Background: The mechanism for selectively targeting membrane proteins to the flagellum of kinetoplastid parasites is unknown.
Results: We have identified a novel protein, KHARON1, which is important for the flagellar targeting of a glucose transporter.
Conclusion: KHARON1 is the first protein identified in Kinetoplastida that targets a membrane protein to the flagellum.
Significance: KHARON1 may be part of a new flagellar targeting pathway.
Keywords: Glucose Transport, Leishmania, Mass Spectrometry (MS), Pathogenesis, Protein Targeting, Flagellar Membrane Protein Targeting, TAP Tagging, Amastigote Viability, Glucose Transporter
Abstract
The LmxGT1 glucose transporter is selectively targeted to the flagellum of the kinetoplastid parasite Leishmania mexicana, but the mechanism for targeting this and other flagella-specific membrane proteins among the Kinetoplastida is unknown. To address the mechanism of flagellar targeting, we employed in vivo cross-linking, tandem affinity purification, and mass spectrometry to identify a novel protein, KHARON1 (KH1), which is important for the flagellar trafficking of LmxGT1. Kh1 null mutant parasites are strongly impaired in flagellar targeting of LmxGT1, and trafficking of the permease was arrested in the flagellar pocket. Immunolocalization revealed that KH1 is located at the base of the flagellum, within the flagellar pocket, where it associates with the proximal segment of the flagellar axoneme. We propose that KH1 mediates transit of LmxGT1 from the flagellar pocket into the flagellar membrane via interaction with the proximal portion of the flagellar axoneme. KH1 represents the first component involved in flagellar trafficking of integral membrane proteins among parasitic protozoa. Of considerable interest, Kh1 null mutants are strongly compromised for growth as amastigotes within host macrophages. Thus, KH1 is also important for the disease causing stage of the parasite life cycle.
Introduction
Eukaryotic cilia and flagella, which are similar in structure and are highly conserved from protozoa to humans, play central roles as sensory organelles that transmit information about the extracellular environment to the cell interior (1–4). Specialized membrane proteins are targeted to the cilia and flagella where they function in sensing and signal transduction (5–8). Because cilia and flagella serve as a platform for sensing and signaling, there is great interest in elucidating the mechanisms that target integral membrane proteins to these organelles.
Kinetoplastid parasites such as Leishmania and Trypanosoma species, which cause devastating diseases that afflict an estimated 60 million people worldwide (9), are flagellated protozoa that constitute attractive model systems for studying flagellar targeting mechanisms. Analysis of individual membrane proteins (reviewed in Ref. 6) and of the trypanosome flagellar membrane proteome (10) have identified flagellar membrane proteins likely to be involved in signal transduction, including potential kinases, adenylate cyclases, and Ca2+ channels. Indeed, the Trypanosoma cruzi Flagellar Ca2+ Binding Protein (TcFCaBP) is localized specifically to the parasite flagellar membrane and has been suggested to have a role in Ca2+-dependent signal transduction (11–13). As demonstrated some years ago, the adenylate cyclase ESAG-4 is also targeted to the flagellar membrane in Trypanosoma brucei (14). Additionally, the aquaglyceroporin channel of Leishmania major (LmjAQP1) is specifically targeted to the flagellar membrane where it is involved in detection of extracellular osmotic gradients and osmotaxis (15). Furthermore, the L. mexicana glucose transporter LmxGT1 is also selectively localized to the flagellar membrane where it may act as a glucose sensor (16, 17).
The cis-acting flagellar targeting signal of LmxGT1 has been described previously (17). Three critical residues within the N-terminal hydrophilic domain, Asn95-Pro96-Met97, are required for efficient flagellar targeting of this permease. However, comparative analysis of the LmxGT1 targeting motif with other known ciliary targeting signals (18, 19) has not identified a similar motif among other eukaryotes, supporting the possibility that novel flagellar targeting machinery may exist in these ancient protozoa. Thus, identification of proteins that interact with the flagellar targeting domain of LmxGT1 and are critical for flagellar trafficking may reveal new flagellar targeting components.
Here we report the identification of a protein, designated KHARON1 (KH1),4 which recognizes the flagellar targeting domain of LmxGT1 and is required for efficient targeting of this permease to the flagellar membrane. A combination of immunolocalization, subcellular fractionation, and ultrastructural studies of tagged KH1 revealed that it is associated with the proximal region of the flagellar axoneme located adjacent to the flagellar pocket of Leishmania promastigotes. Remarkably, KH1 also appears to be important for the viability of the disease causing amastigote stage of the parasite within phagolysosomal vesicles of mammalian host macrophages, raising the possibility that KH1 may be critical for Leishmania pathogenesis. Furthermore, these studies underscore the likely role of the “residual” flagellum of the intracellular amastigote in critical interactions with the host macrophage. KH1 is the first protein identified in kinetoplastid protozoa that functions to selectively target an integral membrane protein to the flagellar compartment. Further characterization of KH1 function may help elucidate novel aspects of flagellar targeting pathways and pathogenesis in kinetoplastid parasites.
EXPERIMENTAL PROCEDURES
Parasite Cultures and Transfections
Wild type Leishmania mexicana promastigotes were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Scientific Hyclone, Logan, UT), 0.1 mm xanthine, and 5 μg/ml of hemin. Parasite lines carrying episomal expression vectors were cultured in the same medium with 100 μg/ml of G418 (Invitrogen), 80 μg/ml of hygromycin B (InvivoGen, San Diego, CA). Δlmxkh1 (Δkh1) null mutants were maintained in RPMI and RPMI supplemented with 50 μg/ml of puromycin and 50 μg/ml of phleomycin (InvivoGen). All cultures were maintained at 26 °C. Leishmania promastigotes were transfected according to previously described electroporation techniques using a Bio-Rad Gene Pulser Xcell (16, 20).
Creation of the Tandem Affinity Tagged LmxGT1 Fusion Proteins
The His6-biotinylation motif-His6 (HBH) affinity tag was amplified from a previously described source (21) using forward primer: 5′-CTAGATCTAGCGGCAGCGGCAGCGGCCATCATCACCACCATCATGCTGGAAAGGC-3′ to include a 3xSG linker (underlined) anterior to the tag and reverse primer: 5′-CGTAGATCTTCAGTGGTGATGATGGTGGTGAACGCCGATCTTGATTAGACC-3′. The tag was cloned into the BglII site of the Leishmania pX63NEORI expression vector (22). Subsequently, the open reading frame of LmxGT1 was amplified and cloned into the BamHI and EcoRI sites to generate the LmxGT1::HBH gene fusion. The LmxGT1(Δ84–100)::HBH fusion protein was created in the same manner using template DNA from the previously generated (17) Δ84–100 deletion mutant. All primer sequences are available upon request. DNA constructs were sequenced at the OHSU sequencing core to verify for accuracy.
In Vivo Cross-linking and Cell Lysis
L. mexicana promastigotes were grown to a density of ∼5 × 106 cells/ml. Approximately 1 × 109 cells were used per sample for experiments employing whole cells, and twice as many were used for experiments using membrane preparations. Cells were collected, washed once with phosphate-buffered saline (PBS: 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4), and resuspended in PBS. For cross-linking with 1% formaldehyde (Ultra Pure EM grade, Polysciences, Warrington, PA), cells were incubated at 25 °C for 10 min followed by another 5 min after the addition of glycine to a final concentration of 125 mm (to stop the cross-linking reaction). PBS was added in place of formaldehyde for non-cross-linked control samples. Cells were washed once with PBS, then resuspended in 1 ml of Buffer 1 (8 m urea, 300 mm NaCl, 0.5% Nonidet P-40, 50 mm NaH2PO4, 50 mm Tris, pH 7.0) on ice. Samples were sonicated on ice at maximum amplitude for 3 × 15 s with 1 min between pulses. For samples where membrane preparations were required, cells were collected, washed, and resuspended in membrane preparation buffer (0.1 m KH2PO4, pH 7.4, 10% glycerol) containing Complete Mini protease inhibitors (Roche Applied Science). Samples were sonicated as described above and cleared by centrifugation at 16,000 × g for 20 min at 4 °C. The supernatants were transferred into Beckman Polyallomer tubes and centrifuged at 124,000 × g using a TLA-45 rotor for 2 h at 4 °C. Membrane pellets were resuspended in Buffer 1.
Tandem Affinity Purification
The cell lysates were cleared by centrifugation at 16,000 × g for 20 min at 4 °C. Cleared supernatants were incubated with 750 μl of HisPurTM Cobalt Resin (Thermo Scientific Pierce, Rockford, IL) on a rocker for 45 min at room temperature. The resins were washed with 5 × 1 ml of Buffer 1 (5 min each), 5 × 1 ml of Buffer 1 at pH 6.4 (5 min each), and 5 × 1 ml of Buffer 1 pH 6.4 with 10 mm imidazole (5 min each). Protein complexes were eluted with 1 ml of Buffer 2 (Elution buffer: 45 mm NaH2PO4, 8 m urea, 270 mm NaCl, 150 mm imidazole). The eluates were incubated with 400 μl of Pierce® High Capacity Streptavidin-agarose Resin (Thermo Scientific Pierce) on a rocker for 16 h at 4 °C. Streptavidin columns were washed with 5 × 1 ml of Buffer 3 (8 m urea, 200 mm NaCl, 100 mm Tris, pH 8.0) containing 0.2% SDS (5 min each), 5 × 1 ml of Buffer 3 with 2.0% SDS (5 min each), 5 × 1 ml of Buffer 3 with no SDS (5 min each), 2 × 1 ml of Buffer A (200 mm NaCl, 100 mm Tris, pH 7.0), and 2 × 1 ml of Tris, pH 8.0. Samples were stored in 500 μl of Tris, pH 8.0, at 4 °C.
Trypsin Digestion
The beads were suspended in 500 μl of 100 mm Tris buffer, pelleted by centrifugation at 5,000 × g for 5 min, resuspended in 100 μl of ammonium bicarbonate buffer, and 1 μg of trypsin (Sigma) was added. After incubation overnight at 37 °C with shaking, the sample containing the suspended beads, digested proteins, and an additional 100 μl wash of water were transferred to 0.45-μm spin filters (Millipore, Burlington, MA) and beads were removed by brief centrifugation. The filtrate was dried by vacuum centrifugation, dissolved in 40 μl of 5% formic acid, and peptides identified by LC-MS/MS analysis.
LC-MS/MS Analysis
After trypsin digestion, the proteins present in the cross-linked and non-cross-link control fractions were identified by MS/MS by the Oregon Health & Science University Proteomics Shared Resource. Tryptic peptides were injected into a trap cartridge in 0.1% formic acid, placed inline with a Zorbax SB-C18 column, and separated using a 2–30% acetonitrile gradient. Eluted peptides were analyzed using a LTQ Velos linear ion trap mass spectrometer (Thermo Scientific) and identified by comparing MS/MS spectra to theoretical fragmentation spectra of peptides generated from the L. mexicana protein database using Sequest software (version 27, Thermo Scientific). Subsequently, the relative levels of specific proteins in the cross-linked and non-cross-linked samples were estimated by “spectral counting,” the quantification of the relative number of spectral counts obtained for peptides in each sample. For comparison of the two samples, the spectral counts of the HBH tag present in both samples were employed for normalization. Putative LmxGT1 binding partners were identified as proteins with high peptide counts in the cross-linked sample and zero counts in the non-cross-linked samples when using wild type LmxGT1::HBH bait protein, and not identified in experiments where the LmxGT1(Δ84–100)::HBH mutant was used.
Generation of Kharon1 (Δkh1) Null Mutants
The LmxM36.5850 (Kharon1) open reading frame (ORF) was genetically removed by two rounds of homologous recombination. Sequences surrounding the Kharon1 ORF were cloned using the following primers containing SfiI sites (underlined) compatible with a previously described method to rapidly generate knock-out constructs (23): 5′-flanking: forward, 5′-GAGGCCACCTAGGCCCGTGTGGACAACTGCAATGGCGGTGAAC-3′; reverse, 5′-GAGGCCACGCAGGCCGCGGCGAAAGACTTCTGTGTGGTGATGCCAGA-3′; 3′-flanking: forward, 5′-GAGGCCTCTGTGGCCGAGGCAGCACCGCCCCTGTTAGCTGATG-3′; reverse, 5′-GAGGCCTGACTGGCCCGAATCGTCGTTGGTATGCGCAAAGACGACAG-3′. Mutant clones were isolated based on the ability of transformants to grow on agar plates containing puromycin (50 μg/ml) and phleomycin (50 μg/ml) and analyzed by Southern blots to verify the absence of the Kharon1 coding region. Two independent clones were analyzed and both showed similar LmxGT1::GFP targeting defects. A rescue construct containing the Kharon1 ORF was cloned into the KpnI and EcoRV sites of a modified pX72-Hyg vector (24). Primer sequences are available upon request. All constructs were verified by sequencing at the Oregon Health & Science University DNA Sequencing Core.
Generation of LmxGT1::HA3 and Flagellar Preparations
The LmxGT1 ORF was amplified with primers that contained a SmaI site for the forward primer and an XbaI site for the reverse primer. The amplified product was then subcloned into a modified pX63NEO-3HA expression vector (D. Rodriguez-Contreras), encoding an in-frame HA3 tag downstream of SmaI and XbaI restriction sites, to generate an ORF encoding LmxGT1::HA3. DNA constructs were sequenced at the Oregon Health & Science University sequencing core.
To isolate L. mexicana promastigote flagella, ∼4 × 109 cells were washed and resuspended in Buffer S1 (25 mm Tris-HCl, 0.2 mm EDTA, 5 mm MgCl2, 0.32 m sucrose, pH 7.4) with HaltTM Protease Inhibitor Mixture (Thermo Scientific Pierce). Cells were sonicated with 3 pulses lasting for 1 s at 25% amplitude with 1-min rest between pulses using a Sonic Dismembrator 505 (Fisher Scientific, Waltham, MA). Detachment of flagella was monitored using phase-contrast microscopy. Cell bodies were separated from flagella by centrifugation at 700 × g for 15 min. Flagella were pelleted at 6,800 × g for 20 min, resuspended in Buffer S2 (25 mm Tris-HCl, 0.2 mm EDTA, 5 mm MgCl2, protease inhibitor), and layered on top of a 0.8 m sucrose cushion (in Buffer S2). After centrifugation at 1,080 × g for 20 min, the top layer was removed and layered on top of a discontinuous sucrose gradient (1.65 and 1.85 m) in Buffer S2. The samples were centrifuged at 133,000 × g for 3 h at 4 °C. The flagellar fraction at the interface between the two sucrose layers was collected, pelleted at 143,000 × g for 45 min, and resuspended in Buffer S1. A small fraction of collected flagella was used for protein quantification using a Bio-Rad DCTM Protein Assay Kit (Bio-Rad). The equivalent of 5 μg of protein were loaded into each lane in the experiments shown in Fig. 3D.
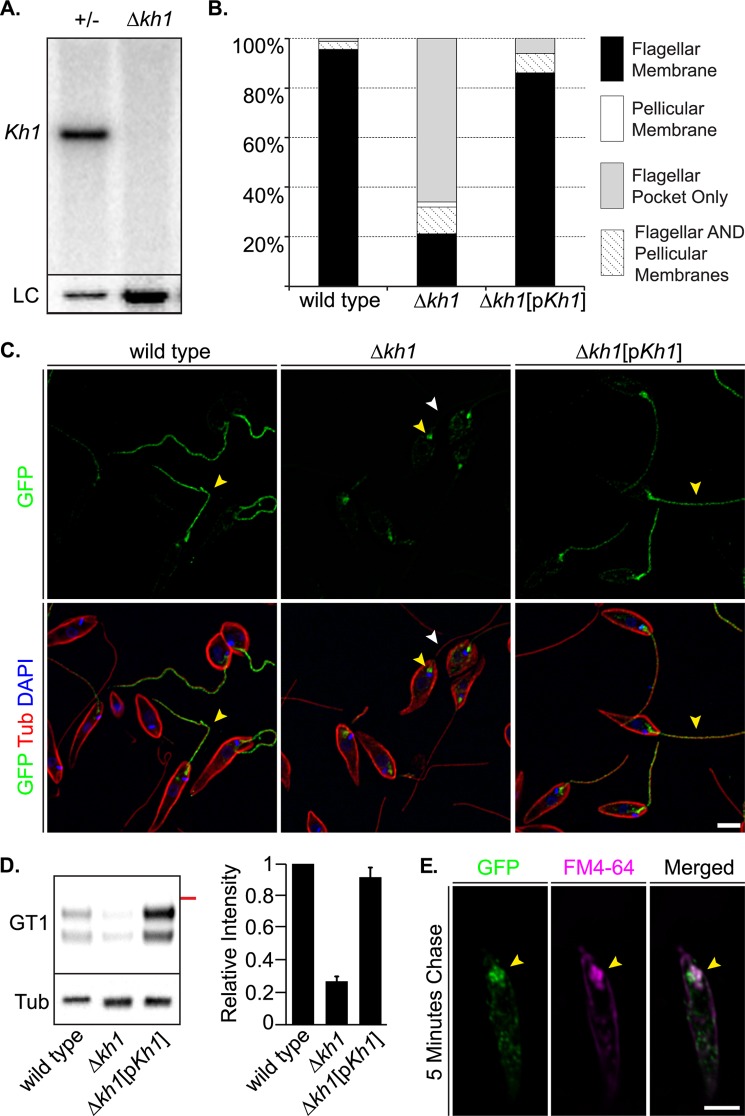
FIGURE 3.
Localization of LmxGT1::GFP in Δkh1 mutants. A, Southern blot of genomic DNA digested with BglII and EcoRI probed for the Kh1 ORF. Left lane is heterozygote (+/−) and right lane is the Δkh1 null mutant. The blot was stripped and re-probed for LmxGT2 as a loading control (LC). More DNA was loaded in the right lane. B, quantification of LmxGT1::GFP localization in wild type, Δkh1 mutant, and Kh1 add-back cell lines (Δkh1[pKh1]). C, immunofluorescence images showing LmxGT1::GFP localization in wild type, Δkh1 mutant, and Kh1 add-back cell lines. Yellow arrowheads show LmxGT1::GFP. White arrowheads indicate absence of GFP signal on the flagellum. Scale bar = 3 μm. D, Western blot (left) and relative quantification (right) of LmxGT1::HA3 in flagellar fractions. Wild type, Δkh1 mutant, and Δkh1[pKh1] flagella were analyzed by Western blot probed with an anti-HA antibody. The red line on the right of the blot represents a 70-kDa molecular mass marker. The blot was also probed with anti-tubulin antibody (Tub), and the intensity of the tubulin signal was used for normalization among lanes (right). Results are plotted as the mean ± S.D. of signal intensity from 2 replicate measurements. E, FM4-64 labeling of Δkh1 mutant cells expressing LmxGT1::GFP following a 5-min chase period. Markers as indicated in each panel. Yellow arrowheads indicate where LmxGT1::GFP and FM4-64 signals overlap. Scale bar = 3 μm.
Generation of HA3- and GFP-tagged Proteins
A 3xFLAG::3xHA (referred to as HA3 throughout the text for all HA-tagged proteins) tag was amplified from a previously described vector (25) and cloned into the SmaI (N terminus tag) or BglII (C terminus tag) site of the modified pX72-Hyg vector as described above. The KH1::HA3-tagged protein was generated by cloning the Kh1 ORF into the KpnI and EcoRV sites of the vector describe above. HA3::KH1 was made in a similar manner with the exception that the reverse primer contains a stop codon (TAG) and the vector for N terminus tag was used. All other 3xFLAG::3xHA-tagged proteins described in this article were tagged on the C terminus. The LmxMIT::GFP fusion protein was generated by cloning the LmxMIT ORF into the BamHI and EcoRV sites of pXG-′GFP+ (26). The LmjAQP1 ORF was amplified using previously described primers (27) with KpnI and EcoRV restriction sites and cloned into the pX72-Hyg vector described above. All primer sequences are available upon request.
Generation of Transgenic Parasites Expressing Tagged KH1 from the Endogenous Kh1 Gene Locus
To monitor the localization of KH1 expressed at near endogenous levels, the Kh1 ORF was tagged at its C terminus with HA3, the Thosea asigna virus 2A (TaV2A) peptide, Renilla luciferase (Luc), and the blasticidin resistance (BSD) protein, in that order, and then flanked by sequences immediately upstream and downstream of the Kh1 ORF in the L. mexicana genome. The linearized construct was transfected into wild type L. mexicana promastigotes, and transgenic parasites containing the integrated transgene replacing one Kh1 allele were selected on agar containing 80 μg/ml of blasticidin. Because the TaV2A peptide induces a co-translational intra-ribosomal cleavage during synthesis of the fusion protein (28), the KH1::HA3::TaV2A fusion protein (without the attached Luc::BSD fusion protein) is expressed in all transgenic parasites from one allele of the endogenous Kh1 gene locus encompassing the correct 5′ and 3′ flanking sequences. The KH1::HA3::TaV2A fusion protein was detected with anti-HA and anti-TaV2A antiserum described below. The development of this methodology will be described elsewhere.5
Molecular Markers and Immunodetection
For immunofluorescence, parasites were collected, washed once with PBS, and attached to poly-l-lysine-treated coverslips for 20 min. Cells were fixed and permeabilized with methanol at −20 °C for 8 min. Fixed cells were washed 3 × 5 min with PBS, then blocked with 5% normal goat serum for 30 min. Parasites were incubated with primary antibodies for 1 h at room temperature, washed 5 × 10 min with PBS, and then incubated with secondary antibodies for 1 h. After secondary antibody incubation, cells were washed as described above. Coverslips were mounted onto microscope slides using DAPI Gold Prolong reagent (Molecular Probes, Eugene, OR). Antibodies, dilutions, and sources are as follows: rabbit GFP, 1:1000 (Molecular Probes); rabbit HA, 1:1000 (Sigma); mouse α-tubulin, 1:2000 (Sigma); rabbit LmjAQP1, 1:200 (15); rabbit 2A (TaV2A), 1:500 (Millipore, Burlington, MA); anti-rabbit IgG Alexa-488 and anti-mouse IgG Alexa-594, 1:1000 (Molecular Probes).
For FM4-64 labeling, parasites were collected and washed once with cold PBS. Cells were incubated on ice with FM4-64FX (Molecular Probes) at 5 μg/ml for 2 min. After labeling, cells were pelleted in a microcentrifuge at 5,000 × g for 15 s and washed once with cold PBS. Cells were incubated at 26 °C in serum-free RPMI for a chase period of 5 min, washed once with cold PBS, and then fixed with 4% ultra-pure formaldehyde and attached to poly-l-lysine-treated coverslips for 30 min at room temperature. After fixation, parasites were washed 3 × 5 min with PBS, and incubated in 5% normal goat serum in PBS containing 0.01% saponin. Primary and secondary antibody incubations were also performed in PBS containing 0.01% saponin.
For immunoblotting, promastigotes at early-mid log density were collected and washed with PBS. Cells were lysed in 1× lithium dodecyl sulfate sample buffer (Invitrogen) containing 5 mm DTT and loaded onto 4–12% polyacrylamide gels for electrophoresis (Invitrogen). Proteins were then transferred onto nitrocellulose membranes, blocked with 3% BSA or 5% milk in PBS containing 0.1% Tween 20, and probed with primary antibodies in blocking solution. Primary antibodies were used as described above. Anti-mouse and anti-rabbit HRP-conjugated secondary antibodies were used at 1:20,000 (Thermo Scientific Pierce) with 5% milk in PBS containing 0.1% Tween 20.
Cellular Fractionations
For the experiment in Fig. 5F, cells were pelleted and washed in Buffer S (25 mm Tris-HCl, 0.2 mm EDTA, 5 mm MgCl2, 0.32 m sucrose, pH 7.4) with Halt Protease Inhibitor Mixture (Thermo Scientific Pierce). Cells were sonicated in Buffer S using 5 pulses of 5 s each at maximum amplitude, and checked for cell breakage by phase-contrast microscopy. Pellet and supernatant were separated by centrifugation at 15,000 × g for 20 min at 4 °C. Detergent extractions were performed using PBS buffer containing 1% Triton X-100, 25 mm HEPES, pH 7.5, 1 mm EDTA, and Halt Protease Inhibitor Mixture for 30 min at 25 °C. Insoluble and soluble fractions were separated by centrifugation at 15,000 × g for 20 min at 4 °C. For all experiments, the same proportion of each fraction was used in SDS-gel electrophoresis.
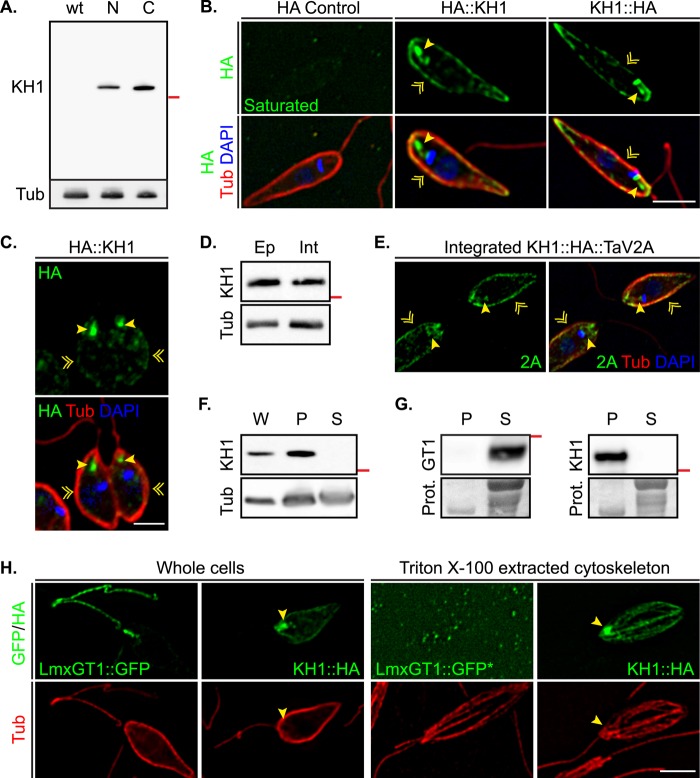
FIGURE 5.
Localization of HA-tagged KH1. A, Western blot of HA-tagged KH1 proteins probed with anti-HA antibodies. Left lane wt, HA tag only; middle lane N, HA3::KH1; right lane C, KH1::HA3. The red line on the right of the blot represents a 70-kDa molecular mass marker. The membrane was stripped and re-probed with anti-tubulin antibodies as a protein loading control. B, immunolocalization of HA3::KH1and KH1::HA3. HA control are parasites transfected with an episomal vector expressing HA3 alone. Yellow arrowheads indicate HA-tagged KH1 proteins at the base of the flagellum. Yellow chevrons indicate localization on the cell periphery. Scale bar = 3 μm. C, HA3::KH1 localization in dividing cells. Yellow indicators are as described in B. Scale bar = 3 μm. D, Western blot of HA3::KH1 expressed from an episomal vector (Ep) and integrated KH1::HA3::TaV2A (Int) probed with anti-HA antibodies. The red line on the right of the blot represents a 70-kDa molecular mass marker. E, immunolocalization of integrated KH1::HA3::TaV2A using anti-2A antibodies (2A). Control for the 2A antibody using wild type cells show no specific staining on the PPM or at the base of the flagellum. Yellow indicators are as described in B. Scale bar = 3 μm. F, Western blot of HA3::KH1 in various cellular fractions. W, whole cells; P, pellet; S, supernatant. Whole cells and fractions from sonicated cells probed with anti-HA antibodies. The red line on the right of the blot represents a 70-kDa molecular mass marker. The membrane was stripped and re-probed with anti-tubulin antibody as a loading control. G, Western blots of fractions from Triton X-100 extractions at 25 °C. Left, GT1, fractions from LmxGT1::HBH expressing parasites were probed with anti-His antibodies. Right, KH1, fractions from HA3::KH1 expressing cells were probed with anti-HA antibodies. The red line on the right of each blot represents a 70-kDa molecular mass marker. Panels marked Prot. show a cropped portion of the membrane stained with Ponceau-S as a protein control. P and S indicate pellet and supernatant, respectively. H, Triton X-100-extracted cytoskeletons of promastigotes expressing both LmxGT1::GFP and KH1::HA3. Yellow arrowheads indicate enrichment of KH1::HA3 at the base of the flagellum. * indicates overexposure in the GFP channel. Scale bar = 3 μm.
L. mexicana promastigote cytoskeletons were obtained by extraction with 1% Triton X-100 in PBS for 30 min at 25 °C, pelleted, and washed once in PBS. Resuspended cytoskeletons were attached to poly-l-lysine coverslips, fixed, and stained as described above.
Deconvolution Microscopy
Fluorescence images were captured on a Deltavision Image Restoration System (Applied Precision, Issaquah, WA) consisting of a Nikon Eclipse TE2000 microscope base, mercury light source, Applied Precision light homogenizer and Nanoposition XYZ stage, and a Kodak CH350 CCD. Cells were imaged at room temperature through a 60 × 1.40NA Nikon objective using SoftWoRx acquisition software version 5.0.0-R6 (Applied Precision, Issaquah, WA). Images were deconvolved in SoftWoRx and then analyzed and processed using ImageJ (NIH, Bethesda, MD). Figures were constructed using Adobe Illustrator CS3 (Adobe Corporation, San Jose, CA).
Electron Microscopy
The immuno-EM experiments were performed at the Molecular Microbiology Imaging Facility of Washington University School of Medicine (St. Louis, MO). For immunolocalization at the ultrastructural level, parasites were fixed in 4% paraformaldehyde, 0.05% glutaraldehyde in 100 mm PIPES, 0.5 mm MgCl2, pH 7.2, for 1 h at 4 °C. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 m sucrose, 20% polyvinylpyrrolidone in PIPES/MgCl2 at 4 °C. Samples were trimmed, frozen in liquid nitrogen, and sectioned with a Leica Ultracut UCT7 cryo-ultramicrotome (Leica Microsystems Inc., Bannockburn, IL). Sixty-nm sections were blocked with 5% FBS, 5% normal goat serum for 30 min and subsequently incubated with rabbit anti-GFP followed by secondary anti-rabbit antibody conjugated to 18 nm colloidal gold (Jackson ImmunoResearch Laboratories Inc., West Grove, PA). Sections were washed in PIPES buffer followed by a water rinse, and stained with 0.3% uranyl acetate, 2% methylcellulose. Samples were viewed with a JEOL 1200EX transmission electron microscope (JEOL USA Inc., Peabody, MA) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques, Woburn, MA). All labeling experiments were conducted in parallel with controls omitting the primary antibody. These controls were consistently negative at the concentration of colloidal gold conjugated secondary antibodies used in these studies.
Macrophage Infections
The human acute leukemia monocyte cell line (THP-1) was cultivated in RPMI 1640 medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Thermo Scientific Hyclone, Logan, UT), 25 mm HEPES, 1% l-glutamine, 50 mm glucose, 5 mm sodium pyruvate, and 1% streptomycin/penicillin at 37 °C and 5% CO2. The cultures were diluted every 3 days to prevent cell count from exceeding 1 × 106 cells/ml. Cells were kept for a maximum of 20 subcultured dilution cycles.
THP-1 cells were differentiated with 100 ng/ml of phorbol 12-myristate 13-acetate (Sigma) for 48 h at 37 °C, 5% CO2. Differentiated THP-1 cells are adherent and were seeded in 4-well Lab-TekII Chamber Slides (Nalge Nunc International, Rochester, NY) at a confluence of 3 × 105 cells/well. L. mexicana promastigotes at stationary phase were added to the plates (1:10 macrophage/parasite ratio) and incubated for 4 h, 1 day, 3 days, 5 days, and 7 days at 37 °C, 5% CO2. At each time point, slides were stained using the HEMA3 STAT PACK staining kit as described by the manufacturer (Fisher Scientific). Infected macrophages were examined using a Nikon Eclipse 50i microscope equipped with a ×100 1.25NA oil objective (Nikon Instruments, Melville, NY), and the number of parasites/100 macrophages were determined by counting 300 cells in each of the triplicate experiments per round of infection. Images were captured using a white iPhone 4S equipped with an 8-megapixel iSight camera (Apple Corp., Cupertino, CA).
RESULTS AND DISCUSSION
Tandem Affinity Purification (TAP)-tagged LmxGT1::HBH Localizes Correctly to the Flagellum and Is Biotinylated
To elucidate the molecular mechanism underlying LmxGT1 flagellar trafficking, and identify new components of the flagellar targeting pathway in kinetoplastid parasites, we employed a TAP tagging strategy (21) that combines in vivo formaldehyde cross-linking and purification under fully denaturing conditions to enrich for proteins that specifically interact with LmxGT1, whereas reducing the nonspecific background. The TAP tag consists of a hexa-histidine motif (H), followed by a peptide that is a substrate for endogenous biotinylation (B), followed by another hexa-histidine motif (H), i.e. the HBH tag, fused to the C terminus of LmxGT1 to generate the LmxGT1::HBH fusion protein (Fig. 1A). LmxGT1::HBH was targeted correctly to the flagellar membrane as shown by immunofluorescence microscopy employing anti-His antibodies (Fig. 1B). Additionally, fluorescence microscopy using Alexa 488-conjugated streptavidin revealed that LmxGT1::HBH was spontaneously biotinylated by an endogenous biotin ligase in Leishmania (Fig. 1B). These results confirm that LmxGT1::HBH can be used to identify proteins that interact with LmxGT1.
FIGURE 1.
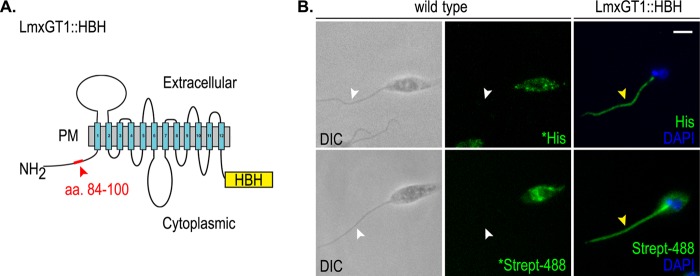
Targeting of LmxGT1::HBH. A, schematic diagram of LmxGT1::HBH. The 12 plasma membrane (PM) spanning domains are numbered. Red box and arrowhead indicate amino acids 84–100 on the cytosolic N terminus. The yellow box labeled HBH indicates the tandem affinity tag. B, immunofluorescence of LmxGT1::HBH. Antibodies and markers used are indicated in panels. An * indicates overexposure compared with other panels to emphasize the absence of flagellar staining in the wild type parasites. White arrowheads indicate the absence of signal on the flagellum. Yellow arrowheads indicate LmxGT1::HBH on the flagellum. DIC indicates differential interference contrast microscopy. Scale bar = 3 μm.
TAP Tagging and Mass Spectrometry Identify KHARON1 as an Interacting Partner Required for LmxGT1 Flagellar Targeting
To identify potential interacting proteins required for LmxGT1 flagellar trafficking, we cross-linked proteins in vivo in L. mexicana promastigotes expressing LmxGT1::HBH followed by TAP and identification of cross-linked affinity purified proteins by tandem mass spectrometry (MS/MS) (Fig. 2A). In addition, we performed experiments using a HBH tagged version of the LmxGT1(Δ84–100) deletion mutant in which the flagellar targeting signal has been deleted. This mutant is defective in flagellar trafficking and targets instead to the pellicular plasma membrane (PPM) surrounding the cell body (17). To identify proteins involved in flagellar targeting, peptides identified among the TAP products of LmxGT1::HBH were compared with those obtained from the non-flagellar LmxGT1(Δ84–100)::HBH. Peptides associated with LmxGT1::HBH but not with LmxGT1(Δ84–100)::HBH should represent proteins that interact with LmxGT1 containing the intact flagellar targeting domain but cannot interact with the non-flagellar deletion mutant.
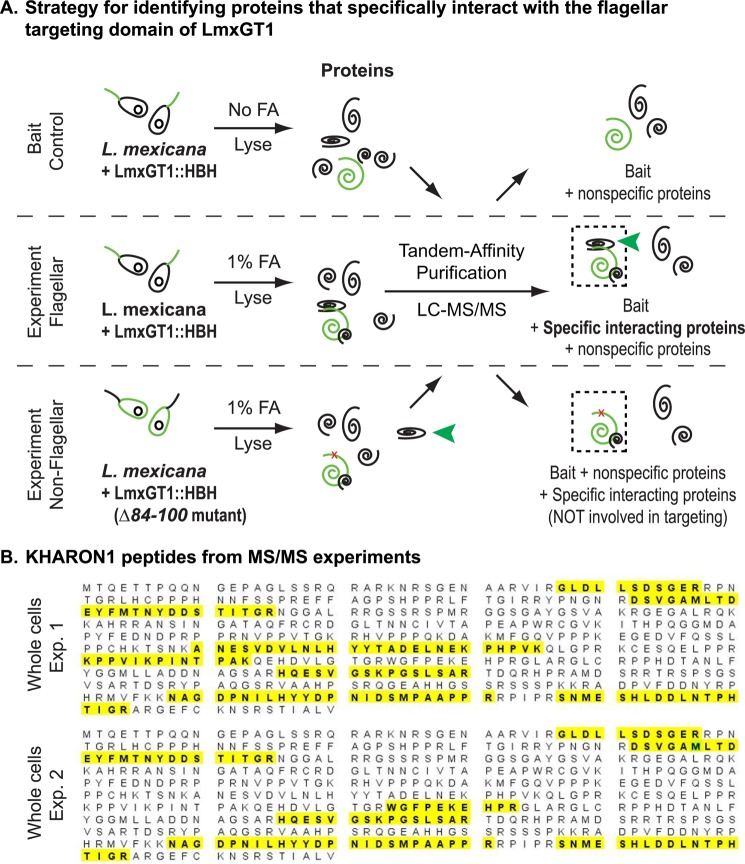
FIGURE 2.
Strategy for identifying proteins that specifically interact with the flagellar targeting domain of LmxGT1. A, three samples were analyzed: Bait Control in which parasites expressing LmxGT1::HBH were not cross-linked with formaldehyde (FA); Experiment Flagellar, in which LmxGT1::HBH expressing parasites were FA cross-linked; and Experiment Non-Flagellar, in which parasites expressing LmxGT1(Δ84–100)::HBH were FA cross-linked. Each sample was lysed and proteins that had been cross-linked to either wild type LmxGT1::HBH or LmxGT1(Δ84–100)::HBH were subjected to tandem affinity purification, and the TAP purified products were analyzed by tandem mass spectrometry (MS/MS). The green spiral represents LmxGT1::HBH, and the black spirals are proteins that are either specifically cross-linked or not cross-linked to LmxGT1::HBH. The dashed boxes indicate complexes of proteins that are cross-linked to LmxGT1::HBH and purified by TAP. The red x in LmxGT1::HBH in the bottom panel (Experiment Non-Flagellar) indicates the Δ84–100 deletion mutant that does not target to the flagellum. The green arrowhead indicates proteins that are specifically bound to the flagellar targeting domain of LmxGT1::HBH. Peptides identified from these three samples were compared to identify those that were present only in the Experiment Flagellar sample but not in the other two samples. B, peptides identifying KH1 from MS/MS spectra of the Experimental Flagellar sample are highlighted on the KH1 coding sequence.
Employing this subtractive approach, we identified a protein that we have designated KH1, encoded by gene LmxM36.5850 in the L. mexicana genome (29), which specifically interacts with the flagellar targeting domain of LmxGT1 based on the following criteria. First, peptides from KH1, covering ∼20% of the protein sequence (Fig. 2B), were reproducibly identified in the cross-linked sample but not in the non-cross-linked sample in three independent experiments using HBH-tagged wild type LmxGT1. Second, KH1 was not identified when the LmxGT1(Δ84–100)::HBH deletion mutant was employed in the TAP experiments. These results suggest that KH1 interacts with wild type flagellar LmxGT1 but not with a mutant LmxGT1 that is targeted to the PPM.
KH1 is annotated in the TritrypDB Kinetoplastid Genomics Resources as a “hypothetical conserved protein” of unknown function. Analysis using BLAST (30), PSI-BLAST (31), and HMMER (32) revealed that KH1 is conserved in all Kinetoplastid parasites whose genomes have been sequenced, but this ORF was not identified in other organisms. KH1 may be specific to kinetoplastid parasites, or it may be highly divergent from orthologs outside of the order Kinetoplastida and therefore cannot be detected by current search algorithms. Thus, we currently consider KH1 as a kinetoplastid-specific protein.
KH1 Is Important for Flagellar Targeting of LmxGT1
To test whether KH1 is involved in targeting LmxGT1 to the flagellar membrane, we generated a Δlmxkh1 null mutant (Δkh1 hereafter) using targeted gene replacement (23, 33) and verified their null genotype by Southern blot (Fig. 3A). In wild type cells, LmxGT1::GFP was targeted exclusively to the flagellar membrane in over 98% of cells with detectable GFP signal (n = 210) (Fig. 3, B and C). In contrast, LmxGT1::GFP was found in the flagellum in only ∼30% of Δkh1 null mutant cells (n = 241) (Fig. 3, B and C). Approximately 66% of Δkh1 cells expressing LmxGT1::GFP showed localization at the base of the flagellum (Fig. 3, B and C). However, when the Kh1 ORF was expressed from an episomal expression vector in Δkh1 mutant cells, LmxGT1::GFP was correctly targeted to the flagellar membrane in ∼85% of cells assayed (n = 139) (Fig. 3, B and C). To provide a biochemical confirmation of these results, flagella were isolated from wild type, Δkh1, and Kh1 complemented cell lines expressing LmxGT1::HA3 and analyzed by Western blotting using an anti-HA antibody (Fig. 3D). Only ∼25% of LmxGT1-HA3 protein present in wild type flagella was detectable in isolated flagella from the Δkh1 null mutant, but almost all of this signal was restored in the add-back line (Fig. 3D). Thus the quantification of residual LmxGT1::HA3 detected in the flagella of Δkh1 null mutants was essentially the same value when measured by Western blotting of isolated flagella (Fig. 3D) or by immunofluorescence of whole parasites (Fig. 3B). These results confirm that KH1 plays a critical role in targeting of LmxGT1 to the flagellar compartment.
To determine whether the localization of LmxGT1 at the base of the flagellum reflects association with the flagellar pocket (FP), Δkh1 mutant cells expressing LmxGT1::GFP were labeled with the lipophilic dye FM4-64. This dye first intercalates into the plasma membrane, but after a chase period, it accumulates at the FP and then subsequently enters the early and late endosomes (34, 35). In cells treated with FM4-64 and then incubated for a chase period of 5 min in serum-free medium lacking FM4-64, the FM4-64 signal showed significant co-localization with LmxGT1::GFP at the base of the flagellum (Fig. 3E).
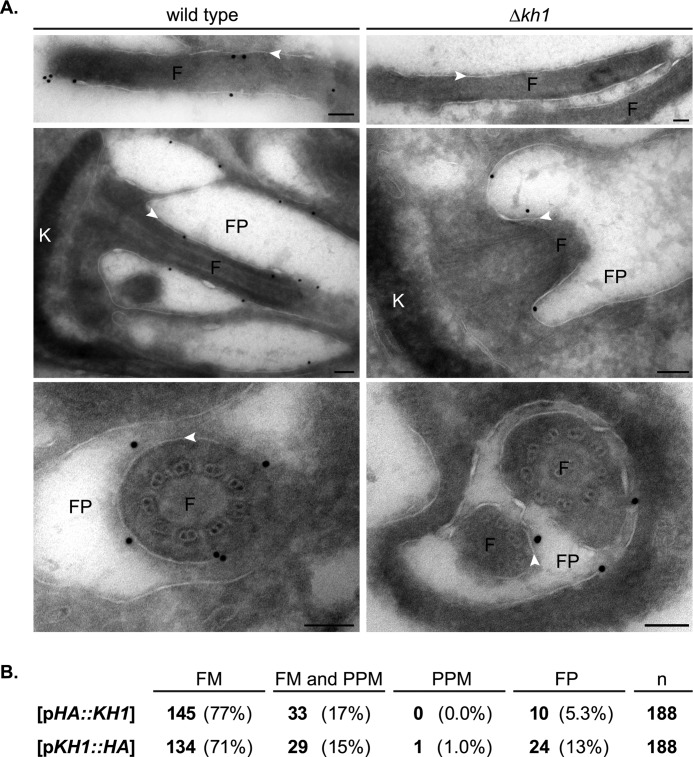
To obtain high resolution localization for LmxGT1::GFP in Δkh1 null mutants, the location of this fusion protein was examined by immuno-EM. Indeed in Δkh1 mutants, LmxGT1::GFP was present on the FP membrane and the component of the flagellar membrane (FM) that is within the FP, but it is not on the FM outside the flagellar pocket (Fig. 4A, right). In contrast, in wild type parasites this fusion protein was located on FP membranes and on the FM both inside and outside of the FP (Fig. 4A, left). Hence, LmxGT1::GFP was impaired in movement into the flagellar compartment outside of the FP in Δkh1 null mutants.
FIGURE 4.
Immuno-EM localization of LmxGT1::GFP in Δkh1 null mutants. A, immuno-EM of LmxGT1::GFP in wild type and Δkh1 mutants. White arrowheads indicate flagellar membrane. Top and middle panels show a longitudinal section through the flagellum and FP region, respectively. Bottom panels show a cross-section through the flagellar pocket region. F, flagellum; FP, flagellar pocket; K, kDNA. Each scale bar = 100 nm. B, table showing quantification of LmxGT1::GFP localization in Δkh1 mutants rescued with HA-tagged KH1. The numbers in bold represent parasites determined to fall under each category, and the same statistics are represented as a percentage in parentheses. n, total number of cells expressing GT1::GFP examined.
Taken together, these results show that KH1 is likely required for efficient sorting of LmxGT1::GFP from the flagellar pocket to the flagellar compartment. Indeed, the reason for naming this protein KHARON1 is that it mediates transit of cargo across a putative barrier between the FP and the external flagellum, analogous to the mythological Kharon who ferries souls across the River Styx that separated the living from the dead.
KH1 Associates with the Cytoskeleton and Flagellar Axoneme at the Base of the Flagellum within L. mexicana Promastigotes
To determine where KH1 is localized within L. mexicana promastigotes, we generated N- and C-terminal HA3-tagged KH1 proteins designated HA3::KH1 and KH1::HA3. Immunoblots of cells expressing tagged KH1 proteins revealed a single band at ∼75 kDa, whereas no signal was detected in cells expressing only the epitope tag (Fig. 5A). Furthermore, tagged KH1 proteins appeared to be functional, as Δkh1 mutant cells expressing either HA3::KH1 or KH1::HA3 from an episomal vector showed correct flagellar targeting of LmxGT1::GFP in over 70% of cells examined (Fig. 4B). The ability to rescue the Δkh1 null mutant suggests that tagged KH1 proteins localize correctly within the cell.
In cells expressing tagged KH1 proteins, the HA positive signal is found at the base of the flagellum and surrounding the cell body (Fig. 5B). In contrast, parasites expressing only the HA epitope tag showed no specific staining with anti-HA antibodies (Fig. 5B). In dividing cells, KH1 can be found at the base of both flagellar structures (Fig. 5C). Furthermore, KH1 tagged with the viral TaV2A peptide (KH1::HA3::TaV2A) and integrated into the endogenous Kh1 locus (“Experimental Procedures”) revealed similar levels of expression (Fig. 5D) and localization within the cell (Fig. 5E), although the proportion of KH1::HA3::TaV2A on the cell periphery was higher than in cells expressing tagged KH1 from an episomal vector.
Subcellular fractionation suggests that KH1 associates with the cytoskeleton (Fig. 5, F–H). KH1 remained in the low speed pellet in parasites lysed either by sonication (Fig. 5F) or Triton X-100 (Fig. 5G), and KH1::HA3 was detected in pelleted and resuspended cytoskeletons from detergent-extracted parasites (Fig. 5H). Cytoskeleton association is further supported by ultrastructural studies using immuno-EM, which revealed HA3::KH1 localization on the basal body, the proximal flagellar axoneme, and subpellicular microtubules surrounding the cell body (Fig. 6). Together, these results suggest that an interaction between KH1 and LmxGT1 within the proximal flagellar structure may facilitate LmxGT1 entry into the flagellar compartment outside of the FP.
FIGURE 6.
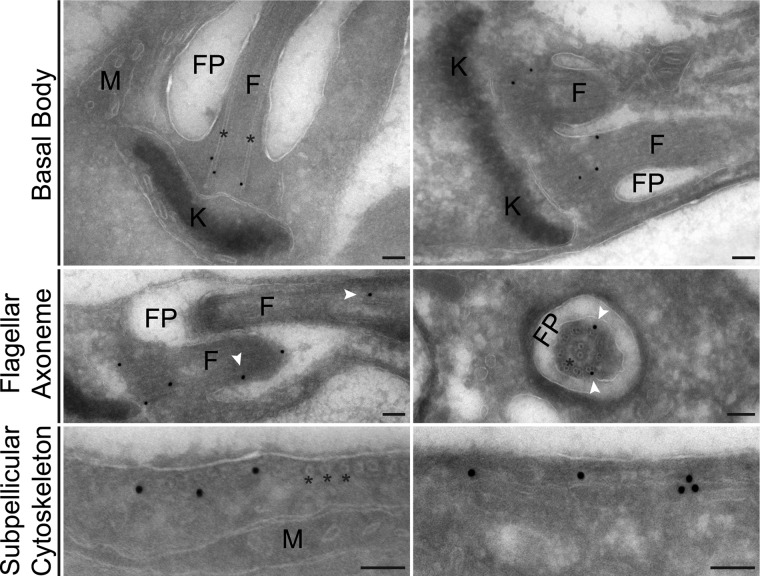
Immuno-EM of HA3::KH1 within L. mexicana promastigotes. Immuno-EM showing examples of HA3::KH1 association with the cytoskeletal structure. F, flagellum; K, kDNA; M, mitochondrion. * indicates microtubules. Top and middle: longitudinal sections through the region at the base of the flagellum and a cross-section of the FP (middle right). White arrowheads indicate KH1 association with the flagellar axoneme. Bottom panels show examples of KH1 association with the subpellicular cytoskeleton. Each scale bar = 100 nm.
KH1 Is Not Required for PPM Targeting of LmxGT2 and LmxMIT
Because sorting of membrane proteins likely takes place at the flagellar pocket (6), and because KH1 can also be found on subpellicular microtubules, it is possible that KH1 also functions to sort non-flagellar polytopic membrane proteins to the PPM. To test this potential role, we examined localization of the glucose transporter LmxGT2::GFP and the myo-inositol transporter LmxMIT::GFP by immunofluorescence microscopy in wild type and Δkh1 cells. LmxGT2::GFP was targeted correctly to the PPM in both wild type and Δkh1 cells (wild type, 99%, n = 150; Δkh1, 98%, n = 171) (Fig. 7A). Likewise, LmxMIT::GFP was targeted to the PPM in wild type and Δkh1 null mutants (wild type, 95%, n = 189; Δkh1, 99%, n = 172) (Fig. 7B). These results indicate that targeting of LmxGT2::GFP and LmxMIT::GFP to the PPM is not dependent on recognition by KH1, and it is unlikely that KH1 performs dual sorting functions.
FIGURE 7.
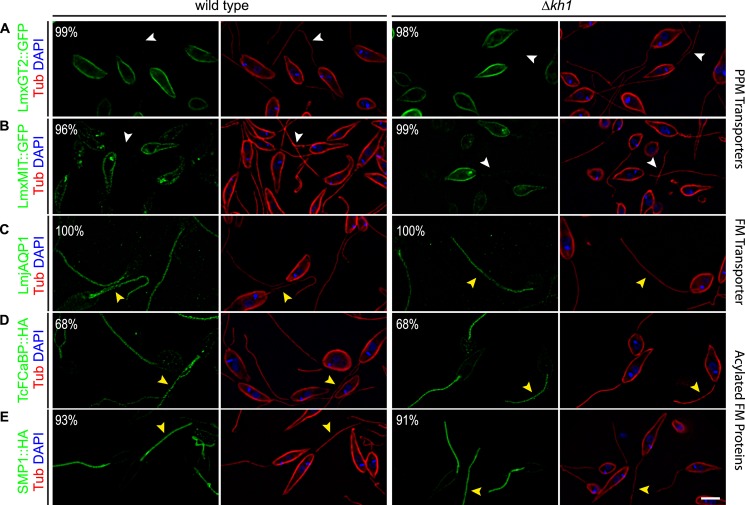
Targeting of other membrane proteins in Δkh1 mutant cells. Immunofluorescence of PPM proteins and other proteins known to specifically target to the FM. Green is GFP, HA, or LmjAQP1; red is tubulin; blue is DAPI. White arrowheads indicate lack of flagellar localization. Yellow arrowheads indicate flagellar localization. Percentage shown in panels represent the fraction of cells with localization as shown. Scale bar = 3 μm.
Flagellar Targeting of LmjAQP1 Is Not Affected in Δkh1 Null Mutants
One other flagellar polytopic membrane protein is the L. major aquaporin channel AQP1 (15). To determine whether LmjAQP1 is targeted to the flagellum in a KH1-dependent manner, the LmjAQP1 coding sequence was expressed from an episomal vector in wild type L. mexicana and Δkh1 null mutants. LmjAQP1 was detected in the flagellum of both wild type and Δkh1 null mutants (wild type, 100%, n = 118; Δkh1, 100%, n = 137) using a previously described polyclonal antibody (15) (Fig. 7C). Therefore, flagellar targeting of LmjAQP1 does not require KH1.
LmjAQP1 and LmxGT1 share no sequence similarity that would suggest a conserved flagellar targeting mechanism. Indeed, a recent study shows that phosphorylation of LmjAQP1 is sufficient to undo its FM membrane specificity and redistributes this protein to the PPM (36). However, we cannot exclude the possibility that KH1 may be an adaptor for a subset of FM proteins, including LmxGT1, which links them to a core FM targeting machinery. In this case, the core complex could target other proteins, such as LmjAQP1, to the FM.
KH1 Is Not Required for Targeting the Acylated TcFCaBP and SMP1 to the Flagella
Several FM proteins identified in kinetoplastid parasites are dually acylated near their N termini and require these acylations for trafficking to the FM (37), but these proteins do not encompass transmembrane segments. To determine whether acylated flagellar proteins are trafficked to the FM in a KH1-dependent manner, we examined the dually acylated TcFCaBP from T. cruzi, which is targeted correctly to the flagellar membrane in Leishmania (38, 39), and SMP1 from L. mexicana (40). In both wild type and Δkh1 backgrounds, TcFCaBP::HA3 was targeted correctly to the flagellum in the majority of cells assayed (wild type: 68%, n = 175; Δkh1: 69%, n = 145) (Fig. 7D), with another 32% of cells showing localization on both the FM and PPM (not shown). Similarly, L. mexicana SMP1 was also correctly targeted to the flagellar membrane in wild type as well as Δkh1 null mutants (wild type: 93%, n = 124; Δkh1: 92%, n = 116) when the epitope-tagged LmxSMP1::HA3 was examined (Fig. 7E). These results indicate that KH1 is not required for flagellar targeting of acylated proteins such as TcFCaBP and SMP1 in Leishmania. It is possible that dually acylated proteins traffic to the flagellar membrane by a different mechanism such as association with lipid raft microdomains (12, 41).
KH1 Is Critical for Viability of Intracellular Amastigotes
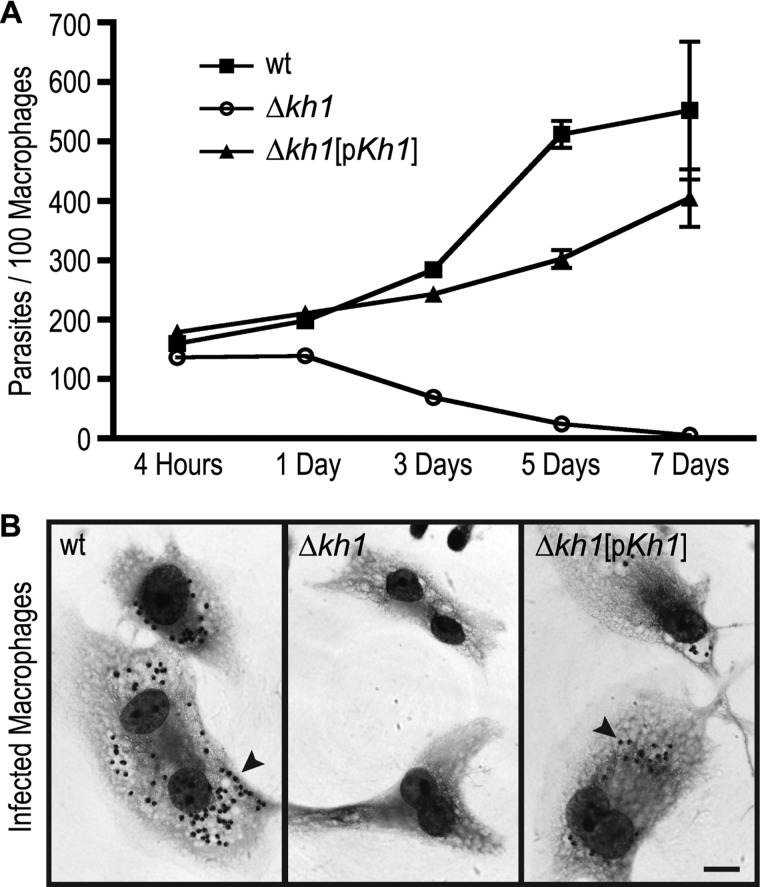
Although the Δkh1 null mutant is not compromised in viability in the promastigote stage employed to generate this mutant, we wished to determine whether it might have a phenotype in disease causing amastigotes that live inside phagolysosomal vesicles of mammalian macrophages. Infections of differentiated THP-1 macrophages (42) revealed that although wild type L. mexicana replicated inside the host cells, Δkh1 null mutants entered macrophages as well as wild type parasites but died off following establishment of the initial infection (Fig. 8). The Δkh1[pKh1] add-back line showed partial restoration of the wild type phenotype maintaining their initial number and then slowly replicating. These results imply that KH1 plays a critical role in the disease causing stage of the parasite life cycle.
FIGURE 8.
Infections of macrophages with Δkh1 null mutants. Growth of wild type, Δkh1, and Δkh1[pKh1] cell lines in THP-1 macrophages. Filled squares are wt; open circles are Δkh1 null mutants; and filled triangles are Δkh1[pKh1] cells. The graph shows average ± S.D. (n = 9) quantified at each time point. B, sample images of macrophage infected by each cell line 7 days post-infection. Black arrowheads indicate amastigotes. Scale bar = 10 μm.
We do not yet know the precise function of KH1 in amastigotes, especially because the LmxGT1 protein is not expressed in this life cycle stage.6 Thus, the impairment of amastigote growth cannot be ascribed to failure to target LmxGT1 properly within amastigotes. However, Gluenz and colleagues (43, 44) have recently emphasized that the short amastigote flagellum may play important roles in the interaction of the parasite with the host cell. In particular, this flagellum has the morphology of a sensory cilium and may be involved in monitoring the environment of the macrophage phagolysomal compartment. Strikingly, these authors also identified a junction that forms between the tip of the amastigote flagellum and the macrophage parasitophorous vacuole in 65% of amastigotes examined. They further proposed that the tip of the flagellum might contain receptors engaged in signal transduction, and/or it might serve as a conduit for secretion of parasite molecules into the macrophage. If components of the amastigote flagellar membrane necessary for junction formation depend upon KH1 for trafficking to the tip, the Δkh1 null mutant may be deficient in formation of structures required for signaling, delivery of parasite “effector” molecules, or both. This deficiency could be fatal to intracellular amastigotes. Hence, a high priority for future studies is to determine whether Δkh1 amastigotes, which retain partial viability at 1–3 days post-infection, fail to form junctions between their flagella and the parasitophorous vacuole membrane. Additionally, it will be important to define the ability of the Δkh1 mutant to mediate disease in the murine model of cutaneous leishmaniasis (45).
Model for Flagellar Targeting by KH1
We propose that KH1 recognizes the flagellar targeting domain of LmxGT1 in the region of the FP and mediates its transit from the FP and proximal flagellar membranes into the external FM. This model is supported by (i) the observation that KH1 cross-linked to wild type LmxGT1 but not to the Δ(84–100) mutant in which critical flagellar targeting sequence, including the NPM motif, has been deleted; (ii) LmxGT1 failed to enter efficiently into the external FM in Δkh1 null mutants and is largely retained in the FP, (iii) LmxGT1 encompassing mutations in the NPM motif is also stalled in the FP (17); and (iv) HA-tagged KH1 is located on the proximal flagellar axoneme within the FP but not on the component of the axoneme that extends beyond the FP.
Although a FM barrier has been predicted to exist in kinetoplastid parasites (37), neither the precise nature of the putative FP to FM barrier nor the mechanism whereby KH1 may overcome this barrier is understood at present. Morphological structures that could potentially play such a barrier role include the FP collar and collarette that have been identified by electron tomography in T. brucei (46). In both mammalian cells and Caenorhabditis elegans, multisubunit complexes such as Tectonic complex (47), B9 complex (48), and the MKS/MKSR/NPHP complex (49) form a ciliary gate that prevents diffusion of non-ciliary proteins into the ciliary membrane. In contrast, the mammalian protein Septin 2, a membrane-associated GTPase, is required for formation of a ciliary diffusion barrier that retains ciliary membrane proteins within the cilium (50). It is possible that KH1 either circumvents similar barriers to flagellar entry or contributes to a barrier preventing exit from the flagellar membrane in L. mexicana.
It is also noteworthy that LmxGT1::GFP exhibits punctate localization on the flagellum (Fig. 3C), a pattern that has been noticed for some flagellar membrane proteins in T. brucei (10). This punctate localization could reflect a significant aspect of distribution within the FM after flagellar entry, such as association with the intraflagellar transport machinery (51) or partitioning into lipid raft domains (52, 53). Additionally, when LmxGT1 was extracted with Triton X-100, it appeared as a single band on SDS-PAGE (Fig. 5G). In contrast, two bands (∼15 kDa difference in apparent molecular mass) were apparent when samples were treated with SDS-containing lysis buffer and immediately heated to 70 °C (Fig. 3D). The “doublet” may indicate the presence of a post-translational modification of LmxGT1 that is reversed when lysis is performed under non-denaturing conditions. At present, we do not know what this potential modification may be or whether it is important for function or targeting of LmxGT1.
Searches employing PSI-BLAST (31) and HMMER (54) did not identify any significant homology between KH1 and other proteins with known function, and all paralogs were hypothetically conserved proteins present in the genome databases of kinetoplastid parasites. Secondary structure algorithms such as NORSp (55) predict that KH1 does not have a defined secondary structure, and may be unstructured in the absence of binding partners. However, the FTwin algorithm on the REPPER server (56) identified hydrophobic repeats with a repeat unit of ∼3.4 residues, between amino acids 250 and 350, which could represent a potential coiled-coil domain, and these hydrophobic repeats are also conserved in the T. brucei ortholog of KH1, Tb927.10.8940, which is only 18.7% identical to KH1. These predictions raise the possibility that KH1 interacts with other partners using this putative coiled-coil domain and/or that other regions of the protein only assume a defined tertiary structure in the presence of other binding partners.
The potential for KH1 being one component of a larger complex is further highlighted by the preliminary observation that the protein migrates as a band of ∼1 MDa when parasite lysates are analyzed by blue native (57) gel electrophoresis (not shown). KH1 may be a core component of this complex or an adaptor that mediates interaction of the complex with a subset of flagellar membrane proteins. In the latter case, other adaptors might link different FM proteins to the larger KHARON complex. It is difficult to resolve this issue with the limited number of known membrane proteins that are selectively targeted to the flagellum in Leishmania. Thus, another question is whether KH1 is involved in flagellar trafficking of other integral or polytopic FM proteins, and the availability of the Δkh1 null mutant will be valuable in addressing a potentially larger role for KH1 in assembly of the FM proteome.
Finally, the existence of homologous Kharon1 genes within the kinetoplastid parasites suggests that similar FM targeting machinery is likely operative among all these species of parasite. Proteomic studies of the trypanosome flagellum also indicate that the T. brucei ortholog of KH1, Tb927.10.8940, is associated with the flagellar “matrix” or cytoskeleton (10), consistent with the results of this study. Of considerable interest, in a genome wide RNAi study Tb927.10.8940 was scored as an essential gene for viability of both procyclic and bloodstream forms of T. brucei parasites (58). This observation implies that the KH1 targeting pathway engages various flagellar membrane proteins in trypanosomes as well as Leishmania, and that this pathway can be critical for the survival of the parasite in the disease causing stage, the L. mexicana amastigote and the T. brucei bloodstream form.
Acknowledgment
We thank Dr. Rita Mukhopadhyay for the LmjAQP1 antibody and for a protocol for isolation of flagella from Leishmania promastigotes.
This work was supported, in whole or in part, by National Institutes of Health Grant AI25920 (to S. M. L.) and American Heart Association Postdoctoral Fellowship 11POST7440105 and National Institutes of Health NRSA F32 Fellowship 1F32AI096854 (to K. D. T.).
P. Yates, manuscript in preparation.
D. Rodriguez-Contreras, unpublished data.
- KH1
- KHARON1
- HBH
- hexa-histidine-biotinylation motif-hexa-histidine tandem affinity tag
- HA3 or 3HA
- 3X hemagluttinin epitope tag
- FM
- flagellar membrane
- FP
- flagellar pocket
- PPM
- pellicular plasma membrane
- TAP
- tandem affinity purification.
REFERENCES
- 1. Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. (2009) The primary cilium as a complex signaling center. Curr. Biol. 19, R526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bloodgood R. A. (2010) Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell Sci. 123, 505–509 [DOI] [PubMed] [Google Scholar]
- 3. Ginger M. L., Portman N., McKean P. G. (2008) Swimming with protists. Perception, motility and flagellum assembly. Nat. Rev. Microbiol. 6, 838–850 [DOI] [PubMed] [Google Scholar]
- 4. Pazour G. J., Witman G. B. (2003) The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 15, 105–110 [DOI] [PubMed] [Google Scholar]
- 5. Clapham D. E. (2003) TRP channels as cellular sensors. Nature 426, 517–524 [DOI] [PubMed] [Google Scholar]
- 6. Maric D., Epting C. L., Engman D. M. (2010) Composition and sensory function of the trypanosome flagellar membrane. Curr. Opin. Microbiol. 13, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marshall W. F., Nonaka S. (2006) Cilia. Tuning in to the cell's antenna. Curr. Biol. 16, R604–614 [DOI] [PubMed] [Google Scholar]
- 8. Singla V., Reiter J. F. (2006) The primary cilium as the cell's antenna. Signaling at a sensory organelle. Science 313, 629–633 [DOI] [PubMed] [Google Scholar]
- 9. WHO W. H. O. (2013) Neglected tropical diseases. www.who.int/neglected_diseases/diseases/en
- 10. Oberholzer M., Langousis G., Nguyen H. T., Saada E. A., Shimogawa M. M., Jonsson Z. O., Nguyen S. M., Wohlschlegel J. A., Hill K. L. (2011) Independent analysis of the flagellum surface and matrix proteomes provides insight into flagellum signaling in mammalian-infectious Trypanosoma brucei. Mol. Cell. Proteomics 10, M111.010538-1–M111.010538-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buchanan K. T., Ames J. B., Asfaw S. H., Wingard J. N., Olson C. L., Campana P. T., Araújo A. P., Engman D. M. (2005) A flagellum-specific calcium sensor. J. Biol. Chem. 280, 40104–40111 [DOI] [PubMed] [Google Scholar]
- 12. Emmer B. T., Maric D., Engman D. M. (2010) Molecular mechanisms of protein and lipid targeting to ciliary membranes. J. Cell Sci. 123, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Engman D. M., Krause K. H., Blumin J. H., Kim K. S., Kirchhoff L. V., Donelson J. E. (1989) A novel flagellar Ca2+-binding protein in trypanosomes. J. Biol. Chem. 264, 18627–18631 [PubMed] [Google Scholar]
- 14. Paindavoine P., Rolin S., Van Assel S., Geuskens M., Jauniaux J. C., Dinsart C., Huet G., Pays E. (1992) A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol. Cell. Biol. 12, 1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Figarella K., Uzcategui N. L., Zhou Y., LeFurgey A., Ouellette M., Bhattacharjee H., Mukhopadhyay R. (2007) Biochemical characterization of Leishmania major aquaglyceroporin LmAQP1. Possible role in volume regulation and osmotaxis. Mol. Microbiol. 65, 1006–1017 [DOI] [PubMed] [Google Scholar]
- 16. Burchmore R. J., Rodriguez-Contreras D., McBride K., Merkel P., Barrett M. P., Modi G., Sacks D., Landfear S. M. (2003) Genetic characterization of glucose transporter function in Leishmania mexicana. Proc. Natl. Acad. Sci. U.S.A. 100, 3901–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tran K. D., Rodriguez-Contreras D., Shinde U., Landfear S. M. (2012) Both sequence and context are important for flagellar targeting of a glucose transporter. J. Cell Sci. 125, 3293–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pazour G. J., Bloodgood R. A. (2008) Targeting proteins to the ciliary membrane. Curr. Top. Dev. Biol. 85, 115–149 [DOI] [PubMed] [Google Scholar]
- 19. Nachury M. V., Seeley E. S., Jin H. (2010) Trafficking to the ciliary membrane. How to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 26, 59–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson K. A., Beverley S. M. (2003) Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol. Biochem. Parasitol. 128, 217–228 [DOI] [PubMed] [Google Scholar]
- 21. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions. Application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 22. Valdés R., Vasudevan G., Conklin D., Landfear S. M. (2004) Transmembrane domain 5 of the LdNT1.1 nucleoside transporter is an amphipathic helix that forms part of the nucleoside translocation pathway. Biochemistry 43, 6793–6802 [DOI] [PubMed] [Google Scholar]
- 23. Fulwiler A. L., Soysa D. R., Ullman B., Yates P. A. (2011) A rapid, efficient and economical method for generating leishmanial gene targeting constructs. Mol. Biochem. Parasitol. 175, 209–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nasser M. I., Landfear S. M. (2004) Sequences required for the flagellar targeting of an integral membrane protein. Mol. Biochem. Parasitol. 135, 89–100 [DOI] [PubMed] [Google Scholar]
- 25. Tran K. D., Miller M. R., Doe C. Q. (2010) Recombineering Hunchback identifies two conserved domains required to maintain neuroblast competence and specify early-born neuronal identity. Development 137, 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ha D. S., Schwarz J. K., Turco S. J., Beverley S. M. (1996) Use of the green fluorescent protein as a marker in transfected Leishmania. Mol. Biochem. Parasitol. 77, 57–64 [DOI] [PubMed] [Google Scholar]
- 27. Gourbal B., Sonuc N., Bhattacharjee H., Legare D., Sundar S., Ouellette M., Rosen B. P., Mukhopadhyay R. (2004) Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J. Biol. Chem. 279, 31010–31017 [DOI] [PubMed] [Google Scholar]
- 28. Heras S. R., Thomas M. C., García-Canadas M., de Felipe P., García-Pérez J. L., Ryan M. D., López M. C. (2006) L1Tc non-LTR retrotransposons from Trypanosoma cruzi contain a functional viral-like self-cleaving 2A sequence in-frame with the active proteins they encode. Cell Mol. Life Sci. 63, 1449–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B. P., Carrington M., Depledge D. P., Fischer S., Gajria B., Gao X., Gardner M. J., Gingle A., Grant G., Harb O. S., Heiges M., Hertz-Fowler C., Houston R., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Logan F. J., Miller J. A., Mitra S., Myler P. J., Nayak V., Pennington C., Phan I., Pinney D. F., Ramasamy G., Rogers M. B., Roos D. S., Ross C., Sivam D., Smith D. F., Srinivasamoorthy G., Stoeckert C. J., Jr., Subramanian S., Thibodeau R., Tivey A., Treatman C., Velarde G., Wang H. (2010) TriTrypDB. A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38, D457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 31. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eddy S. R. (1998) Profile hidden Markov models. Bioinformatics 14, 755–763 [DOI] [PubMed] [Google Scholar]
- 33. Cruz A., Beverley S. M. (1990) Gene replacement in parasitic protozoa. Nature 348, 171–173 [DOI] [PubMed] [Google Scholar]
- 34. McConville M. J., Mullin K. A., Ilgoutz S. C., Teasdale R. D. (2002) Secretory pathway of trypanosomatid parasites. Microbiol Mol. Biol. Rev. 66, 122–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng Z., Butler K. D., Tweten R. K., Mensa-Wilmot K. (2004) Endosomes, glycosomes, and glycosylphosphatidylinositol catabolism in Leishmania major. J. Biol. Chem. 279, 42106–42113 [DOI] [PubMed] [Google Scholar]
- 36. Mandal G., Sharma M., Kruse M., Sander-Juelch C., Munro L. A., Wang Y., Vilg J. V., Tamás M. J., Bhattacharjee H., Wiese M., Mukhopadhyay R. (2012) Modulation of Leishmania major aquaglyceroporin activity by a mitogen-activated protein kinase. Mol. Microbiol. 85, 1204–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fridberg A., Buchanan K. T., Engman D. M. (2007) Flagellar membrane trafficking in kinetoplastids. Parasitol. Res. 100, 205–212 [DOI] [PubMed] [Google Scholar]
- 38. Godsel L. M., Engman D. M. (1999) Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J. 18, 2057–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maric D., McGwire B. S., Buchanan K. T., Olson C. L., Emmer B. T., Epting C. L., Engman D. M. (2011) Molecular determinants of ciliary membrane localization of Trypanosoma cruzi flagellar calcium-binding protein. J. Biol. Chem. 286, 33109–33117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tull D., Vince J. E., Callaghan J. M., Naderer T., Spurck T., McFadden G. I., Currie G., Ferguson K., Bacic A., McConville M. J. (2004) SMP-1, a member of a new family of small myristoylated proteins in kinetoplastid parasites, is targeted to the flagellum membrane in Leishmania. Mol. Biol. Cell 15, 4775–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Denny P. W., Gokool S., Russell D. G., Field M. C., Smith D. F. (2000) Acylation-dependent protein export in Leishmania. J. Biol. Chem. 275, 11017–11025 [DOI] [PubMed] [Google Scholar]
- 42. Tsuchiya S., Yamabe M., Yamaguchi Y., Kobayashi Y., Konno T., Tada K. (1980) Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26, 171–176 [DOI] [PubMed] [Google Scholar]
- 43. Gluenz E., Ginger M. L., McKean P. G. (2010) Flagellum assembly and function during the Leishmania life cycle. Curr. Opin. Microbiol. 13, 473–479 [DOI] [PubMed] [Google Scholar]
- 44. Gluenz E., Höög J. L., Smith A. E., Dawe H. R., Shaw M. K., Gull K. (2010) Beyond 9+0. Noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J. 24, 3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sacks D. L., Melby P. C. (2001) Animal models for the analysis of immune responses to leishmaniasis. Curr. Protoc. Immunol. 28, 19.2.1–19.2.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lacomble S., Vaughan S., Gadelha C., Morphew M. K., Shaw M. K., McIntosh J. R., Gull K. (2009) Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 122, 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garcia-Gonzalo F. R., Corbit K. C., Sirerol-Piquer M. S., Ramaswami G., Otto E. A., Noriega T. R., Seol A. D., Robinson J. F., Bennett C. L., Josifova D. J., García-Verdugo J. M., Katsanis N., Hildebrandt F., Reiter J. F. (2011) A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chih B., Liu P., Chinn Y., Chalouni C., Komuves L. G., Hass P. E., Sandoval W., Peterson A. S. (2012) A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol. 14, 61–72 [DOI] [PubMed] [Google Scholar]
- 49. Williams C. L., Li C., Kida K., Inglis P. N., Mohan S., Semenec L., Bialas N. J., Stupay R. M., Chen N., Blacque O. E., Yoder B. K., Leroux M. R. (2011) MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J. Cell Biol. 192, 1023–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., Spiliotis E. T., Nelson W. J. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P. (2008) Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell 19, 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Denny P. W., Field M. C., Smith D. F. (2001) GPI-anchored proteins and glycoconjugates segregate into lipid rafts in Kinetoplastida. FEBS Lett. 491, 148–153 [DOI] [PubMed] [Google Scholar]
- 53. Denny P. W., Smith D. F. (2004) Rafts and sphingolipid biosynthesis in the kinetoplastid parasitic protozoa. Mol. Microbiol. 53, 725–733 [DOI] [PubMed] [Google Scholar]
- 54. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 55. Liu J., Rost B. (2003) NORSp. Predictions of long regions without regular secondary structure. Nucleic Acids Res. 31, 3833–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gruber M., Söding J., Lupas A. N. (2005) REPPER. Repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 33, W239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wittig I., Schägger H. (2008) Features and applications of blue-native and clear-native electrophoresis. Proteomics 8, 3974–3990 [DOI] [PubMed] [Google Scholar]
- 58. Alsford S., Turner D. J., Obado S. O., Sanchez-Flores A., Glover L., Berriman M., Hertz-Fowler C., Horn D. (2011) High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]