Background: DC-SIGNR, a C-type lectin that promotes infection of pathogens such as HIV, is a promising drug target.
Results: The carbohydrate recognition domain of DC-SIGNR is highly dynamic, displaying unique binding modes for individual glycans.
Conclusion: More complex, disease-associated glycans have binding modes different from those of smaller glycans previously studied.
Significance: Understanding ligand-binding properties and solution dynamics of DC-SIGNR will facilitate therapeutic design.
Keywords: Calcium-binding Proteins, Glycobiology, HIV, Lectin, Nuclear Magnetic Resonance, C-type Lectin, Glycan Binding Affinities, gp120 Man9GlcNAc2, Oligosaccharide Interactions, Solution NMR Dynamics
Abstract
The C-type lectin DC-SIGNR (dendritic cell-specific ICAM-3-grabbing non-integrin-related; also known as L-SIGN or CD299) is a promising drug target due to its ability to promote infection and/or within-host survival of several dangerous pathogens (e.g. HIV and severe acute respiratory syndrome coronavirus (SARS)) via interactions with their surface glycans. Crystallography has provided excellent insight into the mechanism by which DC-SIGNR interacts with small glycans, such as (GlcNAc)2Man3; however, direct observation of complexes with larger, physiological oligosaccharides, such as Man9GlcNAc2, remains elusive. We have utilized solution-state nuclear magnetic resonance spectroscopy to investigate DC-SIGNR binding and herein report the first backbone assignment of its active, calcium-bound carbohydrate recognition domain. Direct interactions with the small sugar fragments Man3, Man5, and (GlcNAc)2Man3 were investigated alongside Man9GlcNAc derived from recombinant gp120 (present on the HIV viral envelope), providing the first structural data for DC-SIGNR in complex with a virus-associated ligand, and unique binding modes were observed for each glycan. In particular, our data show that DC-SIGNR has a different binding mode for glycans on the HIV viral envelope compared with the smaller glycans previously observed in the crystalline state. This suggests that using the binding mode of Man9GlcNAc, instead of those of small glycans, may provide a platform for the design of DC-SIGNR inhibitors selective for high mannose glycans (like those on HIV). 15N relaxation measurements provided the first information on the dynamics of the carbohydrate recognition domain, demonstrating that it is a highly flexible domain that undergoes ligand-induced conformational and dynamic changes that may explain the ability of DC-SIGNR to accommodate a range of glycans on viral surfaces.
Introduction
Calcium-dependent carbohydrate-binding proteins of the C-type lectin family play a large role in the mammalian immune system (1) and have been shown to be responsible for pathogen recognition and neutralization, cell-cell adhesion, and receptor-mediated endocytosis (2). C-type lectins recognize glycan structures with high selectivity via calcium-dependent carbohydrate recognition domains (CRDs)4 (3). The C-type lectin DC-SIGNR (dendritic cell-specific ICAM-3-grabbing non-integrin-related; also known as L-SIGN or CD299) is a type II transmembrane protein that recognizes high mannose N-linked oligosaccharides on viral envelopes and host glycoproteins (4). Although DC-SIGNR is known to bind high mannose ligands, little is known about this molecule's biological function, partly due to difficulties studying its specialized and often inaccessible cell types in which it is natively expressed. Furthermore, functional orthologues of DC-SIGNR in model species such as mice are not clear cut, thus restricting the value and meaning of gene-targeting studies, such as knock-out animals. In humans, it is expressed on specialized endothelia found in liver sinusoids, lymph nodes, and placental capillaries, suggesting important roles in leukocyte adhesion and migration (5). Expression has also been found on precursor lung epithelial cells (6), a site where there is potential exposure to airborne viruses. Despite the lack of understanding of its biological function, important disease associations have been reported for DC-SIGNR, such as vertical transmission of human immunodeficiency virus (HIV) (7) and severe acute respiratory syndrome coronavirus (SARS) infection (8). Recent work also indicates involvement of DC-SIGNR in respiratory syncytial virus infection (9), influenza (10), and within-host dynamics of Dengue infection (11). The affinity of DC-SIGNR for glycoproteins on the surface of viruses, as well as its localization at the primary sites of virus replication, promotes in trans infection by viruses, such as HIV. Specifically, DC-SIGNR is believed to promote HIV infection by transferring the virus to adjacent CD4+ T-cells, where the HIV glycoprotein gp120 binds to the CD4 receptor on these cells, promoting entry of the virus into host T-cells. This reinforces the important role of this protein in immunity as well as its considerable potential as a drug target.
Therapeutic strategies designed to inhibit or stimulate the function of C-type lectins, such as DC-SIGNR, are scarce, considering the scale of the diseases involved in their biology. Of particular interest is the interaction of the DC-SIGNR CRD (residues 262–400) with Man9GlcNAc2, one of the dominant oligosaccharides present on the HIV envelope glycoprotein gp120. It has been speculated that direct blockade of DC-SIGNR could provide a topical barrier against primary HIV infection. Therefore, a detailed understanding of the interaction between the DC-SIGNR CRD and the oligosaccharides present on viral glycoproteins is valuable for the design of compounds that could act as antiviral drugs.
Thus far, x-ray crystallography has been the primary method employed for atomic level study of the DC-SIGNR CRD structure. To date, four crystal structures have been deposited for the DC-SIGNR CRD: 1) in complex with the branched pentasaccharide (GlcNAc)2Man3 (Protein Data Bank code 1K9J); 2) in the absence of ligand (but with one Ca2+) and containing a portion of the N-terminal α-helical neck region (1XPH); 3) with two repeats of the α-helical neck region and one sodium ion bound (1XAR); and 4) in complex with Lewis-x trisaccharide and containing a portion of the neck (1SL6). The DC-SIGNR CRD adopts a typical “lectin fold” consisting of α-helices and antiparallel β-sheets connected by irregular loops that are stabilized by disulfide bonds and calcium ions (2). The structure of the DC-SIGNR CRD in complex with (GlcNAc)2Man3 provides insight into the CRD structure and potential ligand binding mechanism, notably revealing that an extended binding site exists that is composed of α-helix 2 and a solvent-exposed Phe-325 residue. The C-terminal end of α-helix 2 packs against the loop joining β-sheets 6 and 7, forming a continuous binding surface (see Fig. 1A). The “shelf” formed by α-helix 2 and Phe-325 creates a shape complementary to the Manα1–6Man moiety that forms van der Waals contacts with Phe-325 and hydrogen-bonds with Ser-372. The Phe-325 residue is also thought to be responsible for the selective binding of DC-SIGNR to the outer branched trimannose moiety of high mannose structures (such as Man9GlcNAc2) because it sterically hinders binding to the inner branched mannose (4).
FIGURE 1.
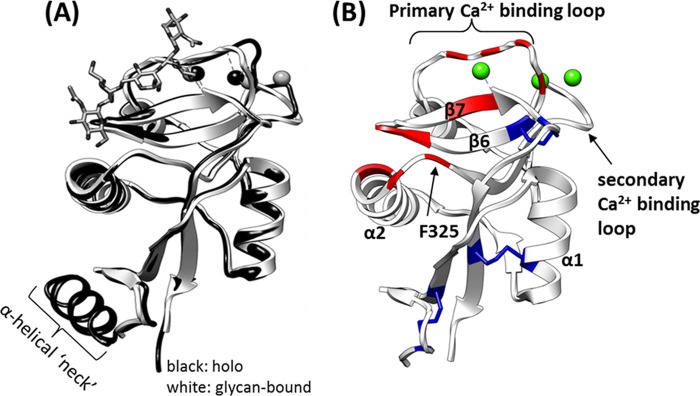
Insight from current crystal structures. A, comparison of holo-structure (Protein Data Bank code 1XPH; black) and (GlcNAc)2Man3-bound (1K9J; white) crystal structure. Ca2+ ions are represented by spheres. All published crystal structures adopt nearly identical conformations, suggesting that the DC-SIGNR CRD adopts the same conformation during crystal formation with or without glycan. B, residues that form direct contacts/bonds with the glycan in the structure of the (GlcNAc)2Man3·CDR complex (1K9J; ligand not shown) are highlighted in red, disulfide bonds are shown in blue, and bound calcium ions are shown in green.
In addition, coordination bonds via the primary Ca2+ binding loop (residues 356–364) and contacts with residues in β-sheets 6 and 7 have been described (4). Regions of interest that form contacts with (GlcNAc)2Man3 are shown in Fig. 1B and listed in supplemental Table S1. However, crystal structures of the DC-SIGNR CRD bound to larger, physiologically relevant oligosaccharides, such as Man9GlcNAc2, have proved to be unattainable thus far. This may be due to as yet uncharacterized conformational/dynamic factors that prohibit crystal growth and diffraction.
High field nuclear magnetic resonance (NMR) studies of the DC-SIGNR CRD have not been reported previously, although NMR studies of ligand interactions with the homologous protein DC-SIGN have started to emerge (12–18). These largely ligand-based studies have been similarly restricted to the use of small glycans and sugar mimetics and have not approached conformational or dynamic properties of DC-SIGN in solution or included larger physiological glycans such as Man9GlcNAc2. As a result, binding of disease-associated ligands such as Man9GlcNAc2 to molecules such as DC-SIGNR and DC-SIGN has been assumed to be consistent with the binding modes observed for smaller glycan fragments co-complexed in the crystalline state (19, 20).
We aimed to increase current understanding of DC-SIGNR-glycan interactions by investigating the binding of the DC-SIGNR CRD to a number of oligosaccharides in solution using heteronuclear solution state NMR techniques that can better deal with issues of dynamics that we surmise to be restricting the rate of progress in DC-SIGNR crystallography. Here we present the backbone assignment of the DC-SIGNR CRD as well as the first structural data for binding of a disease-associated ligand, namely Man9GlcNAc. These results are extended using dynamics measurements (15N T1 and T2), which suggest that the same regions of the DC-SIGNR CRD are highly dynamic in both holo-form and ligand-bound form and interconvert between a number of conformations at similar rates. Our results support the location of the extended binding site observed in the crystal structure of DC-SIGNR CRD-(GlcNAc)2Man3 complex (1K9J). However, our data also demonstrate that DC-SIGNR employs a different binding mode for Man9GlcNAc, suggesting that DC-SIGNR may interact with the HIV glycoprotein gp120 in a way different from that previously observed for smaller glycans. Dynamics data provide new information on the flexible nature of DC-SIGNR and highlight new regions of the protein that are potentially important for ligand recognition.
EXPERIMENTAL PROCEDURES
Expression and Purification of 13C/15N Isotopically Labeled DC-SIGNR CRD
The pT5T overexpression plasmids containing modified cDNA inserts encoding the human DC-SIGNR CRD sequence (Q9H2X3 (CLC4M_HUMAN; residues 262–400); cDNA provided by Elizabeth Soilleux, University of Oxford) were prepared as described previously (21) and subjected to DNA sequencing in order to confirm sequence integrity. The plasmids were then used to transform Escherichia coli strain BL21(DE3), and frozen bacterial stocks were prepared in 15% glycerol and stored at −80 °C. Protein expression was carried out in M9 minimal medium (22) (pH 7.3) containing 50 μg/ml ampicillin, 1 g of [15N]ammonium chloride, and 2 g of [13C]glucose per liter of culture. Two 100-ml starter cultures were inoculated using colonies from an M9/ampicillin plate. Starter cultures were grown at 37 °C with shaking at 200 rpm for 24 h before dilution into 1 liter of M9 minimal medium to a starting A600 nm of ∼0.2. Once an A600 nm = 0.7 was reached, the 1-liter culture was induced with isopropyl-β-d-thiogalactoside to a final concentration of 350 μm. Refolding of the CRD from inclusion bodies was performed as described (21). The protein CRD fragment was purified using a 2-ml mannose-Sepharose column (kindly provided by Dr. Russell Wallis, University of Leicester) equilibrated with 25 mm HEPES, 5 mm CaCl2, 150 mm NaCl, pH 7.8 (loading buffer) as described (21). Protein purity and oligomeric state were assessed using mass spectrometry, SDS-PAGE, and circular dichroism (CD) spectroscopy. The protein predominantly migrated near its monomeric molecular mass of 17.1 kDa (for the isotopically labeled protein) and yielded a CD spectrum that is in good agreement with previous reports (21).
Carbohydrates
All carbohydrate fragments (GlcNAc)2Man3 (M592), Man3 (M336), Man5 (M536)) were purchased from Dextra Laboratories (Reading, UK). The Man9GlcNAc was prepared by digestion of recombinant gp120 with endoglycosidase H using gp120 glycoprotein harvested from 10 μm kifunensine-treated HEK 293T cultures as described (23). The gp120 glycoform was characterized by mass spectrometry as shown in supplemental Fig. S1.
NMR
Purified protein samples were subjected to extensive dialysis into water (1 week with 12 buffer changes) before lyophilization. The protein was then dissolved in 180 μl of 20 mm deuterated HEPES-d18, 20 mm NaCl, pH 6.8, in 10% D2O, 90% H2O to a final concentration of 0.7 mm DC-SIGNR CRD. NMR experiments were carried out at 37 °C on either a 700-MHz Bruker Avance spectrometer fitted with cryoprobe (University of Warwick) or a 5-mm triple resonance cold probe-equipped Varian Unity Inova 800-MHz spectrometer (Henry Wellcome Building for Biomolecular NMR Spectroscopy, University of Birmingham). Proton chemical shifts were referenced against external DSS, whereas nitrogen chemical shifts were referenced indirectly to DSS using the absolute frequency ratio (24). One-dimensional proton spectra were acquired using the pulse sequence described by Liu et al. (25). Two-dimensional 1H–15N HSQC spectra (26) were recorded with 128 increments in the t1 domain and 1024 data points in the t2 dimension. The sweep width was 18.0 ppm in the 1H dimension and 31.8 ppm in the 15N dimension. The triple resonance (1H-13C-15N) experiments (CBCA(CO)NH (27), CBCANH (28), HNCA (29), HN(CO)CA (30), HNCO (29), and HN(CA)CO (31)) were recorded with 128 increments in the t1 domain, 40 increments in the t2 domain, and 2048 increments in the t3 domain. Spectra were processed using Topspin version 2.0 (unless otherwise stated) and analyzed using CCPN Analysis software version 2.1.5 (32, 33) and SPARKY version 3 (34). Secondary structure predictions based on the chemical shift index were carried out using TALOS+ (35).
Unlabeling of specific residues, by the addition of 100 mg of unlabeled amino acid to double-labeled M9 medium, was used to assign difficult residues (such as the highly mobile residues Asn-102 and Asn-103 in the primary calcium binding loop).
Carbohydrate Titrations
Titration experiments were carried out by adding increasing amounts of carbohydrate (0.2, 0.5, 1.0, 2.0, 5.0, 10.0, and 20.0 mm Man3; 1.0, 2.0, 5.0, and 10.0 mm Man5 and (GlcNAc)2Man3; and 0.1, 0.3, 0.7, 1.0, 1.5, 2.0, 5.0, and 10.0 mm Man9GlcNAc) to 0.7 mm [U-15N,13C]DC-SIGNR CRD at pH 6.8 and acquiring a series of two-dimensional 1H-15N HSQC spectra at 37 °C. The pH and temperature were held constant throughout the experiments. The three sugar fragments reached saturation by 10 mm, whereas relaxation properties prevented the determination of Man9GlcNAc saturation.
The total chemical shift perturbation per residue (Δδtotal) was calculated using Equation 1,
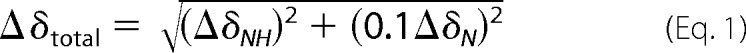 |
where ΔδNH and ΔδN represent the chemical shift differences in the 1H and 15N dimensions, respectively. The weighting factor of 0.1 applied to the nitrogen chemical shift corresponds to the difference in magnetogyric ratios of 15N with 1H nuclei. Although a weighting factor of 0.15 can also be used, Schumann et al. (36) found little difference between the two weighting factors and concluded that either is sufficient. Residues significantly perturbed by the ligand addition were determined by calculating the S.D. of the chemical shift perturbations across all residues for each carbohydrate and using this as a cut-off (36).
To determine the disassociation constant (KD), titration curves were fit to Equation 2, valid for 1:1 complex formation in fast exchange on the NMR chemical shift time scale,
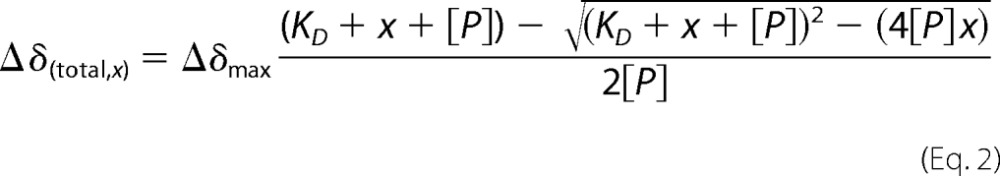 |
where x and [P] represent the ligand and protein concentration, respectively; Δδ(total, x) is the total chemical shift perturbation at ligand concentration x, and Δδmax is the total chemical shift perturbation at saturation of ligand (37, 38). The fit was carried out and analyzed in Origin version 8.5 using the non-linear least squares method.
Backbone Dynamics
15N longitudinal (T1) and transverse (T2) relaxation times were measured using the procedures of Kay et al. (39) and Farrow et al. (40). For T1 measurements, 11 spectra were recorded with relaxation delays between 0.01 and 0.750 s. Matrices of 1024 × 128 complex data points were acquired, using 32 scans per t1 increment and a recycle delay of 3 s. For T2 measurements, 10 spectra were recorded with relaxation delays of 0.0077–0.0850 s. Matrices of 1024 × 128 complex data points were recorded, using 32 (holoprotein) or 64 (Man5-bound protein) scans per t1 increment and a recycle delay of 3 s. Sample heating due to cold probe sensitivity was compensated for by application of continuous wave irradiation to 15N nuclei for a variable time period during the recycle delay (41). Spectral widths of 13,008.1 Hz (1H) and 2500 Hz (15N) were measured. Relaxation data were acquired in an interleaved manner to minimize the effects of sample heating, and three repeat measurements for each of the T1 and T2 data sets were made for determination of peak height uncertainties. Relaxation spectra were processed in NMRpipe (42), and peak heights were calculated and fit to a monoexponential decay in SPARKY 3 (34).
RESULTS
Assignment of Ca2+-bound DC-SIGNR CRD
To investigate the structure, dynamics, and ligand-binding modes of the human DC-SIGNR CRD domain in solution, solution state NMR was used. A 0.7 mm sample of the recombinant 138-residue fragment (containing CRD residues 262–399) was prepared with uniform 13C/15N labeling in the presence of 4 mm Ca2+ (pH 6.8). The purity, secondary structure, and oligomeric state were probed using SDS-PAGE, mass spectrometry, and circular dichroism, and in all cases, we observed pure, monomeric protein. Purification of the CRD using a mannose-Sepharose column served as an effective screen for correct and functional folding of the CRD because incorrectly folded protein fails to bind to immobilized mannose on the column and elutes much earlier.
The 1H-15N HSQC spectrum of the calcium-bound DC-SIGNR CRD is shown in Fig. 2, and the sequential assignment of the N, C, and H nuclei along the protein backbone was achieved using a full suite of triple resonance experiments (see “Experimental Procedures”). The assignments are given in Fig. 2 and supplemental Table S2. Of the 138 amino acids in the CRD, 5 proline residues do not appear in the HSQC spectrum due to their lack of a backbone amide group, and 17 residues (8 at the N terminus and 9 at the C terminus) were also not present. Of the observable residues present in the HSQC, 96% have been unambiguously assigned (80% of the total CRD).
FIGURE 2.
Assignment of DC-SIGNR CRD. Shown are the HSQC spectrum and annotated backbone assignment of holo-DC-SIGNR CRD (20 mm HEPES-d18, 20 mm NaCl, 4 mm CaCl2, pH 6.8, 37 °C), showing 96% of the observed 1H, 15N, and 13C resonances assigned.
The C, H, and N chemical shift assignments (deposited at BMRB, ID 19297) facilitated determination of secondary structure using the TALOS+ program (35) to designate regions containing α-helical/β-sheet structure based on dihedral angle predictions. The secondary structure prediction is in good agreement with crystal data (Protein Data Bank code 1K9J) (supplemental Fig. S2). Therefore, we have used published crystal structures as a foundation for comparison of solution data in this work.
Binding Modes for Three Glycan Fragments in Complex with DC-SIGNR CRD
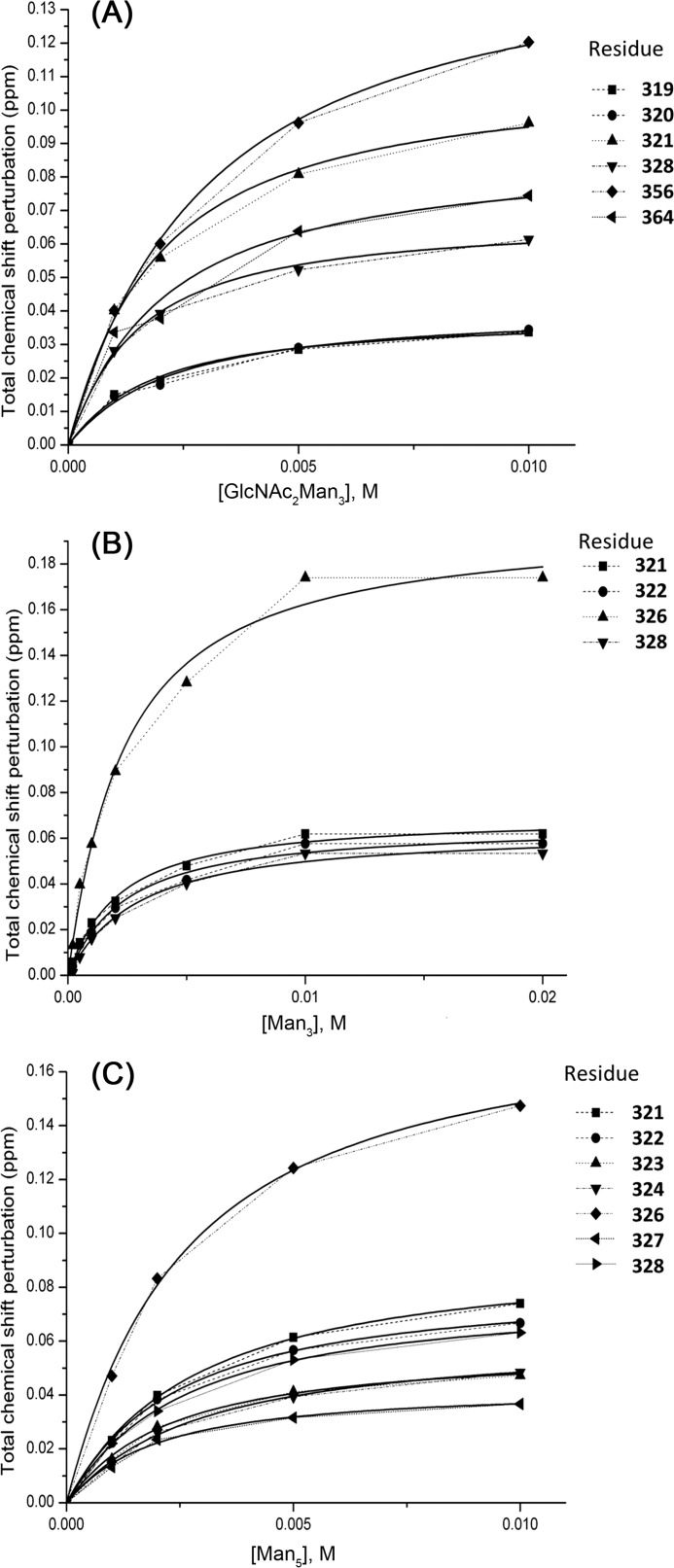
The HSQC spectrum shown in Fig. 2 provides a “map” of the CRD in the calcium-bound state. To investigate a range of glycan-CRD interactions in solution, with an eye toward difficult-to-crystallize glycans, a series of HSQC spectra were acquired upon titration of the oligosaccharide fragment Man3, Man5, or (GlcNAc)2Man3 (see Fig. 3) into the DC-SIGNR CRD. Analysis of (GlcNAc)2Man3, the sugar present in the 1K9J structure, allowed the direct comparison of results in solution with published crystal data. As shown in Fig. 4 and supplemental Figs. S3–S5, binding of all three sugars resulted in significant chemical shift perturbations along the length of the CRD. The S.D. of the chemical shift perturbation across all residues (dashed line in Fig. 4) was used (36) to determine the residues most affected by binding. Perturbations above this threshold were considered significant and are mapped onto the (GlcNAc)2Man3-CRD structure (Protein Data Bank code 1K9J) as shown in Fig. 5.
FIGURE 3.
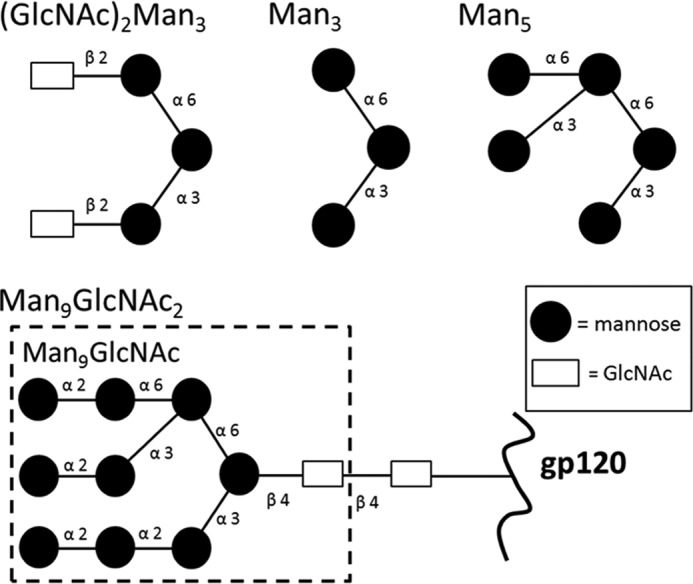
Schematic representation of glycans used in this study. Binding of all four glycans to the DC-SIGNR CRD was measured using chemical shift perturbation analyses.
FIGURE 4.
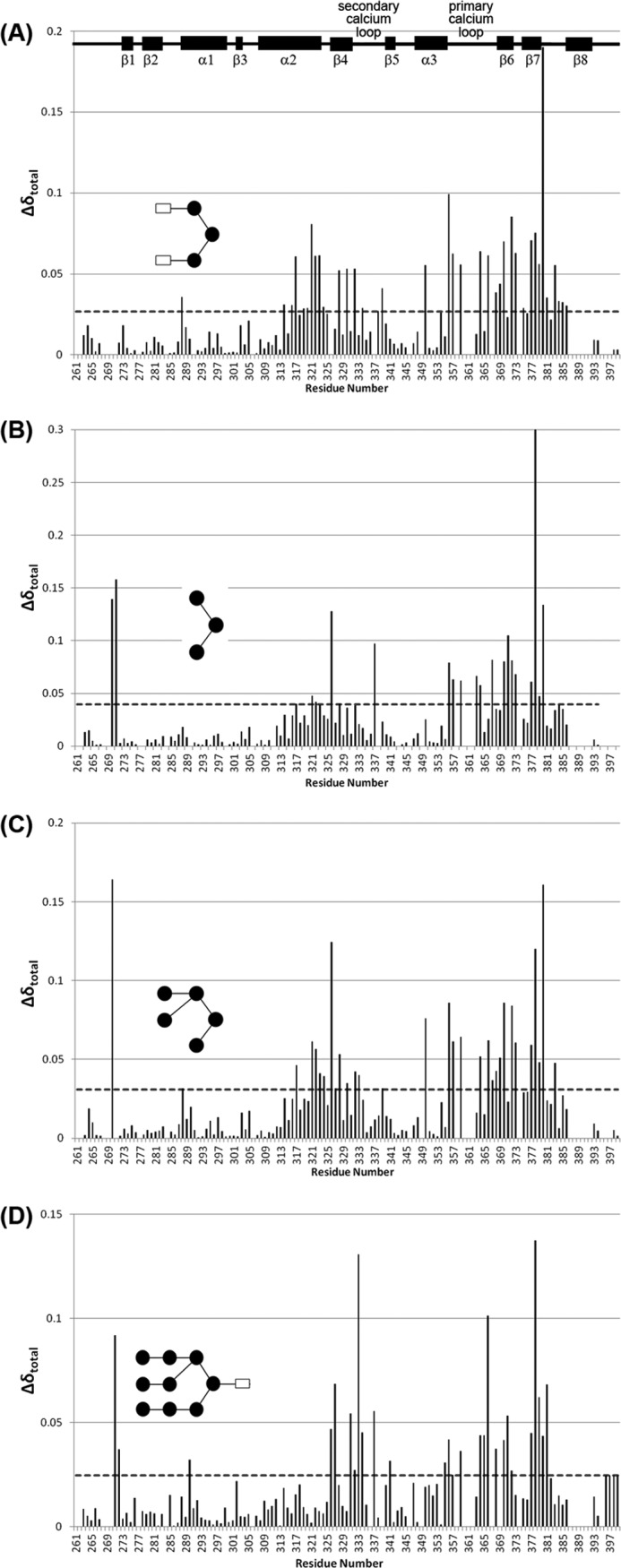
Chemical shift perturbation upon ligand-binding. Total chemical shift perturbation per residue upon the addition of (GlcNAc)2Man3 (A), Man3 (B), Man5 (C), and Man9GlcNAc (D) was calculated according to Equation 1. Dashed horizontal lines represent the 1 × S.D. cut-off used in each data set, above which a change was considered significant.
FIGURE 5.
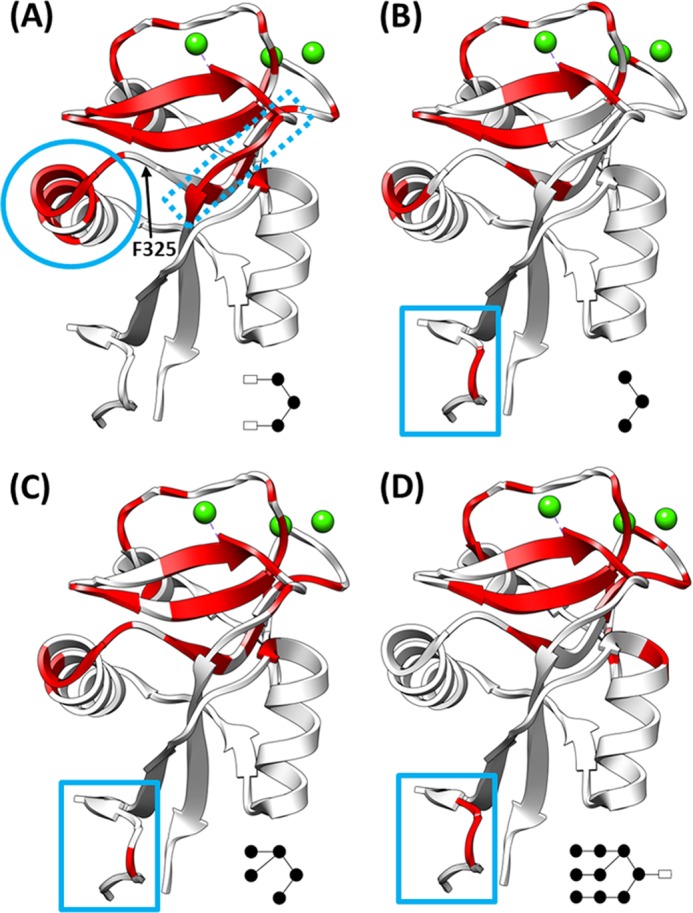
Regions affected by glycan binding. Chemical shifts with perturbations greater than 1 × S.D. upon the addition of 5 mm (GlcNAc)2Man3 (A), Man3 (B), Man5 (C), and Man9GlcNAc (D) are shown in red mapped onto the structure of the (GlcNAc)2Man3·CDR complex (Protein Data Bank code 1K9J). A schematic of the bound glycan is given in the lower right corner of each panel. These maps highlight the conserved binding regions as well as regions unique to each glycan.
Binding of all three sugar fragments was consistent with a principal glycan binding site composed of the primary Ca2+-binding loop (residues 356–364), previously proposed to directly interact with the glycan (3, 4, 19, 20), and residues in β-sheets 6 and 7 at the core of the protein fold (residues 368–379; Fig. 1B).
However, binding of the glycan fragments was not universally consistent with the proposed “shelf” formed by α-helix 2 and Phe-325. The three glycan fragments show marked differences in the degree of perturbation of residues in α-helix 2 (residues 308–323), as shown by the circle in Fig. 5A. Only binding of (GlcNAc)2Man3, the glycan bound in the 1K9J structure, yielded appreciable chemical shift perturbations in α-helix 2 (specifically residues 314–324). Even then, the Phe-325 residue in α-helix 2 shown to form a direct contact with (GlcNAc)2Man3 in the crystal, was not perturbed significantly in solution. Even fewer perturbations in α-helix 2 were observed upon binding of Man3 and Man5, and neither sugar induced any perturbation of the Phe-325 peak. However, Man3 and Man5 binding significantly affect Ser-326, adjacent to Phe-325, whereas (GlcNAc)2Man3 binding does not.
The chemical shift perturbations observed here in solution also highlighted changes in additional regions of the CRD, distal to the proposed glycan binding site. Man3 and Man5 binding induced perturbation of residues 270 and 271 at the N terminus of the CRD (solid box in Fig. 5, B and C), whereas (GlcNAc)2Man3 binding exhibited unique perturbations in the loop region consisting of residues 382–385 (Fig. 5A, dashed box). This suggests that perturbations in this region may be due to the GlcNAc moieties, possibly in conjunction with their β-(1,4) linkages to the trimannose core. These residues were not highlighted in previous glycan-binding studies, suggesting that a conformational or dynamics change is taking place in this region.
More broadly, when our results are compared with the existing structural data, the number of chemical shifts affected by ligand binding in all cases is greater than was expected based on the size of the canonical binding site in the crystal (see summary in supplemental Table S1). This, along with the fact that there are chemical shift perturbations distal to the canonical glycan binding surface, suggests that significant changes in conformation and/or dynamics occur upon ligand binding in solution. This is in contrast to results obtained from crystallography, which yield virtually identical average structures for the free and ligand-bound states (4, 43) (see Fig. 1A for an overlay of two representative structures for these opposing states).
Binding of Man9GlcNAc Derived from HIV gp120 to the DC-SIGNR CRD
To extend current knowledge of the DC-SIGNR CRD to binding of more complex, physiologically important, and/or disease-associated ligands and provide a better understanding of DC-SIGNR-HIV interactions, similar titration experiments were carried out using Man9GlcNAc derived from the gp120 protein of HIV. Man9GlcNAc is very closely related to Man9GlcNAc2 on HIV gp120, differing by a single GlcNAc unit at the reducing terminus, which would be anchored to the polypeptide backbone and hence less likely to play a crucial role in DC-SIGNR binding (Fig. 3).
Similar to the glycan fragments, chemical shift perturbation data upon Man9GlcNAc binding (Figs. 4D and 5D and supplemental Fig. S6) is consistent with a principal binding site containing residues in β-sheets 6 and 7 and the primary calcium binding loop. The same effects on distal regions of the CRD (e.g. N-terminal residues 270–271; solid box in Fig. 5D) are also observed.
Man9GlcNAc binding results in no chemical shift changes for residues in the region thought to compose a “shelf” formed by α-helix 2 and Phe-325. This is interesting because all crystal structures (on small fragments of DC-SIGNR and the homologous protein DC-SIGN) highlight this shelf as forming part of the extended binding site. The NMR data presented here show that, in solution, α-helix 2 is involved in binding to (GlcNAc)2Man3 (and possibly to the smaller glycan fragments), which is in good agreement with 1K9J structure. However, α-helix 2 is not involved in binding to Man9GlcNAc, suggesting that it has a mode of binding to the CRD different from that of (GlcNAc)2Man3 and that DC-SIGNR may interact with the HIV glycoprotein gp120 in a way different from that observed in the(GlcNAc)2Man3-CRD complex. Ongoing work to determine a high resolution solution structure of DC-SIGNR CRD bound to this ligand will shed further light upon this.
Ligand Binding Affinities in Solution
NMR titration data were also used to provide detailed affinity information. Supplemental Figs. S3–S5 show the HSQC spectra of the CRD acquired at increasing concentrations of (GlcNAc)2Man3, Man3, or Man5, respectively. The three sugar fragments behaved similarly upon titration, displaying linear chemical shift perturbations and no line broadening as ligand concentration was increased. This behavior is characteristic of fast exchange between free and bound protein on the NMR chemical shift time scale and suggests that the interaction between the CRD and sugar fragments is weak. This weak binding of our small sugar fragments is unsurprising in light of several reports that DC-SIGNR binds preferentially to larger, highly branched oligosaccharides (4, 19, 43). Fitting the chemical shift perturbations to a 1:1 binding model (see “Experimental Procedures”), which produced the best fits to the data, provides estimates of the dissociation constants for each sugar (Fig. 6). Table 1 shows that (GlcNAc)2Man3, Man3, and Man5 all bind with similar, weak affinities. KD values ranged from 1.57 to 2.2 mm. This value is considerably weaker than binding of similar simple sugars to other lectin CRD domains, such as galectin-1 (lactose bound with a KD of 40 μm (44) to 520 μm (45)), galectin-3 (lactose bound with a KD of 231 μm (46)), BclA (methyl-α-d-mannoside bound with a KD of 2.75 μm (47)), and the asialoglycoprotein receptor (binding constants of 66–539 μm were reported for a variety of simple sugars (48)). However, these KD values are in line with the IC50 values of the analogous protein DC-SIGN for fucose (1.2 mm) and mannose (1.8 mm) (16).
FIGURE 6.
Affinity of glycans for CRD. Shown is a fit of chemical shift perturbations versus glycan concentration to a single site binding model for (GlcNAc)2Man3 (A), Man3 (B), and Man5 (C). Only data derived from residues near α-helix 2 are shown.
TABLE 1.
Dissociation constants calculated from chemical shift perturbations
| Glycan | KD |
|---|---|
| mm | |
| (GlcNAc)2Man3 | 1.57 ± 0.46 |
| Man3 | 2.04 ± 0.54 |
| Man5 | 2.20 ± 0.43 |
Unlike the glycan fragments, binding of Man9GlcNAc caused substantial broadening and disappearance of a number of CRD peaks (supplemental Fig. S6), consistent with intermediate exchange on the NMR chemical shift time scale (49, 50). This suggests higher affinity binding of Man9GlcNAc compared with the sugar fragments (because we move from fast exchange to intermediate exchange as the lifetime of a complex is increased) and supports previous studies reporting a Ki value of 200 μm (21) for the DC-SIGNR·Man9GlcNAc2 complex. Studies of other lectins (e.g. galectin-1) also report increased affinity for larger, more complex glycans (45). However, due to the severe line broadening at the highest Man9GlcNAc concentrations (5 mm), an accurate KD could not be estimated using NMR under these conditions.
NMR Dynamics
The high degree of dynamics in the DC-SIGNR CRD was first suspected after measurement of the HSQC spectrum of the Ca2+-free (apo) form (supplemental Fig. S7), in which variable signal intensities prevented further study using three-dimensional NMR methods. Weak or missing signals suggest that, in its Ca2+-free form, the protein can exchange between an ensemble of conformational states with a rate corresponding to the intermediate exchange regime on the NMR chemical shift time scale. The addition of Ca2+ to the CRD improved the spectrum; however, the variable signal intensity was only satisfactorily minimized after also raising the temperature from 25 to 37 °C, highlighting the intrinsically dynamic nature of the DC-SIGNR CRD.
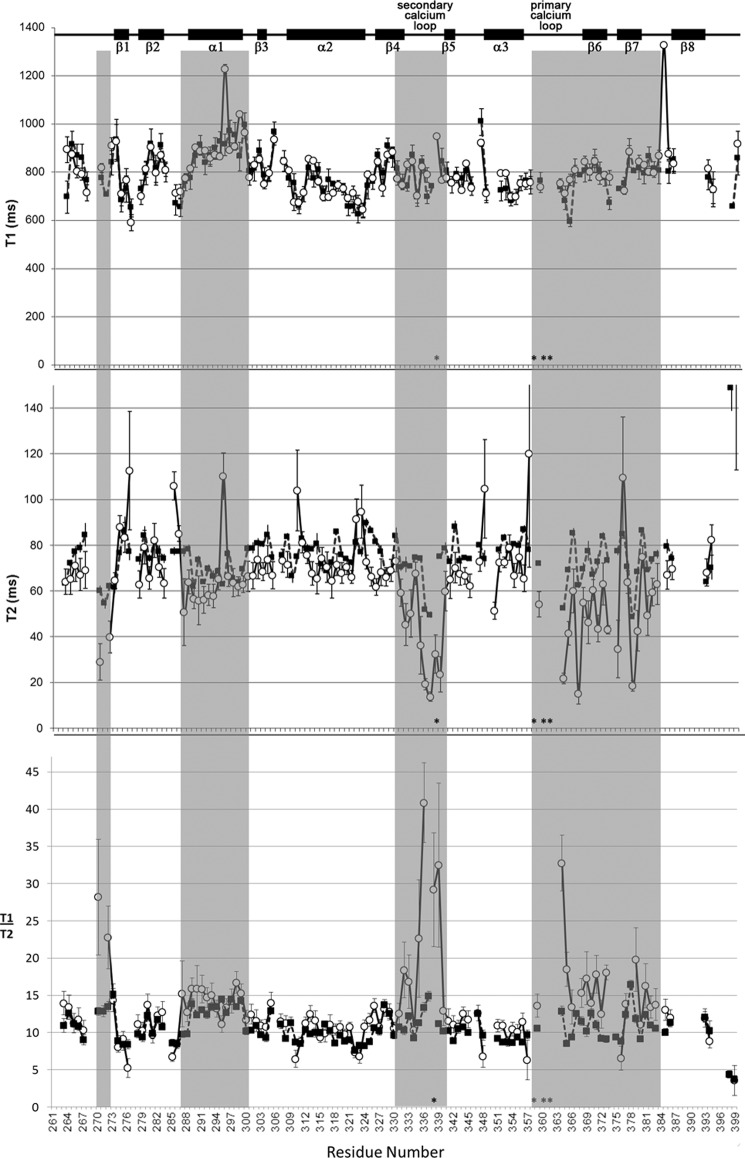
Per residue 15N T1 and T2 relaxation times were measured for the CRD in the absence (holo) and presence of 10 mm Man5 (Fig. 7 and Table 2) to map regions of the CRD where dynamics is altered upon ligand binding. Man5 was selected for this study because binding of Man9GlcNAc produced severely exchange-broadened spectra, preventing accurate measurement of relaxation parameters. The T1 data are very similar for the holo-state and Man5-bound state, showing a similar trend across the protein, and average values of 788.26 ± 22.4 ms (holo) and 806.22 ± 40.35 ms (Man5-bound). Larger differences were observed in the transverse relaxation time constants (T2). For the holo-CRD, although most of the T2 values fall near the average (76.27 ± 2.27 ms), residues in both Ca2+ binding loops, in β-sheets 6 and 7, and at the N terminus (see shaded regions in Fig. 7) display significantly shorter T2 relaxation times, suggesting that these regions are undergoing motions on the micro- to millisecond time scale due to conformational exchange processes (51). A similar trend is seen in the T2 data for the Man5-bound CRD but with a slightly lower average T2 (66.54 ± 8.23 ms) and more pronounced reduction of T2 values for the shaded regions in Fig. 7. These data suggest that micro- to millisecond motions present in the holo-form of the CRD still persist upon glycan binding, albeit at a slightly increased rate.
FIGURE 7.
Dynamics of the CRD. Shown is a plot of per residue values for 15N T1 (top), 15N T2 (middle), and 15N T1/T2 (bottom) for holo-DC-SIGNR CRD (solid squares) and Man5-bound (open circles) DC-SIGNR CRD. Asterisks along the bottom of each panel denote residues that are not observed due to fast relaxation or exchange.
TABLE 2.
Average relaxation parameters of holo-CRD and Man5-bound CRD
| Holo-CRD | Man5-bound CRD | |
|---|---|---|
| Average T1 (ms) | 788.26 ± 22.4 | 806.22 ± 40.35 |
| Average T2 (ms) | 76.27 ± 2.27 | 66.54 ± 8.23 |
| Average T1/T2 | 10.67 ± 0.43 | 13.19 ± 1.75 |
| Rotational correlation time (τc) (ns) | 10.4 ± 0.4 | 12.5 ± 1.36 |
The asterisks in Fig. 7 indicate residues in the Ca2+ binding loops whose signals are so severely broadened as to be unobservable in these experiments, supporting the rapid relaxation of these regions. Specifically, these residues included Glu-359, Asn-361, and Asn-362 in the primary Ca2+ binding loop, which make up the EPN motif conserved among all mannose-binding C-type lectins (52). This binding at the primary Ca2+ site is well characterized (it is a distinguishing feature of C-type lectin binding), and it has been confirmed that the EPN sequence is responsible for mannose specificity (52). In addition to the EPN motif, residues across the entire primary Ca2+ binding loop and β-sheets 6 and 7 display enhanced transverse relaxation in both the holo-form and ligand-bound form. For the holo-form, the increased exchange contribution is possibly driven by the kinetics of Ca2+ binding, confirming that these regions are near the calcium binding sites. A further reduction in T2 values of residues in the primary calcium binding loop are observed upon the addition of Man5. Because there is little change in the average T2 value between holo-form and Man5-bound form, this enhanced relaxation is probably due to an increased conformational exchange contribution as a result of Man5 binding kinetics or hindered motions as a result of Man5 interacting with the principal binding site. This supports previous reports that this is the site of key CRD-mannose interactions. Interestingly, no dynamics changes were observed for residues in α-helix 2 (thought to form the extended glycan-binding “shelf”) upon the addition of Man5.
The relaxation data presented here also highlighted new regions in the CRD that have not yet been implicated in binding to sugars, namely the secondary calcium binding loop and α-helix 1. Residues all along the length of α-helix 1 show a subtle but significant reduction in T2 (as compared with the average) in the holo-CRD and a further reduction upon binding of Man5. α-Helix 1 is positioned toward the N terminus of the CRD, where we also see increased transverse relaxation rates for residues 269–272. The secondary calcium binding site also responds to glycan binding, despite the fact that this region has not (to our knowledge) been implicated in glycan binding previously. In the holoprotein, the reduced T2 values in this region (Thr-337 was broadened beyond detection) were attributed to slow internal motions of the loop (on the micro- to millisecond time scale) upon binding of Ca2+. The further enhancement in transverse relaxation rates upon ligand binding is less obvious because this region of the CRD has not been shown previously to form part of the extended glycan binding site.
The average T2 value decreased significantly from 76.27 to 66.54 ms upon ligand binding. We have attributed this reduction in T2 to slower tumbling of the ligand-bound protein compared with the holoprotein in solution. This was confirmed by using the T1/T2 ratio (Fig. 7) to estimate the overall rotational correlation time (τc) by first excluding residues that contained values more than one S.D. from the average (and thus experience a significant contribution from either chemical exchange or internal motion (53)) and then calculating as described (54). A value for the relaxation-derived τc of 10.4 ± 0.4 ns was obtained for the holo-CRD, which is only slightly longer than the expected value of 8.55 ns obtained using the general rule of 0.5 ns τc per 1 kDa of molecular mass (53–55). This deviation from the ideal value is not large enough to infer oligomerization of the CRD, but it may reflect a non-spherical shape of the monomeric protein. There is a ∼17% increase in the rotational correlation time from 10.4 to 12.5 ns upon binding of Man5 to the CRD (Table 2). Although we acknowledge that a small (∼2% as estimated using a published model (56)) increase in solution viscosity upon the addition of 10 mm Man5 may contribute to this change, and the additional size imparted by the bound sugar may also yield a very small increase, these two factors are unlikely to fully account for the increase in rotational correlation time. Likewise, binding-induced aggregation would result in a much larger increase in τc, suggesting that the CRD adopts a more “open” conformation as a result of Man5 binding.
In broad terms, comparison of the relaxation and chemical shift perturbation data demonstrate that, although several regions in the CRD display micro- to millisecond time scale dynamics that persist upon glycan binding, many more residues display chemical shift perturbations. This suggests that there is a ligand-induced conformational change in the CRD. The extent of this conformational change warrants further investigation because thus far, no conformational changes have been observed in any published crystal structures for DC-SIGNR CRD upon ligand binding.
DISCUSSION
Complex carbohydrate binding events that occur within the human immune system are vital to healthy immune function and proper host responses to a wide variety of pathogens. Greater understanding of this essential glycoimmunology promises to provide important insights into major world health risks, such as HIV, tuberculosis, and Gram-negative multiresistance diseases. C-type lectins represent some of the most important receptors for complex carbohydrates, and their roles in contributing to sophisticated pathogen recognition and cellular response mechanisms are only just beginning to emerge. Structural studies have provided insights into the mechanisms via which C-type lectins assemble and bind to their targets with specificity, displaying a range of strategies, including oligomerization, monosaccharide selectivity, and, in some cases, extended binding sites incorporating multiple protein-glycan contacts. However, characterization of the interaction of larger, disease-associated glycans with C-type lectins (especially HIV-derived Man9GlcNAc with the human C-type lectin DC-SIGNR) has not been reported thus far.
Here we have described the first solution state NMR backbone assignment of the carbohydrate recognition domain of human DC-SIGNR and have used this spectrum as a platform upon which to characterize the solution state binding properties of a variety of glycan ligands, including Man9GlcNAc. We have also used solution NMR methods to begin to characterize the molecular dynamics of the CRD in its free and ligand-bound states for the first time. These data have revealed several interesting properties of the DC-SIGNR CRD summarized below.
Different Binding Modes and Affinities for Small Glycans versus Man9GlcNAc
The C-type lectin family of proteins is striking in that substantial portions of the C-type lectin domain do not adopt regular secondary structure, and typically the ligand binding properties of the C-type lectin domains are located within these nonregular regions. Furthermore, it has been shown that a number of transmembrane human C-type lectins are capable of binding multiple ligands via discrete binding sites and can transduce different intracellular signals through the same receptor molecule, depending upon the type of ligand engaged at the extracellular face (57, 58). Another key feature of the C-type lectin family is its enormous potential for ligand binding diversity, brought about largely through the ability of the C-type lectin domain scaffold to accommodate a substantial variety of nonregular polypeptide loops at several distinct regions within the domain fold (59). It is very likely that, for these regions, the C-type lectin family has evolved into a range of homologous proteins with a very broad spectrum of ligand specificities, including targets of both exogenous and endogenous origin.
The different binding modes for the four glycans studied here, as indicated by four unique patterns of chemical shift perturbations, support the structural plasticity proposed for C-type lectins, which allows them to accommodate a wide range of diverse ligands and heterogeneously glycosylated surfaces (60). All four glycans caused perturbation of protein regions near the principal glycan binding site, namely the primary Ca2+-binding loop and β-sheets 6 and 7. However, each glycan had a unique set of additional perturbations in α-helices 1 and 2, the N terminus of the CRD, the loop region consisting of residues 382–385, and the secondary Ca2+-binding loop. For example, the majority of residues in α-helix 2 were preferentially engaged during binding of (GlcNAc)2Man3 (circled in Fig. 5A), which supported its role as a critical region (along with Phe-325) in forming a “shelf” complementary to (GlcNAc)2Man3 in the binding site. However, this region does not appear to interact with Man9GlcNAc. Overall, the data suggest that the use of small glycans as models for binding of larger, branched physiological ligands should be treated with caution and demonstrate that solution state NMR is highly accommodating, informative, and essential for the design of drug molecules that could inhibit binding of large, disease-associated carbohydrates.
The NMR data presented here also allowed us to report the first dissociation constants for direct binding of the three glycan fragments to the DC-SIGNR CRD. The three glycan fragments displayed 1:1 binding to a single binding site with similar, weak affinities. Man9GlcNAc has a higher affinity for the CRD, as indicated by severe broadening of selected NMR signals unique to this ligand, characteristic of intermediate exchange on the NMR time scale and longer lifetimes for the complex. This higher affinity may explain the fact that Man9GlcNAc (the largest of the ligands tested) yielded the smallest number of chemical shift perturbations (i.e. the binding site in the DC-SIGNR CRD may have evolved around this ligand and does not need to rearrange significantly in order to accommodate it).
NMR Dynamics Reveal a High Degree of Flexibility for the DC-SIGNR CRD and Suggest New Binding Regions
Although several structural analyses of mammalian C-type lectins have revealed substantial spatial information on glycan ligand binding, the level of dynamics data relating to these carbohydrate-binding proteins is surprisingly limited. Given the considerable structural diversity of C-type lectins in nature, especially within regions of nonregular secondary structure, it follows that diversity in the dynamic characteristics of these proteins may play an important role in defining ligand interactions and specificity. Previous studies on the tunicate C-type lectin TC14 have shown that the nonregular sequences in the C-type lectin domain are rigid (61). However, just as primary sequences and ligand specificity for the C-type lectin family are many and varied, so too could be the dynamic properties of the assorted domain family members.
Our data indicate that, unlike TC14, DC-SIGNR shows considerable flexibility within its nonregular sequences, and this may contribute to its ability to interact with large, flexible glycans and transduce intracellular signals. The apo-CRD appeared to be very dynamic, probably exchanging between a broad ensemble of conformations.
Binding of Ca2+ and Man5 leads to enhanced transverse relaxation rates in the primary Ca2+-binding loop and β-sheets 6 and 7, known sites of key CRD-mannose interactions. This rapid relaxation could suggest direct binding and thus more hindered (yet persistent) micro- to millisecond motions in these regions or an enhanced conformational/chemical exchange contribution. This exchange could be compatible with the association/dissociation kinetics of Ca2+ ions or Man5, although there are no existing data in this area. Cis-trans isomerization about the peptide bond of the conserved proline (Pro-360) in the EPN motif (61–65) has been reported for several other C-type lectins in their apo-form, and this could also result in the conformational exchange observed in apo-DC-SIGNR CRD.
Relaxation data also highlighted Ca2+- and Man5-induced changes in the CRD distal to the proposed glycan binding site, namely in the secondary Ca2+-binding loop, α-helix 1, and the N terminus (most pronounced for Asp-271). Previous studies have suggested that Ca2+ binding in the secondary loop is enhanced by glycan binding (43). This type of behavior could explain the reduction in T2 as we go from holo-form to Man5-bound form, with enhanced Ca2+ binding in the secondary loop (in the presence of Man5) leading to a more stabilized loop structure. The secondary Ca2+-binding loop lies in close proximity to the proposed binding site, and given the structural plasticity proposed for C-type lectins, which allows them to accommodate a wide range of diverse ligands, it is also possible that Man5 has a mode of binding to the DC-SIGNR CRD different from that shown for (GlcNAc)2Man3, which includes the secondary Ca2+ binding loop.
The N terminus of the CRD connects to the α-helical neck in the full-length protein, and others have proposed that this region forms a flexible “hinge” (43, 66, 67), allowing the CRD to sample multiple orientations with respect to the neck. Taking this into account, one tentative explanation for the enhanced relaxation in α-helix 1 and the N terminus is that binding of Ca2+ and Man5 increasingly reduces the rate of conformational interconversion of this region. Such dynamic behavior may influence ligand-induced conformational changes throughout the entire DC-SIGNR molecule, including the neck and cytoplasmic region. Alternatively, ligand binding could alter the orientation of the CRD with respect to the neck, promoting multivalent binding by adjacent CRDs. Structural analyses of the human C-type lectin CLEC5A allude to similar possibilities that dynamic changes in the CRD, attributable to distal glycan binding, could contribute to the transmission of conformational information and signaling beyond the target binding site to the intracellular regions of the native polypeptide (68). In the case of DC-SIGNR, a receptor previously believed to be involved primarily in adhesion, evidence of signaling activity has been demonstrated in the context of respiratory syncytial virus glycoprotein binding (9).
Although dynamics data could not be acquired in the case of Man9GlcNAc, binding of this glycan was unique in that NMR spectra displayed severe line broadening, which could be due to extensive micro- to millisecond dynamics in the CRD (compared with Man5-associated CRD). It follows that the larger, higher affinity Man9GlcNAc could restrict the motions of the CRD more than the smaller, low affinity Man5; however, more work is needed to improve the solution behavior of this complex.
Together, the dynamics and chemical shift perturbation data suggest that more residues are affected by ligand binding than can be explained by direct interaction of the protein with the oligosaccharides. Our interpretation of these data leads us to portray the DC-SIGNR CRD as a highly flexible, dynamic domain that can interconvert between a number of conformations over a range of time scales. However, the crystal structures for free and ligand-bound DC-SIGNR CRD (Fig. 1A) do not suggest conformational rearrangement upon ligand binding. Future work will involve solving the solution structures of the holo-state and ligand-bound state in an effort to characterize these conformational changes. An alternative interpretation is that glycans bind to multiple binding sites or experience multivalent binding. For the homologous protein DC-SIGN, crystallography has revealed multiple binding modes for the smaller glycans Man2 and Man6. However, only a single binding mode was observed in crystals of (GlcNAc)2Man3 with DC-SIGNR, and no non-linear chemical shift perturbations or broadening (which would result from multiple binding sites and/or modes) was observed after the addition of the three sugar fragments. The linear chemical shift perturbations also fit very well to a one-site binding model. Man9GlcNAc binding did result in broadening of signals; therefore, we cannot rule out multiple binding modes for this ligand, but taken together with the rest of the data, we conclude that significant changes (both dynamic and structural) are taking place in the CRD as a result of ligand binding in solution that cannot be sampled in the crystal structures. This inherent flexibility may enhance the ability of DC-SIGNR to accommodate a variety of ligands, including those of the HIV envelope.
Acknowledgments
We thank Prof. M. Overduin (Henry Wellcome Building for Biomolecular NMR Spectroscopy, University of Birmingham) and Dr. I. Prokes (Warwick Chemistry) for NMR assistance, Dr. R. Wallis for mannose-Sepharose columns, Snezana Vasiljevic and Camille Bonomelli (University of Oxford) for technical assistance, and Dr. C. Scanlan for guidance on glycan preparation.

This article contains supplemental Tables S1 and S2 and Figs. S1–S7.
The chemical shifts and 15N relaxation data have been deposited in the BioMagnetic Resonance Bank under BMRB accession number 19297 (www.bmrb.wisc.edu/).
- CRD
- carbohydrate recognition domain
- SARS
- severe acute respiratory syndrome coronavirus
- HSQC
- heteronuclear single quantum correlation.
REFERENCES
- 1. Weis W. I., Taylor M. E., Drickamer K. (1998) The C-type lectin superfamily in the immune system. Immunol. Rev. 163, 19–34 [DOI] [PubMed] [Google Scholar]
- 2. Drickamer K. (1997) Making a fitting choice. Common aspects of sugar-binding sites in plant and animal lectins. Structure 5, 465–468 [DOI] [PubMed] [Google Scholar]
- 3. Drickamer K. (1999) C-type lectin-like domains. Curr. Opin. Struct. Biol. 9, 585–590 [DOI] [PubMed] [Google Scholar]
- 4. Feinberg H., Mitchell D. A., Drickamer K., Weis W. I. (2001) Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 294, 2163–2166 [DOI] [PubMed] [Google Scholar]
- 5. Soilleux E. J., Barten R., Trowsdale J. (2000) DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J. Immunol. 165, 2937–2942 [DOI] [PubMed] [Google Scholar]
- 6. Chen Y., Chan V. S., Zheng B., Chan K. Y., Xu X., To L. Y., Huang F. P., Khoo U. S., Lin C. L. (2007) A novel subset of putative stem/progenitor CD34+Oct-4+ cells is the major target for SARS coronavirus in human lung. J. Exp. Med. 204, 2529–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boily-Larouche G., Iscache A. L., Zijenah L. S., Humphrey J. H., Mouland A. J., Ward B. J., Roger M. (2009) Functional genetic variants in DC-SIGNR are associated with mother-to-child transmission of HIV-1. PLoS One 4, e7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan V. S., Chan K. Y., Chen Y., Poon L. L., Cheung A. N., Zheng B., Chan K. H., Mak W., Ngan H. Y., Xu X., Screaton G., Tam P. K., Austyn J. M., Chan L. C., Yip S. P., Peiris M., Khoo U. S., Lin C. L. (2006) Homozygous L-SIGN (CLEC4M) plays a protective role in SARS coronavirus infection. Nat. Genet. 38, 38–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johnson T. R., McLellan J. S., Graham B. S. (2012) Respiratory syncytial virus glycoprotein G interacts with DC-SIGN and L-SIGN to activate ERK1 and ERK2. J. Virol. 86, 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Londrigan S. L., Turville S. G., Tate M. D., Deng Y. M., Brooks A. G., Reading P. C. (2011) N-Linked glycosylation facilitates sialic acid-independent attachment and entry of influenza A viruses into cells expressing DC-SIGN or L-SIGN. J. Virol. 85, 2990–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dejnirattisai W., Webb A. I., Chan V., Jumnainsong A., Davidson A., Mongkolsapaya J., Screaton G. (2011) Lectin switching during dengue virus infection. J. Infect. Dis. 203, 1775–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mari S., Serrano-Gómez D., Cañada F. J., Corbí A. L., Jiménez-Barbera J. (2004) 1D saturation transfer difference NMR experiments on living cells. The DC-SIGN/oligomannose interaction. Angew Chem. Int. Ed. Engl. 44, 296–298 [DOI] [PubMed] [Google Scholar]
- 13. Reina J. J., Sattin S., Invernizzi D., Mari S., Martínez-Prats L., Tabarani G., Fieschi F., Delgado R., Nieto P. M., Rojo J., Bernardi A. (2007) 1,2-Mannobioside mimic. Synthesis, DC-SIGN interaction by NMR and docking, and antiviral activity. Chemmedchem 2, 1030–1036 [DOI] [PubMed] [Google Scholar]
- 14. Angulo J., Díaz I., Reina J. J., Tabarani G., Fieschi F., Rojo J., Nieto P. M. (2008) Saturation transfer difference (STD) NMR spectroscopy characterization of dual binding mode of a mannose disaccharide to DC-SIGN. Chembiochem 9, 2225–2227 [DOI] [PubMed] [Google Scholar]
- 15. Reina J. J., Díaz I., Nieto P. M., Campillo N. E., Páez J. A., Tabarani G., Fieschi F., Rojo J. (2008) Docking, synthesis, and NMR studies of mannosyl trisaccharide ligands for DC-SIGN lectin. Org Biomol. Chem 6, 2743–2754 [DOI] [PubMed] [Google Scholar]
- 16. Timpano G., Tabarani G., Anderluh M., Invernizzi D., Vasile F., Potenza D., Nieto P. M., Rojo J., Fieschi F., Bernardi A. (2008) Synthesis of novel DC-SIGN ligands with an α-fucosylamide anchor. Chembiochem 9, 1921–1930 [DOI] [PubMed] [Google Scholar]
- 17. Guzzi C., Angulo J., Doro F., Reina J. J., Thépaut M., Fieschi F., Bernardi A., Rojo J., Nieto P. M. (2011) Insights into molecular recognition of Lewis(X) mimics by DC-SIGN using NMR and molecular modelling. Org Biomol. Chem. 9, 7705–7712 [DOI] [PubMed] [Google Scholar]
- 18. Prost L. R., Grim J. C., Tonelli M., Kiessling L. L. (2012) Noncarbohydrate glycomimetics and glycoprotein surrogates as DC-SIGN antagonists and agonists. ACS Chem. Biol. 7, 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo Y., Feinberg H., Conroy E., Mitchell D. A., Alvarez R., Blixt O., Taylor M. E., Weis W. I., Drickamer K. (2004) Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 11, 591–598 [DOI] [PubMed] [Google Scholar]
- 20. Feinberg H., Castelli R., Drickamer K., Seeberger P. H., Weis W. I. (2007) Multiple modes of binding enhance the affinity of DC-SIGN for high mannose N-linked glycans found on viral glycoproteins. J. Biol. Chem. 282, 4202–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mitchell D. A., Fadden A. J., Drickamer K. (2001) A novel mechanism of carbohydrate recognition by the C-type lectins DC-SIGN and DC-SIGNR. Subunit organization and binding to multivalent ligands. J. Biol. Chem. 276, 28939–28945 [DOI] [PubMed] [Google Scholar]
- 22. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Dunlop D. C., Bonomelli C., Mansab F., Vasiljevic S., Doores K. J., Wormald M. R., Palma A. S., Feizi T., Harvey D. J., Dwek R. A., Crispin M., Scanlan C. N. (2010) Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology 20, 812–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) H-1, C-13 and N-15 chemical-chift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 25. Liu M. L., Mao X. A., Ye C. H., Huang H., Nicholson J. K., Lindon J. C. (1998) Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J. Magn. Reson. 132, 125–129 [Google Scholar]
- 26. Davis A. L., Keeler J., Laue E. D., Moskau D. (1992) Experiments for recording pure-absorption heteronuclear correlation spectra using pulsed field gradients. J. Magn. Reson. 98, 207–216 [Google Scholar]
- 27. Grzesiek S., Bax A. (1992) An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Reson. 99, 201–207 [Google Scholar]
- 28. Wittekind M., Mueller L. (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α-carbon and β-carbon resonances in proteins. J. Magn. Reson. Ser. B. 101, 201–205 [Google Scholar]
- 29. Kay L. E., Ikura M., Tschudin R., Bax A. (1990) 3-Dimensional triple-resonance NMR-spectroscopy of isotopically enriched proteins. J. Magn. Reson. 89, 496–514 [DOI] [PubMed] [Google Scholar]
- 30. Bax A., Ikura M. (1991) An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the α-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J. Biomol. NMR 1, 99–104 [DOI] [PubMed] [Google Scholar]
- 31. Clubb R. T., Thanabal V., Wagner G. (1992) A constant-time 3-dimensional triple-resonance pulse scheme to correlate intraresidue H-1(N), N-15, and C-13′ chemical-shifts in N-15-C-13-labeled proteins. J. Magn. Reson. 97, 213–217 [Google Scholar]
- 32. Fogh R., Ionides J., Ulrich E., Boucher W., Vranken W., Linge J. P., Habeck M., Rieping W., Bhat T. N., Westbrook J., Henrick K., Gilliland G., Berman H., Thornton J., Nilges M., Markley J., Laue E. (2002) The CCPN project. An interim report on a data model for the NMR community. Nat. Struct. Biol. 9, 416–418 [DOI] [PubMed] [Google Scholar]
- 33. Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) The CCPN data model for NMR spectroscopy. Development of a software pipeline. Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 34. Kneller D. G., Goddard T. D. (1993) UCSF Sparky: An NMR Display, Annotation and Assignment Tool, University of California, San Francisco [Google Scholar]
- 35. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS plus. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schumann F. H., Riepl H., Maurer T., Gronwald W., Neidig K. P., Kalbitzer H. R. (2007) Combined chemical shift changes and amino acid specific chemical shift mapping of protein-protein interactions. J. Biomol. NMR 39, 275–289 [DOI] [PubMed] [Google Scholar]
- 37. Sudmeier J. L., Evelhoch J. L., Jonsson N. B. H. (1980) Dependence of NMR lineshape analysis upon chemical rates and mechanisms. Implications for enzyme histidine titrations. J. Magn. Reson 40, 377–390 [Google Scholar]
- 38. Fielding L. (2007) NMR methods for the determination of protein-ligand dissociation constants. Prog. Nucl. Magn. Reson. Spectrosc. 51, 219–242 [Google Scholar]
- 39. Kay L. E., Nicholson L. K., Delaglio F., Bax A., Torchia D. A. (1992) Pulse sequences for removal of the effects of cross-correlation between dipolar and chemical-shift anisotropy relaxation mechanism on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 97, 359–375 [Google Scholar]
- 40. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and a phosphopeptide-complexed Src homology-2 domain studied by N-15 NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 41. Demers J. P., Mittermaier A. (2009) Binding mechanism of an SH3 domain studied by NMR and ITC. J. Am. Chem. Soc. 131, 4355–4367 [DOI] [PubMed] [Google Scholar]
- 42. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NmrPipe. A multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 43. Snyder G. A., Colonna M., Sun P. D. (2005) The structure of DC-SIGNR with a portion of its repeat domain lends insights to modeling of the receptor tetramer. J. Mol. Biol. 347, 979–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nesmelova I. V., Ermakova E., Daragan V. A., Pang M., Menéndez M., Lagartera L., Solís D., Baum L. G., Mayo K. H. (2010) Lactose binding to galectin-1 modulates structural dynamics, increases conformational entropy, and occurs with apparent negative cooperativity. J. Mol. Biol. 397, 1209–1230 [DOI] [PubMed] [Google Scholar]
- 45. Miller M. C., Nesmelova I. V., Platt D., Klyosov A., Mayo K. H. (2009) The carbohydrate-binding domain on galectin-1 is more extensive for a complex glycan than for simple saccharides. Implications for galectin-glycan interactions at the cell surface. Biochem. J. 421, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Diehl C., Engström O., Delaine T., Håkansson M., Genheden S., Modig K., Leffler H., Ryde U., Nilsson U. J., Akke M. (2010) Protein flexibility and conformational entropy in ligand design targeting the carbohydrate recognition domain of galectin-3. J. Am. Chem. Soc. 132, 14577–14589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lameignere E., Malinovská L., Sláviková M., Duchaud E., Mitchell E. P., Varrot A., Sedo O., Imberty A., Wimmerová M. (2008) Structural basis for mannose recognition by a lectin from opportunistic bacteria Burkholderia cenocepacia. Biochem. J. 411, 307–318 [DOI] [PubMed] [Google Scholar]
- 48. Onizuka T., Shimizu H., Moriwaki Y., Nakano T., Kanai S., Shimada I., Takahashi H. (2012) NMR study of ligand release from asialoglycoprotein receptor under solution conditions in early endosomes. FEBS J. 279, 2645–2656 [DOI] [PubMed] [Google Scholar]
- 49. Keeler J. H. (2005) Understanding NMR Spectroscopy, John Wiley, Chichester, UK [Google Scholar]
- 50. Lian L.-Y., Roberts G. C. K. (2011) Protein NMR Spectroscopy: Practical Techniques and Applications, John Wiley & Sons, Inc., New York [Google Scholar]
- 51. Csizmok V., Felli I. C., Tompa P., Banci L., Bertini I. (2008) Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J. Am. Chem. Soc. 130, 16873–16879 [DOI] [PubMed] [Google Scholar]
- 52. Drickamer K. (1992) Engineering galactose-binding activity into a C-type mannose-binding protein. Nature 360, 183–186 [DOI] [PubMed] [Google Scholar]
- 53. Clore G. M., Driscoll P. C., Wingfield P. T., Gronenborn A. M. (1990) Analysis of the backbone dynamics of interleukin-1 β using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry 29, 7387–7401 [DOI] [PubMed] [Google Scholar]
- 54. Kay L. E., Torchia D. A., Bax A. (1989) Backbone dynamics of proteins as studied by N-15 inverse detected heteronuclear NMR-spectroscopy. Application to staphylococcal nuclease. Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 55. Copié V., Battles J. A., Schwab J. M., Torchia D. A. (1996) Secondary structure of β-hydroxydecanoyl thiol ester dehydrase, a 39-kDa protein, derived from Hα, Cα, Cβ, and CO signal assignments and the chemical shift index. Comparison with the crystal structure. J. Biomol. NMR 7, 335–340 [DOI] [PubMed] [Google Scholar]
- 56. Chirife J., Buera M. P. (1997) A simple model for predicting the viscosity of sugar and oligosaccharide solutions. J. Food Eng. 33, 221–226 [Google Scholar]
- 57. Hibbert R. G., Teriete P., Grundy G. J., Beavil R. L., Reljic R., Holers V. M., Hannan J. P., Sutton B. J., Gould H. J., McDonnell J. M. (2005) The structure of human CD23 and its interactions with IgE and CD21. J. Exp. Med. 202, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gringhuis S. I., den Dunnen J., Litjens M., van der Vlist M., Geijtenbeek T. B. (2009) Carbohydrate-specific signalling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori. Nat. Immunol. 10, 1081–1088 [DOI] [PubMed] [Google Scholar]
- 59. Drickamer K., Taylor M. E. (2005) Targeting diversity. Nat. Struct. Mol. Biol. 12, 830–831 [DOI] [PubMed] [Google Scholar]
- 60. Bonomelli C., Doores K. J., Dunlop D. C., Thaney V., Dwek R. A., Burton D. R., Crispin M., Scanlan C. N. (2011) The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS ONE 6, e23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Poget S. F., Freund S. M., Howard M. J., Bycroft M. (2001) The ligand-binding loops in the tunicate C-type lectin TC14 are rigid. Biochemistry 40, 10966–10972 [DOI] [PubMed] [Google Scholar]
- 62. Ng K. K., Park-Snyder S., Weis W. I. (1998) Ca2+-dependent structural changes in C-type mannose-binding proteins. Biochemistry 37, 17965–17976 [DOI] [PubMed] [Google Scholar]
- 63. Pavlícek J., Sopko B., Ettrich R., Kopecký V., Jr., Baumruk V., Man P., Havlícek V., Vrbacký M., Martínková L., Kren V., Pospísil M., Bezouska K. (2003) Molecular characterization of binding of calcium and carbohydrates by an early activation antigen of lymphocytes CD69. Biochemistry 42, 9295–9306 [DOI] [PubMed] [Google Scholar]
- 64. Nielbo S., Thomsen J. K., Graversen J. H., Jensen P. H., Etzerodt M., Poulsen F. M., Thøgersen H. C. (2004) Structure of the plasminogen kringle 4 binding calcium-free form of the C-type lectin-like domain of tetranectin. Biochemistry 43, 8636–8643 [DOI] [PubMed] [Google Scholar]
- 65. Ho M. R., Lou Y. C., Wei S. Y., Luo S. C., Lin W. C., Lyu P. C., Chen C. (2010) Human RegIV protein adopts a typical C-type lectin fold but binds mannan with two calcium-independent sites. J. Mol. Biol. 402, 682–695 [DOI] [PubMed] [Google Scholar]
- 66. Yu Q. D., Oldring A. P., Powlesland A. S., Tso C. K., Yang C., Drickamer K., Taylor M. E. (2009) Autonomous tetramerization domains in the glycan-binding receptors DC-SIGN and DC-SIGNR. J. Mol. Biol. 387, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Leckband D. E., Menon S., Rosenberg K., Graham S. A., Taylor M. E., Drickamer K. (2011) Geometry and adhesion of extracellular domains of DC-SIGNR neck length variants analyzed by force-distance measurements. Biochemistry 50, 6125–6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Watson A. A., Lebedev A. A., Hall B. A., Fenton-May A. E., Vagin A. A., Dejnirattisai W., Felce J., Mongkolsapaya J., Palma A. S., Liu Y., Feizi T., Screaton G. R., Murshudov G. N., O'Callaghan C. A. (2011) Structural flexibility of the macrophage dengue virus receptor CLEC5A. Implications for ligand binding and signaling. J. Biol. Chem. 286, 24208–24218 [DOI] [PMC free article] [PubMed] [Google Scholar]