Background: Plant cytoplasmic GAPDH undergoes nitrosylation and glutathionylation.
Results: GSNO triggers mainly nitrosylation of Arabidopsis GAPDH, which can be denitrosylated by GSH but not by thioredoxins.
Conclusion: The extent of GAPDH nitrosylation is dependent on the [GSH]/[GSNO] ratio but independent of the [GSH]/[GSSG] ratio.
Significance: The mechanisms of GAPDH nitrosylation are delineated, and the prominent role of glutathione is established.
Keywords: Arabidopsis, Glutathionylation, Nitrosylation, Redox Signaling, Thioredoxin, Denitrosylation, Glutathione, Glyceraldehyde-3-phosphate Dehydrogenase
Abstract
Nitrosylation is a reversible post-translational modification of protein cysteines playing a major role in cellular regulation and signaling in many organisms, including plants where it has been implicated in the regulation of immunity and cell death. The extent of nitrosylation of a given cysteine residue is governed by the equilibrium between nitrosylation and denitrosylation reactions. The mechanisms of these reactions remain poorly studied in plants. In this study, we have employed glycolytic GAPDH from Arabidopsis thaliana as a tool to investigate the molecular mechanisms of nitrosylation and denitrosylation using a combination of approaches, including activity assays, the biotin switch technique, site-directed mutagenesis, and mass spectrometry. Arabidopsis GAPDH activity was reversibly inhibited by nitrosylation of catalytic Cys-149 mediated either chemically with a strong NO donor or by trans-nitrosylation with GSNO. GSNO was found to trigger both GAPDH nitrosylation and glutathionylation, although nitrosylation was widely prominent. Arabidopsis GAPDH was found to be denitrosylated by GSH but not by plant cytoplasmic thioredoxins. GSH fully converted nitrosylated GAPDH to the reduced, active enzyme, without forming any glutathionylated GAPDH. Thus, we found that nitrosylation of GAPDH is not a step toward formation of the more stable glutathionylated enzyme. GSH-dependent denitrosylation of GAPC1 was found to be linked to the [GSH]/[GSNO] ratio and to be independent of the [GSH]/[GSSG] ratio. The possible importance of these biochemical properties for the regulation of Arabidopsis GAPDH functions in vivo is discussed.
Introduction
The crucial importance of redox post-translational modifications (RPTMs)3 in cell signaling and regulation and their association with many diseases is largely recognized (1–7). Reactive oxygen species (ROS) and reactive nitrogen species (RNS) act as signaling molecules to transfer extracellular or intracellular information and elicit specific responses. ROS and RNS can alter the function of proteins by triggering diverse RPTMs on several types of residues (6, 8). Cysteine is the major target of reactive species as its thiol side chain can undergo diverse types of reversible modifications, including oxidoreduction of intra- or interprotein disulfides (SS), nitrosylation (SNO), glutathionylation (SSG), and other types of S-thiolation (e.g. cysteinylation), sulfhydration (SSH), or sulfenylation (SOH). In addition, cysteine residues can be irreversibly oxidized by sulfinylation (SO2H) or sulfonylation (SO3H). These modifications are mainly controlled by small oxidoreductases of the thioredoxin (TRX) and glutaredoxin (GRX) families.
Nitric oxide (NO) is a highly reactive molecule implicated in many signaling processes. NO can be generated by NO synthases (NOS), which convert l-Arg to NO and l-citrulline (three isoforms of NOS are present in mammals), or by the breakdown of nitrite or other compounds to NO, e.g. through cytochrome c oxidase in mitochondria or nitrate reductase in plants (9–11). NO is involved in a post-translational modification named nitrosylation consisting of the formation of nitrosothiols by reaction of NO-related species with protein thiols (12). Several hundred targets of nitrosylation have been identified in bacteria, yeast, plants, and animals, suggesting a role for this modification in many cellular processes (13, 14).
S-Nitrosylation appears to be implicated in a broad spectrum of human diseases, including cancer, diabetes, and several neurodegenerative, cardiovascular, muscular, or respiratory diseases (1, 15–17). Although the enzymes controlling NO production in plants remain to be characterized (18), numerous plant proteins appear to undergo nitrosylation under biotic or abiotic stress conditions (13, 19–22). Nitrosylation appears crucial for plant immune response by controlling the activity and/or subcellular localization of the transcription factors NPR1 (Non-expresser of Pathogenesis-related genes 1) and TGA1 but also the salicylic acid-binding protein 3 (SABP3) (23–25). Nitrosylation also modulates cell death during the hypersensitive response to pathogens through regulation of NADPH oxidase (26) or peroxiredoxin IIE (27).
The extent of nitrosylation of a given cysteine residue is governed by the equilibrium between nitrosylation and denitrosylation reactions. Therefore, protein nitrosylation is dependent on the activity of NO-producing enzymes. Nitrosylation can be triggered chemically by RNS such as the NO radical (NO•), the nitrosonium cation (NO+), the nitroxyl anion (NO−), or peroxynitrite (ONOO−) or by trans-nitrosylation reactions mediated by small nitrosothiols such as nitrosoglutathione (GSNO) or by other nitrosylated proteins (12, 28, 29). Indeed, several nitrosylated proteins are able to act as cysteine trans-nitrosylases by transferring their NO group to target proteins as shown for hemoglobin (30), thioredoxin (31–33), caspase-3 (34), cyclin-dependent kinase 5 (35), or GAPDH (36). Some proteins also mediate transfer of NO from an intrinsically bound heme iron or other transition metal complexes to cysteine thiols (16). These metal-to-Cys nitrosylases mainly catalyze intramolecular NO group transfer (auto-S-nitrosylation, e.g. hemoglobin) or intermolecular reaction with glutathione to form GSNO (e.g. cytochrome c).
GSNO is the main NO reservoir of the cell and a major trans-nitrosylating molecule. GSNO is formed by reaction of glutathione, the main small soluble thiol of the cell, with RNS. GSNO not only induces protein trans-nitrosylation but also protein glutathionylation (37, 38). GSNO concentration is mainly controlled by GSNO reductase, an enzyme widely expressed in prokaryotes and eukaryotes (19, 39, 40). GSNO reductase catalyzes the NADH-dependent reduction of GSNO and, in the presence of GSH, gives rise to GSSG and ammonia. GSNO reductase controls the cellular levels of GSNO and therefore influences indirectly the level of nitrosylated proteins. Several other enzymes have also been suggested to control GSNO levels, including protein-disulfide reductase (41) and thioredoxin (42, 43). Similarly, flavohemoglobins in bacteria and fungi control NO accumulation and also affect indirectly nitrosylation levels (44–46).
The reactions of denitrosylation are much less documented. Two main mechanisms appear to be involved. The first one involves reaction with glutathione that leads to GSNO formation that can be subsequently reduced by GSNO reductase. Consistently, the absence of GSNO reductase leads to a strong increase of protein nitrosylation in mice, yeast, and plants (19, 39, 47). Most protein nitrosothiols (75–80%) were shown to be susceptible to reduction by GSH in rat brain extracts or in rat spinal cords (48, 49). The second mechanism involves mammalian cytoplasmic and mitochondrial TRX, which were shown to denitrosylate numerous proteins (43, 50–54). Other enzymes such as protein-disulfide isomerase, xanthine oxidase, carbonyl reductase, glutathione peroxidase, and superoxide dismutase have been suggested to act as denitrosylases but remain to be confirmed (28).
GAPDH, a glycolytic enzyme, is the most widely used model protein to investigate the molecular mechanisms of RPTMs. Indeed, GAPDH contains a highly reactive catalytic cysteine that undergoes multiple types of RPTM, including SNO, SSG, SSH, SOH, SO2H, or SO3H. All of these modifications inhibit the glycolytic activity of GAPDH, which can therefore be used to measure precisely the extent and the reversibility of a given RPTM. In animal cells, redox modifications of GAPDH can redirect the protein to new and completely unrelated functions. Indeed, this glycolytic enzyme was demonstrated to play a role in several cellular processes such as receptor-mediated signaling, intracellular membrane trafficking, maintenance of DNA integrity, transcriptional and post-transcriptional regulation, oxidative stress response, and apoptosis (55, 56). Several of these functions involve re-localization of GAPDH to the nucleus and are triggered or regulated by redox modifications of the catalytic cysteine. For instance, upon apoptotic stimulation, nitrosylation of GAPDH triggers its binding to the E3 ubiquitin ligase SIAH1, which possesses a nuclear localization signal and mediates translocation of GAPDH to the nucleus (57). The translocation of the GAPDH-SIAH1 complex to the nucleus ultimately leads to apoptosis through several mechanisms involving not only Siah1-dependent ubiquitination and degradation of nuclear proteins but also GAPDH-mediated regulation of gene expression. Indeed, nuclear GAPDH controls the regulation of nuclear genes such as p53, PUMA, BAX, and p21 through interaction with the nuclear acetyltransferases p300 and CBP (58). Moreover, nitrosylated GAPDH was also shown to trans-nitrosylate several nuclear proteins (HDAC, SIRT-1, and DNA-PK (36)).
Plants contain several isoforms of GAPDH encoded by gapA, gapB, and gapC genes. Indeed, in addition to glycolytic GAPDH isoforms (cytoplasmic GAPC1, GAPC2, and plastidic GAPCP1, GAPCP2), A2B2-GAPDH and A4-GAPDH are located in chloroplast stroma and participate in the Calvin-Benson cycle. These chloroplastic GAPDH are likely regulated by different types of RPTMs. We have recently employed two different forms of GAPDH from Arabidopsis thaliana (hereafter referred to as Arabidopsis) as tools to analyze the enzymes and mechanisms controlling glutathionylation/deglutathionylation reactions. Notably, A4-GAPDH and GAPC1 are very sensitive to oxidation and are rapidly irreversibly inactivated by sulfinylation and/or sulfonylation in the presence of ROS such as H2O2 (59, 60). However, both types of enzymes can be protected from irreversible oxidative inactivation by glutathionylation of the catalytic cysteine, a modification occurring by reaction of GSH with the sulfenic acid formed on the catalytic cysteine of A4-GAPDH and GAPC1 after primary oxidation by H2O2 (59, 60). Because of the stable nature of glutathione-protein-mixed disulfide, glutathionylated forms of A4-GAPDH and GAPC1 were used to delineate the mechanisms involved in deglutathionylation reactions catalyzed by oxidoreductases like GRXs and TRXs (60–62).
By contrast, the molecular mechanisms of plant GAPDH nitrosylation remain much less documented. Several proteomic studies have identified cytoplasmic and chloroplastic GAPDH from Arabidopsis as potential targets of nitrosylation (27, 63), and the modification was confirmed in vitro for the Arabidopsis GAPC isoforms (64). More generally, the mechanisms of nitrosylation and denitrosylation remain poorly studied in plants. In addition, it is not known whether plant TRXs can catalyze protein denitrosylation.
In this study, we have employed glycolytic GAPDH isoform 1 from A. thaliana (AtGAPC1) as a tool to investigate the molecular mechanisms of nitrosylation and denitrosylation using a combination of approaches, including activity assays, the biotin switch technique, site-directed mutagenesis, and mass spectrometry. Nitrosylation reactions were induced chemically with a strong NO donor or by trans-nitrosylation with GSNO. The ability of GSNO to trigger both nitrosylation and glutathionylation was investigated, and the site of modification was demonstrated experimentally. The ability and the efficiency of glutathione and different types of plant TRXs to catalyze protein denitrosylation were also examined. Nitrosylation and glutathionylation of GAPC1 were found to be alternative and not inter-convertible RPTMs of the same catalytic cysteine. Denitrosylation of GAPC1 was specifically performed by GSH and found to be directly controlled by the [GSH]/[GSNO] ratio but independent from the [GSH]/[GSSG] ratio.
EXPERIMENTAL PROCEDURES
Materials and Enzymes
PD-10 and NAP-5 columns were obtained from GE Healthcare. N-[6-(Biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide (HPDP-biotin) was purchased from Pierce. GSNO (nitrosoglutathione) and DEA-NONOate (DEA-NO, diethylamine NONOate (diazeniumdiolate) diethylammonium salt) were purchased from Sigma, and their concentration in solution was determined spectrophotometrically using molar extinction coefficients of 920 m−1 cm−1 at 335 nm (GSNO) and 6700 m−1 cm−1 at 250 nm (DEA-NO). All other chemicals were obtained from Sigma. Glutathione reductase (GR) and 3-phosphoglycerate kinase from S. cerevisiae were purchased from Sigma. Recombinant wild-type TRXh1 from C. reinhardtii was prepared as described previously (65). Recombinant NADPH TRX reductase b from A. thaliana and poplar recombinant h-type TRXs (isoforms h 1, h 2, h 3, and h 5) were expressed and purified as described previously (66–69).
Expression and Purification
Cytoplasmic GAPC1 from A. thaliana and the cysteine mutants GAPC1-C149S (C149S) and GAPC1-C153S (C153S) were expressed and purified according to Ref. 60. The subunit molecular mass and purity of the proteins were analyzed by SDS-PAGE (12% gel) and Coomassie Blue staining after desalting on PD-10 columns equilibrated with 50 mm potassium phosphate buffer (pH 7.5). The protein concentration (on a subunit basis) of recombinant proteins was determined spectrophotometrically using a molar extinction coefficient at 280 nm of 40,910 m−1 cm−1. The resulting homogeneous proteins were stored at −20 °C.
Activity Assays
GAPC1 activity was monitored spectrophotometrically at 340 nm and 25 °C by following the reverse (gluconeogenic) reaction assay. The reaction was measured in an assay mixture containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 5 mm MgCl2, 3 mm 3-phosphoglycerate, 5 units/ml of Saccharomyces cerevisiae 3-phosphoglycerate kinase, 2 mm ATP, and 0.2 mm NADH.
Inactivation of GAPC1 by GSNO and DEA-NO Treatments
Before each treatment, wild-type GAPC1 and its cysteine variants were reduced in the presence of 10 mm dithiothreitol (DTT) for 30 min at room temperature (25 °C). DTT was subsequently removed by desalting on NAP-5 columns equilibrated with 50 mm BisTris buffer (pH 7.0). Reduced GAPC1 (2.0 μm or 78 μg/ml final concentration) was incubated in 50 mm BisTris buffer (pH 7.0) with different concentrations of GSNO (0.1, 0.5, and 1 mm) or DEA-NO (0.1, 0.5, and 1 mm) in the presence or absence of 0.14 mm NAD+. At the indicated times, an aliquot of the sample (5–10 μl) was withdrawn for the assay of enzyme activity. The reversibility of GAPC1 inactivation (10 min of incubation with 0.5 mm GSNO or 0.5 mm DEA-NO) was assessed by measuring GAPDH activity after incubation for 15 min in the presence of 20 mm DTT.
MALDI-TOF Mass Spectrometry
Wild-type GAPC1 and the C149S mutant were treated for 30 min either with 1 mm GSNO or with 50 μm H2O2 and 0.5 mm GSH. The MALDI-TOF mass spectrometry analyses were performed before and after incubation with 20 mm DTT for 30 min. The samples were mixed with a saturated solution of sinapinic acid in 30% acetonitrile containing 0.3% trifluoroacetic acid, deposited onto the sample plate, and allowed to dry. Mass determination of WT and C149S GAPC1 was performed as described previously (70).
Reactivation of Nitrosylated GAPC1
Nitrosylated GAPC1 (GAPC1-SNO) was prepared by incubating the enzyme for 30 min in the presence of 1 mm DEA-NO at room temperature and then desalted on a NAP-5 column equilibrated with 50 mm Tris-HCl (pH 7.5) and 1 mm EDTA. Reactivation treatments of GAPC1-SNO were performed in Tris-HCl (pH 7.5) and 1 mm EDTA with 20 mm DTT or in the presence of TRXh isoforms supplemented with 0.22 μm A. thaliana NTRb and 0.2 mm NADPH. Control samples were analyzed under similar conditions by omitting TRXs. The reactivation of GAPC1 was also carried out in the presence of 2 mm GSH at different pH values (7.0, 7.5, and 7.9) or in the presence of 2 mm GSH (pH 7.5) alone or supplemented with 0.5 mm GSSG or with the GR system (6 μg/ml yeast GR and 0.2 mm NADPH). At the indicated times, aliquots (5–10 μl) were withdrawn to assay enzyme activity.
Biotin Switch Technique (BST) to Analyze GAPC1 Nitrosylation/Denitrosylation
The extent of GAPC1 nitrosylation was assessed by the biotin switch assay adapted from Ref. 71. After nitrosylation or denitrosylation treatments, proteins (1 mg/ml) were precipitated with 4 volumes of cold acetone at −20 °C during 30 min and pelleted by centrifugation at 4 °C for 5 min at 15,000 × g. The pellet was resuspended in TENS buffer (30 mm Tris-HCl (pH 7.9), 1 mm EDTA, 100 mm NaCl, and 1% SDS) supplemented with a mixture of alkylating reagents (10 mm iodoacetamide, 10 mm N-ethylmaleimide), to allow blocking of free thiols. After 30 min of incubation at 25 °C under shaking, the samples were precipitated as described above to remove unreacted alkylating reagents. After resuspension in TENS buffer, proteins were incubated in the presence of 40 mm ascorbate and 1 mm HPDP-biotin for 30 min. This step allows reduction of S-nitrosylated cysteines and their derivatization with biotin. Proteins were precipitated to remove unreacted labeling compounds, pelleted by centrifugation as described above, and resuspended in TENS buffer. All steps were performed in darkness until proteins were biotinylated with HPDP-biotin. After final precipitation, proteins were resuspended and quantified using the bicinchoninic acid assay. Proteins were then separated by nonreducing SDS-PAGE, transferred onto nitrocellulose membranes, and analyzed by Western blotting using a primary anti-biotin antibody (1:5,000 dilution; Sigma) and an anti-mouse secondary antibody coupled to peroxidase (1:10,000 dilution; Sigma). Signals were visualized by enhanced chemiluminescence, as described previously (60). A mirror SDS-polyacrylamide gel was prepared from the same samples and stained with Coomassie Brilliant Blue to assess equal loading. All BST assays included a negative control where ascorbate was omitted to prevent reduction of S-nitrosothiols and subsequent biotinylation. This control without ascorbate allows us to assess the efficiency of the initial thiol-blocking step.
Reactivation of GSNO-treated GAPC1 in the Presence of Variable GSH Amounts
GAPC1 (5 μm) was treated with 2 mm GSNO in 50 mm Tris-HCl (pH 7.5) and 1 mm EDTA, and after 30 min of incubation, various amounts of GSH were added to achieve the following [GSH]/[GSNO] ratios: 25, 10, 1, and 0.1. After 10 min of incubation, an aliquot of the samples was withdrawn for the assay of enzyme activity monitored as described above.
Reactivation of Nitrosylated GAPC1 in the Presence of Various [GSH]/[GSSG] and [GSH]/[GSNO] Ratios
GAPC1-SNO was prepared as described above and then desalted on a NAP-5 column equilibrated with 50 mm Tris-HCl (pH 7.5) and 1 mm EDTA. Reactivation treatments were carried out by incubating GAPC1-SNO (5 μm) in 50 mm Tris-HCl (pH 7.5) and 1 mm EDTA in the presence of 2 mm GSH supplemented with variable amounts of GSSG (0.02, 0.08, 0.2, and 2 mm). After 10 min of incubation, an aliquot of the samples was withdrawn for the assay of enzyme activity monitored as described above.
Redox Titrations of GAPC1 in the Presence of Different [GSH]/[GSNO] or [GSH]/[GSSG] Ratios
Reduced GAPC1 (5 μm) was incubated in 50 mm Tris-HCl (pH 7.5) and 1 mm EDTA in the presence of different [GSH]/[GSNO] or [GSH]/[GSSG] ratios at a total concentration of 2 mm. After reaching the equilibrium (30 min), aliquots were withdrawn to assay enzyme activity monitored as described above.
Replicates
All the results reported are representative of at least three independent experiments and expressed as mean ± S.D.
RESULTS
GAPC1 Is Reversibly Inhibited by DEA-NO and GSNO
In A. thaliana, cytoplasmic glyceraldehyde-3-phosphate dehydrogenase is represented by two isozymes, GAPC1 (At3g04120) and GapC2 (At1g13440), that are 98% identical and most probably share common biochemical properties. We have recombinantly expressed and purified GAPC1 from A. thaliana, an enzyme strictly specific for NADH and containing a strictly conserved catalytic cysteine (60). Although this catalytic cysteine is the 155th residue of Arabidopsis GAPC1 sequence, it is named Cys-149 here for congruence with the first solved GAPDH structure (from Bacillus stearothermophilus, Protein Data bank code 2dbv (72)) in which catalytic cysteine is numbered Cys-149.
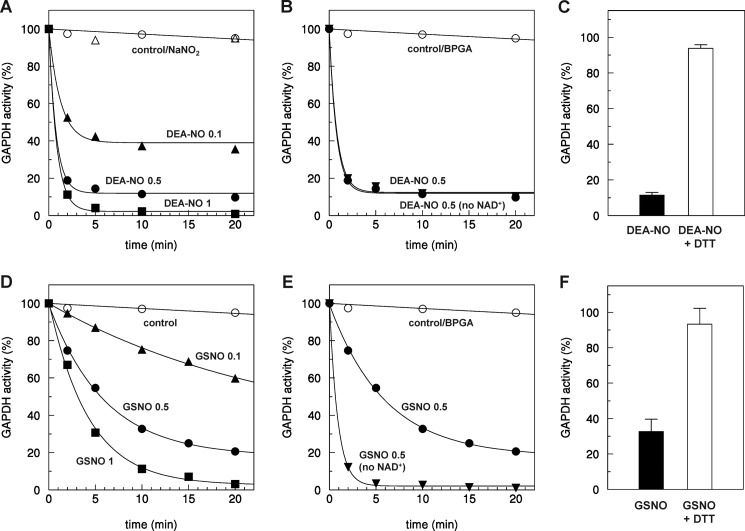
To investigate the regulation of GAPC1 by nitrosylation, the purified enzyme was incubated with different types of NO donors. DEA-NONOate (DEA-NO), dissolved in 10 mm NaOH, behaves as a strong chemical NO donor upon transfer to neutral pH. Typical DEA-NO decomposition rates of 28.8 ± 2.3 μm/min were optically determined in 0.2 mm DEA-NO solutions at neutral pH. In the presence of DEA-NO at pH 7.0, GAPC1 activity was strongly inhibited, although the activity remained high in the buffer alone or in the presence of sodium nitrite (NaNO2), a product of the spontaneous reaction of NO with O2, commonly used for GSNO synthesis (Fig. 1A). The rate and extent of inhibition were dependent on DEA-NO concentration, and GAPC1 was fully inactivated after 5 min of incubation with 1 mm DEA-NO. Lower concentrations of DEA-NO caused rapid but only partial inhibition of GAPC1 (Fig. 1A), probably reflecting the rapid consumption of NO by oxygen. The cofactor NAD, which was present in all incubation media, did not protect GAPC1 from DEA-NO-dependent inhibition (Fig. 1B). By contrast, the substrate 1,3-bisphosphoglycerate, which covalently binds to catalytic Cys-149, allowed full protection of the enzyme suggesting that Cys-149 may be the target of DEA-NO (Fig. 1B). Reaction of DEA-NO with cysteine residues can lead to S-nitrosylation, a modification that can be reversed by the strong chemical reductant DTT. Consistently, the inhibition caused by 10 min of incubation with 0.5 mm DEA-NO was nearly completely reversed by DTT (Fig. 1C).
FIGURE 1.
Inactivation of AtGAPC1 by DEA-NO and GSNO. A, time-dependent inactivation of GAPC1 in the presence of DEA-NO. Reduced GAPC1 (2 μm) was incubated either with buffer (open circles) or NaNO2 (open triangles) as control or with DEA-NO at different concentrations (0.1 mm, closed up-triangles; 0.5 mm, closed circles; 1 mm, closed squares). B, kinetics of inactivation of GAPC1 in the presence of 0.5 mm DEA-NO supplemented or not with 0.14 mm NAD+. C, reversibility of DEA-NO treatment. Reduced GAPC1 was incubated for 10 min with 0.5 mm DEA-NO (black bar). The reversibility of GAPC1 inactivation was assessed by incubation for 15 min in the presence of 20 mm DTT (white bars). NADH-dependent GAPC1 activity was then determined. D, time-dependent inactivation of GAPC1 in the presence of GSNO. Reduced GAPC1 (2 μm) was incubated with buffer as control (open circles) or with GSNO at different concentrations (0.1 mm, closed up-triangles; 0.5 mm, closed circles; 1 mm, closed squares). E, kinetics of inactivation of GAPC1 in the presence of 0.5 mm GSNO supplemented or not with 0.14 mm NAD+. F, reversibility of GSNO treatment. Reduced GAPC1 was incubated for 10 min with 0.5 mm GSNO (black bar). The reversibility of GAPC1 inactivation was assessed by incubation for 15 min in the presence of 20 mm DTT (white bars). NADH-dependent GAPC1 activity was then determined. A, B, D, and E, aliquots of the incubation mixtures were withdrawn at the indicated time points, and the remaining NADH-dependent activity was determined. Data represent the mean percentage (n = 3) of maximal control activity. C and F, NADH-dependent activities of GAPC1 are expressed as mean percentages (n = 3) of the GAPC1 activity measured after 10 min of incubation under control conditions. BPGA, 1,3-bisphosphoglycerate. Except for C and F, S.D. was omitted for clarity.
We also analyzed the effect of the trans-nitrosylating agent GSNO on GAPC1 activity. GSNO was found to inhibit GAPC1 activity, and the inactivation was dependent on GSNO concentration (Fig. 1D). The inactivation appeared slower with GSNO compared with DEA-NO. This lower efficiency was most probably due to the presence of the NAD cofactor, bound to the active site, which partially protected the enzyme from GSNO-dependent inhibition. Indeed, in the absence of NAD, 0.5 mm GSNO led to complete inhibition of GAPC1 after 5 min of incubation. This partial protection is consistent with a higher steric hindrance of the NAD cofactor for GSNO compared with NO (Fig. 1E). GAPC1 incubated for 10 min in the presence of 0.5 mm GSNO could be largely reversed by DTT. All these results are consistent with both DEA-NO and GSNO triggering inhibition of GAPC1 through S-nitrosylation.
GAPC1 Is Nitrosylated on Catalytic Cys-149
The BST allows replacement of labile nitrosothiols by a biotin moiety (71). BST consists of three major steps. 1) All free thiols are initially blocked. 2) Nitrosothiols are specifically reduced with ascorbate. 3) Newly exposed thiols are derivatized with HPDP-biotin. Omission of ascorbate in the second step can be used as control. BST allows direct visualization of protein nitrosylation by Western blot using anti-biotin antibodies.
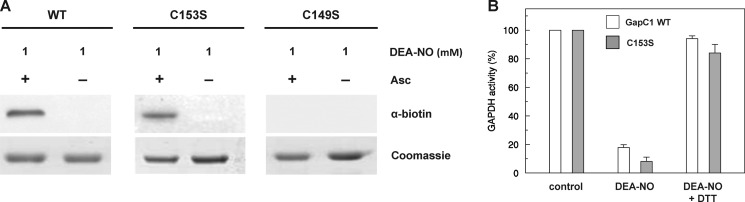
We employed BST to confirm GAPC1 nitrosylation. GAPC1 was incubated for 30 min with 1 mm DEA-NO and analyzed by BST followed by anti-biotin Western blots after separation by nonreducing SDS-PAGE. For wild-type (WT) GAPC1, a strong nitrosylation signal dependent on the presence of ascorbate was observed (Fig. 2A). Arabidopsis GAPC1 only contains two cysteines that are both strictly conserved, catalytic Cys-149 and the closely located Cys-153. Cys-149 is absolutely required for GAPDH catalysis, which implies the acylation of the thiolate Cys-149 by the substrate as an essential step of the catalytic cycle (73). However, Cys-153 has apparently no role in catalysis, despite its close proximity with catalytic Cys-149 (60).
FIGURE 2.
Nitrosylation of GAPC1 and its cysteine variants in the presence of DEA-NO. A, WT GAPC1, C153S, or C149S mutants (25 μm each) were treated for 30 min in the presence of 1 mm DEA-NO, and nitrosylation was visualized using the biotin switch technique followed by anti-biotin Western blots as described under “Experimental Procedures.” The Coomassie Brilliant Blue staining of the gel shows equal loading in each lane. Asc, ascorbate. B, WT GAPC1 or C153S mutants (25 μm) were treated with 1 mm DEA-NO, and after 30 min the residual NADH-dependent activity was assayed. The reversibility of the reaction was assessed by 15 min of treatment with 20 mm DTT. Data are represented as mean percentage ± S.D. (n = 3) of the GapC activity measured after 30 min of incubation under control conditions.
BST was used to analyze the nitrosylation of WT GAPC1 compared with the site-specific mutants C149S and C153S. Whereas no signal could be detected with the C149S mutant, strong and comparable signals were detected with the WT enzyme and the C153S mutant (Fig. 2A). The C149S variant is “virtually” inactive (60) and could not be used for activity assays. By contrast, activity assays were performed on the WT enzyme and the C153S mutant. As shown in Fig. 2B, both WT GAPC1 and the C153S mutant could be inhibited by DEA-NO and reactivated by DTT. Because the C149S mutant is not nitrosylated and the C153S GAPC1 cannot be distinguished from the WT enzyme in BST and activity assays, we conclude that catalytic Cys-149 is the only target of nitrosylation in WT GAPC1.
GSNO Inhibition of GAPC1 Activity Is Essentially Due to Trans-nitrosylation
GSNO can react with cysteine residues to induce not only nitrosylation but also glutathionylation. GAPC1 activity is known to be reversibly inhibited by glutathionylation of catalytic Cys-149 through a mechanism involving reaction of GSH with Cys-149 sulfenate generated by reaction of Cys-149 thiolate with H2O2 (60). As shown in this study for nitrosylated GAPC1, the inhibition of GAPC1 activity by glutathionylation can be reversed by DTT (60). Therefore, it is not possible to distinguish nitrosylation from glutathionylation based on activity measurements because both modifications inhibit GAPC1 activity and are reversed by DTT.
The ability of GSNO to induce GAPC1 nitrosylation was analyzed using BST and anti-biotin Western blots (Fig. 3). After incubation of GAPC1 for 30 min in the presence of 1 mm GSNO, a strong signal, which is absent without ascorbate, was observed. This suggests that GAPC1 undergoes nitrosylation in the presence of GSNO. However, this does not exclude the possibility that GAPC1 also undergoes glutathionylation concomitantly with nitrosylation. Indeed, with the BST assay being qualitative but not quantitative, it is not possible to determine whether the nitrosylation signal detected corresponds to the total pool of GAPC1 molecules present in the samples or only to a fraction of this pool. It is possible that GSNO induces a partial nitrosylation of GAPC1 molecules (even a minor fraction) and leads to glutathionylation of the remaining GAPC1 enzymes. This situation would be consistent with the BST and the activity assays reported above because GAPC1 would be fully and reversibly inhibited by GSNO due to both nitrosylation and glutathionylation.
FIGURE 3.
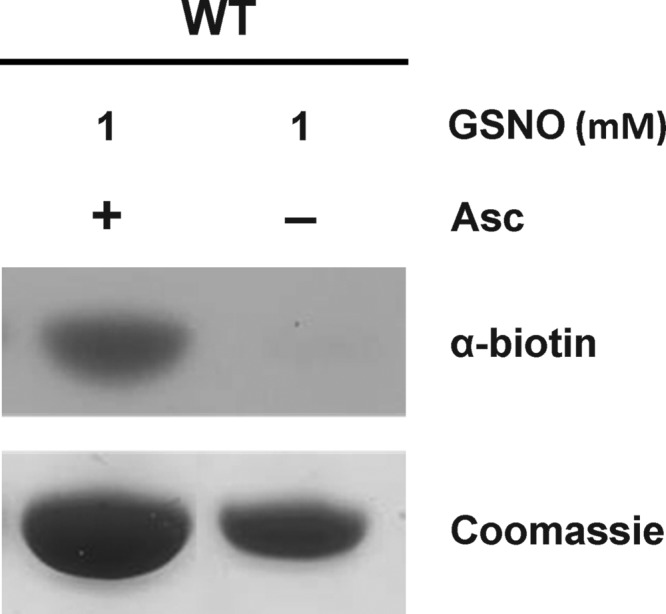
Nitrosylation of GAPC1 in the presence of GSNO. GAPC1 (25 μm) was treated (30 min) with 1 mm GSNO, and nitrosylation was visualized using the biotin switch technique followed by anti-biotin Western blots as described under “Experimental Procedures.” The Coomassie Brilliant Blue staining of the gel shows almost equal loading in each lane. Asc, ascorbate.
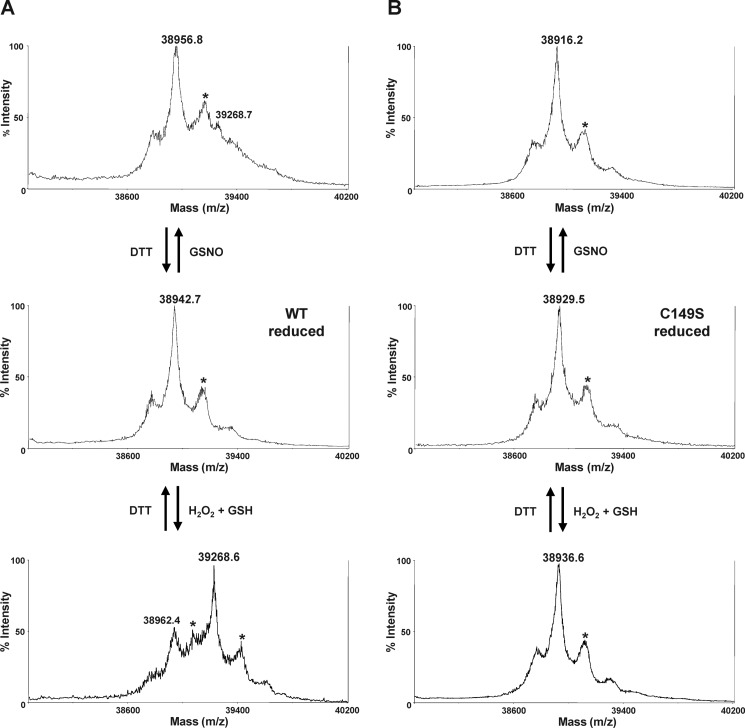
The ability of GSNO to trigger GAPC1 glutathionylation was investigated by MALDI-TOF mass spectrometry. Glutathionylation typically results in a 305-Da shift of the mass of the protein that can be usually observed by MALDI-TOF mass spectrometry. By contrast, the labile SNO bond is lost during the laser-induced ionization process (74), and therefore the nitrosylated protein is not distinguishable from its unmodified form. MALDI-TOF analysis was performed on WT GAPC1 and the C149S mutant after incubation with 1 mm GSNO for 30 min (Fig. 4). The same proteins were also treated with 50 μm H2O2 and 0.5 mm GSH, conditions known to trigger GAPC1 glutathionylation. After GSNO treatment of WT GAPC1, a minor peak (39,268 Da), shifted by ∼305 Da compared with the unmodified/nitrosylated protein, was observed (Fig. 4A, upper and middle panels). The control glutathionylation treatment induced a shift of the main peak of WT GAPC1 to the same size of 39,268 Da corresponding to the glutathionylated form (Fig. 4A, lower panel). In both cases, the shift could be reversed by DTT treatment. This reversible mass increase is consistent with the formation of one glutathione adduct per GAPDH monomer. This indicates that GSNO may induce glutathionylation of GAPC1, although much less efficiently than treating with H2O2 and GSH (glutathionylation treatment). Integration of the area of the minor peak detected after GSNO treatment indicated that glutathionylated GAPC1 represents less than 10% of the total GAPC1 pool. Therefore, the strong inhibition exerted by GSNO on GAPC1 activity (Fig. 1) must depend on effects other than glutathionylation, i.e. nitrosylation (Fig. 2). By contrast with WT GAPC1, the spectra recorded for unmodified or GSNO-treated C149S mutants were comparable. A similar spectrum was also measured after the glutathionylation treatment of C149S (Fig. 4B), confirming that this mutant could not be glutathionylated.
FIGURE 4.
MALDI-TOF mass spectrometry analysis. Mass spectra of wild-type GAPC1 (WT) (A) and C149S mutant (B) treated with 1 mm GSNO for 30 min at room temperature were performed before (upper panels) and after (medium panels) a treatment in the presence of 20 mm DTT (30 min). The lower panels show the spectra of WT and C149S subjected to a typical glutathionylation treatment in the presence of 50 μm H2O2 and 0.5 mm GSH for 30 min at room temperature as described in Ref. 60. The differences between mass peaks of GSNO-treated GAPC1 WT and C149S mutant before (38,942.7 and 38,929.5 Da, respectively) and after DTT treatment (38,956.8 and 38,916.2 Da, respectively) are within the experimental error (0.1%) of the instrument. The peaks marked with asterisks correspond to GAPC1 (WT and C149S mutant) with a sinapinic acid adduct.
Altogether, the results reported above indicate the following: (i) GAPC1 activity is fully inhibited by GSNO; (ii) the inhibition is fully reversed by DTT; (iii) GAPC1 is nitrosylated on Cys-149 based on the BST assay, and (iv) GSNO triggers glutathionylation of the same cysteine in a small fraction of GAPC1, as revealed by mass spectrometry. Therefore, it can be concluded that GSNO triggers both trans-nitrosylation and glutathionylation of GAPC1 catalytic Cys-149, with nitrosylated GAPC1 representing by far the major final product of the reaction.
GAPC1 Can Be Denitrosylated by GSH but Not by Thioredoxin
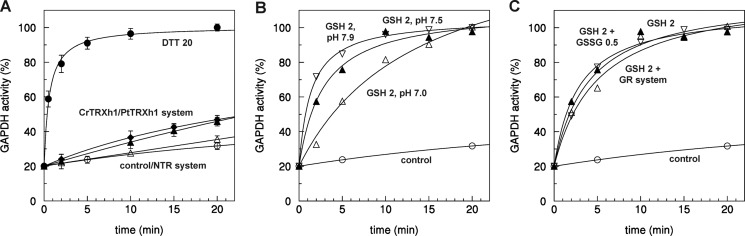
Two main mechanisms, involving either GSH or TRX, have been proposed to control protein denitrosylation reactions (28). We therefore investigated the contribution of these two mechanisms in the control of GAPC1 denitrosylation. GAPC1 was nitrosylated with DEA-NO and desalted to remove unreacted NO donors. In these conditions, the activity of GAPC1-SNO was inhibited by 80% and fully recovered in the presence of 20 mm DTT (Fig. 5A). The partial residual activity in these samples compared with previous assays (Fig. 1) was reminiscent of the desalting step and was most probably linked to changes of protein/NO donor ratio and to the instability of the nitrosothiol bond (e.g. photolysis, trace metals, etc.) during desalting. We first investigated the ability of different cytoplasmic h-type TRXs from both poplar and Chlamydomonas reinhardtii to catalyze GAPC1 denitrosylation. DTT, even at low concentrations, could not be used as a reductant for TRXs due to its fast reaction with GAPC1-SNO. Therefore, we rather used a physiological reducing system for TRXs composed of NADPH and A. thaliana NADPH-thioredoxin reductase b (NTR system). The NTR reducing system alone (Fig. 5A) or in the presence of poplar TRXh2, TRXh3, or TRXh5 (data not shown) had no effect on GAPC1 activity. A very slow reactivation could be observed with the NTR system in the presence of TRXh1 from either poplar or Chlamydomonas (Fig. 5A). By contrast, treatment of GAPC1-SNO with a physiological concentration of GSH (2 mm) allowed a rapid and complete recovery of GAPDH activity comparable with the reactivation kinetics observed with DTT (Fig. 5B). The efficiency of GSH-mediated reactivation decreased with decreasing pH values between 8 and 7. This result is consistent with the estimated pKa value of GSH at 8.7 (75) and indicates that the thiolate form of GSH is involved in the reaction. The full reactivation by GSH of nitrosylated GAPC1 (Fig. 5B) clearly indicates that the GSH thiolate (more abundant at pH 7.9 than 7.0) attacks the N nucleus (and not the S nucleus) of the S-nitrosocysteine. As a result, the GAPC1 cysteine is reduced, and enzyme activity is restored, whereas GSNO is released. Otherwise, if the GSH thiolate attacked the S nucleus of the nitrosocysteine, GAPC1 would be consequently glutathionylated and, as such, would be insensitive to GSH reactivation, as demonstrated previously (60). Hence, full reactivation by GSH of nitrosylated GAPC1 can only be explained by GSH directly removing NO from the catalytic Cys-149.
FIGURE 5.
Kinetics of reactivation of nitrosylated GAPC1. A, nitrosylated GAPC1 was incubated with buffer as control (open circles), with 20 mm DTT (closed circles), with NTR system alone (0.2 mm NADPH and 0.22 μm A. thaliana NTRb, open up-triangles), or in the presence of Chlamydomonas (Cr) TRXh1 (10 μm, closed up-triangles) or poplar (Pt) TRXh1 (10 μm, closed diamonds). Error bars represent average ± S.D. (n = 3). When error bars are not visible, they are within the symbol. B, nitrosylated GAPC1 was incubated with buffer as control (open circles) or in the presence of 2 mm GSH at pH 7.0 (open up-triangles), pH 7.5 (closed up-triangles), or pH 7.9 (open down-triangles). C, nitrosylated GAPC1 was incubated with buffer as control (open circles), with 2 mm GSH alone (closed up-triangles), or in the presence of 0.5 mm GSSG (open down-triangles) or GR system (0.2 mm NADPH and 6 μg/ml glutathione reductase; open up-triangles). For all panels, aliquots of the incubation mixtures were withdrawn at the indicated time points, and the NADH-dependent activity was determined. Data represent the mean percentage (n = 3) of maximal control activity (100% corresponds to the mean maximal reactivation measured after treatment with 20 mm DTT). Except for A, S.D. was omitted for the sake of clarity.
GAPC1 reactivation with 2 mm GSH was not influenced by the addition of glutathione reductase and NADPH (which keeps glutathione fully reduced during the incubation) or, on the contrary, by GSSG (Fig. 5C). These results suggest that GSH-mediated denitrosylation may not be influenced by the presence of GSSG.
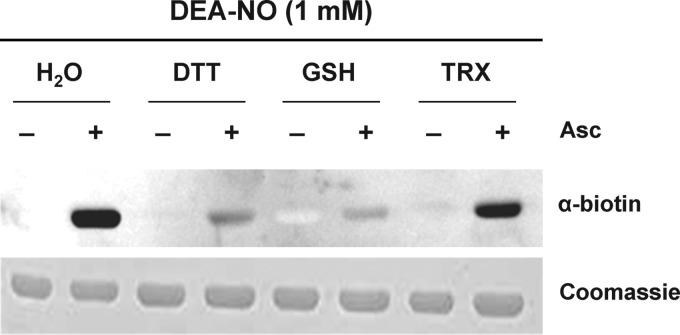
The denitrosylation of GAPC1-SNO by TRX and GSH was also analyzed with the BST assay (Fig. 6). A strong nitrosylation signal was detected for untreated GAPC1-SNO, although only a faint signal remained after treatment of the nitrosylated protein with DTT or GSH. By contrast, the signal was only slightly decreased with the NTR system supplemented with Chlamydomonas TRXh1, consistently with activity assays.
FIGURE 6.
Denitrosylation of GAPC1 in the presence of reducing systems. GAPC1 (25 μm) was treated with 1 mm DEA-NO (30 min) and desalted on NAP-5 columns equilibrated with 50 mm Tris-HCl, 1 mm EDTA. Subsequently, the samples were incubated for 30 min in the presence of buffer, 20 mm DTT, 2 mm GSH, or TRX system (0.2 mm NADPH, 0.22 μm A. thaliana NTRb, and 10 μm Chlamydomonas TRXh1) and subjected to the biotin switch assay. Biotinylated proteins were visualized using anti-biotin Western blots as described under “Experimental Procedures.” The Coomassie Brilliant Blue staining of the gel shows equal loading in each lane. Asc, ascorbate.
All of these results indicate that plant cytoplasmic TRXs are not efficient catalysts of GAPC1 denitrosylation. In contrast, GSH induces rapid denitrosylation of GAPC1 under physiologically relevant concentrations and redox states of the glutathione pool.
GSH-dependent Denitrosylation Is Linked to the [GSH]/[GSNO] Ratio and Independent of the [GSH]/[GSSG] Ratio
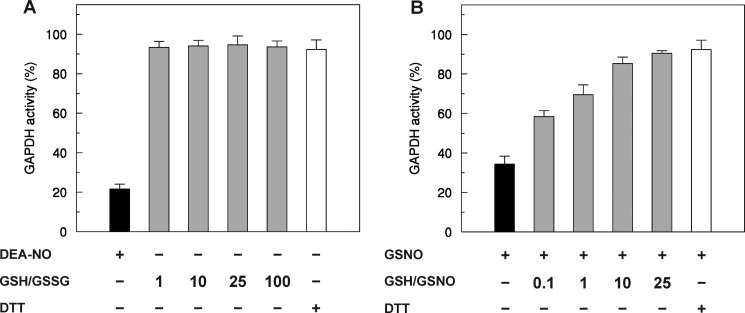
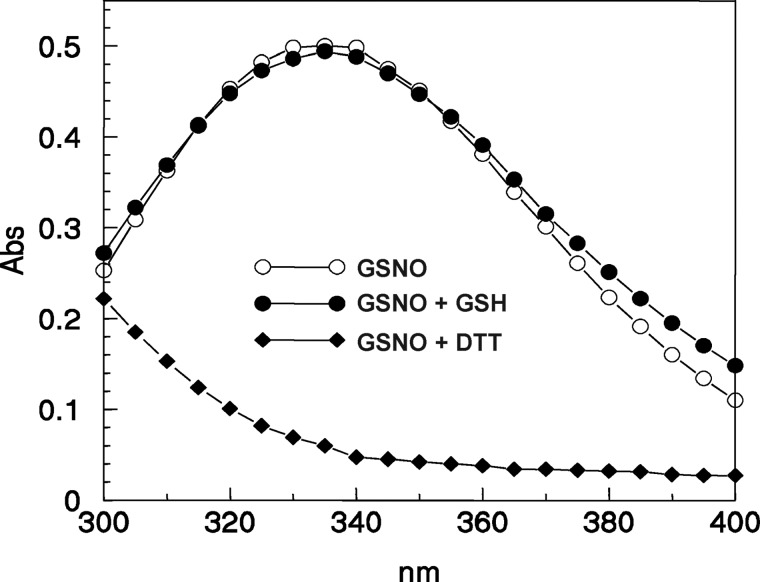
To determine the dependence of GSH-mediated denitrosylation of GAPC1 on the glutathione redox state, GAPC1-SNO was incubated in the presence of different [GSH]/[GSSG] ratios using physiological glutathione concentrations (Fig. 7A). GAPC1 is a protein found within the nucleocytoplasmic compartment, which constitutes a reducing environment where the [GSH]/[GSSG] ratio is maintained in the 10:1 to 100:1 range in diverse organisms. The most oxidizing environment is generally found in the endoplasmic reticulum where the ratio is estimated to be close to 1:1. In vitro, the activity of GAPC1-SNO was efficiently and fully recovered in the presence of GSH under all conditions, even when the [GSH]/[GSSG] ratio was set to 1:1, a very unlikely redox state of the glutathione pool in the nucleocytoplasmic compartment, even under artificial oxidative stress (76–78). These results indicate that GSH-mediated GAPC1 denitrosylation is totally independent of the [GSH]/[GSSG] ratio. This conclusion is consistent with the fact that GSSG is not involved in the GSH-mediated denitrosylation reaction that most probably yields GAPC1-SH and GSNO. By contrast, the recovery of GAPC1-SNO activity increased with increasing [GSH]/[GSNO] ratios (Fig. 7B). Although GSNO (S atom) may react with GSH to release GSSG- and NO-related species, this reaction is very slow (second order rate constant of 0.01 m−1 s−1 at 37 °C) (79). Consistently, we observed no significant decrease of the typical absorbance of GSNO at 335 nm in the presence of GSH, although the absorbance rapidly decreased in the presence of DTT (Fig. 8). However, GSH may also react with the N atom of GSNO, but in this case the products of the reaction (GSNO and GSH) would correspond to the substrates (GSH and GSNO), thus with no effects on [GSH]/[GSNO] ratios. Therefore, we conclude that GSH-dependent denitrosylation of GAPC1-SNO is truly correlated with the [GSH]/[GSNO] ratio. Little is known about GSNO levels and compartmentalization in plant or animal tissues. In a recent study on pepper plants, GSH/GSNO ratios were found in the same order as GSH/GSSG ratios (i.e. 102) in whole organs (roots, stem, leaves; see Ref. 40).
FIGURE 7.
Effect of the [GSH]/[GSSG] and [GSH]/[GSNO] ratios on the reactivation of GAPC1-SNO. A, nitrosylated GAPC1 (black bar) was incubated in the presence of 2 mm GSH supplemented with variable amounts of GSSG (0.02, 0.08, 0.2, and 2 mm) (gray bars). After 10 min of incubation, an aliquot of the samples was withdrawn for the assay of enzyme activity. B, reduced GAPC1 was treated with 2 mm GSNO (black bar), and after 30 min of incubation, various amounts of GSH were added to obtain the indicated [GSH]/[GSNO] ratios (gray bars). After 10 min of incubation, an aliquot of the samples was withdrawn for the assay of enzyme activity. For both panels, the reactivations in the presence of 20 mm DTT are shown (white bars). Data are represented as mean percentage ± S.D. (n = 3) of the initial activity of reduced GAPC1 measured before the nitrosylation treatment.
FIGURE 8.
Stability of GSNO in the presence of DTT or GSH. UV-visible spectra (300–400 nm) were recorded after 30 min of incubation in Tris-HCl (pH 7.5), 1 mm EDTA buffer of 0.54 mm GSNO alone (open circles) or in the presence of 2 mm GSH (closed circles) or 10 mm DTT (closed diamonds).
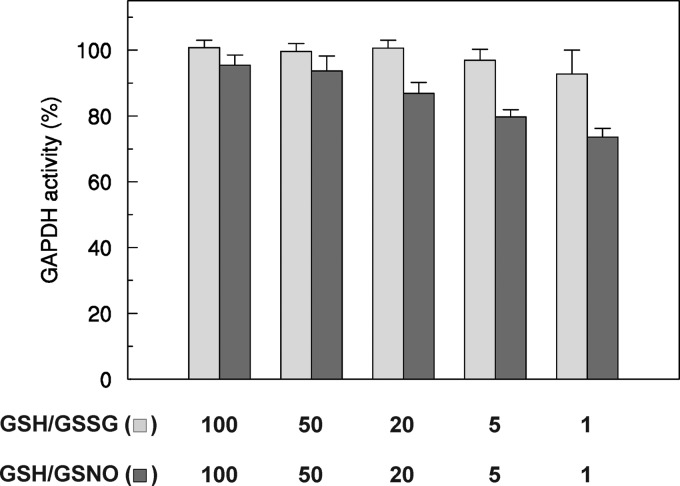
The effects of different [GSH]/[GSSG] and [GSH]/[GSNO] ratios on reduced GAPC1 were also analyzed (Fig. 9). Incubation with the physiological [GSH]/[GSSG] ratio in the 100:1 to 1:1 range had no significant effect on GAPDH activity, although the enzyme was progressively inhibited with decreasing [GSH]/[GSNO] ratios. Overall, these results suggest that GSSG-mediated glutathionylation of GAPC1 does not occur at physiological [GSH]/[GSSG] ratios, although the extent of GAPC1 nitrosylation may be influenced by [GSH]/[GSNO] ratios in the same range (Fig. 9).
FIGURE 9.
Redox titrations of GAPC1 in the presence of various [GSH]/[GSNO] or [GSH]/[GSSG] ratios. Reduced GAPC1 was incubated in the presence of varying [GSH]/[GSNO] (light gray bars) or [GSH]/[GSSG] (dark gray bars) ratios at a total concentration of 2 mm. After equilibrium in the redox buffers was reached (30 min), the remaining NADH-dependent activity was measured. Data are represented as mean percentage ± S.D. (n = 3) of the initial activity of reduced GAPC1 measured before the treatments.
DISCUSSION
This study confirmed that Arabidopsis GAPC1 can be reversibly inactivated by nitrosylation of its catalytic Cys-149. This modification occurs by reaction of reduced GAPC1 (GAPC1-S−) with NO donors or through GSNO-mediated trans-nitrosylation (Fig. 10). This result is consistent with proteomic analyses and targeted studies that identified this enzyme as a nitrosylated protein in Arabidopsis and other species (63, 80–83) and with an in vitro analysis of RPTMs controlling GAPC1 and GapC2 from Arabidopsis (64). In the latter study, the activity of nitrosylated GAPDH was not fully restored by reducing treatment in the presence of DTT suggesting that NO donors might also induce partial irreversible inactivation of Arabidopsis cytoplasmic GAPDH. We show here that NO donors fully inhibit the enzyme activity due essentially to nitrosylation of Cys-149 and that this inactivation is fully reversed by DTT. These biochemical properties of Arabidopsis cytoplasmic GAPDH are similar to those reported for mammalian GAPDH. Therefore, these results open the possibility that Arabidopsis GAPC1 has the ability to participate in nonglycolytic functions, as shown previously for its mammalian counterpart that participates in different regulatory and signaling functions triggered by RPTM of its catalytic cysteine and subsequent nuclear relocalization. Different plant GAPDH isoforms, including Arabidopsis GAPC1, may localize in the nucleus under certain conditions (64, 83–85). Although the function of GAPC1 in the nucleus is still unclear, nuclear relocalization is stimulated by stress conditions that also induce RPTMs of active site GAPDH cysteine (83, 85). Therefore, also in plants, nuclear GAPDH is likely to play completely different roles than catalyzing a glycolytic reaction.
FIGURE 10.
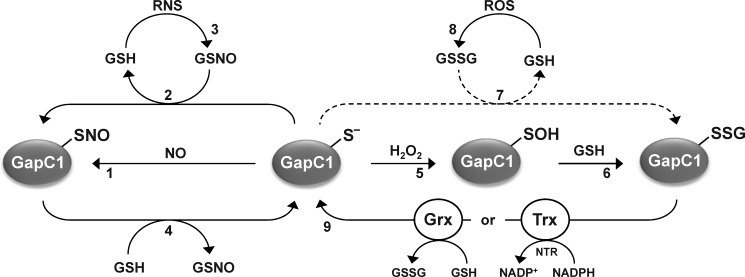
Reactions leading to (de)nitrosylation and (de)glutathionylation of AtGAPC1. Reduced AtGAPC1 (GAPC1-S−) undergoes S-nitrosylation in the presence of NO donors (1) or trans-nitrosylation by GSNO with concomitant release of GSH (2). GSNO can be generated by the reaction of GSH with RNS (3). Denitrosylation of nitrosylated GAPC1 (GAPC1-SNO) is not catalyzed by plant cytoplasmic TRXs, but it is efficiently catalyzed by GSH with formation of GSNO (4), hence controlled by the [GSH]/[GSNO] ratio. AtGAPC1 can undergo primary oxidation to sulfenic acid (GAPC1-SOH) in the presence of H2O2 (5) that can subsequently react with GSH leading to the glutathionylated form (GAPC1-SSG) (6). In principle, GAPC1 glutathionylation may also be performed by GSSG with release of GSH (7), but this reaction does not occur under physiological [GSH]/[GSSG] ratios (dashed arrow). GSSG can be formed by the reaction of GSH with ROS (8). Deglutathionylation of GAPC1-SSG can be catalyzed by either TRX system (i.e. TRX plus NADPH and NADPH:thioredoxin reductase (NTR)) or GRX system (GRX plus GSH) (9). Therefore, glutathionylation of GAPC1 is indirectly controlled by either [NADPH]/[NADP+] and/or [GSH]/[GSSG] ratios.
GSNO was found to induce both nitrosylation and glutathionylation of GAPC1, although nitrosylation was largely prominent. Mammalian GAPDH was also shown to undergo both nitrosylation and glutathionylation in the presence of GSNO and to be preferentially nitrosylated (37, 86). In addition to GAPDH, several enzymes have been reported to undergo both modifications upon GSNO treatment. Although most enzymes are preferentially nitrosylated by GSNO, several enzymes such as creatine kinase (87), cytoplasmic branched chain aminotransferase (88), or papain (37) are mostly glutathionylated in the presence of this compound. Chloroplastic GRXS12 is even exclusively glutathionylated in the presence of GSNO (38, 61). The structural determinants accounting for these preferences remain unclear but likely rely on the local environment of the target cysteine, its influence on the interaction with GSNO, and on the relative stabilities of the nitrosothiol bond and the glutathionylated cysteine. That trans-nitrosylation is the preferred reaction between GAPC1 and GSNO is also consistent with the fact that GSH can denitrosylate GAPC1 with total recovery of enzyme activity, i.e. without glutathionylating GAPC1. Indeed, denitrosylation by GSH is the reverse reaction of nitrosylation by GSNO (Fig. 10), and it makes sense that both reactions may occur in the same local environment (i.e. the active site of GAPC1).
Nitrosylation and glutathionylation are two types of RPTMs that in the case of GAPC1 appear to be alternative and differently regulated. NO or GSNO induce nitrosylation of catalytic Cys-149, but nitrosylation of GAPC1 is not an intermediate step toward glutathionylation, because denitrosylation by GSH leaves the protein reduced (Fig. 10). However, sulfenation of Cys-149 by H2O2 is a mandatory step of GSH-dependent glutathionylation, a modification that requires GRX or TRX systems for being reverted (Fig. 10) (60). Therefore, in the case of GAPC1, nitrosylation and glutathionylation are not easily interchangeable and may well play different roles in vivo; the former is induced by RNS and directly controlled by the [GSH]/[GSNO] ratio, and the latter is induced by ROS (e.g. H2O2) and indirectly controlled by [GSH]/[GSSG] and [NADPH]/[NADP+] ratios via GRX and TRX systems, respectively (Fig. 10).
Both nitrosylation and denitrosylation of purified GAPC1 were found to be dependent on the [GSH]/[GSNO] ratio, which is therefore likely a major factor controlling the abundance of GAPC1-SNO (Fig. 10). However, considering the physiological concentrations of GSH in the millimolar range and a [GSH]/[GSNO] ratio on the order of 100:1 (as determined in whole organs of pepper plants (40)), accumulation of GAPC1-SNO would only be possible at cell locations where local concentrations of GSNO are strongly increased at the expense of GSH, such as in the vicinity of NO-producing enzymes or in sites where GSNO reductase, the major enzyme controlling the levels of GSNO in vivo, may be inhibited (19, 40). Indeed, it has emerged that nitrosylation exhibits a high degree of spatiotemporal precision partly due to interaction of target proteins with sources of the NO group, including NO synthases and GSNO (12, 16).
Nitrosylation was found involved in the signaling role of GAPDH in mammals, where nitrosylated GAPDH interacts with other proteins such as the SIAH1 ubiquitin ligase and is consequently transferred to the nucleus (57). In the nucleus, GAPDH could function as a trans-nitrosylase (36), and mechanisms may exist to stabilize GAPDH nitrosothiol under the highly reducing conditions of the nucleocytoplasmic environment. Animal GAPDH was indeed found resistant to GSH-mediated denitrosylation in vivo or in total extracts (48). A possible mechanism of protection could be promoted by interaction of nitrosylated GAPDH with other proteins. The binding of partner proteins could shield access to the catalytic cysteine or could induce a conformational change of GAPDH resulting in the protection of the nitrosocysteine from GSH. In the former case, denitrosylation of GAPDH would depend on the mechanisms controlling the stability of the complex between GAPDH-SNO and other proteins. It is even possible that TRX plays a role in these reactions, a hypothesis that would fit with previous observations reporting GSH-dependent denitrosylation of GAPDH in conditions where the complex is not formed and TRX-dependent denitrosylation when the complex is present (37, 53, 89).
We have clearly shown here that plant cytoplasmic TRXs are not efficient catalysts of GAPC1 denitrosylation, whereas GSH is an excellent denitrosylating agent. This indicates that GAPC1 is not a good model enzyme for the analysis of TRX-dependent denitrosylation in plants but does not necessarily preclude a role of plant TRXs as denitrosylases whose targets remain to be identified. Further studies will be required to determine the in vivo and in planta stability of nitrosylated GAPDH in Arabidopsis, but it should be noted that stability of the nitrosocysteine is not necessarily an essential requisite for a role of GAPC1 in signaling. In fact, nitrosylation might trigger nuclear relocalization of GAPC1 (e.g. via partner recruitment), and the role played by GAPC1 in the nucleus might be independent from its nitrosylated state. For instance, both animal and plant GAPDHs display a secondary activity of uracil glycosylase, an activity that prevents mutagenesis by eliminating uracil from DNA molecules (90, 91).
Arabidopsis GAPDH isoforms are several, and they are controlled by different types of RPTMs as follows: intramolecular disulfide bond controlled by ferredoxin-dependent TRX for A2B2-GAPDH (92, 93) and A4-GAPDH complexed with the regulatory protein CP12 (94); glutathionylation controlled by either only GRX or both TRX and GRX for A4-GAPDH and GapC, respectively (60–62); and nitrosylation controlled by GSH and GSNO for GAPC (this study). The possibility to alter GAPDH function by different types of RPTM and the fact that these RPTMs are controlled by distinct redox couples (reduced/oxidized ferredoxin for A2B2-GAPDH and CP12 disulfides; [NADPH]/[NADP+] and/or [GSH]/[GSSG] for glutathionylation; [GSH]/[GSNO] for nitrosylation) likely contributes to the functional flexibility of GAPDH allowing this enzyme of primary carbon metabolism to participate in diverse cellular processes and pathways that remain to be established in plants. GAPDH is probably one example among hundreds of moonlighting proteins that remain to be identified and characterized.
Acknowledgments
We thank Gaia Tioli and Roberto Orru for experimental support.
This work was supported in part by Grants PRIN 2008 (to P. T.) and PRIN 2009 (to M. Z.), ANR Grant 12-BSV5-0019 REDPRO2 (to C. H. M. and S. D. L.), a “Research in Paris” fellowship from Ville de Paris (to M. Z.), and by LABEX DYNAMO ANR-11-0011.
- RPTM
- redox post-translational modification
- BST
- biotin switch technique
- DEA-NO
- diethylamine NONOate
- GR
- glutathione reductase
- GSNO
- S-nitrosoglutathione
- TRX
- thioredoxin
- ROS
- reactive oxygen species
- RNS
- reactive nitrogen species
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GRX
- glutaredoxin
- HPDP-biotin
- N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)propionamide
- NTR
- NADPH:thioredoxin reductase.
REFERENCES
- 1. Foster M. W., Hess D. T., Stamler J. S. (2009) Protein S-nitrosylation in health and disease: a current perspective. Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giustarini D., Dalle-Donne I., Tsikas D., Rossi R. (2009) Oxidative stress and human diseases: origin, link, measurement, mechanisms, and biomarkers. Crit. Rev. Clin. Lab. Sci. 46, 241–281 [DOI] [PubMed] [Google Scholar]
- 3. Trushina E., McMurray C. T. (2007) Oxidative stress and mitochondrial dysfunction in neurodegenerative diseases. Neuroscience 145, 1233–1248 [DOI] [PubMed] [Google Scholar]
- 4. Xiong Y., Uys J. D., Tew K. D., Townsend D. M. (2011) S-Glutathionylation: From molecular mechanisms to health outcomes. Antioxid. Redox Signal. 15, 233–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dietz K. J. (2008) Redox signal integration: from stimulus to networks and genes. Physiol. Plant. 133, 459–468 [DOI] [PubMed] [Google Scholar]
- 6. Forman H. J., Maiorino M., Ursini F. (2010) Signaling functions of reactive oxygen species. Biochemistry 49, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Foyer C. H., Noctor G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905 [DOI] [PubMed] [Google Scholar]
- 8. Hancock J. T. (2009) The role of redox mechanisms in cell signalling. Mol. Biotechnol. 43, 162–166 [DOI] [PubMed] [Google Scholar]
- 9. Castello P. R., Woo D. K., Ball K., Wojcik J., Liu L., Poyton R. O. (2008) Oxygen-regulated isoforms of cytochrome c oxidase have differential effects on its nitric oxide production and on hypoxic signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 8203–8208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spadaro D., Yun B. W., Spoel S. H., Chu C., Wang Y. Q., Loake G. J. (2010) The redox switch: dynamic regulation of protein function by cysteine modifications. Physiol. Plant. 138, 360–371 [DOI] [PubMed] [Google Scholar]
- 11. Sun J., Murphy E. (2010) Protein S-nitrosylation and cardioprotection. Circ. Res. 106, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 13. Astier J., Rasul S., Koen E., Manzoor H., Besson-Bard A., Lamotte O., Jeandroz S., Durner J., Lindermayr C., Wendehenne D. (2011) S-Nitrosylation: an emerging post-translational protein modification in plants. Plant Sci. 181, 527–533 [DOI] [PubMed] [Google Scholar]
- 14. Seth D., Stamler J. S. (2011) The SNO-proteome: causation and classifications. Curr. Opin. Chem. Biol. 15, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akhtar M. W., Sunico C. R., Nakamura T., Lipton S. A. (2012) Redox regulation of protein function via cysteine S-nitrosylation and its relevance to neurodegenerative diseases. Int. J. Cell Biol. 2012, 463756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anand P., Stamler J. S. (2012) Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J. Mol. Med. 90, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lima B., Forrester M. T., Hess D. T., Stamler J. S. (2010) S-Nitrosylation in cardiovascular signaling. Circ. Res. 106, 633–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moreau M., Lindermayr C., Durner J., Klessig D. F. (2010) NO synthesis and signaling in plants–where do we stand? Physiol. Plant. 138, 372–383 [DOI] [PubMed] [Google Scholar]
- 19. Feechan A., Kwon E., Yun B. W., Wang Y., Pallas J. A., Loake G. J. (2005) A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102, 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lindermayr C., Durner J. (2009) S-Nitrosylation in plants: pattern and function. J. Proteomics 73, 1–9 [DOI] [PubMed] [Google Scholar]
- 21. Yu M., Yun B. W., Spoel S. H., Loake G. J. (2012) A sleigh ride through the SNO: regulation of plant immune function by protein S-nitrosylation. Curr. Opin. Plant Biol. 15, 424–430 [DOI] [PubMed] [Google Scholar]
- 22. Astier J., Besson-Bard A., Lamotte O., Bertoldo J., Bourque S., Terenzi H., Wendehenne D. (2012) Nitric oxide inhibits the ATPase activity of the chaperone-like AAA+ ATPase CDC48, a target for S-nitrosylation in cryptogein signalling in tobacco cells. Biochem. J. 447, 249–260 [DOI] [PubMed] [Google Scholar]
- 23. Lindermayr C., Sell S., Müller B., Leister D., Durner J. (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. (2008) Plant immunity requires conformational changes (corrected) of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Y. Q., Feechan A., Yun B. W., Shafiei R., Hofmann A., Taylor P., Xue P., Yang F. Q., Xie Z. S., Pallas J. A., Chu C. C., Loake G. J. (2009) S-Nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J. Biol. Chem. 284, 2131–2137 [DOI] [PubMed] [Google Scholar]
- 26. Yun B. W., Feechan A., Yin M., Saidi N. B., Le Bihan T., Yu M., Moore J. W., Kang J. G., Kwon E., Spoel S. H., Pallas J. A., Loake G. J. (2011) S-Nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268 [DOI] [PubMed] [Google Scholar]
- 27. Romero-Puertas M. C., Campostrini N., Mattè A., Righetti P. G., Perazzolli M., Zolla L., Roepstorff P., Delledonne M. (2008) Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8, 1459–1469 [DOI] [PubMed] [Google Scholar]
- 28. Benhar M., Forrester M. T., Stamler J. S. (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 29. Hogg N. (2002) The biochemistry and physiology of S-Nitrosothiols. Annu. Rev. Pharmacol. Toxicol. 42, 585–600 [DOI] [PubMed] [Google Scholar]
- 30. Pawloski J. R., Hess D. T., Stamler J. S. (2001) Export by red blood cells of nitric oxide bioactivity. Nature 409, 622–626 [DOI] [PubMed] [Google Scholar]
- 31. Mitchell D. A., Marletta M. A. (2005) Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nat. Chem. Biol. 1, 154–158 [DOI] [PubMed] [Google Scholar]
- 32. Mitchell D. A., Morton S. U., Fernhoff N. B., Marletta M. A. (2007) Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc. Natl. Acad. Sci. U.S.A. 104, 11609–11614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu C., Liu T., Chen W., Oka S., Fu C., Jain M. R., Parrott A. M., Baykal A. T., Sadoshima J., Li H. (2010) Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol. Cell. Proteomics 9, 2262–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nakamura T., Wang L., Wong C. C., Scott F. L., Eckelman B. P., Han X., Tzitzilonis C., Meng F., Gu Z., Holland E. A., Clemente A. T., Okamoto S., Salvesen G. S., Riek R., Yates J. R., 3rd, Lipton S. A. (2010) Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell 39, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qu J., Nakamura T., Cao G., Holland E. A., McKercher S. R., Lipton S. A. (2011) S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proc. Natl. Acad. Sci. U.S.A. 108, 14330–14335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kornberg M. D., Sen N., Hara M. R., Juluri K. R., Nguyen J. V., Snowman A. M., Law L., Hester L. D., Snyder S. H. (2010) GAPDH mediates nitrosylation of nuclear proteins. Nat. Cell Biol. 12, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Giustarini D., Milzani A., Aldini G., Carini M., Rossi R., Dalle-Donne I. (2005) S-Nitrosation versus S-glutathionylation of protein sulfhydryl groups by S-nitrosoglutathione. Antioxid. Redox Signal. 7, 930–939 [DOI] [PubMed] [Google Scholar]
- 38. Zaffagnini M., Bedhomme M., Marchand C. H., Couturier J. R., Gao X. H., Rouhier N., Trost P., Lemaire S. P. (2012) Glutaredoxin S12: unique properties for redox signaling. Antioxid. Redox Signal. 16, 17–32 [DOI] [PubMed] [Google Scholar]
- 39. Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J. S. (2001) A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410, 490–494 [DOI] [PubMed] [Google Scholar]
- 40. Airaki M., Sánchez-Moreno L., Leterrier M., Barroso J. B., Palma J. M., Corpas F. J. (2011) Detection and quantification of S-nitrosoglutathione (GSNO) in pepper (Capsicum annuum L.) plant organs by LC-ES/MS. Plant Cell Physiol. 52, 2006–2015 [DOI] [PubMed] [Google Scholar]
- 41. Sliskovic I., Raturi A., Mutus B. (2005) Characterization of the S-denitrosation activity of protein-disulfide isomerase. J. Biol. Chem. 280, 8733–8741 [DOI] [PubMed] [Google Scholar]
- 42. Nikitovic D., Holmgren A. (1996) S-Nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271, 19180–19185 [DOI] [PubMed] [Google Scholar]
- 43. Stoyanovsky D. A., Tyurina Y. Y., Tyurin V. A., Anand D., Mandavia D. N., Gius D., Ivanova J., Pitt B., Billiar T. R., Kagan V. E. (2005) Thioredoxin and lipoic acid catalyze the denitrosation of low molecular weight and protein S-nitrosothiols. J. Am. Chem. Soc. 127, 15815–15823 [DOI] [PubMed] [Google Scholar]
- 44. Foster M. W., Liu L., Zeng M., Hess D. T., Stamler J. S. (2009) A genetic analysis of nitrosative stress. Biochemistry 48, 792–799 [DOI] [PubMed] [Google Scholar]
- 45. Liu L., Zeng M., Hausladen A., Heitman J., Stamler J. S. (2000) Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. U.S.A. 97, 4672–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Forrester M. T., Foster M. W., Benhar M., Stamler J. S. (2009) Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic. Biol. Med. 46, 119–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu L., Yan Y., Zeng M., Zhang J., Hanes M. A., Ahearn G., McMahon T. J., Dickfeld T., Marshall H. E., Que L. G., Stamler J. S. (2004) Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell 116, 617–628 [DOI] [PubMed] [Google Scholar]
- 48. Paige J. S., Xu G., Stancevic B., Jaffrey S. R. (2008) Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem. Biol. 15, 1307–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Romero J. M., Bizzozero O. A. (2009) Intracellular glutathione mediates the denitrosylation of protein nitrosothiols in the rat spinal cord. J. Neurosci. Res. 87, 701–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benhar M., Forrester M. T., Hess D. T., Stamler J. S. (2008) Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science 320, 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Benhar M., Thompson J. W., Moseley M. A., Stamler J. S. (2010) Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry 49, 6963–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Forrester M. T., Thompson J. W., Foster M. W., Nogueira L., Moseley M. A., Stamler J. S. (2009) Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 27, 557–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu C., Parrott A. M., Liu T., Jain M. R., Yang Y., Sadoshima J., Li H. (2011) Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach. J. Proteomics 74, 2498–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sengupta R., Holmgren A. (2013) Thioredoxin and thioredoxin reductase in relation to reversible S-nitrosylation. Antioxid. Redox Signal. 18, 259–269 [DOI] [PubMed] [Google Scholar]
- 55. Sirover M. A. (2011) On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 1810, 741–751 [DOI] [PubMed] [Google Scholar]
- 56. Tristan C., Shahani N., Sedlak T. W., Sawa A. (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 23, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 58. Sen N., Hara M. R., Kornberg M. D., Cascio M. B., Bae B. I., Shahani N., Thomas B., Dawson T. M., Dawson V. L., Snyder S. H., Sawa A. (2008) Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat. Cell Biol. 10, 866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., Miginiac-Maslow M., Noctor G., Trost P., Lemaire S. D. (2007) The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 274, 212–226 [DOI] [PubMed] [Google Scholar]
- 60. Bedhomme M., Adamo M., Marchand C. H., Couturier J., Rouhier N., Lemaire S. D., Zaffagnini M., Trost P. (2012) Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 445, 337–347 [DOI] [PubMed] [Google Scholar]
- 61. Couturier J., Koh C. S., Zaffagnini M., Winger A. M., Gualberto J. M., Corbier C., Decottignies P., Jacquot J. P., Lemaire S. D., Didierjean C., Rouhier N. (2009) Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site. J. Biol. Chem. 284, 9299–9310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gao X. H., Zaffagnini M., Bedhomme M., Michelet L., Cassier-Chauvat C., Decottignies P., Lemaire S. D. (2010) Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii: kinetics and specificity in deglutathionylation reactions. FEBS Lett. 584, 2242–2248 [DOI] [PubMed] [Google Scholar]
- 63. Lindermayr C., Saalbach G., Durner J. (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Holtgrefe S., Gohlke J., Starmann J., Druce S., Klocke S., Altmann B., Wojtera J., Lindermayr C., Scheibe R. (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 133, 211–228 [DOI] [PubMed] [Google Scholar]
- 65. Goyer A., Decottignies P., Lemaire S., Ruelland E., Issakidis-Bourguet E., Jacquot J. P., Miginiac-Maslow M. (1999) The internal Cys-207 of sorghum leaf NADP-malate dehydrogenase can form mixed disulphides with thioredoxin. FEBS Lett. 444, 165–169 [DOI] [PubMed] [Google Scholar]
- 66. Jacquot J. P., Rivera-Madrid R., Marinho P., Kollarova M., Le Maréchal P., Miginiac-Maslow M., Meyer Y. (1994) Arabidopsis thaliana NADPH thioredoxin reductase. cDNA characterization and expression of the recombinant protein in Escherichia coli. J. Mol. Biol. 235, 1357–1363 [DOI] [PubMed] [Google Scholar]
- 67. Gelhaye E., Rouhier N., Jacquot J. P. (2003) Evidence for a subgroup of thioredoxin h that requires GSH/Grx for its reduction. FEBS Lett. 555, 443–448 [DOI] [PubMed] [Google Scholar]
- 68. Gelhaye E., Rouhier N., Laurent P., Sautière P. E., Martin F., Jacquot J. P. (2002) Isolation and characterization of an extended thioredoxin h from poplar. Physiol. Plant 114, 165–171 [DOI] [PubMed] [Google Scholar]
- 69. Rouhier N., Unno H., Bandyopadhyay S., Masip L., Kim S. K., Hirasawa M., Gualberto J. M., Lattard V., Kusunoki M., Knaff D. B., Georgiou G., Hase T., Johnson M. K., Jacquot J. P. (2007) Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc. Natl. Acad. Sci. U.S.A. 104, 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sicard-Roselli C., Lemaire S., Jacquot J. P., Favaudon V., Marchand C., Houée-Levin C. (2004) Thioredoxin Ch1 of Chlamydomonas reinhardtii displays an unusual resistance toward one-electron oxidation. Eur. J. Biochem. 271, 3481–3487 [DOI] [PubMed] [Google Scholar]
- 71. Jaffrey S. R., Snyder S. H. (2001) The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 72. Biesecker G., Harris J. I., Thierry J. C., Walker J. E., Wonacott A. J. (1977) Sequence and structure of d-glyceraldehyde-3-phosphate dehydrogenase from Bacillus stearothermophilus. Nature 266, 328–333 [DOI] [PubMed] [Google Scholar]
- 73. Talfournier F., Colloc'h N., Mornon J. P., Branlant G. (1998) Comparative study of the catalytic domain of phosphorylating glyceraldehyde-3-phosphate dehydrogenases from bacteria and archaea via essential cysteine probes and site-directed mutagenesis. Eur. J. Biochem. 252, 447–457 [DOI] [PubMed] [Google Scholar]
- 74. Kaneko R., Wada Y. (2003) Decomposition of protein nitrosothiols in matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. J. Mass Spectrom. 38, 526–530 [DOI] [PubMed] [Google Scholar]
- 75. Srinivasan U., Mieyal P. A., Mieyal J. J. (1997) pH profiles indicative of rate-limiting nucleophilic displacement in thioltransferase catalysis. Biochemistry 36, 3199–3206 [DOI] [PubMed] [Google Scholar]
- 76. Gallogly M. M., Mieyal J. J. (2007) Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 77. Starke D. W., Chen Y., Bapna C. P., Lesnefsky E. J., Mieyal J. J. (1997) Sensitivity of protein sulfhydryl repair enzymes to oxidative stress. Free Radic. Biol. Med. 23, 373–384 [DOI] [PubMed] [Google Scholar]
- 78. Wang J., Boja E. S., Tan W., Tekle E., Fales H. M., English S., Mieyal J. J., Chock P. B. (2001) Reversible glutathionylation regulates actin polymerization in A431 cells. J. Biol. Chem. 276, 47763–47766 [DOI] [PubMed] [Google Scholar]
- 79. Broniowska K. A., Diers A. R., Hogg N. (2013) S-Nitrosoglutathione. Biochim. Biophys. Acta 1830, 3173–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tanou G., Job C., Rajjou L., Arc E., Belghazi M., Diamantidis G., Molassiotis A., Job D. (2009) Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60, 795–804 [DOI] [PubMed] [Google Scholar]
- 81. Tanou G., Filippou P., Belghazi M., Job D., Diamantidis G., Fotopoulos V., Molassiotis A. (2012) Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 72, 585–599 [DOI] [PubMed] [Google Scholar]
- 82. Abat J. K., Mattoo A. K., Deswal R. (2008) S-Nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata–ribulose-1,5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J. 275, 2862–2872 [DOI] [PubMed] [Google Scholar]
- 83. Wawer I., Bucholc M., Astier J., Anielska-Mazur A., Dahan J., Kulik A., Wysłouch-Cieszynska A., Zareba-Kozioł M., Krzywinska E., Dadlez M., Dobrowolska G., Wendehenne D. (2010) Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 429, 73–83 [DOI] [PubMed] [Google Scholar]
- 84. Anderson L. E., Ringenberg M. R., Carol A. A. (2004) Cytosolic glyceraldehyde-3-P dehydrogenase and the B subunit of the chloroplast enzyme are present in the pea leaf nucleus. Protoplasma 223, 33–43 [DOI] [PubMed] [Google Scholar]
- 85. Vescovi M., Zaffagnini M., Festa M., Trost P., Lo Schiavo F., Costa A. (2013) Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 162, 333–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mohr S., Hallak H., de Boitte A., Lapetina E. G., Brüne B. (1999) Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 274, 9427–9430 [DOI] [PubMed] [Google Scholar]
- 87. Konorev E. A., Kalyanaraman B., Hogg N. (2000) Modification of creatine kinase by S-nitrosothiols: S-nitrosation versus S-thiolation. Free Radic. Biol. Med. 28, 1671–1678 [DOI] [PubMed] [Google Scholar]
- 88. Coles S. J., Easton P., Sharrod H., Hutson S. M., Hancock J., Patel V. B., Conway M. E. (2009) S-Nitrosoglutathione inactivation of the mitochondrial and cytosolic BCAT proteins: S-nitrosation and S-thiolation. Biochemistry 48, 645–656 [DOI] [PubMed] [Google Scholar]
- 89. Chakravarti R., Stuehr D. J. (2012) Thioredoxin-1 regulates cellular heme insertion by controlling S-nitrosation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 287, 16179–16186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Mazzola J. L., Sirover M. A. (2003) Subcellular alteration of glyceraldehyde-3-phosphate dehydrogenase in Alzheimer's disease fibroblasts. J. Neurosci. Res. 71, 279–285 [DOI] [PubMed] [Google Scholar]
- 91. Wang X., Sirover M. A., Anderson L. E. (1999) Pea chloroplast glyceraldehyde-3-phosphate dehydrogenase has uracil glycosylase activity. Arch. Biochem. Biophys. 367, 348–353 [DOI] [PubMed] [Google Scholar]
- 92. Fermani S., Sparla F., Falini G., Martelli P. L., Casadio R., Pupillo P., Ripamonti A., Trost P. (2007) Molecular mechanism of thioredoxin regulation in photosynthetic A2B2-glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 104, 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Marri L., Zaffagnini M., Collin V., Issakidis-Bourguet E., Lemaire S. D., Pupillo P., Sparla F., Miginiac-Maslow M., Trost P. (2009) Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol. Plant 2, 259–269 [DOI] [PubMed] [Google Scholar]
- 94. Fermani S., Trivelli X., Sparla F., Thumiger A., Calvaresi M., Marri L., Falini G., Zerbetto F., Trost P. (2012) Conformational selection and folding-upon-binding of intrinsically disordered protein CP12 regulate photosynthetic enzymes assembly. J. Biol. Chem. 287, 21372–21383 [DOI] [PMC free article] [PubMed] [Google Scholar]