.7 and Nav1.2, respectively. No specific binding was observed in WT HEK293 cells. Incubation of cells stably expressing Nav1.7 with 125I-HwTX-IV resulted in time-dependent association and dissociation of the radioligand (kobs of 1.13 × 10−4 s−1; association rate constant, kon, of 1.17 × 104 m−1 s−1; and dissociation rate constant, koff, of 6.65 × 10−5 s−1 (n = 3; Fig. 3, c and d)). The pKd value derived from these kinetic data was 8.2, in reasonable agreement with the corresponding value from the saturation experiment. In competition studies, specific binding of synthetic 125I-HwTX-IV to Nav1.7 and Nav1.2 was inhibited by unlabeled synthetic HwTX-IV with pKi values of 7.4 ± 0.03 and 7.6 ± 0.08 (n > 7), respectively, in good agreement with the pKd values described above (Fig. 3, e and f). Hill coefficients were −0.95 ± 0.1 (n = 8) and −0.7 ± 0.05 (n = 7), respectively. The HwTX-IV pKi values determined in the binding assay, however, were slightly lower than the respective pIC50 values determined in the QPatch functional assay.
FIGURE 3.
Binding of 125I-HwTX-IV to HEK-293 cells stably expressing Nav1.7 or Nav1.2. a and b, saturation studies. HEK-293 cells stably expressing Nav1.7 (a) and Nav1.2 (b) were incubated with increasing concentrations of 125I-HwTX-IV. Total binding (●), nonspecific binding determined in the presence of 1 μm HwTX-IV (■), and specific binding (▴), defined as the difference between total and nonspecific binding, are presented. Specific binding of 125I-HwTX-IV displayed pKd values of 7.5 ± 0.1 and 7.8 ± 0.06 and Bmax values of 19,122 ± 2,586 and 12,343 ± 778.2 binding sites/cell for Nav1.7 and Nav1.2, respectively. c, association kinetics. Nav1.7 cells were incubated with 125I-HwTX-IV for the indicated times at 37 °C, and specific binding is plotted against time, yielding a kobs of 1.13 × 10−4 s−1. d, dissociation kinetics. Dissociation of 125I-HwTX-IV, initiated by dilution, followed monoexponential kinetics with a dissociation rate constant, koff, of 6.65 × 10−5 s−1. An association rate constant of kon, of 1.17 × 104 m−1 s−1, and a pKd value of 8.2 were derived from the kinetic data. e and f, competition binding. Cells were incubated with 4 nm 125I-HwTX-IV in the presence or absence of increasing concentrations of HwTX-IV for 3 h at 37 °C. Specific binding data were fit to a single-site inhibition model, yielding pKi values of 7.4 and 7.6 for Nav1.7 (e) and Nav1.2 (f), respectively. n >3 for all experiments. Values shown are means ± S.E.
Inhibition constants for recombinant HwTX-IV and the 30 single amino acid mutants were determined in 125I-HwTX-IV competition binding assays in cells expressing either Nav1.7 or Nav1.2, and the results are summarized in Table 2. As for parent HwTX-IV, pKi values determined for the alanine mutants in the binding assay were slightly lower than the respective pIC50 values determined in the QPatch functional assay. Notably, there were no examples where pKi values were higher than the respective QPatch pIC50 values. All mutants that exhibited a marked loss in activity versus Nav1.7 and Nav1.2 in QPatch (F6A, P11A, D14A, L22A, S25A, W30A, K32A, and Y33A) also exhibited a >10-fold loss of activity in the respective 125I-HwTX-IV binding assays. The binding assays also confirmed the preferential loss of Nav1.2 activity with R26A and K27A. In addition, R29A also resulted in preferential loss of Nav1.2 binding, whereas E4A showed more activity versus Nav1.7 and Nav1.2 in both binding assays.
Molecular Dynamics Simulations
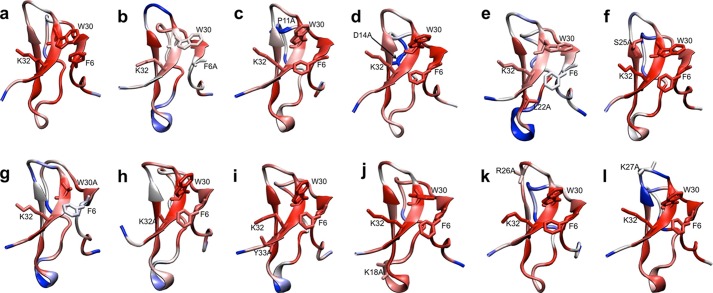
To help understand whether changes in toxin activity or selectivity could be explained by changes in peptide stability/flexibility, we conducted a series of molecular dynamics (MD) simulations, with particular emphasis on the possible effects of the alanine mutations that lead to significant loss of activity (F6A, P11A, D14A, L22A, S25A, W30A, K32A, and Y33A) or changes in channel selectivity (K18A, R26A, and K27A). The previously generated NMR structure for HwTX-IV (PDB code 1MB6) was used as a template for building the various alanine mutant peptides, and each toxin variant was subjected to 50 ns of molecular dynamics simulations (see supplemental Video 1). Static representations of the molecular dynamic simulations are shown in Fig. 4. For each mutant, CA RMSD values were calculated for each residue and colored on a gradient from 0.5 Å in red to 2.2 Å in blue. The average CA RMSD of native HwTX-IV peptide was only 1.0 Å, indicating a highly rigid peptide, stabilized by an antiparallel β-sheet, three disulfide bonds, and π-π interactions between Trp-30 and Phe-6 (which can be observed to flip between a face-to-face interaction and an edge-to-face interaction (Fig. 4a and supplemental Video 1)). MD simulations revealed that only F6A (Fig. 4b), L22A (Fig. 4e), and W30A (Fig. 4g) had a modest influence on core stability. Other loss of function mutants exerted little to no effect on core stability. However, most loss of function mutants as well as the mutants that differentially affected Nav1.7 and Nav1.2 activity influenced the flexibility of the loop regions to some extent. For example, F6A (Fig. 4b), L22A (Fig. 4e), W30A (Fig. 4g), and Y33A (Fig. 4i) appeared to increase the flexibility of loop 3, whereas P11A (Fig. 4c) and D14A (Fig. 4d) showed a clear increase in loop 2 flexibility. R26A (Fig. 4k) and K27A (Fig. 4l) were found to increase loop 4 flexibility. S25A (Fig. 4f), K32A (Fig. 4h), and K18A (Fig. 4j), on the other hand, did not seem to have any significant impact on peptide flexibility.
FIGURE 4.
Molecular dynamics simulations of selected HwTX-IV mutants. Various HwTX-IV alanine mutants were colored by average per residue CA RMSD calculated from their respective molecular dynamics simulations (50 ns each). The CA RMSDs are colored on a gradient from 0.5 Å in red to 2.2 Å in blue. The average CA RMSD of native HwTX-IV (a) was only 1.0 Å, indicating a highly rigid peptide (see also supplemental Video 1). MD simulations revealed that alanine substitutions at Phe-6 (b), Leu-22 (e), and Trp-30 (g) had a modest influence on core stability. Other selected alanine substitutions exerted little to no effect on core stability, but many influenced the flexibility of the loop regions. For example, F6A (b), L22A (e), W30A (g), and Y33A (i) appeared to increase the flexibility of loop 3, whereas P11A (c) and D14A (d) increased loop 2 flexibility. R26A (k) and K27A (l) were found to increase loop 4 flexibility. S25A (f), K32A (h), and K18A (j), on the other hand, did not seem to have any significant impact on peptide flexibility.
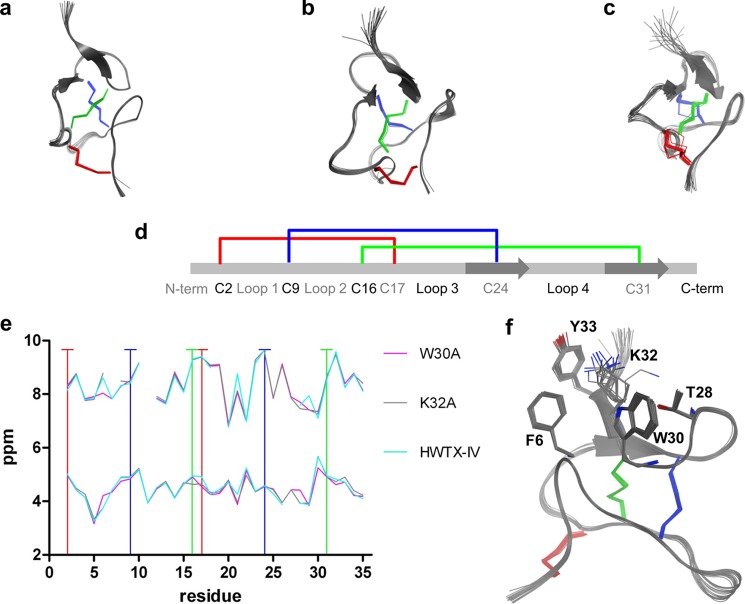
NMR
To gain insight into the structural features of recombinant HwTX-IV and two inactive mutants, W30A and K32A, we determined the NMR structures of the three toxins (Fig. 5). 1H chemical shifts and CYANA statistics for the three structures are shown in supplemental Tables S1 and S2, respectively. Our solution structure of recombinant HwTX-IV (Fig. 5a) confirms some of the key structural features described previously (37), including the presence of a twisted, anti-parallel β-sheet formed by residues Leu-22, Val-23, Cys-24, and Ser-25 and Arg-29, Trp-30, Cys-31, Lys-32, and Tyr-33 and an ICK structure formed by three disulfide bridges, between Cys-2 and Cys-17, Cys-9 and Cys-24, and Cys-16 and Cys-31. The toxin structure can be described one-dimensionally: N terminus, Cys-2, loop 1, Cys-9, loop 2, Cys-16, Cys-17, loop 3, β-strand 1/Cys-17, loop 4, β-strand 2/Cys-31, C terminus (Fig. 5d).
FIGURE 5.
NMR structures of HwTX-IV, W30A, and K32A. Ensembles of the 20 lowest energy structures of HwTX-IV (a), W30A (b), and K32A (c) are shown. Disulfide side chains are highlighted in color. d, a “one-dimensional” representation of the ICK fold found in HwTX-IV and analogs. Loops, β-sheets, and disulfide pattern are shown. e, backbone 1H chemical shifts. Top, amide proton (N-H); bottom, α-carbon proton (HA). Chemical shifts show low RMSD except where indole (Trp30)-derived chemical shift anisotropy is present. Vertical lines, cysteine residues. f, HwTX-IV ensemble highlighting 5 key residues. See the supplemental material for 1H chemical shifts (supplemental Table S1) and NMR structure statistics (supplemental Table S2).
Despite the complete loss of activity measured in the QPatch and binding assays, but largely in keeping with the molecular dynamic simulations, W30A and K32A exhibit a global structure similar to that of WT recombinant HwTX-IV (Fig. 5, b and c). The chemical shifts observed for amide protons and α-carbon protons along the length of the peptide backbone did not differ significantly between WT and W30A and K32A mutants, indicating that the surrounding molecular contexts of equivalent residues between structures are not significantly different (Fig. 5e). Although interproton NOESYs and backbone chemical shift values indicate that W30A, K32A, and the wild type peptides have very similar global folds and structure, local differences are apparent near the solvent-exposed face of the twisted β-sheet. These differences include observation of strong ring current anisotropy within a 5-Å radius of the Trp-30 indole of the K32A and wild type peptides, potential cation-π interaction between Lys-32 and Tyr-33, and minor differences in loop conformations (Fig. 5f). Coordinates and chemical shifts have been deposited in the PDB and BMRB: HWTX-IV (2M4X and 19026), W30A (2M4Z and 19030), and K32A (2M50 and 19032).
Homology Modeling and Docking
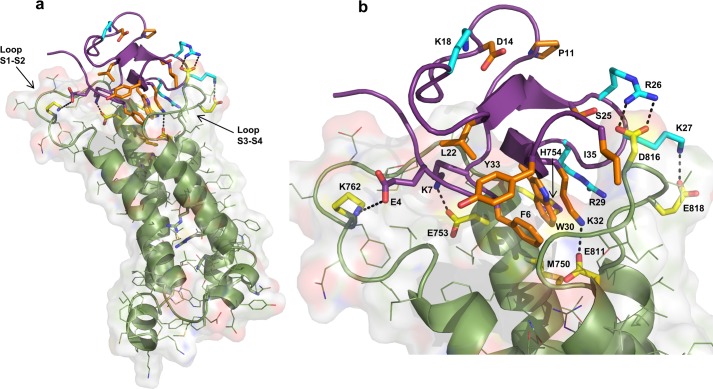
In order to explore the potential specific interactions made between HwTX-IV and Nav1.7, a homology model of the Nav1.7 domain II voltage sensor domain (S1–S4) was constructed, using NavAB as a template (34). The model was further refined to produce a resting state structure, using the crystal structure of Kv1.2 in the ground state as a guide (35). HwTX-IV was manually docked onto the homology model using our alanine scan SAR data and published channel mutation data to optimize the fit (8, 18, 19). These channel mutation data suggest that HwTX-IV binds in the DII voltage sensor domain and makes important interactions with the channel S1-S2 and S3-S4 loops (specifically with residues Glu753, Glu811, Asp816, and Glu818). The resulting docked structure is presented in Fig. 6, with the hydrophobic patch composed of Trp30 and Phe6, along with the basic Lys32 residue, oriented in the groove formed by the Nav1.7 S1-S2 and S3-S4 loops. The docked model predicts that the Trp30 and Phe6 hydrophobic patch interacts with a corresponding hydrophobic residue, Met750, in the channel groove. Charged interactions along the edge of the S1-S2 loop and S3-S4 loop allow the HwTX-IV to orient itself in the binding site. Specifically, on the S1-S2 loop, charge-charge interactions are made between Lys7 and Glu753 and between Glu4 and Lys762 of HwTX-IV and the Nav channel, respectively. Likewise, a series of charge-charge interactions (between Arg26 and Asp816, Lys27 and Glu818, and Lys32 and Glu811) between the HwTX-IV and the S3-S4 Nav loop are observed.
FIGURE 6.
Homology/docking model of HwTX-IV with Nav1.7 domain II voltage sensor. a, according to the model, HwTX-IV binds in the cleft between the S1-S2 and S3-S4 loops of the domain II voltage-sensing domain. b, detailed view of proposed peptide-channel interactions described under “Results” and “Discussion.” Channel residues are highlighted in yellow. Peptide residues that contribute to affinity are orange. Peptide residues that show selective interactions are shown in cyan. Peptide residues shown in purple may contribute to binding but have minimal impact when mutated to alanine.
DISCUSSION
Using a combination of complementary techniques, including a novel recombinant peptide expression system and a novel radioligand binding assay, we have identified the key toxin residues responsible for the interaction between HwTX-IV and the VGSCs, Nav1.7 and Nav1.2. Our results provide the most comprehensive SAR evaluation for a family 1 VGSC-blocking toxin described to date. Furthermore, our results provide the first systematic evaluation of the determinants of toxin selectivity between two closely related VGSCs. Our data show that Phe6, Pro11, Asp14, Leu22, Ser25, Trp30, Lys32, and Tyr33 are important for toxin activity (binding and inhibition). It is interesting to note that Trp30 and Lys32 are conserved across two of the best characterized families of sodium channel-blocking toxins, namely NaSpTx families 1 and 3 (15), but do not feature in other classes of spider toxins. These observations underscore the importance of Trp30 and Lys32 and suggest that these residues may be a universal determinant of sodium channel activity in these toxin families. Phe6, Lys18, Arg26, and Lys27 appear to make isoform-specific interactions. These novel findings may provide a basis for the rational design of toxin-based peptides with improved VGSC potency and/or selectivity.
Alanine Perturbation Maps
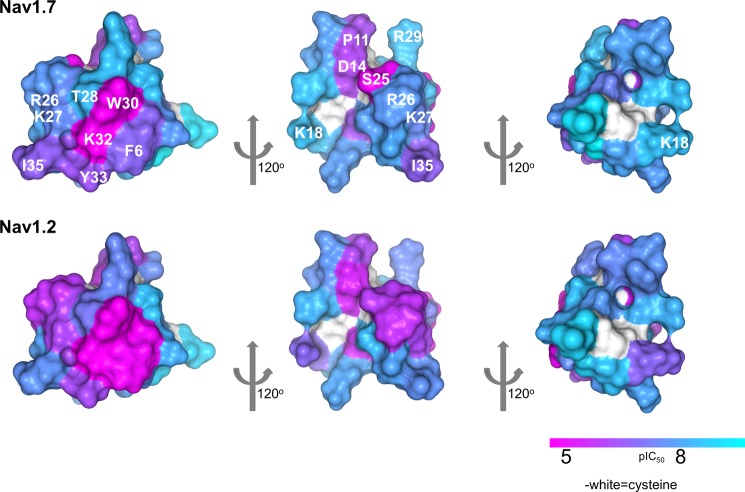
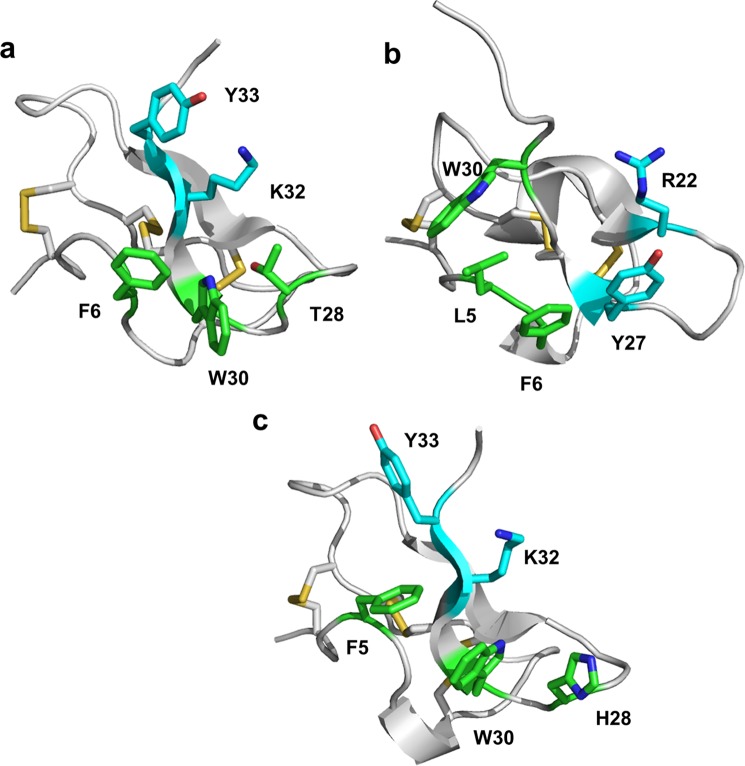
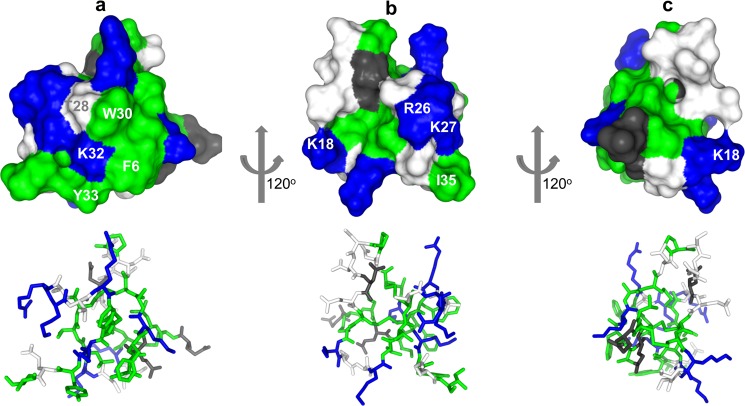
The complete loss of activity and binding to sodium channels by the W30A and K32A variants led us to question whether this was the result of a loss of binding energy or simply a disruption of the peptide folding. The NMR structure determination of HwTX-IV and of the W30A and K32A mutants, however, allowed us to eliminate misfolding as a possibility, suggesting that these and probably other residues make specific interactions with the sodium channels. Nav1.7 and Nav1.2 pIC50 values were mapped onto the surface of HwTX-IV as a color gradient from cyan (>8) to magenta (<5). Fig. 7 illustrates three different views of the toxin by successive 120º rotations and highlights the close proximity of Phe6, Trp30, Lys32, and Tyr33 and the deleterious effect on affinity toward both Nav1.2 and Nav1.7 when these residues are mutated to alanine (colors other than cyan indicate significant decrease in activity). These residues, along with Thr28, belong to β-strand 2, loop 1, and loop 4, creating a polar-aryl face. This aspect of the toxin we refer to as the affinity interface, which may represent a pharmacophore for VGSC interaction (Fig. 9). The affinity interface is characterized by an arrangement of hydrophobic and basic residues (often described as a hydrophobic patch, or protrusion) surrounded by a basic ring of residues (Fig. 8) that probably forms the bulk of toxin interactions with the voltage sensor of DII and is reminiscent of similar surfaces in other toxins that are known to directly interact with voltage-gated ion channels (38–40). Lys32 is geometrically poised for a cation-π interaction between the protonated side chain and the aromatic π electrons of Tyr33. Interestingly, this functional dyad, or pairing of a hydrophobic and a basic residue in close (6–7 Å) spatial proximity, has been described previously as being important for the activity of scorpion-derived K+ channel pore-blocking peptides, suggesting that peptides with disparate mechanisms of action across different channel types utilize a common structural binding motif (41). The central residue of the hydrophobic component of the affinity interface, Trp30, is sandwiched between and tightly coupled to two neighbors, Thr-28 and Phe6 (Figs. 5f and 9). The close coupling between the side chains of this triad manifests itself in the NMR spectrum as a large chemical shift anisotropy from the ring current exerted by the aromatic indole. The α and β carbon hydrogens are most affected, with their chemical shifts often shifted ∼1 ppm upfield, indicative of close spatial proximity. π-Stacking between Trp30 and Phe6 aromatic rings is also apparent in supplemental Video 1, where the movement of the two residues can be seen to be tightly correlated. Similar ring current-derived shielding is apparent in the related family 1 toxins hainantoxin-IV and phrixotoxin-I (20, 42). A similar “aromatic sandwich” of residues critical for affinity, proximal to a π-cation functional dyad pairing, is observed in the Kv2.1 gating modifier, SGTx, which suggests that these two features may represent a common motif for peptide-mediated inhibition of voltage-gated ion channel voltage sensors in general (Fig. 9) (40). It is well known that cation-π interactions on the surface of proteins have a stabilizing effect on structure; however, we conclude based on our NMR data that mutations in Trp30 and Lys32 do not experience any significant structural destabilization. It is also well established that desolvation penalties can destabilize electrostatic interactions at protein-protein interaction interfaces (43, 44) and that the desolvation penalty of a cation-π interaction is significantly less than that of a normal electrostatic interaction (45). Given the importance of Lys32 as an anchoring interaction for huwentoxin-IV binding and activity, it is tempting to speculate that the role of the cation-π interaction is to mitigate the energetic cost that would otherwise be associated with desolvation of Lys32 and thus strengthen the overall equilibrium constant more in favor of binding (anchored by Lys32–Glu811 according to our model). Further, the proximity of the aromatic sandwich to the functional dyad (Fig. 9) seems to suggest that a redundancy in local π system availability intramolecularly may be a common mechanism employed by peptide toxins to cloak a residue that will be involved in an anchoring charge-charge interaction in the complex, keeping it largely free from solvation prior to voltage sensor binding. The rigidity of the aromatic sandwich and the resulting imperfect geometry of the cation-π interaction both there and within the functional dyad may be indicative that multiple, weak cation-π interactions are available to Lys32, sufficient to preclude solvation but not so strong as to thermodynamically preclude formation of the proper charge pairing upon complex formation.
FIGURE 7.
Alanine perturbation maps. A color gradient is applied to the molecular surface of HwTX-IV, highlighting alanine mutant differences (pIC50 values) from HwTX-IV for Nav1.7 (top) and Nav1.2 (bottom). For clarity, residues are labeled in the top panel only.
FIGURE 9.
The functional dyad (cyan) and the aromatic sandwich (green) combine to form the affinity interface in three different inhibitory cysteine knot peptides that act as gating modifiers of distinct types of voltage-gated ion channels. a, huwentoxin-IV; b, SGTx (PDB code 1LA4), an inhibitor of the voltage-gated potassium channel, Kv2.1 (40); c, hainantoxin-IV (PDB code 1NIY) (20). Lys32 in huwentoxin-IV and hainantoxin-IV and Arg22 in SGTx are known to be important for overall activity of the respective peptides (21, 40). The redundancy of π systems available for cation-π interactions may help to mitigate the energetic desolvation penalty that would otherwise be incurred upon complex formation between the peptide and voltage sensor.
FIGURE 8.
Surface and stick representations of HwTX-IV colored by residue type: hydrophobic (green), basic (blue), neutral polar (white), and acidic (gray). a, the affinity interface characterized by hydrophobic and basic residues. b, the selectivity interface, characterized by a mosaic distribution of residue properties. c, a surface relatively tolerant (outside of Lys18) to alanine mutation, perhaps indicating a surface not directly involved in binding to the channel.
Fig. 8b reveals residues lending marked differences in affinity toward the channel subtypes. This face of the molecule illustrates the contrast between Nav1.2 and Nav1.7 when mutating three basic residues, Lys18, Arg26, and Lys27. HwTX-IV affinity for Nav1.7 is much less sensitive to mutations at these positions than Nav1.2. It is our assumption that these residues interact with corresponding regions on the respective channels that contain lower sequence homology. This aspect of the toxin we refer to as the selectivity interface. In contrast to the affinity interface, the selectivity interface is a mosaic of many different physicochemical properties. All four general property types are represented: hydrophobic, basic, acidic, and neutral polar. Although all property types exist in the interface, it is primarily basic residues located on loops 3 and 4 that confer selectivity between Nav1.2 and Nav1.7. Basicity at the Lys18, Arg26, and Lys27 positions appears to be a requirement for affinity toward Nav1.2. It is unclear where on the channel the selectivity interface interacts, but acidic residues located on the DII S3-S4 linker of Nav1.7 along with residues in the pore domain of DIII may be implicated. Future studies will include testing these HwTX-IV mutants on additional Nav subtypes with the intent of producing “global” sodium channel selectivity profiles and selectivity interface maps.
Fig. 8c illustrates a surface relatively tolerant (outside of Lys18) to alanine mutation, perhaps indicating a surface not directly involved in binding to the channel (i.e. a solvent- or membrane-exposed surface) made up of residues from the N-terminal, loop 2, and loop 3 regions.
Toxin Dynamics
ICK folds are inherently stable and exhibit little flexibility. NMR-derived relaxation and order parameters have shown related venom peptides to be very rigid over a picosecond to nanosecond time frame (38). Underlying this stability is the high ratio of cysteine to non-cysteine residues. It is common for venom peptides to contain six or more cysteines, resulting in a high number of disulfide bonds over an average peptide length of 35 residues. These folds impart not only conformational stability but metabolic and thermal stability as well. It was our intent to examine, via MD simulations, what role peptide flexibility has on affinity with and selectivity between Nav1.2 and Nav1.7. Simulations on a variety of HwTX-IV mutants over a 50-ns trajectory revealed only very subtle differences in core structure when compared with HwTX-IV. Perhaps not surprisingly, greater variation, as measured by α-carbon RMSDs, was shown to exist in the loop regions. Although the MD simulations revealed modest increases in loop 2, 3, or 4 flexibility in specific mutants, little correlation exists between loss of function and changes in loop flexibility. We do not rule out the potential importance peptide dynamics and conformational flexibility/rigidity have for binding affinity; however, our findings suggest that side chain-specific interactions, between toxin and channel, play a greater role.
HwTX-IV-Domain II Voltage Sensor Interactions
Docking of HwTX-IV with the domain II voltage sensor predicts that HwTX-IV binds in the cleft formed by the DII S1-S2 and S3-S4 loops, where Trp30 may make a hydrophobic interaction with Met750 in Nav1.7 (Met776 in Nav1.2) and a non-bonded van der Waals contact with His754 (Fig. 6b). In addition, our docking model suggests a likely interaction of basic Lys32 with acidic Glu811 in Nav1.7 (Glu837 in Nav1.2), an attractive model given the highly disruptive nature of the K32A and E811Q mutations individually compared with other peptide and channel mutations (19). Interestingly, according to our model, P11A, D14A, L22A, and K18A are located on loops 2 and 3, facing away from the groove formed by the Nav1.7 S1-S2 and S3-S4 loops. By analogy to the recently described binding site for β-scorpion toxins, it is possible that these residues may make important contacts with the pore module of the adjacent DIII (46). More subtle and primarily subtype-specific reductions in activity were observed with K18A, R26A, K27A, and R29A. Arg26, Lys27, and Arg29 are located on loop 4 of HwTX-IV, and our model predicts that some of these amino acids may interact with acidic residues, such as Asp816 and Glu818, located on the DII S3-S4 linker of Nav1.7, that have previously been identified as important for HwTX-IV activity (19). Two of these basic residues (Lys27 and Arg29) have also been shown to be important for block of VGSCs in rat DRG neurons by the closely related spider toxin, hainantoxin IV (μ-Theraphotoxin-Hhn1b) (20). These observations may indicate that hainantoxin-IV and HwTX-IV interact with VGSCs in a similar manner. In contrast to all of the mutations described that result in loss of sodium channel blocking activity, mutation of Glu4 to alanine results in a small but consistent increase in activity. If the modeled interaction with Lys762 is correct, this behavior of E4A mutants could be due to loss of a competing electrostatic repulsion with nearby Glu759 and Glu760. Alternatively, the proposed interaction between Glu4 and Lys762 may represent a thermodynamic local minimum, the loss of which allows the peptide to find a still more favorable binding mode. In our model, the predominant interactions are contributed by the S3-S4 loop with supporting interactions from the S1-S2 loop. This is similar to the model proposed for β-scorpion toxin interaction, CssIV, where the S3-S4 loop provides the interaction hot spot for binding with supporting interactions coming through the S1-S2 loop (47). In that case, the toxin locks the channel in an activated state. This may suggest that the key to locking helix 4 into a given position is primarily dependent on interactions with the S3-S4 loop of domain II. Overall, our results indicate that HwTX-IV makes numerous interactions with Nav1.7 channel residues in the S1-S2 and S3-S4 linkers, hydrophobic residues at the top of S1 in domain II, and, potentially, in yet to be defined residues in the adjacent pore loop of domain III (46).
The mechanism of action of HwTX-IV is thought to involve binding to the voltage sensor of domain II and trapping of the voltage sensor in the inward, closed conformation (5, 8). In order to determine if different interactions are responsible for binding and voltage sensor trapping, we compared the activity of the panel of alanine mutants in the functional QPatch assay (where activity is a function of affinity and efficacy) with activity in a novel radioligand binding assay (measure of binding affinity only). Our results show that binding and functional activity were generally well correlated. There were no examples where binding was retained but efficacy was lost, suggesting that binding and voltage sensor trapping may represent a single event. Most alanine mutants were slightly less potent in the binding assay than in the functional assay. The reasons for this are not entirely clear, but this may indicate that binding is somewhat voltage-dependent, as suggested by the unbinding of HwTX-IV following strong depolarization (8). Interestingly, the magnitude of the difference between functional activity and binding affinity varied between mutants, possibly suggesting that different mutants exhibit varying degrees of voltage dependence.
VGSCs have been implicated in a variety of disorders, including arrhythmia, epilepsy, and pain (10, 12, 13). Nav1.7, in particular, has attracted much attention recently because it appears to play a critical role in human pain sensation. Gain-of-function mutations in Nav1.7 have been linked to the episodic painful conditions primary erythermalgia (48) and paroxysmal extreme pain disorder (49), whereas loss-of-function mutations of Nav1.7 in humans cause congenital indifference to pain, a rare autosomal recessive disorder characterized by a complete indifference or insensitivity to painful stimuli (50–52). Nav1.2 is also of medical importance, with mutations in Nav1.2 being linked to epilepsy and autism (53, 54). Our results provide the most comprehensive evaluation of the SAR for a family 1 VGSC-blocking toxin described to date, providing unique insight into the residues required for both Nav1.7 and Nav1.2 affinity and selectivity. These experimental findings, coupled with the predictions of our model, provide a structural framework for the interaction between HwTX-IV and VGSCs and may provide a basis for rational design of toxin-based VGSC-blocking peptides with improved potency and/or selectivity for the treatment of human diseases.

This article contains supplemental Tables S1 and S2 and Video 1.
The atomic coordinates and structure factors (codes 2M4X, 2M4Z, and 2M50) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- VGSC
- voltage-gated sodium channels
- ICK
- inhibitory cysteine knot
- SAR
- structure-activity relationship(s)
- HwTX-IV
- huwentoxin-4
- HSQC
- heteronuclear single quantum correlation
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation
- CA
- α-carbon.
REFERENCES
- 1. King G. F. (2004) The wonderful world of spiders. Preface to the special Toxicon issue on spider venoms. Toxicon 43, 471–475 [DOI] [PubMed] [Google Scholar]
- 2. Escoubas P. (2006) Molecular diversification in spider venoms. A web of combinatorial peptide libraries. Mol. Divers. 10, 545–554 [DOI] [PubMed] [Google Scholar]
- 3. Herzig V., Wood D. L., Newell F., Chaumeil P. A., Kaas Q., Binford G. J., Nicholson G. M., Gorse D., King G. F. (2011) ArachnoServer 2.0, an updated online resource for spider toxin sequences and structures. Nucleic Acids Res. 39, D653–D657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood D. L., Miljenović T., Cai S., Raven R. J., Kaas Q., Escoubas P., Herzig V., Wilson D., King G. F. (2009) ArachnoServer: a database of protein toxins from spiders. BMC Genomics 10, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosmans F., Rash L., Zhu S., Diochot S., Lazdunski M., Escoubas P., Tytgat J. (2006) Four novel tarantula toxins as selective modulators of voltage-gated sodium channel subtypes. Mol. Pharmacol. 69, 419–429 [DOI] [PubMed] [Google Scholar]
- 6. King G. F., Escoubas P., Nicholson G. M. (2008) Peptide toxins that selectively target insect Na(V) and Ca(V) channels. Channels 2, 100–116 [DOI] [PubMed] [Google Scholar]
- 7. Middleton R. E., Warren V. A., Kraus R. L., Hwang J. C., Liu C. J., Dai G., Brochu R. M., Kohler M. G., Gao Y. D., Garsky V. M., Bogusky M. J., Mehl J. T., Cohen C. J., Smith M. M. (2002) Two tarantula peptides inhibit activation of multiple sodium channels. Biochemistry 41, 14734–14747 [DOI] [PubMed] [Google Scholar]
- 8. Xiao Y., Bingham J. P., Zhu W., Moczydlowski E., Liang S., Cummins T. R. (2008) Tarantula huwentoxin-IV inhibits neuronal sodium channels by binding to receptor site 4 and trapping the domain ii voltage sensor in the closed configuration. J. Biol. Chem. 283, 27300–27313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xiao Y., Liang S. (2003) Inhibition of neuronal tetrodotoxin-sensitive Na+ channels by two spider toxins. Hainantoxin-III and hainantoxin-IV. Eur. J. Pharmacol. 477, 1–7 [DOI] [PubMed] [Google Scholar]
- 10. Yogeeswari P., Ragavendran J. V., Thirumurugan R., Saxena A., Sriram D. (2004) Ion channels as important targets for antiepileptic drug design. Curr. Drug Targets 5, 589–602 [DOI] [PubMed] [Google Scholar]
- 11. Noble D. (2002) Unraveling the genetics and mechanisms of cardiac arrhythmia. Proc. Natl. Acad. Sci. U.S.A. 99, 5755–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cannon S. C. (2000) Spectrum of sodium channel disturbances in the nondystrophic myotonias and periodic paralyses. Kidney Int. 57, 772–779 [DOI] [PubMed] [Google Scholar]
- 13. Wood J. N., Boorman J. P., Okuse K., Baker M. D. (2004) Voltage-gated sodium channels and pain pathways. J. Neurobiol. 61, 55–71 [DOI] [PubMed] [Google Scholar]
- 14. Saez N. J., Senff S., Jensen J. E., Er S. Y., Herzig V., Rash L. D., King G. F. (2010) Spider-venom peptides as therapeutics. Toxins 2, 2851–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Klint J. K., Senff S., Rupasinghe D. B., Er S. Y., Herzig V., Nicholson G. M., King G. F. (2012) Spider-venom peptides that target voltage-gated sodium channels. Pharmacological tools and potential therapeutic leads. Toxicon 60, 478–491 [DOI] [PubMed] [Google Scholar]
- 16. Liang S. P., Zhang D. Y., Pan X., Chen Q., Zhou P. A. (1993) Properties and amino acid sequence of huwentoxin-I, a neurotoxin purified from the venom of the Chinese bird spider Selenocosmia huwena. Toxicon 31, 969–978 [DOI] [PubMed] [Google Scholar]
- 17. Xiao Y., Luo X., Kuang F., Deng M., Wang M., Zeng X., Liang S. (2008) Synthesis and characterization of huwentoxin-IV, a neurotoxin inhibiting central neuronal sodium channels. Toxicon 51, 230–239 [DOI] [PubMed] [Google Scholar]
- 18. Xiao Y., Blumenthal K., Jackson J. O., 2nd, Liang S., Cummins T. R. (2010) The tarantula toxins ProTx-II and huwentoxin-IV differentially interact with human Nav1.7 voltage sensors to inhibit channel activation and inactivation. Mol. Pharmacol. 78, 1124–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao Y., Jackson J. O., 2nd, Liang S., Cummins T. R. (2011) Common molecular determinants of tarantula huwentoxin-IV inhibition of Na+ channel voltage sensors in domains II and IV. J. Biol. Chem. 286, 27301–27310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li D., Xiao Y., Xu X., Xiong X., Lu S., Liu Z., Zhu Q., Wang M., Gu X., Liang S. (2004) Structure-activity relationships of hainantoxin-IV and structure determination of active and inactive sodium channel blockers. J. Biol. Chem. 279, 37734–37740 [DOI] [PubMed] [Google Scholar]
- 21. Liu Y., Li D., Wu Z., Li J., Nie D., Xiang Y., Liu Z. (2012) A positively charged surface patch is important for hainantoxin-IV binding to voltage-gated sodium channels. J. Pept. Sci. 18, 643–649 [DOI] [PubMed] [Google Scholar]
- 22. Liu Y., Beck E. J., Flores C. M. (2011) Validation of a patch clamp screening protocol that simultaneously measures compound activity in multiple states of the voltage-gated sodium channel Nav1.2. Assay Drug Dev. Technol. 9, 628–634 [DOI] [PubMed] [Google Scholar]
- 23. Roland B. L., Sutton S. W., Wilson S. J., Luo L., Pyati J., Huvar R., Erlander M. G., Lovenberg T. W. (1999) Anatomical distribution of prolactin-releasing peptide and its receptor suggests additional functions in the central nervous system and periphery. Endocrinology 140, 5736–5745 [DOI] [PubMed] [Google Scholar]
- 24. Bax A., Davis D. G. (1985) MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 65, 355–360 [Google Scholar]
- 25. Jeener J., Meier B. H., Bachmann P., Ernst R. R. (1979) Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 71, 4546–4553 [Google Scholar]
- 26. Cavanagh J., Fairbrother W., Palmer A. G., III, Skleton N. J. (2007) Protein NMR Spectroscopy: Principles and Practice, 2nd Ed., pp. 535–673, Academic Press, Inc., New York [Google Scholar]
- 27. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 29. Berjanskii M. V., Neal S., Wishart D. S. (2006) PREDITOR. A web server for predicting protein torsion angle restraints. Nucleic Acids Res. 34, W63–W69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheung M. S., Maguire M. L., Stevens T. J., Broadhurst R. W. (2010) DANGLE. A Bayesian inferential method for predicting protein backbone dihedral angles and secondary structure. J. Magn. Reson. 202, 223–233 [DOI] [PubMed] [Google Scholar]
- 31. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mackerell A. D., Jr., Feig M., Brooks C. L., 3rd. (2004) Extending the treatment of backbone energetics in protein force fields. Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 [DOI] [PubMed] [Google Scholar]
- 33. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 34. Payandeh J., Scheuer T., Zheng N., Catterall W. A. (2011) The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khalili-Araghi F., Jogini V., Yarov-Yarovoy V., Tajkhorshid E., Roux B., Schulten K. (2010) Calculation of the gating charge for the Kv1.2 voltage-activated potassium channel. Biophys. J. 98, 2189–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spassov V. Z., Yan L., Szalma S. (2002) Introducing an implicit membrane in generalized born/solvent accessibility continuum solvent models. J. Phys. Chem. B 106, 8726–8738 [Google Scholar]
- 37. Peng K., Shu Q., Liu Z., Liang S. (2002) Function and solution structure of huwentoxin-IV, a potent neuronal tetrodotoxin (TTX)-sensitive sodium channel antagonist from Chinese bird spider Selenocosmia huwena. J. Biol. Chem. 277, 47564–47571 [DOI] [PubMed] [Google Scholar]
- 38. Lee S., Milescu M., Jung H. H., Lee J. Y., Bae C. H., Lee C. W., Kim H. H., Swartz K. J., Kim J. I. (2010) Solution structure of GxTX-1E, a high-affinity tarantula toxin interacting with voltage sensors in Kv2.1 potassium channels. Biochemistry 49, 5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Takahashi H., Kim J. I., Min H. J., Sato K., Swartz K. J., Shimada I. (2000) Solution structure of hanatoxin1, a gating modifier of voltage-dependent K+ channels. Common surface features of gating modifier toxins. J. Mol. Biol. 297, 771–780 [DOI] [PubMed] [Google Scholar]
- 40. Wang J. M., Roh S. H., Kim S., Lee C. W., Kim J. I., Swartz K. J. (2004) Molecular surface of tarantula toxins interacting with voltage sensors in Kv channels. J. Gen. Physiol. 123, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mouhat S., De Waard M., Sabatier J. M. (2005) Contribution of the functional dyad of animal toxins acting on voltage-gated Kv1-type channels. J. Pept. Sci. 11, 65–68 [DOI] [PubMed] [Google Scholar]
- 42. Chagot B., Escoubas P., Villegas E., Bernard C., Ferrat G., Corzo G., Lazdunski M., Darbon H. (2004) Solution structure of phrixotoxin 1, a specific peptide inhibitor of Kv4 potassium channels from the venom of the theraphosid spider Phrixotrichus auratus. Protein Sci. 13, 1197–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fiorucci S., Zacharias M. (2010) Prediction of protein-protein interaction sites using electrostatic desolvation profiles. Biophys. J. 98, 1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burgoyne N. J., Jackson R. M. (2006) Predicting protein interaction sites. Binding hot-spots in protein-protein and protein-ligand interfaces. Bioinformatics 22, 1335–1342 [DOI] [PubMed] [Google Scholar]
- 45. Gallivan J. P., Dougherty D. A. (2000) A computational study of cation-π interactions vs. salt bridges in aqueous media. Implications for protein engineering. J. Am. Chem. Soc. 122, 870–874 [Google Scholar]
- 46. Zhang J. Z., Yarov-Yarovoy V., Scheuer T., Karbat I., Cohen L., Gordon D., Gurevitz M., Catterall W. A. (2012) Mapping the interaction site for a β-scorpion toxin in the pore module of domain III of voltage-gated Na+ channels. J. Biol. Chem. 287, 30719–30728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang J. Z., Yarov-Yarovoy V., Scheuer T., Karbat I., Cohen L., Gordon D., Gurevitz M., Catterall W. A. (2011) Structure-function map of the receptor site for β-scorpion toxins in domain II of voltage-gated sodium channels. J. Biol. Chem. 286, 33641–33651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Y., Wang Y., Li S., Xu Z., Li H., Ma L., Fan J., Bu D., Liu B., Fan Z., Wu G., Jin J., Ding B., Zhu X., Shen Y. (2004) Mutations in SCN9A, encoding a sodium channel α subunit, in patients with primary erythermalgia. J. Med. Genet. 41, 171–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fertleman C. R., Baker M. D., Parker K. A., Moffatt S., Elmslie F. V., Abrahamsen B., Ostman J., Klugbauer N., Wood J. N., Gardiner R. M., Rees M. (2006) SCN9A mutations in paroxysmal extreme pain disorder. Allelic variants underlie distinct channel defects and phenotypes. Neuron 52, 767–774 [DOI] [PubMed] [Google Scholar]
- 50. Cox J. J., Reimann F., Nicholas A. K., Thornton G., Roberts E., Springell K., Karbani G., Jafri H., Mannan J., Raashid Y., Al-Gazali L., Hamamy H., Valente E. M., Gorman S., Williams R., McHale D. P., Wood J. N., Gribble F. M., Woods C. G. (2006) An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Goldberg Y. P., MacFarlane J., MacDonald M. L., Thompson J., Dube M. P., Mattice M., Fraser R., Young C., Hossain S., Pape T., Payne B., Radomski C., Donaldson G., Ives E., Cox J., Younghusband H. B., Green R., Duff A., Boltshauser E., Grinspan G. A., Dimon J. H., Sibley B. G., Andria G., Toscano E., Kerdraon J., Bowsher D., Pimstone S. N., Samuels M. E., Sherrington R., Hayden M. R. (2007) Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin. Genet. 71, 311–319 [DOI] [PubMed] [Google Scholar]
- 52. Ahmad S., Dahllund L., Eriksson A. B., Hellgren D., Karlsson U., Lund P. E., Meijer I. A., Meury L., Mills T., Moody A., Morinville A., Morten J., O'donnell D., Raynoschek C., Salter H., Rouleau G. A., Krupp J. J. (2007) A stop codon mutation in SCN9A causes lack of pain sensation. Hum. Mol. Genet. 16, 2114–2121 [DOI] [PubMed] [Google Scholar]
- 53. Meisler M. H., O'Brien J. E., Sharkey L. M. (2010) Sodium channel gene family. Epilepsy mutations, gene interactions and modifier effects. J. Physiol. 588, 1841–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sanders S. J., Murtha M. T., Gupta A. R., Murdoch J. D., Raubeson M. J., Willsey A. J., Ercan-Sencicek A. G., DiLullo N. M., Parikshak N. N., Stein J. L., Walker M. F., Ober G. T., Teran N. A., Song Y., El-Fishawy P., Murtha R. C., Choi M., Overton J. D., Bjornson R. D., Carriero N. J., Meyer K. A., Bilguvar K., Mane S. M., Sestan N., Lifton R. P., Günel M., Roeder K., Geschwind D. H., Devlin B., State M. W. (2012) De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]