Background: Claudin pore domain contains a highly conserved aromatic residue.
Results: Cation selectivity of claudin-2 and claudin-10b was impaired by substitution of residues lacking an aromatic group.
Conclusion: The aromatic residue confers cation selectivity by cation-π interaction and restricting the pore diameter.
Significance: This advances our understanding of the paracellular ion selectivity mechanism.
Keywords: Epithelial Cell, Ion Channels, Membrane Proteins, Membrane Transport, Phosphotyrosine Signaling, Sodium Channels, Tight Junctions, Tyrosine, Paracellular Transport
Abstract
In tight junctions, both claudin-2 and claudin-10b form paracellular cation-selective pores by the interaction of the first ECL 1 with permeating ions. We hypothesized that a highly conserved aromatic residue near the pore selectivity filter of claudins contributes to cation selectivity by cation-π interaction with the permeating cation. To test this, we generated MDCK I Tet-off cells stably transfected with claudin-2 Tyr67 mutants. The Y67L mutant showed reduced cation selectivity compared with wild-type claudin-2 due to a decrease in Na+ permeability, without affecting the Cl− permeability. The Y67A mutant enlarged the pore size and further decreased the charge selectivity due to an increase in Cl− permeability. The Y67F mutant restored the Na+ permeability, Cl− permeability, and pore size back to wild-type. The accessibility of Y67C to methanethiosulfonate modification indicated that its side chain faces the lumen of the pore. In claudin-10b, the F66L mutant reduced cation selectivity, and the F66A mutant lost pore conductance. We conclude that the conserved aromatic residue near the cation pore domain of claudins contributes to cation selectivity by a dual role of cation-π interaction and a luminal steric effect. Our findings provide new insight into how ion selectivity is achieved in the paracellular pore.
Introduction
Epithelial cells are connected via multiple junctional complexes. The tight junction separates the apical and basolateral membrane domains and acts as the paracellular barrier, while remaining selectively permeable to ions and water. In tight junctions, the first ECL1 of claudin forms the paracellular pore or barrier (1). Both claudin-2 and claudin-10b can form paracellular cation pores with PNa+/PCl− of 6 to 8 (2–4). The pore diameter of claudin-2 is estimated to be 6.5–7.5 Å (2, 5). The primary determinant of claudin-2 ion charge selectivity is an aspartate residue in ECL1 (Asp65) (2, 6). When all three negatively charged residues in the claudin-2 ECL 1, including Asp65, were mutated to neutral amino acids, the pore became less cation-selective. However, it remained four times more selective to Na+ than to Cl− (2). This observation led us to postulate that other mechanisms may also play a role in cation selectivity such as cation interaction with polar residues (e.g. carbonyl oxygen, as is the case in the KcsA potassium channel (7)), or cation-π interactions. The latter possibility prompted us to search for a conserved aromatic residue near Asp65 and Ile66, where the cation-selective filter is located (2, 8). We found position 67 of claudin-2 and position 66 of claudin-10b to have an aromatic residue that is highly conserved in all of the classic claudins (tyrosine in claudin-2 and phenylalanine in claudin-10b). The goal of this study was to assess the role of this aromatic residue in cation pore-forming claudins.
We hypothesized that Tyr67 (claudin-2) and Phe66 (claudin-10b) may interact with permeating cations through cation-π interaction. Cation-π interaction is defined as the interaction between positively charged molecules and negatively charged π electrons on the benzene ring of the aromatic amino acid side chain. Cation-π interaction has been identified in the nicotinic receptor ligand binding site (9) as well as in the binding site for tetraethylammonium in potassium channel (10). To test whether Tyr67 or Phe66 interacts with the permeating cations through cation-π interaction, we mutated this aromatic residue to leucine, a bulky and hydrophobic residue without the benzene ring. By eliminating the cation-π interaction, both claudin-2 Y67L and claudin-10b F66L were predicted to be less cation-selective than its respective wild-type protein.
The aromatic residue may also have a role via its steric effect. Its bulky benzene group could have a mechanical effect to modulate protein conformation and hence function. In the ATP-sensitive K channel, a pore-lining phenylalanine gates the channel by steric hindrance (11), and in the KcsA channel, activation and inactivation are mechanically coupled by a phenylalanine residue (12). To test whether the conserved aromatic residue exerts a steric effect, we substituted Tyr67 (claudin-2) or Phe66 (claudin-10b) with an alanine, a smaller hydrophobic residue. Our findings suggest that the conserved aromatic residue confers cation selectivity in cation pore-forming claudins by interacting with the permeating cation both via cation-π interaction and by restricting the pore size via its steric effect.
EXPERIMENTAL PROCEDURES
Generation and Screening of MDCK I Tet-off Claudin-2 and Claudin-10b Cell Lines
MDCK I Tet-off cells expressing wild-type claudin-2, wild-type claudin-10b, claudin-2 mutants (Y67L, Y67A, Y67C, D65N/Y67L, Y67F), and claudin-10b mutants (F66L, F66A) were generated by methods described previously (13). In short, the mutants of mouse claudin-2 and human claudin-10b were generated by site-directed mutagenesis on the template plasmid, pRevTREP-mouse-claudin2-wt and pRevTREP-human-claudin10b-wt respectively, using the QuikChange kit (Stratagene). These plasmids have been deposited and are available from the PSI:Biology-Materials Repository at DNASU. The plasmids were lipofected into the viral packaging cell line, PT67. Viral particles were collected from the growth medium of PT67 cells and used to transduce MDCK I Tet-off cells. After 7–10 days in a 0.3 mg/ml hygromycin-selective medium, independent clones of MDCK I Tet-off cell lines with transduced constructs were selected using cloning cylinders. To induce protein expression, doxycycline was omitted from the culture medium; otherwise 50 ng/ml doxycycline was included to suppress the protein expression.
Immunoblotting
Protein expression was tested by SDS-PAGE and immunoblotting. Confluent cells grown on tissue culture dishes were mechanically lysed by passing through a 25-gauge needle 10 times in sucrose-histidine lysis buffer containing 0.25 m sucrose, 30 mm histidine, 1 mm EDTA (pH 8), and protease inhibitor (Complete Mini, Roche Diagnostics). Cell lysates were loaded in reducing SDS-PAGE buffer (1% (v/v) 2-mercaptoethanol added) and heated at 75 °C for 10 min. 20 μg of protein samples were loaded on 12% polyacrylamide gel, transferred to a PVDF membrane, blotted with 1:500 mouse anti-claudin-2 antibody (Invitrogen) or 1:500 rabbit anti-claudin-10b antibody (Invitrogen) and then appropriate horseradish peroxidase-conjugated secondary antibodies (GE), detected with the ECL chemiluminescent method (Pierce), and imaged by an ImageQuant LAS-4000 (GE Healthcare).
Immunofluorescent Staining
The cells were plated at a density of 105 cells/1.16 cm2 on 12-well Transwell plates and grown for 7 days. The cells were washed in ice-cold PBS, fixed with 4% paraformaldehyde at 4 °C for 15 min, permeabilized, and blocked in a permeation buffer (0.3% Triton X-100, 1% BSA, and 5% goat serum in PBS) for 1 h. The filters were incubated in primary antibodies (1:500 mouse anti-claudin-2 and 1:500 rabbit anti-ZO-1; or 1:500 rabbit anti-claudin-10b and 1:500 mouse anti-ZO-1) for 2 h at room temperature, washed in PBS, and incubated in secondary antibodies for 1 h. For claudin-2 staining, Alexa Fluor 488-conjugated anti-mouse IgG and Alexa Fluor 555-conjugated anti-rabbit IgG were used (both 1:1000); for claudin-10b staining, Alexa Fluor 555-conjugated anti-mouse IgG and Alexa Fluor 488-conjugated anti-rabbit IgG were used (both 1:1000). The filters were then washed in PBS and mounted in the ProLong anti-fade mounting medium. All of the reagents were from Invitrogen. Slides were imaged by a Leica TCS SP2 multi-photon confocal microscope.
Electrophysiological Studies in Ussing Chamber
Cells were plated at a density of 105 cells/1.16 cm2 on Snapwell filters (Corning) and cultured for 7 days in the presence (Dox+)2 or absence (Dox−) of 50 ng/ml doxycycline. The Ussing chamber setup and liquid junction potential correction method was employed as described previously (13). The conductance and permeability attributed to claudin-2 pore was calculated by subtracting the average value of the uninduced (Dox+) state from the values of the induced (Dox−) state. The standard Ringer solution used at base line contained the following: 150 mm NaCl, 2 mm CaCl2, 1 mm MgCl2, 10 mm glucose, 10 mm Tris-HEPES, pH 7.4. To measure Na+ permeability, the solution in the basolateral chamber was changed to 75 mm NaCl Ringer solution (osmolarity adjusted with mannitol). To measure alkali metal biionic potential, the basolateral medium was changed to 150 mm alkali metal chloride salt. To measure organic cation permeability, the basolateral medium was changed to the solution containing 75 mm organic cation chloride salt and 75 mm NaCl. The organic cations included methylamine, ethylamine, and tetraethylammonium. The ion permeability ratio, β = PCl−/PNa+, was calculated from the Goldman-Hodgkin-Katz voltage equation. The absolute Na+ permeability was estimated by the method devised by Kimizuka and Koketsu (14). The alkali metal permeability was calculated from γ = PM/PNa, where γ was estimated as shown in Equation 1.
The organic cation permeability was calculated from the following equation.
Here, α represents the activity ratio of NaCl in the apical compartment over the basolateral compartment. The activity of NaCl at 150 mm is 0.752 and 0.797 at 75 mm, resulting in α = (150 × 0.752)/(75 × 0.797) = 1.89.
The pore size of the claudin pore was estimated by the method described previously (2). In short, the claudin-2 pore was assumed to be a cylinder of diameter, D, across which cations of diameter, d, diffused. According to the Renkin equation (Equation 3),
 |
the square root of the relative permeability of methylamine, ethylamine, and tetraethylammonium to Na+ is linearly related to the cation diameter. D was estimated from the x intercept of the best-fit line, as determined by linear regression.
Substituted Cysteine Modification
To test the functional effect of covalent modification of the substituted cysteine, (2-(trimethylammonium) ethyl)methanthiosulfonate (MTSET) was added to the cells, and the changes of pore conductance and NaCl dilution potential were measured. The working concentration of MTSET was 1 mm. To avoid hydrolysis, the reagent was freshly dissolved as 100-fold concentrated stock solution immediately before starting the experiment. The stock solution was added simultaneously to the medium of the apical and basolateral chambers and rapidly mixed by gas lifts. The change in conductance was calculated as the percentage change in conductance from pretreatment state to the 5-min post-treatment state. The NaCl dilution potential was measured before and after the treatment.
Cysteine-specific Surface Biotinylation
To test the accessibility of the substituted cysteine, cysteine-specific surface biotinylation was performed. Cells were plated at a density of 5 × 105 cells/well on six-well plates and grown for 6 days. Cells were washed with PBS contained 1 mm CaCl2 and 1 mm MgCl2 (PBS/CM), and a solution of 0.5 ml/well 0.5 mg/ml MTSEA-biotin freshly dissolved in PBS/CM was added. The plate was incubated at room temperature for 10 min and washed three times with ice-cold PBS, and the cells were harvested in radioimmune precipitation assay buffer (50 mm Tris-HCl pH 8, 150 mm NaCl, 0.1% (w/v) SDS, 0.5% (w/v) deoxycholic acid, 1% (v/v) Nonidet P-40). The cell lysate was centrifuged at 16,000 × g for 15 min. The supernatant was added to a 40-μl slurry of streptavidin-coated beads and rotated at 4 °C for 2 h. The beads were then pelleted, and the supernatant was saved for analysis. The beads were washed three times in TBS (50 mm Tris-HCl and 150 mm NaCl), added to 20 μl of 2× reducing SDS-PAGE loading buffer, and heated at 75 °C for 10 min with occasional agitation. Both bead (biotinylated protein fraction) and supernatant (non-biotinylated fraction) samples were then subjected to immunoblotting as described above.
Statistics
The data are presented as means ± S.E.. Statistical significance was determined using unpaired two-tailed Student's t test or one-way analysis of variance test. The p value of multiple comparisons was corrected using the Bonferroni correction. p < 0.05 was considered to be statistically significant.
RESULTS
Generation of Claudin-2 and Claudin-10b Mutants in Stably Transduced MDCK I Tet-off Cells
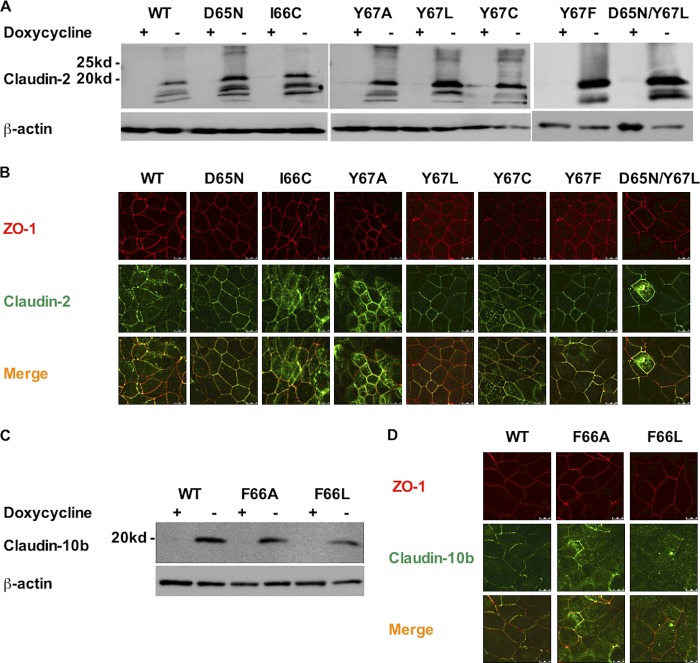
To test the role of the aromatic residue near the pore selective filter, claudin-2 constructs (Y67L, Y67A, Y67C, Y67F, and D65N/Y67L) and claudin-10b constructs (wild-type, F66L, F66A) were transduced into MDCK I Tet-off cells using retroviral transduction, and stably transduced clones were selected. Inducible protein expression was verified by immunoblotting, which showed a characteristic band of both the claudin-2 monomer (Fig. 1A) and the claudin-10b monomer (Fig. 1C) at ∼20 kDa in the absence of doxycycline. There were also multiple bands <20 kDa in the claudin-2 blot, which are not seen in the claudin-2 blot of mouse kidney lysates (data not shown). They are therefore probably proteolysis products, which we often see in overexpressing protein in cells. Immunofluorescent staining of claudins and ZO-1 showed that all the claudin-2 constructs (Fig. 1B) and claudin-10b constructs (Fig. 1D) were localized at the tight junction.
FIGURE 1.
Characterization of stably transduced MDCK I Tet-Off cell lines expressing claudin-2 and claudin-10b constructs. A, immunoblot of protein expression in clones stably transduced with claudin-2 constructs (wild-type (WT), D65N, I66C, Y67L, Y67A, Y67C, Y67F, and D65N/Y67L). C, claudin-10b constructs (WT, F66A, and F66L), grown in the presence (+) or absence (−) of Dox. Cell lysates were subjected to reducing SDS-PAGE and immunoblotted with anti-claudin-2 antibody or anti-claudin-10b antibody. The image was exposed for 15 min. Inducible claudin-2 monomer and claudin-10b monomer expression at ∼20 kDa was seen in all constructs. Shown are localization of claudin-2 (B) and claudin-10b (D) constructs by immunofluorescent staining and confocal microscopy examination. The cells were cultured on Transwells for 6 days and then immunostained for ZO-1 (red) and claudin-2 or claudin-10b (green). All mutants co-localized with ZO-1 at the tight junction.
In Claudin-2, Leucine Substitution Leads to Partial Loss of Cation Selectivity without Affecting the Pore Size
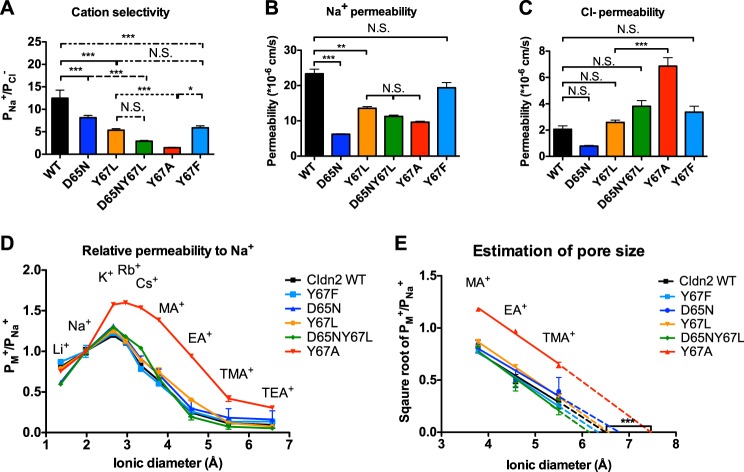
To test whether Tyr67 contributes to cation selectivity by cation-π interaction, we mutated this position to leucine, a bulky and hydrophobic residue without the benzene ring. Y67L was predicted to be less cation-selective than wild-type. Consistent with this, the PNa+/PCl− of claudin-2 Y67L was 5.4 ± 0.3, which was significantly smaller than claudin-2 wild-type (12.5 ± 1.8) (Fig. 2A). If both Asp65 and Tyr67 function as a site that independently confers cation selectivity, the PNa+/PCl− of D65N/Y67L should be further reduced from the single mutants. The PNa+/PCl− of D65N/Y67L was 2.9 ± 0.2, a significant decrease from D65N (8.1 ± 0.5). In addition, it was approximately half of the PNa/PCl ratio of Y67L, although the difference did not reach a level of statistical significance.
FIGURE 2.
Characterization of the electrophysiological properties of claudin-2 constructs. MDCK I Tet-off cells transfected with claudin-2 constructs (WT, D65N, Y67L, D65N/Y67L, Y67A, and Y67F) were plated at 105 cells/1.16 cm2 and grown for 7 days before mounting in Ussing chambers. A, the permeability ratio was calculated as PNa+/PCl−, where PNa+ and PCl− were calculated from NaCl dilution potentials and subtracting the average base-line permeability of uninduced (Dox+) cells from that of induced (Dox−) cells. Shown are Na+ permeability (B) and Cl− permeability (C) of claudin-2 (Cldn2) WT, Y67F, D65N, Y67L, D65N/Y67L, and Y67A. D, the permeability of claudin-2 constructs (WT, Y67F, D65N, Y67L, D65N/Y67L, and Y67A) to alkali metal cations and organic cations relative to their Na+ permeability were plotted against the ionic diameters. E, the relationship between the square roots of the relative permeability of methylamine (MA), ethylamine (EA), and tetramethylammonium (TMA), and ionic diameters were fitted by linear regression, and the pore diameter was estimated as the x intercept of the best-fit line. Data points represent the means of 3–9 filters ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. N.S., non-significant. p values were obtained from one-way analysis of variance test with the Bonferroni's correction.
Compared with wild-type, the partial decrease in the cation selectivity of Y67L was due to a significant decrease in Na+ permeability (Fig. 2B) without affecting the Cl− permeability (Fig. 2C). The PNa+ of D65N/Y67L was less than Y67L, and the PCl− was higher than Y67L. Neither, however, reached a level of statistical significance. The relative permeability of Y67L to alkali metal cations (Fig. 2D, orange line) was identical to wild-type. As shown previously (2), the relative permeability to Li+ (PLi+/PNa+) of D65N (Fig. 2D, blue line) was less than wild-type, indicating that there was loss of a strong intrapore binding site for dehydrated cations. In contrast, the PLi+/PNa+ of Y67L was no different than wild-type. The relative permeability of D65N/Y67L to Li+ was similar to D65N. The phenotype of the double mutation appears to be additive of the effect of the two single mutants, suggesting that Asp65 and Tyr67 are two distinct sites that independently confer cation selectivity. The pore diameter (in Å) of Y67L and D65N/Y67L was estimated to be 6.7 ± 0.2 and 6.1 ± 0.5 (Fig. 2E), respectively, neither of which was significantly different from wild-type (6.6 ± 0.2). In summary, the leucine substitution in the aromatic residue of claudin-2 decreased the cation selectivity due to a decrease in Na+ permeability without affecting Cl− permeability or the pore size.
In Claudin-2, Alanine Substitution Abolishes Cation Selectivity and Enlarges Pore Size
To test whether the aromatic group exerts a steric effect, we substituted an amino acid with a side chain that was as small as possible. Glycine is the smallest amino acid, but it would introduce flexibility to the peptide backbone. Instead, alanine was selected because it has the second smallest side chain and because its substitution is usually well tolerated and only infrequently causes protein misfolding.
Mutation of Tyr67 to alanine eliminates both the benzene ring and the bulky side chain. Thus, Y67A was compared with Y67L to specifically pinpoint the role of steric effects from the bulky side chain. The PNa+/PCl− of Y67A was 1.4 ± 0.1, smaller than the PNa+/PCl− of Y67L (Fig. 2A). The cation selectivity of Y67A approached the ratio of mobilities of these ions in free solution (PNa+/PCl− = 0.7) (15). Thus, Y67A almost completely abolished the cation selectivity of claudin-2. Compared with Y67L and D65N/Y67L, the decrease in the cation selectivity in Y67A was due to a significant increase in Cl− permeability (Fig. 2C) without further affecting Na+ permeability (Fig. 2B). In Y67A, the relative permeability of large alkali metal and organic cations (Fig. 2D, red line) was significantly increased from wild-type. The estimated pore size of Y67A was 7.6 ± 0.1Å (Fig. 2E), which was significantly larger than that of wild-type, D65N, Y67L, and D65N/Y67L.
In summary, alanine substitution almost completely abolished the cation selectivity of claudin-2 due to increase in Cl− permeability without affecting Na+ permeability. The pore size of Y67A was significantly enlarged from Y67L and wild-type, suggesting that Tyr67 restricted the pore size by a steric effect.
In Claudin-2, Substitution of Another Aromatic Residue at Position 67 Partially Restores Cation Selectivity and Pore Size
If cation selectivity is conferred by a bulky aromatic ring at position 67, substitution of phenylalanine at this position should have a similar function. To test this, we made the claudin-2 mutation, Y67F. Y67F partially restored cation selectivity as evidenced by a PNa+/PCl− ratio of 5.9 ± 0.4, which was significantly greater than Y67A, yet still lower than that of wild-type (Fig. 2A). The PNa+ of Y67F was lower than wild-type and the PCl− of Y67F was higher than wild-type, but neither of them reach a level of statistical significance (Fig. 2, B and C). The relative cation permeability curve (Fig. 2D) and the pore size (Fig. 2E) of Y67F were almost identical to wild-type.
In Claudin-2, the Side Chain of Tyr67 Is Accessible from the Pore Lumen
There are two possible side chain conformations that Tyr67 could adopt that would restrict the pore size. The side chain could point directly into the pore lumen. Alternatively, the side chain could be buried inside the protein fold and sterically push the pore-lining residues into the pore lumen. To determine the conformation of the Tyr67 side chain relative to the pore, we generated a Y67C mutant and assessed the accessibility of the substituted cysteine to membrane-impermeable methanethiosulfonate (MTS) reagents. As positive controls, we used cysteine mutants of a known pore-lining residue in claudin-2, Ile66, and of a residue known to face the outside of the pore, Tyr35 (8). If the side chain at position 67 points to the pore lumen, Y67C will be accessible to extracellularly applied MTS reagents.
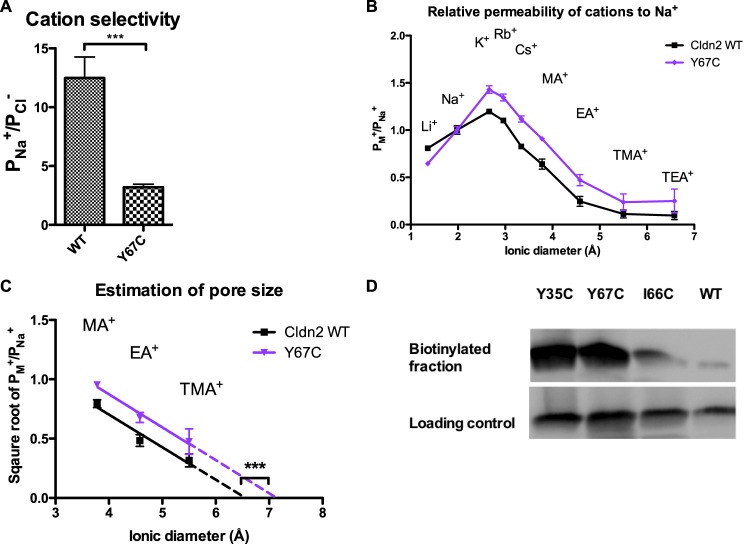
Y67C had lower PNa+/PCl− than wild-type, consistently suggesting cation-π interaction has an important role in cation selectivity (Fig. 3A). Y67C had higher relative permeability of cations larger than Na+ (Fig. 3B), and the estimated pore size of Y67C (7.1 ± 0.4 Å) was significantly increased compared with wild-type (Fig. 3C). Next, we probed the accessibility of the substituted cysteine by MTSEA-biotin. Y67C was biotinylatable (Fig. 3D), suggesting that the amino acid side chain at position 67 was accessible and was not folded within the protein. The biotinylated fraction of Y67C was similar to Y35C in abundance and was much greater than I66C, suggesting that Y67C was more accessible than I66C, likely reflecting the enlarged pore size.
FIGURE 3.
Characterization of the functional and structural properties of claudin-2 Y67C. A, cation selectivity of Y67C. B, the permeability of claudin-2 constructs (WT and Y67C) to alkali metal cations and organic cations relative to their Na+ permeability were plotted against the ionic diameters. C, the square roots of the relative permeability of methylamine (MA), ethylamine (EA), and tetramethylammonium (TMA) were fitted by linear regression, and the pore diameter was estimated as the x-intercept. D, cells expressing claudin-2 (Cldn2) Y35C, Y67C, I66C, and WT were treated with MTSEA-biotin, followed by streptavidin precipitation. The bead fraction and the supernatant fraction were subjected to SDS-PAGE and blotted with anti-claudin-2 antibody. The upper blot shows the biotinylated claudin-2 on the beads. The lower blot shows the non-biotinylated claudin-2 in the supernatant as the loading control. E, conductance inhibition assay by MTSET in Ussing chamber. The change of conductance was calculated as the percentage change in the conductance at 5-min after addition of MTSET to claudin-2 Y67C, compared with pre-treatment. Data points represent the means of 3 filters ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. p values were obtained from one-way analysis of variance test with the Bonferroni's correction.
If the side chain of Tyr67 faces the pore lumen, MTS reagents might be expected to block the pore. However, we found that neither MTSET (Fig. 3E) nor MTSEA-biotin (data not shown) altered the conductance or ion selectivity of Y67C. Furthermore, when we attempted to restore a benzene group to Y67C by adding benzyl MTS, the benzyl MTS treatment neither changed the conductance of Y67C nor the Na+ or Cl− permeability (data not shown), suggesting that the MTS reagents may not mimic the actual Tyr67 side chain confirmation in the wild-type protein.
In summary, similar to alanine, cysteine substitution in Tyr67 enlarged the pore size. Y67C was accessible from the aqueous environment, but MTS reagents were unable to block the pore conductance or change ion selectivity.
Phe66 Is Critical for the Function of Claudin-10b
To determine whether the findings of Tyr67 in claudin-2 can be generalized to other cation pore-forming claudins, we generated MDCK I Tet-off cells expressing claudin-10b wild-type, F66L, and F66A. Phe66 in claudin-10b is the aromatic residue homologous to Tyr67 in claudin-2.
The pore properties of wild-type claudin-10b were consistent with previous findings (Fig. 4) (3, 4). In brief, claudin-10b increased the transepithelial conductance by ∼6.5-fold. It was four times more permeable to Na+, and the order of relative permeability to alkali metal cations was that of Eisenman sequence VIII.
FIGURE 4.
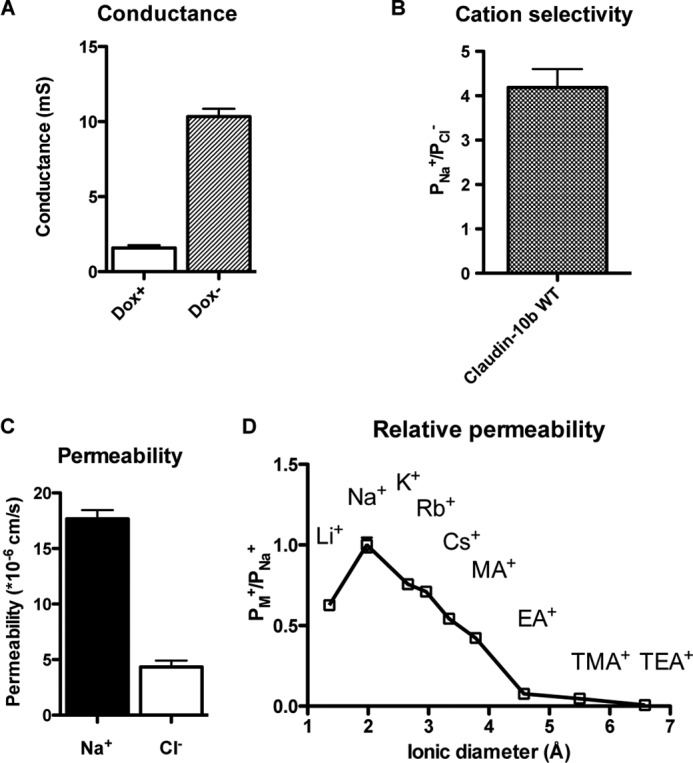
Characterization of the electrophysiological properties of wild-type claudin-10b. MDCK I Tet-off cells transfected with claudin-10b wild-type (WT) were plated at 105 cells/1.16 cm2 and grown for 7 days before mounting in Ussing chamber. A, change of conductance from uninduced state (Dox+) to induced state (Dox−). mS, millisiemens. B, cation selectivity presented as PNa+/PCl−, where PNa+ and PCl− were calculated from NaCl dilution potentials and subtracting the average base-line permeability of the uninduced (Dox+) cells from that of the induced (Dox−) cells. C, Na+ permeability and Cl− permeability. D, the relative permeability of alkali metal cations and organic cations relative to their Na+ permeability were plotted against the ionic diameters. Data points represent the means of three filters ± S.E.
Claudin-10b F66L increased conductance similarly to wild-type claudin-10b (Fig. 5A). Similar to claudin-2 Y67L, the PNa+/PCl− ratio of claudin-10b F66L was 1.3 ± 0.3 (Fig. 5B), which was significantly less than that of wild-type claudin-10b (4.2 ± 0.4). The decrease of cation selectivity of F66L was due to reduced Na+ permeability without changing the Cl− permeability (Fig. 5C). This suggests that the role of the aromatic residue at this site can be generalized to other cation pore-forming claudins.
FIGURE 5.
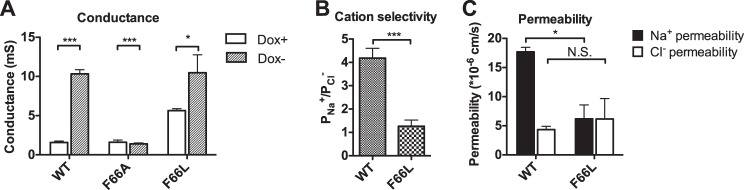
Characterization of the electrophysiological properties of claudin-10b constructs. MDCK I Tet-off cells transduced with claudin-10b constructs (WT, F66L, and F66A) were plated at 105 cells/1.16 cm2 and grown for 7 days before mounting in an Ussing chamber. A, change of conductance from uninduced state (Dox+) to induced state (Dox−). B, the permeability ratio was calculated as PNa+/PCl−, where PNa+ and PCl− were calculated from NaCl dilution potentials and subtracting the average base-line permeability of the uninduced (Dox+) cells from that of the induced (Dox−) cells. C, Na+ permeability and Cl− permeability of claudin-10b WT and F66L. Data points represent the means of three filters ± S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. N.S., non-significant. p values were obtained from unpaired Student's t test.
Interestingly, claudin-10b F66A did not increase the conductance of MDCK I cells at all (1.61 ± 0.28 mS in Dox+ and 1.40 ± 0.11 mS in Dox−), suggesting that it was not functional (Fig. 5A). Thus, it was uninformative to compare the effect of F66A on the pore size and charge selectivity of claudin-10b to the effect of Y67A on claudin-2.
DISCUSSION
Claudin-2 and claudin-10b are cation-selective pores at the tight junction. In claudin-2, mutating all three negatively charged amino acids in the pore-forming first extracellular domain makes the pore become less cation-selective. However, the pore still remains four times more permeable to Na+ than to Cl−, suggesting that other non-charged amino acids may also contribute to the cation selectivity. Tyr67 and Phe66 are conserved aromatic residues in claudin-2 and claudin-10b, respectively, which are located near the pore selectivity filter. We initially hypothesized that Tyr67 (Phe66) contributes to cation selectivity by side chain cation-π interaction with the permeating cation. We found that this aromatic residue in cation claudin pores was required for cation selectivity because of a dual role: facilitating cation permeation by cation-π interaction and preventing anion permeation by a luminal steric effect.
In Claudin-2, Tyr67 Provides the Minor Interaction Site for the Permeating Cation by Cation-π Interaction
Na+ is hydrated in solution. The hydration enthalpy for Na+ is −405 kJ/mol (16). In claudin-2 wild-type, cations permeate through the pore in a partially dehydrated form (2). The majority of the energy for dehydration comes from the electrostatic interaction of the cations with Asp65 within the pore (2). In addition to Asp65, Tyr67 seems to play a role. Y67L decreases Na+ permeability without changing the Cl− permeability, alkali metal cation permeability pattern, or pore size. This suggests that Y67L loses the ability to facilitate Na+ permeation rather than alters the pore conformation. The most likely explanation is that Tyr67 facilitates Na+ permeation by cation-π interaction. Cation-π interaction could provide 16–125 kJ/mol of binding energy (17), and it is generally weaker than electrostatic interaction. The quantitative measurement for the binding energy of Na+ to Asp65 and Tyr67 is not available because the stoichiometry of claudin-2 pore is not known. However, D65N was much less permeable than wild-type for the heavily hydrated cation, Li+ (2), whereas Y67L did not significantly decrease the relative permeability of Li+. This relationship suggests that Asp65 provides the major portion of cation permeation energy cost, and Tyr67 contributes a minor portion of that, in agreement with the magnitude of strength of electrostatic interaction and cation-π interaction. Furthermore, the double mutant D65N/Y67L was less cation selective than D65N, reflecting the additive cation selective effect of Tyr67. Meanwhile, the PLi+/PNa+ of D65N/Y67L was less than Y67L, reflecting the loss of the strong intrapore ion-binding site: Asp65. This suggests that Asp65 and Tyr67 are two distinct sites that independently confer cation selectivity.
In Claudin-2, Tyr67 Restricts the Pore Size by Steric Effect to Prevent Cl− Permeation
The claudin-2 pore is 6.5–7.5 Å in diameter, and the hydrated diameter of Na+ and Cl− is estimated to be 9.4 and 7.8 Å, respectively (18). Because Na+ can be partially dehydrated within the pore, and hence has a smaller hydrated diameter than Cl−, Na+ is more permeable than Cl− in claudin-2 wild-type. In Y67A, the pore is enlarged by 0.8–1.2Å, which allows ions to diffuse without dehydration. Because Cl− is more mobile than Na+ in free diffusion, Y67A increases Cl− permeability disproportionately to Na+ permeability. A similar pore enlarging effect is seen in Y67C, precluding the explanation that the pore enlarging effect is an artifact of the introduced amino acid. Comparing the substitution of alanine with that of leucine at this site, Y67A lacks the bulky side chain. A bulky side chain could potentially exert a steric effect on channel gating (11) and coupling (12). However, the most likely explanation for our results is that a bulky side chain at position 67 restricts the pore size by a steric effect.
In Claudin-2, the Side Chain of Tyr67 Likely Points toward the Pore Lumen
There are two possible side chain conformations for Tyr67 that could restrict the pore size. The side chain could directly protrude into the pore lumen. Less directly, the side chain could fold inside the protein and push the pore-lining residues into the pore lumen. Y67C is structurally accessible to MTSEA-biotin, excluding the possibility that the side chain is folded within. Whether the side chain points toward the pore lumen, as is the case with Ile66, or on the outside surface of the protein, as is the case with Tyr35, is debatable. After MTSEA-biotin exposure, the biotinylated fraction of Y67C is much greater than that of I66C and similar to Y35C. This may be the result of Y67C being on the outside of the protein. However, this interpretation does not explain why the Tyr67 mutants have dramatically altered the pore properties. Moreover, Tyr67 is embedded in the middle of a series of consecutive pore-lining residues: Asp65 (2), Ile66 (8), and Ser68.3 It is unlikely that Y67C faces outside while its two neighboring residues are lining the pore. We therefore conclude that the Tyr67 side chain most likely faces toward the pore lumen, and that the high biotinylation fraction is due to the enlarged pore size and hence increased accessibility to MTSEA-biotin.
In Claudin-10b, Phe66 Is Critical for the Pore Function
Claudin-10b is also a cation pore. In the mutagenesis study of Phe66, the F66L mutation reduced the cation selectivity as Y67L did in claudin-2. Interestingly, the F66A mutant did not enlarge the pore size as Y67A did in claudin-2 but instead disrupted the cation pore function of claudin-10b. This indicates that Phe66 is a critical residue for the function of claudin-10b.
The Dual Role of the Aromatic Residue in the Ion Selectivity Mechanism of Pore-forming Claudins
Fig. 6 shows a homology alignment of part of the first extracellular domain of the major pore-forming claudins and their charge selectivity. All claudins have two conserved extracellular cysteines separated by 8–10 residues. Counting from the second extracellular cysteine, all of the pore claudins have a major charge selectivity site (Asp, Glu, Arg, or Lys) located at the +1 and/or +2 position, and one to two aromatic amino acid residues located within the +2 to +4 positions. In cation-selective pore claudins, the role of the aromatic residue(s) is to enhance the cation selectivity: first, by facilitating Na+ permeation by cation-π interaction and second, by preventing hydrated Cl− permeation by a steric effect. In anion-selective pore claudins, we speculate that the presence of a positively charged binding site overrides the effect of the π electrons and facilitates stabilization of a dehydrated Cl− ion in the pore and hence Cl− permeation. Concurrently, the steric effect prevents the hydrated Na+ ions from permeating.
FIGURE 6.
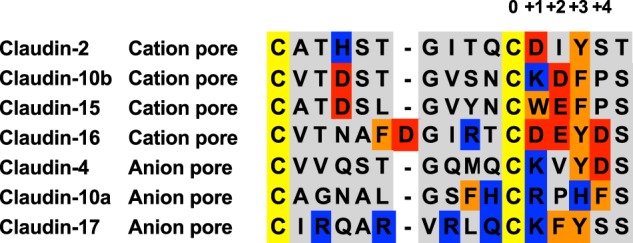
Homology alignment of major pore-forming claudins. Cation-selective pore claudins are as follows: claudin-2 (2), claudin-10b (3, 4, 19), claudin-15 (20), and claudin-16 (21). Anion-selective pore claudins are as follows: claudin-17 (22), claudin-10a (4, 19), and claudin-4 (6). Claudins with inconclusive or controversial selectivity properties such as claudin-7 and claudin-19 are excluded. Displayed here is a homology alignment of the amino acid sequence of the first extracellular domain from the first conserved extracellular cysteine to the fifth residue downstream of the second conserved cysteine. Negatively charged residues are in red, positively charged residues are in blue, and aromatic residues are in orange. The numbers denote relative positions downstream of the second cysteine, where 0 corresponds to the second cysteine, +1 corresponds to Asp65 in claudin-2, and +3 corresponds to Tyr67 in claudin-2 or Phe66 in claudin-10b.
In conclusion, we demonstrate that the conserved aromatic residue located one to two residues downstream of the major charge selective site has a dual role for cation selectivity. It facilitates cation permeation by cation-π interaction and prevents anion permeation by a luminal steric effect. This provides new insight into how ion selectivity is achieved in the paracellular pore.
Acknowledgment
The Confocal Imaging Core at the University of Kansas Medical Center was supported by National Institutes of Health Grant 9P20GM104936 (to Dale Abrahamson).
This work was supported by National Institutes of Health Grants R01DK062283 and U01GM094627 (to A. S. L. Y.).
J. Li, M. Zhuo, L. Pei, and A. S. L. Yu, unpublished results.
- Dox
- doxycycline
- MTSET
- 2-(trimethylammonium) ethyl)methanthiosulfonate
- MTS
- methanethiosulfonate.
REFERENCES
- 1. Colegio O. R., Van Itallie C., Rahner C., Anderson J. M. (2003) Claudin extracellular domains determine paracellular charge selectivity and resistance but not tight junction fibril architecture. Am. J. Physiol. Cell Physiol. 284, C1346–1354 [DOI] [PubMed] [Google Scholar]
- 2. Yu A. S., Cheng M. H., Angelow S., Günzel D., Kanzawa S. A., Schneeberger E. E., Fromm M., Coalson R. D. (2009) Molecular basis for cation selectivity in claudin-2-based paracellular pores: identification of an electrostatic interaction site. J. Gen. Physiol. 133, 111–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenthal R., Milatz S., Krug S. M., Oelrich B., Schulzke J. D., Amasheh S., Günzel D., Fromm M. (2010) Claudin-2, a component of the tight junction, forms a paracellular water channel. J. Cell Sci. 123, 1913–1921 [DOI] [PubMed] [Google Scholar]
- 4. Van Itallie C. M., Rogan S., Yu A., Vidal L. S., Holmes J., Anderson J. M. (2006) Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am. J. Physiol. Renal Physiol. 291, F1288–1299 [DOI] [PubMed] [Google Scholar]
- 5. Van Itallie C. M., Holmes J., Bridges A., Gookin J. L., Coccaro M. R., Proctor W., Colegio O. R., Anderson J. M. (2008) The density of small tight junction pores varies among cell types and is increased by expression of claudin-2. J. Cell Sci. 121, 298–305 [DOI] [PubMed] [Google Scholar]
- 6. Hou J., Renigunta A., Yang J., Waldegger S. (2010) Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc. Natl. Acad. Sci. U.S.A. 107, 18010–18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77 [DOI] [PubMed] [Google Scholar]
- 8. Angelow S., Yu A. S. (2009) Structure-function studies of claudin extracellular domains by cysteine-scanning mutagenesis. J. Biol. Chem. 284, 29205–29217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhong W., Gallivan J. P., Zhang Y., Li L., Lester H. A., Dougherty D. A. (1998) From ab initio quantum mechanics to molecular neurobiology: a cation-π binding site in the nicotinic receptor. Proc. Natl. Acad. Sci. U.S.A. 95, 12088–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guidoni L., Carloni P. (2002) Tetraethylammonium binding to the outer mouth of the KcsA potassium channel: implications for ion permeation. J. Recept. Signal Transduct. Res. 22, 315–331 [DOI] [PubMed] [Google Scholar]
- 11. Rojas A., Wu J., Wang R., Jiang C. (2007) Gating of the ATP-sensitive K+ channel by a pore-lining phenylalanine residue. Biochim. Biophys. Acta 1768, 39–51 [DOI] [PubMed] [Google Scholar]
- 12. Cuello L. G., Jogini V., Cortes D. M., Pan A. C., Gagnon D. G., Dalmas O., Cordero-Morales J. F., Chakrapani S., Roux B., Perozo E. (2010) Structural basis for the coupling between activation and inactivation gates in K+ channels. Nature 466, 272–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu A. S. (2011) Electrophysiological characterization of claudin ion permeability using stably transfected epithelial cell lines. Methods Mol. Biol. 762, 27–41 [DOI] [PubMed] [Google Scholar]
- 14. Kimizuka H., Koketsu K. (1964) Ion transport through cell membrane. J. Theor. Biol. 6, 290–305 [DOI] [PubMed] [Google Scholar]
- 15. Koneshan S., Rasaiah J. C., Lynden-Bell R. M., Lee S. H. (1998) Solvent structure, dynamics, and ion mobility in aqueous solutions at 25 °C. J. Phys. Chem. B 102, 4193–4204 [Google Scholar]
- 16. Wulfsberg G. (2000) Inorganic Chemistry, p. 57, University Science Books, Sausalito, CA [Google Scholar]
- 17. Gallivan J. P., Dougherty D. A. (1999) Cation-π interactions in structural biology. Proc. Natl. Acad. Sci. U.S.A. 96, 9459–9464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kielland J. (1937) Individual activity coefficients of ions in aqueous solutions. J. Am. Chem. Soc. 59, 1675–1678 [Google Scholar]
- 19. Günzel D., Stuiver M., Kausalya P. J., Haisch L., Krug S. M., Rosenthal R., Meij I. C., Hunziker W., Fromm M., Müller D. (2009) Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J. Cell Sci. 122, 1507–1517 [DOI] [PubMed] [Google Scholar]
- 20. Tamura A., Hayashi H., Imasato M., Yamazaki Y., Hagiwara A., Wada M., Noda T., Watanabe M., Suzuki Y., Tsukita S. (2011) Loss of claudin-15, but not claudin-2, causes Na+ deficiency and glucose malabsorption in mouse small intestine. Gastroenterology 140, 913–923 [DOI] [PubMed] [Google Scholar]
- 21. Hou J., Renigunta A., Konrad M., Gomes A. S., Schneeberger E. E., Paul D. L., Waldegger S., Goodenough D. A. (2008) Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J. Clin. Invest. 118, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krug S. M., Günzel D., Conrad M. P., Rosenthal R., Fromm A., Amasheh S., Schulzke J. D., Fromm M. (2012) Claudin-17 forms tight junction channels with distinct anion selectivity. Cell. Mol. Life Sci. 69, 2765–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]