Background: Fibulin-3, -4, and -5 (short fibulins) participate in elastogenesis, although the molecular mechanism remains unknown.
Results: Short fibulins are cleaved by MMPs and bind cells and heparin, and fibulin-4 dimerizes and multimerizes.
Conclusion: Short fibulins have a proteolytically susceptible linker and can bind cells independently of the RGD motif, and fibulin-4 multimerization is crucial for heparin binding.
Significance: The uncovered properties will advance understanding of elastogenesis.
Keywords: Cell Adhesion, Extracellular Matrix, Extracellular Matrix Proteins, Heparin-binding Protein, Matrix Metalloproteinase (MMP), Elastic Fiber, Fibulin, Protein Multimerization
Abstract
Extracellular short fibulins, fibulin-3, -4, and -5, are components of the elastic fiber/microfibril system and are implicated in the formation and homeostasis of elastic tissues. In this study, we report new structural and functional properties of the short fibulins. Full-length human short fibulins were recombinantly expressed in human embryonic kidney cells and purified by immobilized metal ion affinity chromatography. All three fibulins showed various levels of degradation after the purification procedure. N-terminal sequencing revealed that all three fibulins are highly susceptible to proteolysis within the N-terminal linker region of the first calcium-binding epidermal growth factor domain. Proteolytic susceptibility of the linker correlated with its length. Exposure of these fibulins to matrix metalloproteinase (MMP)-1, -2, -3, -7, -9, and -12 resulted in similar proteolytic fragments with MMP-7 and -12 being the most potent proteases. Fibulin-3 proteolysis was almost completely inhibited in cell culture by the addition of 25 μm doxycycline (a broad spectrum MMP inhibitor). Reducible fibulin-4 dimerization and multimerization were consistently observed by SDS-PAGE, Western blotting, and mass spectrometry. Atomic force microscopy identified monomers, dimers, and multimers in purified fibulin-4 preparations with sizes of ∼10–15, ∼20–25, and ∼30–50 nm, respectively. All short fibulins strongly adhered to human fibroblasts and smooth muscle cells. Although only fibulin-5 has an RGD integrin binding site, all short fibulins adhere at a similar level to the respective cells. Solid phase binding assays detected strong calcium-dependent binding of the short fibulins to immobilized heparin, suggesting that these fibulins may bind cell surface-located heparan sulfate.
Introduction
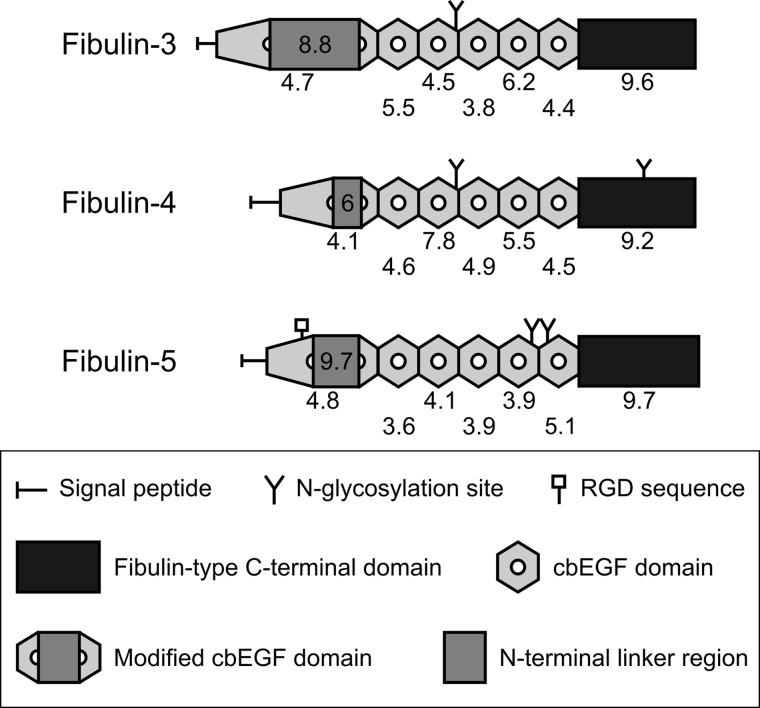
The fibulin family of extracellular matrix glycoproteins comprises eight members, fibulin-1 through -8 (for a review, see Ref. 1). Fibulin-3, -4, and -5 are collectively referred to as the short fibulins, sharing a similar domain structure with calculated molecular masses of 52–56 kDa. The short fibulins are each composed of six tandem calcium-binding epidermal growth factor-like (cbEGF)2 domains followed by a fibulin-type C terminus (see Fig. 1A) (2–4). Characteristic for the short fibulins is an insertion of amino acid residues between the fourth and fifth cysteines known as the N-terminal linker region (5). This insert of unknown function varies in size among human fibulin-3, -4, and -5 with 88, 28, and 44 amino acid residues, respectively. Fibulin-3 contains one and fibulin-4 and -5 contain two conserved sequons for N-linked glycosylation. Only fibulin-5 contains a conserved integrin-binding RGD sequence in its first cbEGF domain preceding the insert, whereas fibulin-3 and -4 are devoid of RGD sequences (5). Electron microscopy has identified the short fibulins as 10–20-nm rodlike monomers with a short globule on one end (3, 5). Jones et al. (6) demonstrated for fibulin-5 a compact dimer in the presence of calcium.
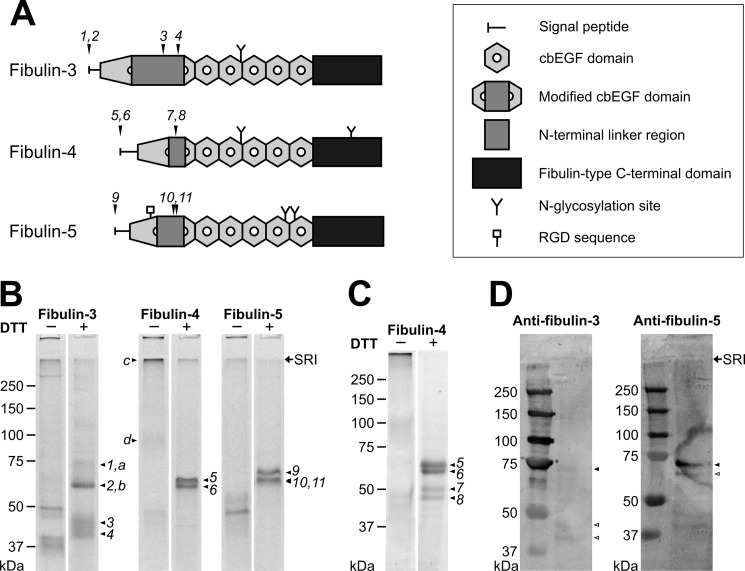
FIGURE 1.
Overview and characterization of fibulin-3, -4, and -5. A, schematic of the domain organization of human fibulin-3, -4, and -5. The numbers above the schematic indicate proteolytic cleavage sites corresponding to the numbered protein bands in B and C. The 28–88-amino acid-long N-terminal linker region between cysteines 4 and 5 of cbEGF1 of each short fibulin is indicated by a gray box. B, analysis of purified recombinant human short fibulins by SDS-PAGE. The samples (10 μg/lane) were separated on a 7.5% polyacrylamide gel under non-reducing (−) or reducing (+) conditions using DTT. C, analysis of an occasionally purified recombinant human fibulin-4 by SDS-PAGE containing two additional breakdown fragments. Numbered protein bands in B and C were analyzed by N-terminal sequencing and are summarized in Table 1 and schematically in A. Protein bands indicated by letters were analyzed by mass spectrometry. D, anti-fibulin-3 and -5 Western blots of guanidine extracts from mouse aorta. Tissue extracts were extracted as described and analyzed on 7.5% polyacrylamide gels under reducing conditions. Proteins were then detected with polyclonal anti-human fibulin-3 and -5 antibodies (1:500) followed by secondary antibody and development as described under “Experimental Procedures.” Faint detection of mouse fibulin-3 and -5 is likely the result of using our anti-human fibulin-3 and -5 antibodies with limited cross-reactivity against mouse. Closed arrowheads indicate bands correlating to full-length proteins, and open arrowheads indicate degradation products. Globular marker proteins are indicated in kDa, and the stacking-resolving interface (SRI) of the gels is indicated with an arrow.
Overlapping and distinct tissue expression patterns have been described for the short fibulins. Fibulin-3 is expressed in diverse tissues, including cartilage, bone, subretinal pigment epithelium, skin, and capillaries. Fibulin-4 and -5 are expressed in blood vessels, lung, skin, and cartilage with fibulin-5 being additionally expressed in the uterus and Bruch membrane (for a review, see Ref. 7).
The importance of the short fibulins in elastic fiber formation and homeostasis became apparent through the generation of knock-out mouse models, which demonstrated various degrees of elastic fiber malformation. Although fibulin-3-null mice survive until adulthood, they exhibit reduced bone density and reproductivity as well as premature aging, inguinal herniation, and uterine and rectal prolapse (8). The herniation and prolapse are likely explained by the observed reduced elastic fibers in the fascia of the fibulin-3-deficient mice. In contrast, human mutations in fibulin-3 have been associated with Doyne honeycomb retinal dystrophy (9). The fibulin-4-null mouse has the most severe phenotype of the short fibulin knock-out mice with perinatal death caused by aortic rupture, emphysematous lung, and aortic aneurysm as well as arterial tortuosity, narrowing, and dilatation (10). Only small aggregates of amorphous elastin instead of fully developed elastic fibers are present in the skin, lung, and blood vessels. In humans, fibulin-4 mutations give rise to cutis laxa characterized by inelastic and saggy skin, vascular tortuosity, increased risk of ascending aortic aneurysms, emphysema, and other clinical symptoms (11). The fibulin-5-null mouse survives until adulthood with thin and fragmented elastic fibers in the ascending aorta, lung, and skin; tortuosity and elongation of the aorta; severe emphysema; loose and inelastic skin; and pelvic organ prolapse during pregnancy (12, 13). Fibulin-5 mutations in humans result in cutis laxa or age-related macular degeneration (14).
The short fibulins are characterized by overlapping and distinct protein and cell binding properties. It has been shown that mouse fibulin-3 and human fibulin-4 and -5 interact with tropoelastin (5). Fibulin-4 and -5 can interact with fibrillin-1 and some members of the lysyl oxidase family of tropoelastin-cross-linking enzymes (5, 12, 13, 15, 16). Of the short fibulins, fibulin-5 cell attachment properties have been explored and attributed to the presence of an RGD site. Specifically, fibulin-5 has been demonstrated to bind human umbilical vein endothelial cells in an RGD-dependent manner; Chinese hamster ovary cells via integrins α9β1, αvβ3, and αvβ5; and primary human aortic smooth muscle cells via integrins α5β1 and α4β1 (4, 12, 17). A mouse knock-in model replacing the RGD with inactive RGE in fibulin-5 forms normal elastic fibers, demonstrating that the RGD cell interaction site is dispensable for elastogenesis (18). It is currently not known whether fibulin-5 and the other short fibulins that do not possess an RGD site can interact with cells through RGD-independent mechanisms.
Proteolytic cleavage of fibulin-5 has been reported in its N-terminal linker region in aged mice (19). The resulting fibulin-5 fragment is no longer able to participate in elastogenesis, suggesting that fibulin-5 cleavage may be involved in the aging of elastic tissues. Whether or not fibulin-3 and -4 are susceptible to proteolytic cleavage in the N-terminal linker region remains to be elucidated.
One of the goals of the present study was to explore the susceptibility of the short fibulins to proteolytic degradation by matrix metalloproteinases (MMPs). The MMP family comprises 23 proteases, either membrane-bound or secreted into the extracellular matrix where they are able to degrade many extracellular matrix proteins, including collagen, elastin, fibronectin, and fibrillin (20–22). This family of endopeptidases is characterized by a Zn2+ ion in the active site of the catalytic domain. MMP-3, -7, -9, and -12 are elastin-degrading enzymes (23). Although MMPs promote extracellular matrix turnover and remodeling, their dysregulation is at the root of numerous pathologies, including thoracic and abdominal aortic aneurysm, emphysema, and cutis laxa (24–27).
Because of the relatively mild elastogenic phenotype of the fibulin-3-null mouse and the involvement of the protein in retinal dystrophy in humans, the role of fibulin-3 in elastogenesis has been poorly studied to date. Based on in vitro binding experiments and in vivo mouse and human data, fibulin-4 and -5 have recently been incorporated into a working model of elastogenesis. Fibulin-4 is hypothesized to bring tropoelastin and pro-lysyl oxidase in close proximity to facilitate initial cross-linking within the coacervate (28). Fibulin-5 is presumed to then bind the partially cross-linked tropoelastin coacervate and direct it to the microfibril for further cross-linking and incorporation into the mature elastic fiber (12, 13, 29).
This study aimed to advance the understanding of the characteristics of the short fibulins with a focus on proteolysis and cell attachment. Human short fibulins were recombinantly expressed and purified, and SDS-PAGE analysis revealed that all three proteins were N-glycosylated and partially cleaved in the N-terminal linker region. MMP-1, -2, -3, -7, -9, and -12 cleaved the short fibulins to fragments almost identical in size and cleavage sites to the fragments obtained upon purification. The short fibulins bound human fibroblasts and smooth muscle cells possibly through heparan sulfate-containing cell surface receptors as all three proteins bound heparin. Furthermore, fibulin-4 monomers, dimers, and multimers were observed by atomic force microscopy, and monomers were separated from multimers by gel filtration chromatography. In contrast to the fibulin-4 monomers and dimers, the multimers strongly bound heparin, suggesting that multimerization may be required for fibulin-4 cell attachment.
EXPERIMENTAL PROCEDURES
All chemicals used were of the highest purity and purchased from Sigma-Aldrich if not otherwise stated.
Antibodies
Polyclonal antisera were produced against fibulin-3, -4, and -5 in rabbit following standard procedures using recombinant full-length proteins as antigen. The specificity and cross-reactivity of each antiserum was verified by Western blotting and ELISA with recombinant short fibulins. The polyclonal antiserum against the C-terminal half of human fibrillin-1 was generated and characterized previously (30). The monoclonal primary antibodies against human fibronectin (Sigma, product F7387) and against a C-terminal polyhistidine sequence (anti-His(C-term), Invitrogen, product R93025) were commercially purchased. The secondary antibody used to detect all polyclonal primary antibodies was goat anti-rabbit antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, product 111-035-003). Conversely, the secondary antibody used to detect all monoclonal primary antibodies was goat anti-mouse antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch Laboratories, product 115-035-062). All primary antibodies were diluted 1:500 for Western blotting and 1:1000 for solid phase and ELISA assays, whereas secondary antibodies were always diluted 1:800.
Cloning Strategy
Full-length human fibulin-3, -4, and -5 constructs were generated using the pcDNA3.1(+) vector (Invitrogen, product V79020), which included the sequence for an N-terminal BM-40 signal peptide for efficient secretion from human embryonic kidney cells (HEK293) and was thus named pDNSP (30). A C-terminal hexahistidine tag sequence was included to allow for protein purification by immobilized metal ion affinity chromatography. Full-length short fibulin cDNA was obtained by reverse transcription of total RNA isolated from human skin fibroblasts (described below).
The expression plasmid for Gln18–Phe493 of human fibulin-3 was generated by subcloning the NheI-XhoI-restricted 1459-bp polymerase chain reaction (PCR) product generated with oligonucleotides 5′-CAGGCTAGCACAGGACACCGAAGAAACCATC-3′ and 5′-TAACTCGAGCTATTAGTGATGGTGATGGTGATGAAATGAAAATGGCCCCACTATTATTG-3′ into the NheI-XhoI-linearized pDNSP plasmid, resulting in the pDNSP-hFBLN3 plasmid. The expressed protein was APLAQ18–F493HHHHHH. The expression plasmid for Gln28–Phe443 of human fibulin-4 was generated by subcloning the NheI-XhoI-restricted 1279-bp PCR product generated with oligonucleotides 5′-CAGGCTAGCTCAGGATTCTGAAGAGCCCGAC-3′ and 5′-TAACTCGAGCTATTAGTGATGGTGATGGTGATGGAAGGTGTAGGCCCCTACAAAG-3′ into the NheI-XhoI-linearized pDNSP plasmid, resulting in the pDNSP-hFBLN4 plasmid. The expressed protein was APLAQ28–F443HHHHHH. The expression plasmid for Gln24–Phe448 of human fibulin-5 was generated by subcloning the NheI-XhoI-restricted 1306-bp PCR product generated with oligonucleotides 5′-CAGGCTAGCACAGGCACAGTGCACGAATGG-3′ and 5′-TAACTCGAGCTATTAGTGATGGTGATGGTGATGGAATGGGTACTGCGACACATATATC-3′ into the NheI-XhoI-linearized pDNSP plasmid, resulting in the pDNSP-hFBLN5 plasmid coding for APLAQ24–F448HHHHHH. The entire coding region of all expression plasmids was verified by DNA sequencing.
Transfection and Cell Culture
The expression plasmids pDNSP-hFBLN3, -4, and -5 were linearized with PvuI, ethanol-precipitated, and redissolved in water. 10 μg of DNA was used for transfection of HEK293 cells with calcium phosphate as described previously (30). The selection procedure was initiated 2 days after transfection with the addition of 750 μg/ml geneticin sulfate (G418; Wisent, product 450-130-QL) to the medium for 1 week and 250 μg/ml G418 thereafter. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Wisent, product 319-005) supplemented with 10% fetal calf serum (FCS; Wisent, product 080150), 100 μg/ml penicillin and streptomycin, and 2 mm glutamine (Wisent, product 450-202-EL). Cells were cultured at 37 °C and 5% CO2. Recombinant protein secretion was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting following trichloroacetic acid precipitation of 1 ml of conditioned medium.
Stably transfected HEK293 cells were plated on eight triple layer flasks (500-cm2 surface area; Nalge Nunc International, product 132913) in DMEM supplemented with 10% FCS, 100 μg/ml penicillin and streptomycin, 2 mm glutamine, and 250 μg/ml G418 to maintain clonal selection. Upon confluence, serum proteins were removed by two washes with 20 mm HEPES, 150 mm NaCl, 2.5 mm CaCl2, pH 7.4. All flasks were then placed on serum-free DMEM, and 500 ml of conditioned medium was collected every 48 h for a total of 10 collections. After each collection, cellular debris was removed from the conditioned medium by centrifugation at 6000 × g for 15 min at 4 °C. 100 μm phenylmethanesulfonyl fluoride (PMSF) was then added to prevent serine protease proteolysis of the collected material, and the collection was stored at −20 °C until protein purification.
Treatment of stably transfected HEK293 cells with doxycycline (DOX; Sigma, product D9891) was attempted to inhibit MMPs to prevent proteolytic cleavage of the recombinant short fibulins. 25 μm DOX supplementation commenced when the cells were transferred onto eight triple layer flasks and was repeated with each collection of serum-free medium every 48 h. As a control, the transfected HEK293 cells on eight triple layer flasks were cultured without DOX.
Human skin fibroblasts were extracted from foreskins of healthy boys (2–5 years of age) following a standard circumcision procedure. This procedure was approved by the Montreal Children's Hospital Research Ethics Committee (PED-06-054), and written consent of each patient's parents was obtained. Cell lines of human fetal lung fibroblasts (ATCC, product CCL-75) and of human umbilical vein smooth muscle cells (ATCC, product CRL-2481) were commercially purchased. Human umbilical artery smooth muscle cells were a generous gift from Dr. Cynthia Goodyer. Cell culture conditions were identical to those used for HEK293 cells but without G418 supplementation. Cells in passages 4–10 were used for all experiments.
Recombinant Protein Purification
Recombinant hexahistidine-tagged short fibulins were purified to homogeneity by immobilized metal ion affinity chromatography with a Ni2+-loaded HisTrap HP 1-ml column (GE Healthcare, product 17-5247-01) using an ÄktaPurifier 10 low pressure liquid chromatography system (GE Healthcare) as follows. 2.5 liters of conditioned medium was concentrated to 50 ml in pressurized ultrafiltration cells (Amicon, products 5124 and 6028) through a 10-kDa cutoff membrane (Millipore, product PLGC07610) over ∼24 h at 4 °C. The resulting 50 ml were dialyzed two times for ∼6 h against 2 liters of 20 mm HEPES, 500 mm NaCl, pH 7.4 (running buffer) at 4 °C. Any remaining precipitate was centrifuged at 11,000 × g for 15 min at 4 °C, and the supernatant was used to fill a 50-ml Superloop (GE Healthcare, product 19-7850-01). The concentrate was loaded at a flow rate of 0.5 ml/min followed by a 5-ml wash with running buffer. Bound protein was eluted from the column at 1 ml/min with a 0–500 mm linear imidazole gradient in 20 mm HEPES, 500 mm NaCl, pH 7.4. 1-ml eluate fractions were collected, and aliquots were analyzed by SDS-PAGE and stained with Coomassie Brilliant Blue. Fractions containing purified short fibulins were pooled and dialyzed two times for ∼6 h against 1 liter of CaCl2-containing Tris-buffered saline (TBS/Ca; 50 mm Tris-HCl, 150 mm NaCl, 2 mm CaCl2, pH 7.4). The BCA protein assay kit (Thermo Fisher Scientific, product 23225) was used to determine the concentration of proteins following the manufacturer's microtiter plate protocol.
N-terminal Protein Sequencing
N-terminal protein sequencing (Edman degradation) was used to identify proteolytic cleavage sites in the recombinant short fibulins upon purification and after treatment with exogenous MMPs. Duplicates containing 20 μg of the protein of interest as determined by BCA assay were separated by SDS-PAGE and transferred onto a methanol-soaked polyvinylidene fluoride membrane (Bio-Rad, product 162-0184) with cold 10 mm tetrasodium borate, pH 8.9 for 1.5 h at 0.4 A. After the transfer, the membrane was stained for 1 min with 1.2% (w/v) Coomassie Brilliant Blue G-250 in 50% methanol, 5% acetic acid, 45% H2O. The blot was destained three times with 50% methanol, 5% acetic acid solution for 3 min followed by a final wash with H2O. The blot was air-dried, and the bands of interest were excised and analyzed by N-terminal protein sequencing as described (22). The resulting protein sequencing chromatograms were analyzed with SequencePro 2.0 software (Applied Biosystems).
Proteolytic Cleavage Assays
MMP-1, -2, -3, -7, -9, and -12 were evaluated for their ability to cleave each of the short fibulins. The MMPs (R&D Systems, products 901-MP, 902-MP, 513-MP, 907-MP, 911-MP, and 917-MP) were activated with 4-aminophenylmercuric acetate (Sigma, product A9563) at 37 °C for 1–32 h according to the manufacturer's instructions. Upon enzyme activation, 10 μg of the fibulins was added, and the mixture was incubated at 37 °C for 24 h. An enzyme-to-protein ratio of 1:100 (w/w) was used for all experiments. The MMPs were deactivated by heat denaturation at 95 °C and reduced with 20 mm dithiothreitol (DTT), and samples were immediately separated by 4–20% gradient SDS-PAGE and stained with colloidal Coomassie Brilliant Blue (Sigma, product G1041).
Mouse Tissue Extraction
Aortas from 25-day-old mice were dissected and homogenized in 50 mm Tris-HCl, pH 7.4, 1 m NaCl, 10 mm EDTA, 2 mm PMSF (200 mg of tissue/ml). After 24 h, the homogenates were centrifuged, and the residual pellets were resuspended in 50 mm Tris-HCl, pH 7.4, 250 mm NaCl, 5 mm CaCl2, 6 m guanidine HCl. After 24 h, the homogenates were centrifuged, and the supernatants were collected. Samples were ethanol-precipitated and separated on a 7.5% polyacrylamide gel (50 μl/lane) under reducing conditions. Proteins were detected with polyclonal anti-human fibulin-3 and -5 primary antibodies (diluted 1:500) followed by secondary antibody and color development.
Mass Spectrometry Analysis
Mass spectrometry was performed to identify protein bands in the purified recombinant short fibulin preparations as well as to identify cleavage sites of fibulin-4 cleaved by MMP-12. 10-μg aliquots of protein with or without MMP-12 were heat-denatured at 95 °C in the presence or absence of 20 mm DTT. The samples were separated by 10% SDS-PAGE and stained with colloidal Coomassie Blue. Protein bands of interest were excised and processed according to standard procedures (31). In brief, all bands were reduced with 10 mm DTT in 100 mm ammonium bicarbonate and alkylated with 55 mm iodoacetamide in 100 mm ammonium bicarbonate. Samples were digested with trypsin (12 ng/μl) overnight at 37 °C. The peptides were extracted from the gel and dried under vacuum. The dried samples were resuspended in the appropriate buffer prior to injection into the Velos Orbitrap (Thermo Fisher Scientific).
Gel Filtration Chromatography
Purified recombinant short fibulins were each loaded onto an analytical Superose 12 column (total column volume, 24 ml; GE Healthcare, product 17-5173-01) to separate high, intermediate, and low molecular weight proteins and protein aggregates in the preparations. 0.7–1.4 mg (total volume, 1.5–2 ml) of purified protein in TBS/Ca was loaded at 0.5 ml/min onto the Superose 12 column equilibrated in TBS/Ca using an ÄktaPurifier 10. 0.5-ml fractions were collected, separated by SDS-PAGE, and stained with Coomassie Brilliant Blue. Fractions containing high, intermediate, and low molecular weight proteins and protein aggregates were pooled based on the elution profile and the Coomassie-stained fraction gels. Protein concentrations (40–170 μg/ml) of the pools were determined by BCA assay, and the pools were stored at −80 °C until further analysis.
Cell Attachment Assay
Short fibulin binding to human skin and lung fibroblasts as well as to human umbilical vein and artery smooth muscle cells was tested to identify differences in cell surface receptor binding between the RGD-containing fibulin-5 and the short fibulins lacking the RGD sequence. Serial dilutions (1:2 starting at 50 μg/ml; 100 μl/well) of short fibulins (and fibronectin, a positive control known to bind multiple integrins and cell surface heparan sulfate proteoglycans) were coated in TBS overnight at 4 °C on 96-well plates (Maxisorp, Nalge Nunc International, product 439454). If not otherwise stated, all subsequent steps were performed at a constant room temperature of 22 °C. The wells were blocked for 1 h with 200 μl of 10 mg/ml heat-denatured (85 °C for 11 min) bovine serum albumin (BSA) in phosphate-buffered saline (PBS; 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4). Confluent human and lung skin fibroblasts as well as human umbilical artery and vein smooth muscle cells were washed twice with PBS and trypsinized with 3 ml of 0.05% trypsin, 0.53 mm EDTA for 3 min at 37 °C. The trypsinization was stopped with 10 ml of FCS-containing DMEM, and the cells were centrifuged and resuspended in cell suspension solution (DMEM without FCS). Cell numbers were determined with a hemocytometer, and cells were diluted to 400,000 cells/ml in cell suspension solution. The plate was washed once with PBS to remove excess blocking solution after which 40,000 cells (100 μl) in cell suspension solution were added to each well and incubated at 37 °C for 1 h. The plate was washed once with 100 μl of PBS in combination with three gentle taps to remove unbound cells. All solutions were added to and removed from wells with a multipipetter with care not to touch or scratch the bottom of the well. Bound cells were fixed with 100 μl/well 5% glutaraldehyde solution in PBS for 20 min. The plate was washed three times with 100 μl of PBS in combination with three gentle taps each time to remove excess glutaraldehyde. Cells were stained with 100 μl of crystal violet solution (0.1% crystal violet in 200 mm MES, pH 6.0) for 1 h. The plate was washed three times with 400 μl/well PBS in combination with three gentle taps each time to remove excess crystal violet. An image of the plate was recorded after which 100 μl of solubilizing solution (10% acetic acid in PBS) was added to each well, and the absorbance was measured at 570 nm in a microplate reader (Beckman Coulter).
Heparin-BSA Coupling
Covalent coupling of heparin derived from porcine intestinal mucosa (Sigma, product H3393) to delipidized BSA (Sigma, product A6003) was performed to allow heparin to be immobilized onto 96-well plates. Heparin alone does not coat these plates efficiently because of its highly negative charge, and coupling to BSA facilitates coating of this glycosaminoglycan. Additionally, three to four heparin molecules are typically coupled per BSA molecule, which mimics the clustering of heparan sulfate chains on cell surface proteoglycans. Heparin is used as a cost-effective model of heparan sulfate as the two share a similar structure of repeating iduronic/glucuronic acid and N-sulfated glucosamine/N-acetylglucosamine disaccharide units. Heparin was cross-linked to BSA following a protocol established previously (32). Heparin-BSA protein concentrations were determined by BCA assay, and the pools were stored at −80 °C until use.
Solid Phase Binding Assay
Solid phase binding assays were performed to assess short fibulin binding to heparin-BSA and tropoelastin following a protocol established previously (30). The tropoelastin was a generous gift from Drs. Ming Miao and Fred Keeley (Hospital for Sick Children, Toronto, Ontario, Canada). Briefly, 10 μg/ml heparin-BSA or tropoelastin in TBS were immobilized per well on a 96-well plate overnight at 4 °C. Three 5-min washes with 200 μl of TBS/Ca containing 0.05% Tween 20 were performed between all subsequent steps to remove unbound ligand or antibody. Nonspecific binding surfaces on the plastic were blocked with blocking buffer (5% (w/v) nonfat milk in TBS/Ca) for 1 h. Serial dilutions (1:2 starting at either 100 μg/ml or at the final concentrations of individual protein pools after gel filtration) of short fibulins in binding buffer (2% (w/v) nonfat milk in TBS/Ca) were incubated for 2 h. Polyclonal primary antibody against each of the short fibulins was diluted in binding buffer (1:1000) and incubated with the bound soluble ligand for 90 min. Secondary goat anti-rabbit antibody conjugated to horseradish peroxidase was diluted in binding buffer (1:800) and added to the wells for a 90-min incubation. The final color reaction was developed with 5-aminosalicylic acid for 2–20 min, and the reaction was stopped with 2 m sodium hydroxide.
Solid phase binding assays investigating calcium dependence of protein-protein interactions were performed as indicated above with the following modifications. Calcium-containing and calcium-free binding assays were performed in parallel on separate 96-well plates. The calcium-containing assay was performed as described above. For the calcium-free assay, 2 mm CaCl2 was replaced by 10 mm EDTA in the washing, blocking, and binding buffers to chelate calcium ions in solution and extract protein-bound calcium. After binding of the soluble ligand and washing with the respective calcium- and EDTA-containing wash buffers, the final washing step in both assays was performed with calcium-containing buffer to saturate the proteins with calcium for subsequent analysis. The primary and secondary antibodies were then added in the presence of 2 mm CaCl2 in both assays to ensure equal antibody binding.
N- and O-Deglycosylation Assays
Short fibulins were deglycosylated at their N-glycosylation sites by peptide:N-glycosidase F (PNGase F; New England Biolabs, product P0704S) and at their O-glycosylation sites by O-glycosidase in conjunction with neuraminidase (New England Biolabs, product E0540S). The manufacturer's protocols were followed with the following adjustments. 10 μg of recombinant fibulin, 1.5 μl of PNGase F, 4 μl of O-glycosidase, and 4 μl of neuraminidase were used. The samples were immediately separated by 4–20% SDS-PAGE and stained with colloidal Coomassie Brilliant Blue.
Atomic Force Microscopy
All experiments, including specimen preparation and AFM measurements, were performed in air at a constant temperature at 22 °C. Diluted fibulin-4 solutions at protein concentrations in the range of 0.2–2 μg/ml were adsorbed onto freshly cleaved muscovite mica (Electron Microscopy Sciences, product 71850-01) substrates for 5 min. The mica surfaces were then quickly dried under dry nitrogen gas and mounted on a stub for AFM scanning. AFM measurements were performed with a multimode scanning probe microscope (Digital Instruments Nano-Scope IIIa, MultiMode SPM, Santa Barbara, CA) operating in the tapping mode. AFM probes were PointProbe-Plus Silicon (typical tip radius at apex, ≤7 nm) and SuperSharpSilicon (typical tip radius at apex, 2 nm) both with a 10–130 newton/m force constant and both from Nanosensors (Neuchatel, Switzerland). Height AFM images were obtained using a scanning rate of 1–2 Hz with 512 lines and 512 sampling per line, providing images of 262,144 pixels. The drive frequency was ∼300 kHz, and the amplitude set point was in the 1.5–2.5-V range with a drive amplitude of 50–500 mV. Measurements were performed with feedback control parameters set in the ranges as follows: integral gain, 0.5–1; proportional gain, 1–2; look-ahead gain, 0–0.8.
Sequence Alignments
Human fibulin-3, -4, and -5 protein sequences were retrieved from the National Center for Biotechnology Information protein database (www.ncbi.nlm.nih.gov/protein/). Sequences NP_001034437.1, NP_058634.4, and NP_006320.2 were used, respectively. Protein sequences were stored in a GCG software package version 11.1 database (Accelrys). Sequences were aligned with ClustalX 2.0 (33).
RESULTS
Short Fibulin Purification and Analysis
Recombinant human full-length short fibulin constructs were generated and transfected into HEK293 cells from which conditioned medium was collected. The secreted histidine-tagged fibulins were purified by immobilized metal ion affinity chromatography, typically yielding 1–1.5 mg of fibulin-3, -4, and -5/2.5 liters of conditioned medium.
The purified fibulins were separated by SDS-PAGE under reducing and non-reducing conditions and stained with Coomassie Brilliant Blue (Fig. 1B). Consistently, under reducing conditions, purified fibulin-3 appeared as four predominant bands (74, 62, 45, and 40 kDa), fibulin-4 appeared as two bands (65 and 61 kDa), and fibulin-5 also appeared as two bands (70 and 65 kDa) (Fig. 1B, labeled with numbers). Occasionally, the fibulin-4 preparations showed, in addition to the double band at 65 and 61 kDa (designated 5 and 6), a second and less intense double band at 51 and 47 kDa (designated 7 and 8) (Fig. 1C). Under non-reducing conditions, some higher molecular mass bands of ∼90 to >250 kDa were observed primarily for fibulin-3 and -4. Occasionally, fibulin-5 showed similar bands in some preparations. Upon reduction, all of these bands shifted to the monomeric position of the respective fibulin. The numbered bands in Fig. 1B were all observed by Western blotting with anti-His(C-term) antibody under either reducing conditions, non-reducing conditions, or both, confirming that the C termini of all three fibulins are intact (data not shown).
The various protein bands observed were further analyzed by N-terminal protein sequencing (Table 1). The 74- and 62-kDa bands of fibulin-3 (bands 1 and 2) were both identified as the intact N terminus after signal peptide cleavage, whereas the 45- and 40-kDa bands (bands 3 and 4) were shown to be cleavage products starting at amino acid residue Ala124 and Ile149, respectively (starting from Met1 of NCBI Reference Sequence NP_001034437.1). Both identified cleavage sites are located in the N-terminal extended linker region within the first, modified cbEGF domain of fibulin-3 (Fig. 1A). For fibulin-4, the 65- and 61-kDa bands (bands 5 and 6) were both determined to correspond to the N terminus of the protein after cleavage of the signal peptide. The 51- and 47-kDa bands (bands 7 and 8) were both determined to be cleaved at Gly93 (starting from Met1 of NCBI Reference Sequence NP_058634.4). This cleavage site is also located in the N-terminal linker region of the first, modified cbEGF domain of fibulin-4 (Fig. 1A). The 70-kDa band of fibulin-5 (band 9) correlates to full-length fibulin-5 after cleavage of the signal peptide, whereas the 65-kDa band (bands 10 and 11) was demonstrated to be a mixture of two fragments cleaved at Ala96 and Ala101, respectively (starting from Met1 of NCBI Reference Sequence NP_006320.2). At least one of these cleaved fibulin-5 fragments was present in each protein preparation, and some preparations contained both fragments. Both of these identified cleavage sites are also located in the N-terminal linker region of the first, modified cbEGF domain of fibulin-5 (Fig. 1A). For all three fibulins, the cleavage products resulting from cleavage within the extended linker regions were not only observed under reducing conditions but also under non-reducing conditions (Fig. 1B). This result demonstrates that the first six cysteine residues in all three fibulins are not disulfide-bonded to cysteine residues C-terminally located to the linker region. This suggests that the first extended cbEGF likely adopts a similar fold compared with other cbEGF domains. Furthermore, similarly sized fibulin-3 and -5 bands were detected in Western blots of mouse aortic extracts, indicating fibulin-3 and -5 cleavage in vivo (Fig. 1D). All cleavage sites observed had a non-polar amino acid residue (Ala, Ile, or Gly) in the P1′ position (Table 1). A ClustalX alignment of the linker insert between cysteines 4 and 5 of cbEGF1 of the three fibulins identified similarities for the cleavage sites between fibulin-3 and -5 (AA↓A), whereas the cleavage site in fibulin-4 was different (Fig. 2). In summary, despite the presence of the serine protease inhibitor PMSF, the results demonstrate that all extended linker regions in the first cbEGF domain represent critically sensitive sites for proteolytic attack.
TABLE 1.
Cleavage sites in purified short fibulin proteins identified by N-terminal sequencing
Shown are the N-terminal amino acid sequences of the purified short fibulin bands numbered in Fig. 1B as determined by N-terminal protein sequencing. NCBI Reference Sequences NP_001034437.1, NP_058634.4, and NP_006320.2 were used to number residues in fibulin-3, -4, and -5, respectively. Eight residues after the cleavage site (P1′) were identified in each N-terminal sequencing reaction, unambiguously identifying the cleavage site (P1′–P8′). Cysteine residues were identified from the stable cysteine-acrylamide compound peak on each chromatogram (47). All identified cleavage sites are found in the N-terminal linker region of the first, modified cbEGF domain of each short fibulin.
| Fibulin | Band number | N-terminal cleavage site |
Position (P1′) | Band size | |
|---|---|---|---|---|---|
| P4–P1 | P1′–P8′ | ||||
| kDa | |||||
| 3 | 1, 2 | APLAQDTE | N terminus | 74, 62 | |
| 3 | ASAA | AVAGPEMQ | 124 | 45 | |
| 4 | DPQR | IPSNPSHR | 149 | 40 | |
| 4 | 5, 6 | APLAQDSE | N terminus | 65, 61 | |
| 7, 8 | NDLH | GEGPPPPV | 93 | 51, 47 | |
| 5 | 9 | APLAQAQC | N terminus | 70 | |
| 10 | YPAA | APPLSAPN | 96 | 65 | |
| 11 | PPLS | APNYPTIS | 101 | 65 | |
FIGURE 2.

ClustalX alignment of the short fibulin N-terminal linker regions. The linker region between cysteine residues 4 and 5 of the first cbEGF domain of fibulin-3, -4, and -5 was included in the ClustalX 2.0 sequence alignment. Cleavage sites identified after the purification procedure are included as filled black arrowheads. Identified MMP cleavage sites are indicated as open arrowheads. At each position, amino acid residues conserved between two or all of the short fibulins are identically colored.
Some of the observed bands were further analyzed by mass spectrometry (Fig. 1B). For fibulin-3 preparations, the 65- and 61-kDa bands (a and b) were both confirmed to be fibulin-3. The high molecular mass bands at >250 and 92 kDa in non-reducing fibulin-4 preparations (c and d) were both identified as fibulin-4. This demonstrates that fibulin-4 forms reducible high molecular mass aggregates.
The short fibulins were functional despite partial proteolysis upon purification. Fibulin-4 and -5 interacted with immobilized tropoelastin in solid phase binding assays with similar binding profiles as reported previously (5, 12, 13, 16) (data not shown). Fibulin-3 also bound to tropoelastin albeit less efficiently than fibulin-4 and -5.
Glycosylation Analysis
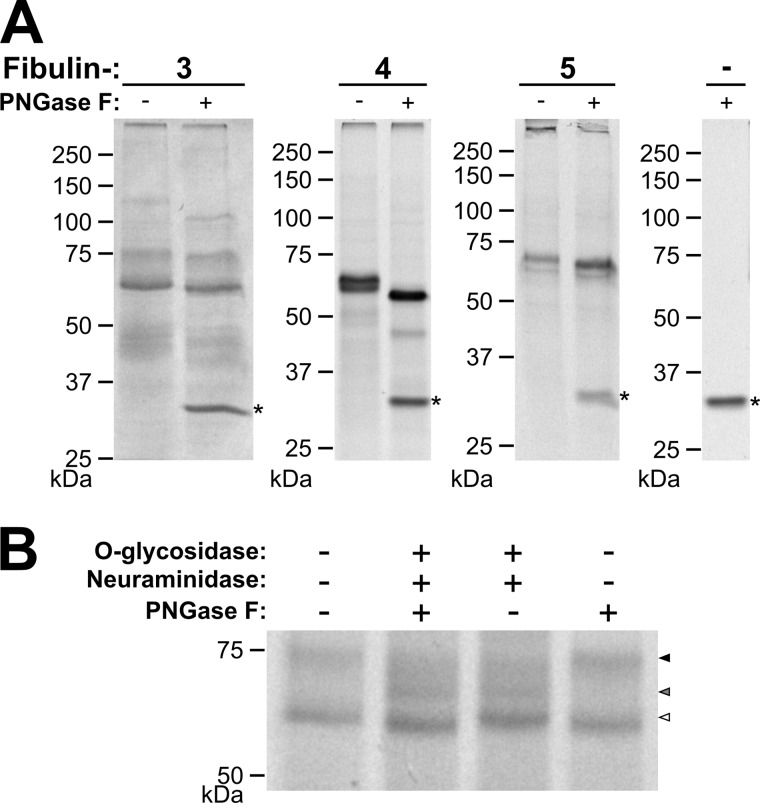
For fibulin-3 and -5, deglycosylation assays with PNGase F resulted in mobility shifts of about 4 kDa of the full length and the proteolytic cleavage products (Fig. 3A). These data indicate that the one N-linked glycosylation site in fibulin-3 and one of the two sites in fibulin-5 are glycosylated. PNGase F treatment of fibulin-4 resulted in a mobility shift of the typical double band pattern at 65 and 61 kDa to a single band of 57 kDa. Similarly, the rare double band observed at 51 and 47 kDa shifted to about 43 kDa after PNGase F treatment. This demonstrates that the double band pattern in the fibulin-4 preparations originates from a differential N-linked glycosylation pattern with either one or two sites occupied.
FIGURE 3.
Short fibulin deglycosylation by PNGase F and O-glycosidase. A, fibulin-3, -4, and -5 (10 μg/lane) were treated with (+) or without (−) PNGase F (1. 5 μl/10 μg of protein), reduced with DTT, and separated by gel electrophoresis. The protein band for PNGase F is indicated by the asterisk. B, fibulin-3 (10 μg/lane) was treated with (+) or without (−) O-glycosidase (4 μl/10 μg of protein), neuraminidase (4 μl/10 μg of protein), and PNGase F (1.5 μl/10 μg of protein); reduced with DTT; and separated by gel electrophoresis. Globular marker proteins are indicated in kDa. The gel was cut between 50 and 75 kDa, removing the two fibulin-3 fragment bands and three enzyme bands, to simplify interpretation of the results. In addition, arrowheads indicate the glycosylation variants of full-length fibulin-3. The black arrowhead indicates the most highly O-glycosylated form, the gray arrowhead indicates the O-glycosylation intermediate, and the white arrowhead indicates the least or non-O-glycosylated form of full-length fibulin-3.
O-Deglycosylation assays with fibulin-3 resulted in the full-length 74-kDa band shifting to an intermediate band closer to the full-length 62-kDa band (Fig. 3B). This mobility shift indicates that the protein has multiple O-linked glycosylation sites at least some of which were removed by the enzymatic treatment.
Degradation by Matrix Metalloproteinases
Based on the absence of efficient protease inhibition by serine protease inhibitors, it was rationalized that MMPs could potentially be responsible for the cleavage of the short fibulins during protein purification. For protease degradation assays, we first attempted to separate the degradation products present after initial chromatographic purification from the full-length proteins by gel filtration chromatography. This procedure enriched full-length fibulin-3 and -4, whereas the degradation products in the fibulin-5 preparation migrated too close to full-length fibulin-5 to be efficiently separated (Fig. 4, right control lanes, and see Fig. 8). MMP-1, -2, -3, -7, -9, and -12 were selected for degradation experiments with the short fibulins based on their relevant tissue distribution.
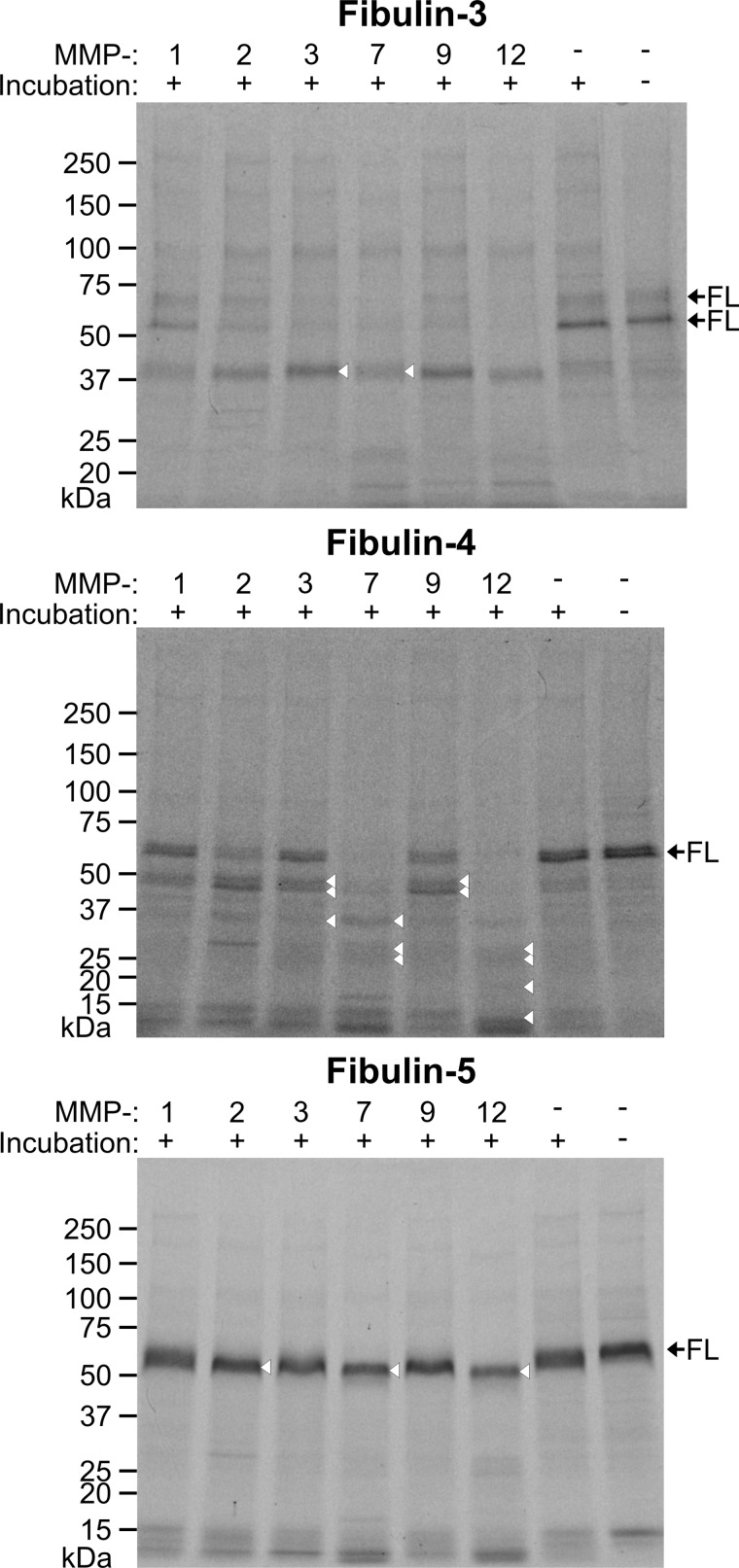
FIGURE 4.
Short fibulin proteolysis by MMPs. To separate degradation products present in short fibulin preparations after immobilized metal ion affinity chromatography, the proteins were passed through an analytical Superose 12 gel filtration column. The fractions with the most full-length fibulins were pooled and used for proteolytic assays with MMPs. 10 μg of fibulin-3, -4, and -5 treated with active MMP-1, -2, -3, -7, -9, and -12 (1:100 (w/w) enzyme-to-protein ratio, 37 °C, 24 h) were analyzed on a 4–20% gradient acrylamide gel and stained with colloidal Coomassie. Fibulins not treated with protease and/or not incubated at 37 °C for 24 h were included as controls. Full-length (FL) short fibulins are indicated by black arrows. Protein bands that were N-terminally sequenced are indicated by white triangles and are listed in Table 2. Globular marker proteins are indicated in kDa. SDS-PAGE was also performed on all six MMPs without the addition of fibulins. The usual 0.1 μg of enzyme/well was loaded, but no protein was detected by colloidal Coomassie staining (data not shown).
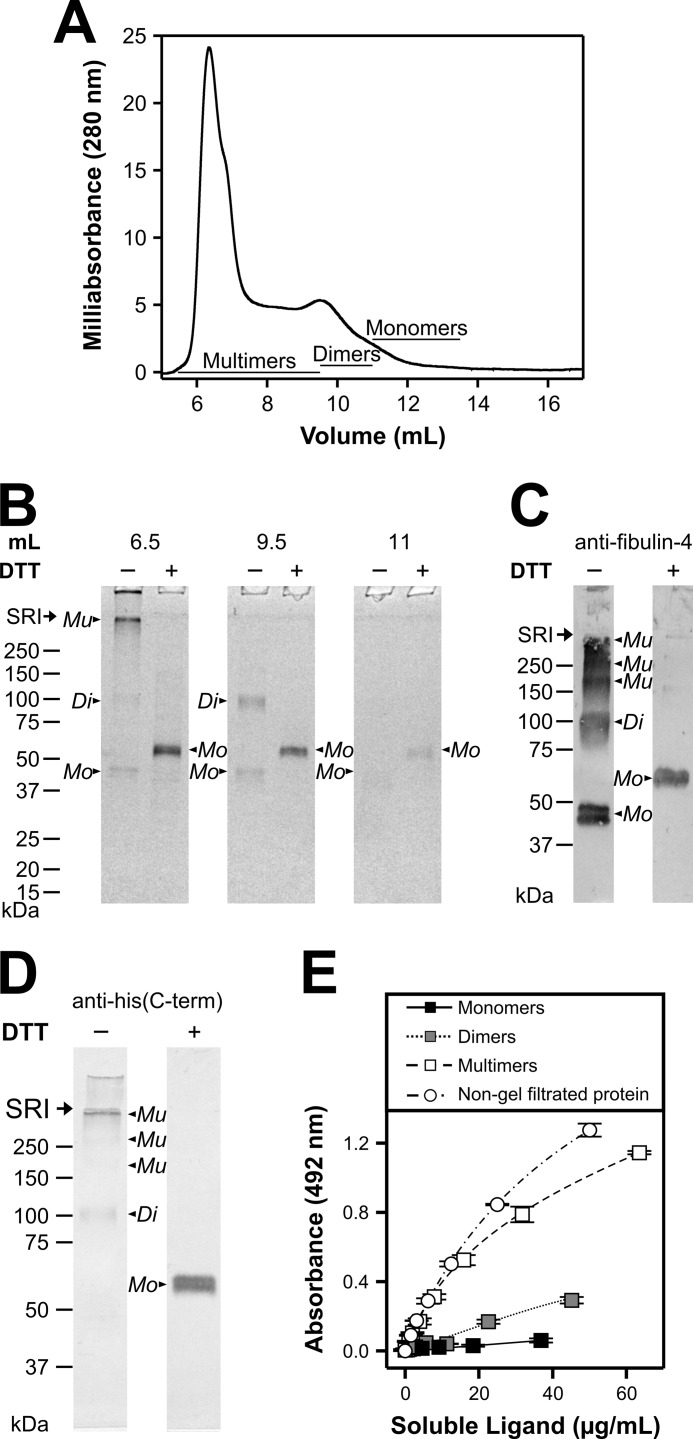
FIGURE 8.
Fibulin-4 multimer and monomer separation by gel filtration chromatography, analysis by Western blotting, and binding to heparin-BSA. A, representative analytical Superose 12 gel filtration chromatogram and corresponding fraction gels of fibulin-4. The y axis denotes the absorbance at 280 nm in milliabsorbance units. The x axis denotes elution volume in milliliters, corresponding to the numbers shown on the SDS-PAGE in B. B, individual fractions of the gel filtration shown in A were subjected to 4–20% SDS-gel electrophoresis under reducing (+) or non-reducing (−) conditions using DTT and subsequently Coomassie-stained. Multimeric (∼5.5–9 ml), dimeric (∼9.5–11 ml), and monomeric (∼11.5–13.5 ml) fibulin-4 was pooled as indicated in A. C and D, Western blot of non-gel-filtrated fibulin-4 separated on a 7.5% polyacrylamide gel under non-reducing (−) or reducing (+) conditions using DTT. Protein was detected with polyclonal anti-human fibulin-4 primary antibody (diluted 1:500) (C) or monoclonal anti-His(C-term) antibody (diluted 1:500) (D) followed by secondary antibody and development as described under “Experimental Procedures.” Note that small differences in protein migration relative to marker proteins in B versus C and D are due to the use of homogenous versus gradient gels. Positions of fibulin-4 multimers (Mu), dimers (Di), and monomers (Mo) are indicated in B and C. Globular marker proteins are indicated in kDa, and the stacking-resolving interface (SRI) of the gel is marked with an arrow. E, shown is a representative typical solid phase binding assay. Heparin-BSA was immobilized (10 μg/ml) along with BSA as a negative control (subtracted to attain the plotted values). Serial dilutions of recombinant fibulin-4 multimers, dimers, and monomers separated by gel filtration chromatography in A were added as soluble ligands at the indicated concentrations in the presence of 2 mm CaCl2. Non-gel-filtrated recombinant human fibulin-4 was used as a positive control. Data sets represent means of duplicates; S.D. (error bars) is indicated.
Each of the tested MMPs cleaved fibulin-3 and -4 to a certain extent as evidenced by the presence of several additional degradation bands in the colloidal Coomassie-stained SDS-PAGE gels (Fig. 4). MMP-7 and -12 were particularly potent and almost always completely degraded full-length fibulin-3 and -4. Short fibulin controls that were neither incubated at 37 °C nor treated with MMPs remained full-length. However, controls incubated for 24 h at 37 °C but not with MMPs already began to break down to cleavage products whose bands were similar in size to those produced by exogenous MMP addition. Fibulin-5 proteolysis by MMPs was not obvious on SDS-PAGE gels alone. However, small full-length fibulin-5 protein mobility shifts and the appearance of low molecular mass bands of about 10–15 kDa were observed with some MMP additions, in particular with MMP-7 and -12.
The major fibulin-3 band at 45 kDa produced by MMP cleavage appeared to be identical for all MMPs and corresponded in mass to the 45-kDa cleavage product band identified in purified fibulin-3 (not treated with enzyme). Analysis by N-terminal protein sequencing confirmed that the MMP-3- and -7-generated band is cleaved one amino acid residue (Val125) after the identified 45-kDa cleavage product in purified, untreated fibulin-3 (Ala124) (Table 2).
TABLE 2.
Short fibulin MMP cleavage sites identified by N-terminal sequencing
Shown are the N-terminal amino acid sequences of the MMP-treated degradation products indicated in Fig. 4 as determined by N-terminal protein sequencing. NCBI Reference Sequences NP_001034437.1, NP_058634.4, and NP_006320.2 were used to number residues in fibulin-3, -4, and -5, respectively. Eight residues after the cleavage site (P1′) were identified in each N-terminal sequencing reaction, unambiguously identifying the cleavage site (P1′–P8′). All identified cleavage sites are found in the N-terminal linker region of the first, modified cbEGF domain of each short fibulin. All sequenced bands of fibulin-4 were cleaved identically at the indicated site (see Fig. 4).
| Fibulin | Cleaved by MMP | N-terminal cleavage site |
Position (P1′) | Band size | |
|---|---|---|---|---|---|
| P4–P1 | P1′–P8′ | ||||
| kDa | |||||
| 3 | 3, 7 | SAAA | VAGPEMQT | 125 | 74 |
| 4 | 3, 7, 9, 12 | VIND | LHGEGPPP | 91 | 65, 61 |
| 5 | 2, 7, 12 | AAPP | LSAPNYPT | 99 | 70 |
Several cleavage products were generated primarily by MMP-7 and -12 treatment of fibulin-4 as judged by the complete degradation of fibulin-4 full length and fragments of >37 kDa. MMP-1, -2, -3, and -9 also degraded fibulin-4 to a typical 51- and 47-kDa double band. This double band is similar in mass to the rare lower double band (Fig. 1C, bands 7 and 8) observed in some purified fibulin-4 preparations not treated with exogenous protease. Analysis by N-terminal protein sequencing revealed that this MMP-generated double band is cleaved two amino acid residues (Leu91) upstream of the identified cleavage site of the double band in purified, untreated fibulin-4 (Gly93) (Table 2). Surprisingly, the cleavage site of this double band was identical to all of the indicated degradation products resulting from MMP-3, -7, -9, and -12 treatment. These data indicate that after initial cleavage within the first cbEGF domain additional, more C-terminally located cleavage events take place.
The starting material for fibulin-5 digestions with MMPs was a typical, two-band purified product with more full-length 70-kDa than cleaved 65-kDa fibulin-5 (Fig. 4, right control lane). As mentioned above, the resolution of SDS-PAGE gels did not suffice in determining whether MMPs cleaved the protein. N-terminal protein sequencing determined that MMP-2, -7, and -12 cleaved fibulin-5 at Leu99, three amino acid residues downstream (Ala96) and two amino acid residues upstream (Ala101) of the identified 65-kDa band of untreated fibulin-5 (Table 2).
In summary, several MMPs were able to cleave the short fibulins within their linker region of the first modified cbEGF domain, similar to what was found during protein purification. This renders the MMPs excellent candidates for physiologically relevant proteases.
Inhibition of Proteolytic Degradation by Doxycycline
Because proteolytic susceptibility of the short fibulins was observed, inhibition of MMP degradation was attempted with a broad spectrum MMP inhibitor, DOX (34). 25 μm DOX was added to the cell culture medium of recombinant short fibulin-expressing HEK293 cells. This procedure significantly prevented proteolytic breakdown of fibulin-3 (Fig. 5). Purified fibulin-3 from the DOX-treated recombinant cells lacked the 45- and 40-kDa cleavage products. The 70-kDa full length and the 65-kDa proteolytic fibulin-5 fragment were observed with or without DOX supplementation (data not shown). As fibulin-4 is regularly purified as the full-length protein, no difference was observed in purified fibulin-4 treated with or without DOX (data not shown). In summary, the data indicate that fibulin-3 is proteolytically processed by a protease in the recombinant medium that is inhibitable by DOX.
FIGURE 5.
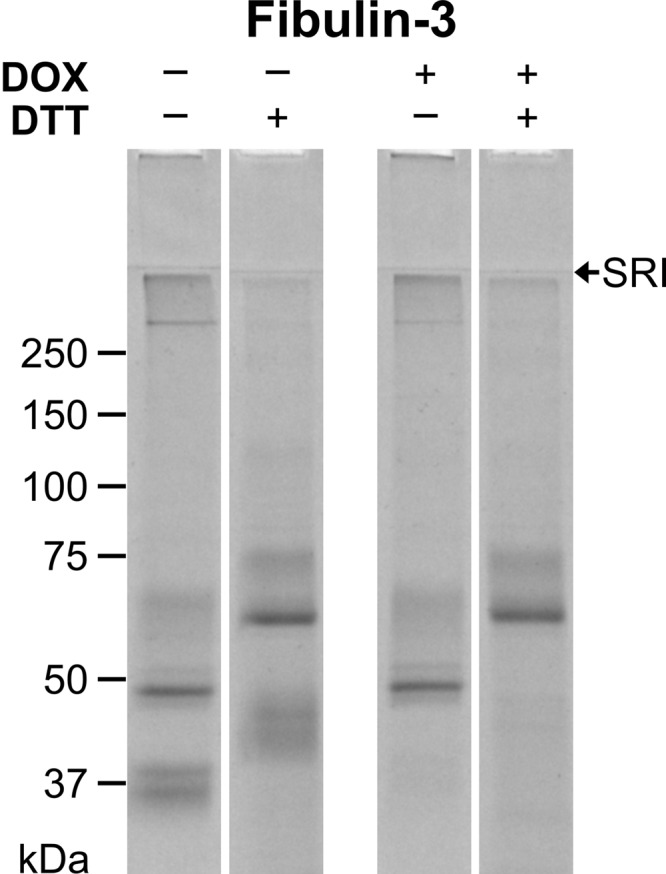
Fibulin-3 purifications with and without DOX treatment. Recombinant HEK293 cells expressing fibulin-3 were treated with or without 25 μm DOX in the cell culture medium. Conditioned medium was collected and purified by metal ion affinity chromatography. The purified proteins (10 μg/lane) were separated on a 7.5% polyacrylamide gel under non-reducing (−) or reducing (+) conditions using DTT and stained with Coomassie Brilliant Blue. SRI indicates the stacking-resolving gel interface. Globular marker proteins are indicated in kDa.
Cell Interactions
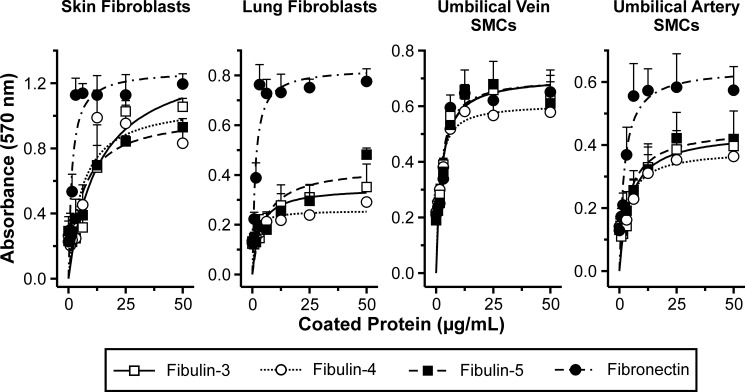
Fibulin-5, the only short fibulin possessing an RGD sequence, has been demonstrated to interact with human umbilical vein endothelial cells in an RGD-dependent manner and to bind a variety of integrins on the cell surface of Chinese hamster ovary and primary human aortic smooth muscle cells (4, 12, 17). Because no such studies have been performed on fibulin-3 and -4, which do not contain an RGD sequence, the cell interaction properties of the short fibulins were explored and compared. Representative skin and lung fibroblasts as well as umbilical vein and umbilical artery smooth muscle cells were used in cell attachment assays. The short fibulins consistently and strongly bound to all four cell types tested in a dose-dependent manner (Fig. 6). The attachment level to the various cell types was similar among fibulin-3, -4, and -5, and saturation was typically achieved at about 25 μg/ml (460 nm) coated fibulin. Short fibulin half-maximal binding was observed at 5.7–13.2 (105–243 nm) and 1.1–5.4 μg/ml (20–99 nm) for skin and lung fibroblasts and at 1.4–2.3 (26–42 nm) and 3.2–4.7 μg/ml (59–87 nm) for umbilical vein and artery smooth muscle cells, respectively. In comparison, fibronectin half-maximal binding was 1.4 μg/ml (6 nm) for fibroblasts and 2.2 μg/ml (10 nm) for smooth muscle cells. Trypsinization time was kept consistent to 3 min as shorter times did not allow proper detachment and separation of cells and longer times compromised the ability of the cells to bind the coated protein. In summary, fibulin-3, -4, and -5 interacted strongly with fibroblasts and smooth muscle cells.
FIGURE 6.
Short fibulin binding to skin and lung fibroblasts as well as to umbilical vein and artery smooth muscle cells. Shown is a representation of a typical crystal violet cell attachment assay. A serial dilution of recombinant short fibulins and fibronectin (positive control) was immobilized at the indicated concentrations. TBS without protein was used as a negative control (plotted at 0 μg/ml). 40,000 cells were added to all wells and incubated for 1 h. Data sets represent means of triplicates; S.D. (error bars) is indicated with bars omitted on one side to improve legibility of the graphs. SMCs, smooth muscle cells.
Interactions with Heparin
Because fibulin-3 and -4 interacted strongly with cells even in the absence of an RGD integrin binding site, it was hypothesized that the proteins contain intrinsic properties to interact with glycosaminoglycans. Covalently coupled heparin-BSA was used as a model of cell surface heparan sulfate as heparin alone does not efficiently coat plastic due to its highly negative charge. In addition, the two glycosaminoglycans share a similar structure, and the clustering of heparan sulfate chains on cell surface proteoglycans is mimicked by coupling three to four heparin molecules per molecule of BSA. This established procedure has been used in many other studies (32, 35–38).
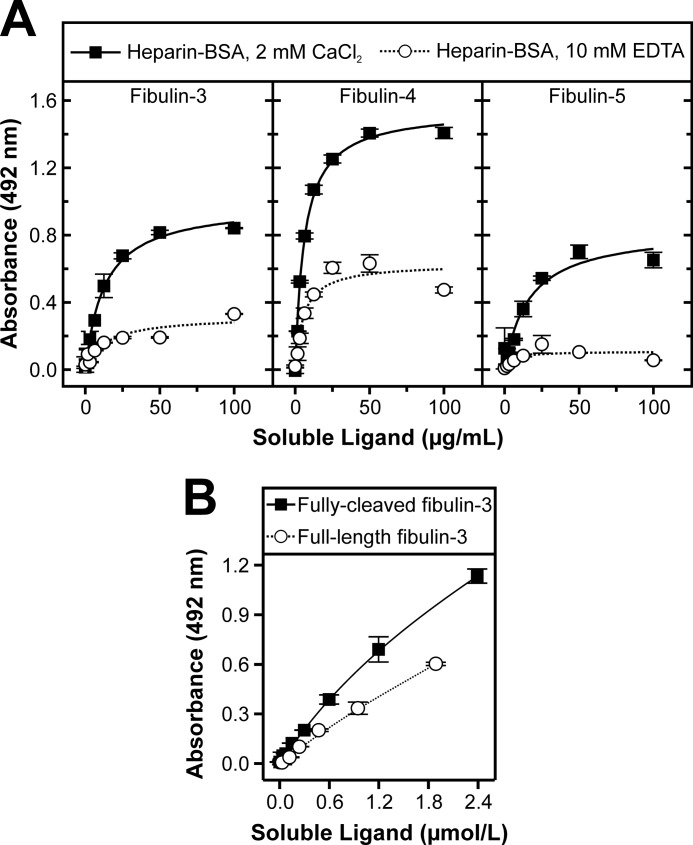
Binding of fibulin-3, -4, and -5 was tested in solid phase binding assays in the presence of calcium or EDTA (Fig. 7A). In the presence of 2 mm calcium, all short fibulins bound strongly to heparin with half-maximal binding at 13, 6, and 16 μg/ml (239, 110, and 294 nm) for fibulin-3, -4, and -5, respectively. Upon calcium chelation with 10 mm EDTA, fibulin-3 and -4 binding to heparin-BSA decreased by about half, whereas fibulin-5 binding was almost completely abolished. To ensure that the observed differences were due to the calcium-loaded and the calcium-free forms of the fibulins and not due to different affinities of these forms for the detection antibodies, a control ELISA confirmed that the detection level of the antibodies did not differ under the experimental conditions. In summary, fibulin-3, -4, and -5 strongly interact with heparin in a calcium-dependent fashion.
FIGURE 7.
Full-length and fully cleaved short fibulin binding to heparin-BSA under calcium-containing and calcium-free conditions. Shown are representative typical solid phase binding assays. Heparin-BSA was immobilized (10 μg/ml) along with BSA as a negative control (subtracted from all values to attain the plotted values). A, serial dilutions of recombinant short fibulins were added as soluble ligands at the indicated concentrations in the presence of 2 mm CaCl2 or 10 mm EDTA. B, serial dilutions of full-length and fully cleaved fibulin-3 were added as soluble ligands at the indicated concentrations in the presence of 2 mm CaCl2. Data sets represent means of duplicates; S.D. (error bars) is indicated.
Full-length fibulin-3, purified after DOX treatment, and fully cleaved fibulin-3 were compared in their ability to bind heparin-BSA (Fig. 7B). Although both proteins bound the glycosaminoglycan, the cleaved form consistently interacted more strongly.
Multimerization of Fibulin-4 and Its Functional Implication
All three fibulins showed various levels of multimerization with fibulin-4 being the most consistent in this regard (see Fig. 1B). Therefore, we analyzed multimerization of fibulin-4 and its functional consequence in more detail. Fibulin-4 purified by metal ion affinity chromatography was further separated by gel filtration chromatography using an analytical Superose 12 gel filtration column (Fig. 8A). Fibulin-4 was separated into multimers (∼5.5–9 ml), dimers (∼9.5–11 ml), and monomers (∼11.5–13.5 ml) (Fig. 8B). Western blotting of the non-gel-filtrated fibulin-4 preparation with anti-fibulin-4 antibody (Fig. 8C) and with an anti-His(C-term) antibody (Fig. 8D) confirmed that the high and intermediate molecular mass proteins were indeed multimers and dimers of fibulin-4, respectively, further confirming the mass spectrometry analyses detailed above. All dimeric and multimeric bands were reduced to the typical monomer bands after reduction (Fig. 8, B, C, and D).
Fibulin-4 multimers, dimers, and monomers separated by gel filtration (Fig. 8B) were tested for their ability to interact with heparin-BSA with respect to each other and to a non-gel-filtrated fibulin-4 control (Fig. 8E). Strong binding to heparin-BSA was determined for the control and the multimer fraction, intermediate binding was determined for the dimer fraction, and no binding was determined for the monomer fraction. These results demonstrate that multimerization provides the structural basis for the interaction with heparin-BSA and thus functionalizes fibulin-4.
Atomic force microscopy identified a heterogeneous population in purified recombinant fibulin-4 preparations (Fig. 9). Three main types of particles were observed, corresponding to monomers (45%), dimers (23%), and multimers (32%), based on shape and size. Elongated crescent-, rod-, or double knob-shaped monomers were observed with a longitudinal end-to-end length of 9–16 nm, a transversal width of 4–8 nm, and a height of 0.3–0.9 nm. Symmetrical or asymmetrical donut-shaped dimers formed from two monomers were found with a circumference of 20–33 nm, a width of 4–9 nm, an outer diameter of 19–26 nm, an inner diameter of 6–10 nm, and a height of 0.6–1 nm. The spheroidal multimers displayed a diameter of 30–50 nm and a height of 3–4 nm.
FIGURE 9.
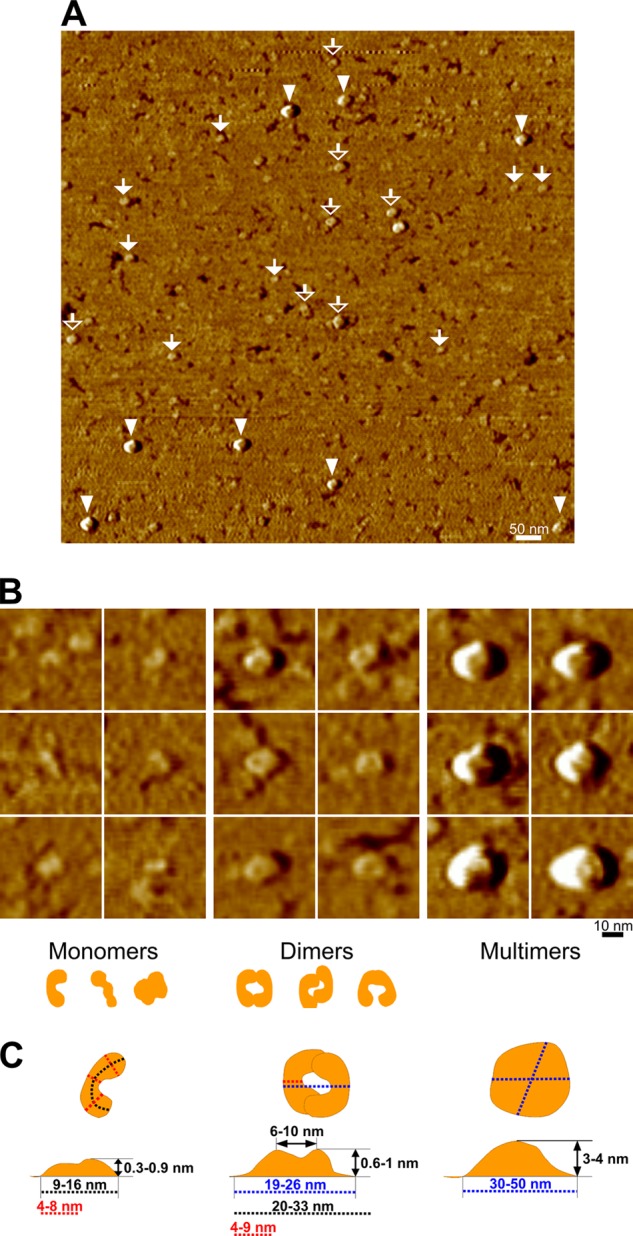
Atomic force microscopy of fibulin-4 monomers, dimers, and multimers. Atomic force microscopy imagery of fibulin-4 molecules adsorbed onto freshly cleaved mica surfaces is shown. A, representative AFM phase overview image of a 1 × 1-μm area showing three distinct populations of molecules attributed to monomers (filled white arrows), dimers (open white arrows), and multimers (white arrowheads). B, close-up of individual fibulin-4 molecules corresponding to monomers, dimers, and multimers detailing the shape and size of each of these populations. All close-up images represent an area of 50 × 50 nm. Schematic drawings of monomers and dimers are included below the panels. C, measurements of the dimensions of fibulin-4 monomers, dimers, and multimers. Note that the molecule height measurements were performed with AFM height images.
DISCUSSION
Fibulin-3, -4, and -5 comprise a subgroup of the larger fibulin family, which is involved at various levels in the formation and homeostasis of elastic tissues. Full understanding of the functional roles of these fibulins in the context of tissue biogenesis and integrity requires knowledge of their molecular properties. The chief aims of this work were to elucidate the proteolytic susceptibility of the short fibulins, their interaction with cells and heparin, and macromolecule formation.
One of the important findings of this study is that all three short fibulins are intrinsically sensitive to proteolytic degradation within their extended linker regions between cysteine residues 4 and 5 of the cbEGF1 domain. This linker region in human fibulins varies in length with 88 (fibulin-3), 28 (fibulin-4), and 44 (fibulin-5) amino acid residues. Proteolytic susceptibility of this region became apparent through (i) the presence of characteristic degradation patterns after purification by immobilized metal ion affinity chromatography and (ii) treatment with candidate MMPs.
After purification, fibulin-3 cleavage products were identified at Ala124 and Ile149, major fibulin-4 degradation products started with Gly93, and Ala96 and Ala101 were found after purification of fibulin-5. No candidate proteases could be identified based on the determined cleavage site sequences. All sites are located in the linker region of cbEGF1. Overall, the level of protease susceptibility clearly correlated with the length of the cbEGF1 linker region. Fibulin-3 was always heavily degraded after standard purification, and fibulin-5 was always somewhat degraded, whereas fibulin-4 only rarely showed the typical proteolytic breakdown products. These findings suggest that in solution the linker region in cbEGF1 protrudes from the domain, rendering it accessible to proteolytic attack, especially in the case of fibulin-3 whose linker region is particularly lengthy. Some similarities of the proteolytic cleavage site sequences and/or positions were observed between fibulin-3 and -5. In both proteins, we identified a susceptible site within an Ala-Ala-Ala sequence, and the position of the second cleavage site close to the end of the linker is similar. The cleavage site identified for fibulin-4 differs in its position and sequence. Overall, it is remarkable that, despite the low conservation of the primary linker sequence between the short fibulins, protease susceptibility in this region appears to be a conserved functional feature.
DOX supplementation of the recombinant cell culture medium successfully prevented fibulin-3 proteolysis in the linker region but had little effect on fibulin-4 and -5 degradation. As DOX is a broad spectrum MMP inhibitor, one or more members of the MMP family may be responsible for the observed fibulin-3 cleavage during purification. DOX is thought to inactivate MMPs by binding to the Zn2+ ion in their active site. However, because the a disintegrin and metalloproteinase (ADAM) enzyme family and the a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) enzyme family also contain a Zn2+ ion in their active site, they may also participate in short fibulin proteolysis.
For fibulin-3 and -4, proteolytic susceptibilities were never described. For fibulin-5, a two-band pattern of recombinant human fibulin-5 has been described previously by two groups (19, 39). Hirai et al. (19) observed that fibulin-5 was preferentially cleaved in aged mice and identified a cleavage site at position Arg77 of recombinant human fibulin-5 that is also located within the N-terminal linker region. Using aprotinin in cell culture, Hirai et al. (19) were able to inhibit fibulin-5 proteolysis, suggesting that a serine protease was involved, although the specific protease could not be identified. Similarly, Kapustin et al. (39) observed fibulin-5 cleavage in cell culture at Ser85 and Ser89 of the N-terminal linker region. Both cleavages were inhibited by aprotinin and were determined to be the result of proteolysis by plasmin. The serine protease inhibitor PMSF was routinely used in all of our recombinant fibulin purifications, explaining why no serine protease cleavage product for fibulin-5 was observed. The 65-kDa cleavage fragment of fibulin-5 is of particular importance as it does not contain the N-terminal RGD site, preventing fibulin-5 binding to integrins. The fibulin-5 fragment remains despite 25 μm DOX supplementation, suggesting that different enzymes cleave fibulin-3 and -5. Despite a partial N-terminal cleavage, fibulin-5 interacted as expected with tropoelastin in solid phase binding assays (40). The fibulin-5 cleavage product described by Hirai et al. (19) is no longer able to participate in elastogenesis. Furthermore, this cleaved fibulin-5 was found in significantly greater quantities in aged as opposed to young mice. Therefore, fibulin-5 proteolysis in the N-terminal linker region may be detrimental to its role in elastogenesis and may also contribute to aging in humans. It will be of interest to investigate whether fibulin-3 and -4 cleavage also occurs in vivo at the identified sites in the N-terminal linker region, particularly in aging mice or humans. We are currently elucidating the consequences of such fragmentation on elastogenesis.
Proteases that cleave the recombinant human short fibulins are presently unknown. We tested various MMPs (MMP-1, -2, -3, -7, -9, and -12) for their ability to hydrolyze short fibulins based on the inhibiting effects of DOX on fibulin-3 degradation and on relevant MMP expression patterns in elastic tissues. In addition, MMP-2, -3, -9, and -12 are able to cleave elastin likely in close vicinity to expressed short fibulins (23, 41). All of the tested MMPs were able to cleave fibulin-3 and -4 to various degrees with MMP-7 and -12 being the most potent. At least MMP-2, -7, and -12 were able to degrade fibulin-5. Identified cleavage products included Val125 (fibulin-3), Leu91 (fibulin-4), and Leu99 (fibulin-5). These sites are exclusively located in the cbEGF1 linker region just one to three amino acid residues away from the cleavage site observed upon recombinant short fibulin purification. Despite one reported MMP consensus cleavage site, it is still not clear whether the MMP family cleaves based on specific amino acid sequence or rather more generally based on the three-dimensional structure of the protein in question (42). MMP cleavage of the short fibulins has never been demonstrated previously. This is an important result as the various pathologies associated with short fibulin deletions and mutations may in part be propagated by the MMPs. The MMPs are associated with some of these conditions, including thoracic aortic aneurysms, pelvic organ prolapse, and lung cancer, and short fibulin expression is sometimes modified by MMPs in these diseases (18, 43–45). The treatment of these diseases with specific (tissue inhibitors of MMPs) or broad spectrum (DOX) MMP inhibitors could also prove to be beneficial.
All predicted N-glycosylation sites are located at some distance from cbEGF1 (see Fig. 1A) and are thus not expected to interfere with proteolytic processing of the linker region. In this study, we determined for fibulin-3 that the one predicted N-glycosylation site is occupied. In addition, similar to what Kobayashi et al. (5) observed for mouse fibulin-3, we hypothesized that one or more of the predicted O-glycosylation sites in human fibulin-3 is occupied based on the two different migration rates of the full-length protein. Indeed, the O-deglycosylation assay demonstrates that the 74-kDa band corresponding to full-length fibulin-3 is O-glycosylated as upon O-deglycosylation this band shifts closer to the 62-kDa full-length band. The latter band is not affected by the addition of O-glycosidase, suggesting that it is not O-glycosylated or that the enzyme is not efficient in cleaving any existing O-linked oligosaccharide chains possibly due to steric hindrance. For fibulin-4, we also observed two species of glycosylated products likely resulting from the occupation of one or both predicted N-glycosylation sites. This may be a consequence of the high level of recombinant fibulin-4 expression in HEK293 cells overloading the glycosylation machinery, or alternatively, it may have a physiological function. Based on the migration shift upon PNGase F treatment, only one of the two N-glycosylation sites present in fibulin-5 appears to be occupied. The close proximity between the two predicted N-glycosylation sites separated by only 12 amino acid residues may prevent glycosylation of both sites through steric hindrance.
Cell attachment assays detected short fibulin binding to human skin and lung fibroblasts as well as to human umbilical artery and vein smooth muscle cells. This demonstrates for the first time that fibulin-3 and -4 are able to interact with cell surface receptors despite the absence of an RGD sequence. In addition, all three short fibulins interacted with heparin (used as a model of heparan sulfate) in the presence of calcium with fibulin-4 binding with the highest affinity. Based on these results, we hypothesize that the short fibulins, in particular fibulin-4, may bind cell surface heparan sulfate, such as that on syndecans or glypicans. Furthermore, when calcium was chelated by EDTA, binding to heparin decreased by about half for fibulin-3 and -4 and was almost completely abolished for fibulin-5.
Of the short fibulin primary sequences, only cbEGF6 of fibulin-5 contains a potential heparin-binding consensus sequence in the XBBXBX form where B represents a basic amino acid residue (46). Therefore, we calculated the isoelectric point of each of the cbEGF and C-terminal domains of the short fibulins using a bioinformatics prediction tool for protein isoelectric points and molecular weights, ExPASy Compute pI/Mw Tool (Fig. 10). According to these calculations, the third cbEGF domain of fibulin-4 and the C-terminal domains of the three short fibulins have relatively basic isoelectric points that are good candidate domains for the potential interactions with negatively charged heparin. In contrast, the partial (fibulin-3 and -4) and full (fibulin-5) calcium dependence of the short fibulin interaction with heparin suggests that there are two heparin binding sites in fibulin-3 and -4 (one in a cbEGF mediated by calcium and one in the C terminus that is calcium-independent) but only one such site in fibulin-5 (in a cbEGF mediated by calcium). This contradiction may indicate that there are shorter sequences within select short fibulin domains that are able to bind heparin. The N-terminal linker regions of the short fibulins have isoelectric points between 6 and almost 10 (Fig. 10). Interestingly, fully cleaved fibulin-3 bound heparin-BSA more strongly than full-length fibulin-3. This result suggests that the N-terminal portion that is cleaved off of the protein does not contain a heparin binding site and that the linker region itself is not cleaved in a region that binds heparin. Furthermore, this result may indicate that upon fibulin-3 cleavage a heparin binding site is unmasked by the removal of the N-terminal moiety or by a more complex conformational change in the protein.
FIGURE 10.
Isoelectric points of domains and regions in human short fibulins. A schematic representation of the human short fibulins with the isoelectric points of each of the cbEGF and C-terminal domains indicated below each domain is shown. The isoelectric points of each of the N-terminal linker regions are indicated within each linker representation.
The generation of the fibulin-5RGE/RGE knock-in mouse, which developed normal elastic fibers, indicated that fibulin-5 integrin binding is dispensable for elastogenesis (18). This result raises the possibility of fibulin-5 cell attachment through cell surface heparan sulfate proteoglycans, which might be necessary for elastic fiber formation. In addition, as fibulin-3 and -4 are able to bind cells in an RGD-independent manner, uncovering which receptors are responsible for their cell interaction could help to define their roles in elastogenesis.
Under non-reducing conditions, SDS-PAGE gels of purified recombinant fibulin-4 consistently showed three protein populations of about 50, 90, and >250 kDa (Fig. 1B). Purified fibulin-3 and -5 sometimes include >250-kDa bands on SDS-PAGE gels. However, these bands are neither as consistent nor as prominent as the high molecular weight bands observed for fibulin-4. These fibulin-4 high molecular weight bands were also detected in Western blots and were all confirmed to be fibulin-4 by mass spectrometry. Upon reduction with DTT, all bands converged, suggesting that the band at 50 kDa corresponds to fibulin-4 monomers, whereas the bands at 90 and >250 kDa correspond to disulfide-bonded dimers and multimers, respectively. Preliminary data produced identical SDS-PAGE gel profiles of fibulin-4 treated with or without 10 mm EDTA, indicating that dimerization and multimerization of the protein is not calcium-dependent (data not shown). Contributions of the hexahistidine tag to fibulin-4 multimerization were excluded as non-tagged fibulin-4 produced similar dimeric and multimeric patterns (data not shown). Atomic force microscopy analysis confirmed a heterogeneous population in the fibulin-4 preparations consisting of monomers, dimers, and multimers. The bent rodlike shape and size of the monomers as visualized here are very similar to those of the fibulin-4 monomers determined previously (3, 5, 16). However, those authors only detected monomeric fibulin-4 by non-reducing SDS-PAGE, rotary shadowing, and equilibrium ultracentrifugation among others most likely because this material was gel-filtrated, separating monomers from the higher molecular weight multimers. Furthermore, multiangle laser light scattering and single particle averaging demonstrated monomeric fibulin-4 in the absence of calcium that mostly dimerized upon addition of calcium (16). In contrast, reducible dimers and multimers make up the majority of the purified fibulin-4 in the present study. Atomic force microscopy revealed symmetrical and asymmetrical donut-shaped fibulin-4 dimers with an open core. This structure is similar to the shape of fibulin-5 analyzed by single particle image processing (6). Here, we demonstrate for the first time that a significant portion (>30%) of the purified fibulin-4 occurs in discrete disulfide-bonded high molecular weight spheroidal multimers with a diameter of 30–50 nm and a height of 3–4 nm. The functional importance of this multimerization is emphasized by the strong interaction between multimeric fibulin-4 and heparin-BSA in contrast to the negligible interaction of monomeric fibulin-4 with heparin. This suggests that multimerization may promote the interaction of fibulin-4 with cell surface proteoglycans, which may in turn be an essential step in the formation of elastic fibers.
In conclusion, this work reveals a number of new and surprising characteristics of the short fibulin proteins. Proteolytic susceptibility of the short fibulin N-terminal linker region was demonstrated as well as the susceptibility of the short fibulins to numerous MMPs commonly found in short fibulin-rich tissues. Short fibulin cell attachment and binding to heparin were also determined, suggesting that the short fibulins, in particular fibulin-3 and -4, which lack an RGD sequence, may bind non-integrin cell surface receptors such as heparan sulfate containing proteoglycans. Lastly, fibulin-4 dimerization and multimerization were demonstrated by Western blotting, mass spectrometry, and atomic force microscopy.
Acknowledgments
We thank Dr. Kurt Dejgaard, Dr. Jing Liu, and Amy Wong for performing the mass spectrometry analysis; Dr. Cynthia Goodyer for providing human umbilical artery smooth muscle cells; Drs. Ming Miao and Fred Keeley for providing human tropoelastin; and Dr. Nicolas Giorgi for performing select preliminary experiments. We also express gratitude to Drs. Dirk Hubmacher and Laetitia Sabatier for many valuable suggestions.
This work was supported by Natural Sciences and Engineering Research Council of Canada Grant RGPIN 375738-09, Canadian Institutes of Health Research Grant MOP-106494, and the Network for Oral and Bone Health Research.
- cbEGF
- calcium-binding epidermal growth factor
- BCA
- bicinchoninic acid
- DOX
- doxycycline
- MMP
- matrix metalloproteinase
- PNGase F
- peptide:N-glycosidase F
- RGD
- arginine-glycine-aspartic acid
- RGE
- arginine-glycine-glutamic acid
- TBS/Ca
- CaCl2-containing Tris-buffered saline
- AFM
- atomic force microscopy.
REFERENCES
- 1. Cooley M. A., Argraves W. S. (2011) in Biology of Extracellular Matrix (Mecham R. P., ed) pp. 337–359, Springer, New York [Google Scholar]
- 2. Lecka-Czernik B., Lumpkin C. K., Jr., Goldstein S. (1995) An overexpressed gene transcript in senescent and quiescent human fibroblasts encoding a novel protein in the epidermal growth factor-like repeat family stimulates DNA synthesis. Mol. Cell. Biol. 15, 120–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giltay R., Timpl R., Kostka G. (1999) Sequence, recombinant expression and tissue localization of two novel extracellular matrix proteins, fibulin-3 and fibulin-4. Matrix Biol. 18, 469–480 [DOI] [PubMed] [Google Scholar]
- 4. Nakamura T., Ruiz-Lozano P., Lindner V., Yabe D., Taniwaki M., Furukawa Y., Kobuke K., Tashiro K., Lu Z., Andon N. L., Schaub R., Matsumori A., Sasayama S., Chien K. R., Honjo T. (1999) DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J. Biol. Chem. 274, 22476–22483 [DOI] [PubMed] [Google Scholar]
- 5. Kobayashi N., Kostka G., Garbe J. H., Keene D. R., Bächinger H. P., Hanisch F. G., Markova D., Tsuda T., Timpl R., Chu M. L., Sasaki T. (2007) A comparative analysis of the fibulin protein family. Biochemical characterization, binding interactions, and tissue localization. J. Biol. Chem. 282, 11805–11816 [DOI] [PubMed] [Google Scholar]
- 6. Jones R. P., Wang M. C., Jowitt T. A., Ridley C., Mellody K. T., Howard M., Wang T., Bishop P. N., Lotery A. J., Kielty C. M., Baldock C., Trump D. (2009) Fibulin 5 forms a compact dimer in physiological solutions. J. Biol. Chem. 284, 25938–25943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Vega S., Iwamoto T., Yamada Y. (2009) Fibulins: multiple roles in matrix structures and tissue functions. Cell. Mol. Life Sci. 66, 1890–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McLaughlin P. J., Bakall B., Choi J., Liu Z., Sasaki T., Davis E. C., Marmorstein A. D., Marmorstein L. Y. (2007) Lack of fibulin-3 causes early aging and herniation, but not macular degeneration in mice. Hum. Mol. Genet. 16, 3059–3070 [DOI] [PubMed] [Google Scholar]
- 9. Stone E. M., Lotery A. J., Munier F. L., Héon E., Piguet B., Guymer R. H., Vandenburgh K., Cousin P., Nishimura D., Swiderski R. E., Silvestri G., Mackey D. A., Hageman G. S., Bird A. C., Sheffield V. C., Schorderet D. F. (1999) A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat. Genet. 22, 199–202 [DOI] [PubMed] [Google Scholar]
- 10. McLaughlin P. J., Chen Q., Horiguchi M., Starcher B. C., Stanton J. B., Broekelmann T. J., Marmorstein A. D., McKay B., Mecham R., Nakamura T., Marmorstein L. Y. (2006) Targeted disruption of fibulin-4 abolishes elastogenesis and causes perinatal lethality in mice. Mol. Cell. Biol. 26, 1700–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hucthagowder V., Sausgruber N., Kim K. H., Angle B., Marmorstein L. Y., Urban Z. (2006) Fibulin-4: a novel gene for an autosomal recessive cutis laxa syndrome. Am. J. Hum. Genet. 78, 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamura T., Lozano P. R., Ikeda Y., Iwanaga Y., Hinek A., Minamisawa S., Cheng C. F., Kobuke K., Dalton N., Takada Y., Tashiro K., Ross J., Jr., Honjo T., Chien K. R. (2002) Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature 415, 171–175 [DOI] [PubMed] [Google Scholar]
- 13. Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., Olson E. N. (2002) Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 415, 168–171 [DOI] [PubMed] [Google Scholar]
- 14. Jones R. P., Ridley C., Jowitt T. A., Wang M. C., Howard M., Bobola N., Wang T., Bishop P. N., Kielty C. M., Baldock C., Lotery A. J., Trump D. (2010) Structural effects of fibulin 5 missense mutations associated with age-related macular degeneration and cutis laxa. Invest. Ophthalmol. Vis. Sci. 51, 2356–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. El-Hallous E., Sasaki T., Hubmacher D., Getie M., Tiedemann K., Brinckmann J., Bätge B., Davis E. C., Reinhardt D. P. (2007) Fibrillin-1 interactions with fibulins depend on the first hybrid domain and provide an adapter function to tropoelastin. J. Biol. Chem. 282, 8935–8946 [DOI] [PubMed] [Google Scholar]
- 16. Choudhury R., McGovern A., Ridley C., Cain S. A., Baldwin A., Wang M.-C., Guo C., Mironov A., Jr., Drymoussi Z., Trump D., Shuttleworth A., Baldock C., Kielty C. M. (2009) Differential regulation of elastic fiber formation by fibulin-4 and -5. J. Biol. Chem. 284, 24553–24567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lomas A. C., Mellody K. T., Freeman L. J., Bax D. V., Shuttleworth C. A., Kielty C. M. (2007) Fibulin-5 binds human smooth-muscle cells through α5β1 and α4β1 integrins, but does not support receptor activation. Biochem. J. 405, 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Budatha M., Roshanravan S., Zheng Q., Weislander C., Chapman S. L., Davis E. C., Starcher B., Word R. A., Yanagisawa H. (2011) Extracellular matrix proteases contribute to progression of pelvic organ prolapse in mice and humans. J. Clin. Investig. 121, 2048–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirai M., Ohbayashi T., Horiguchi M., Okawa K., Hagiwara A., Chien K. R., Kita T., Nakamura T. (2007) Fibulin-5/DANCE has an elastogenic organizer activity that is abrogated by proteolytic cleavage in vivo. J. Cell Biol. 176, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sekhon B. S. (2010) Matrix metalloproteinases—an overview. Res. Rep. Biol. 2010, 1–20 [Google Scholar]
- 21. Ashworth J. L., Murphy G., Rock M. J., Sherratt M. J., Shapiro S. D., Shuttleworth C. A., Kielty C. M. (1999) Fibrillin degradation by matrix metalloproteinases: implications for connective tissue remodelling. Biochem. J. 340, 171–181 [PMC free article] [PubMed] [Google Scholar]
- 22. Kirschner R., Hubmacher D., Iyengar G., Kaur J., Fagotto-Kaufmann C., Brömme D., Bartels R., Reinhardt D. P. (2011) Classical and neonatal Marfan syndrome mutations in fibrillin-1 cause differential protease susceptibilities and protein function. J. Biol. Chem. 286, 32810–32823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Visse R., Nagase H. (2003) Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 92, 827–839 [DOI] [PubMed] [Google Scholar]
- 24. Koullias G. J., Ravichandran P., Korkolis D. P., Rimm D. L., Elefteriades J. A. (2004) Increased tissue microarray matrix metalloproteinase expression favors proteolysis in thoracic aortic aneurysms and dissections. Ann. Thorac. Surg. 78, 2106–2110 [DOI] [PubMed] [Google Scholar]
- 25. Albin R. J., Senior R. M., Welgus H. G., Connolly N. L., Campbell E. J. (1987) Human alveolar macrophages secrete an inhibitor of metalloproteinase elastase. Am. Rev. Respir. Dis. 135, 1281–1285 [DOI] [PubMed] [Google Scholar]
- 26. Gu W., Liu W., Yang X., Yuan X., Tian Y., Meng R., Zhao Q. (2011) Cutis laxa: analysis of metalloproteinases and extracellular matrix expression by immunohistochemistry and histochemistry. Eur. J. Dermatol. 21, 717–721 [DOI] [PubMed] [Google Scholar]
- 27. Vine N., Powell J. T. (1991) Metalloproteinases in degenerative aortic disease. Clin. Sci. 81, 233–239 [DOI] [PubMed] [Google Scholar]
- 28. Horiguchi M., Inoue T., Ohbayashi T., Hirai M., Noda K., Marmorstein L. Y., Yabe D., Takagi K., Akama T. O., Kita T., Kimura T., Nakamura T. (2009) Fibulin-4 conducts proper elastogenesis via interaction with cross-linking enzyme lysyl oxidase. Proc. Natl. Acad. Sci. U.S.A. 106, 19029–19034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Freeman L. J., Lomas A., Hodson N., Sherratt M. J., Mellody K. T., Weiss A. S., Shuttleworth A., Kielty C. M. (2005) Fibulin-5 interacts with fibrillin-1 molecules and microfibrils. Biochem. J. 388, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin G., Tiedemann K., Vollbrandt T., Peters H., Batge B., Brinckmann J., Reinhardt D. P. (2002) Homo- and heterotypic fibrillin-1 and -2 interactions constitute the basis for the assembly of microfibrils. J. Biol. Chem. 277, 50795–50804 [DOI] [PubMed] [Google Scholar]
- 31. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 32. Hubmacher D., Cirulis J. T., Miao M., Keeley F. W., Reinhardt D. P. (2010) Functional consequences of homocysteinylation of the elastic fiber proteins fibrillin-1 and tropoelastin. J. Biol. Chem. 285, 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 34. Franco C., Ho B., Mulholland D., Hou G., Islam M., Donaldson K., Bendeck M. P. (2006) Doxycycline alters vascular smooth muscle cell adhesion, migration, and reorganization of fibrillar collagen matrices. Am. J. Pathol. 168, 1697–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tiedemann K., Bätge B., Müller P. K., Reinhardt D. P. (2001) Interactions of fibrillin-1 with heparin/heparan sulfate: implications for microfibrillar assembly. J. Biol. Chem. 276, 36035–36042 [DOI] [PubMed] [Google Scholar]
- 36. Kett W. C., Osmond R. I., Stevenson S. M., Moe L., Coombe D. R. (2005) Direct detection of the binding of avidin and lactoferrin fluorescent probes to heparinized surfaces. Anal. Biochem. 339, 206–215 [DOI] [PubMed] [Google Scholar]
- 37. Boyle M. J., Richards J. S., Gilson P. R., Chai W., Beeson J. G. (2010) Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood 115, 4559–4568 [DOI] [PubMed] [Google Scholar]
- 38. Xin X., Higai K., Imaizumi Y., Suzuki C., Ito K., Itoh A., Matsumoto S., Azuma Y., Matsumoto K. (2011) Natural killer group 2A (NKG2A) and natural killer group 2C (NKG2C) bind to sulfated glycans and α2,3-NeuAc-containing glycoproteins. Biol. Pharm. Bull. 34, 480–485 [DOI] [PubMed] [Google Scholar]
- 39. Kapustin A., Stepanova V., Aniol N., Cines D. B., Poliakov A., Yarovoi S., Lebedeva T., Wait R., Ryzhakov G., Parfyonova Y., Gursky Y., Yanagisawa H., Minashkin M., Beabealashvilli R., Vorotnikov A., Bobik A., Tkachuk V. (2012) Fibulin-5 binds urokinase-type plasminogen activator and mediates urokinase-stimulated β1-integrin-dependent cell migration. Biochem. J. 443, 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng Q., Davis E. C., Richardson J. A., Starcher B. C., Li T., Gerard R. D., Yanagisawa H. (2007) Molecular analysis of fibulin-5 function during de novo synthesis of elastic fibers. Mol. Cell. Biol. 27, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li D. Q., Meller D., Liu Y., Tseng S. C. (2000) Overexpression of MMP-1 and MMP-3 by cultured conjunctivochalasis fibroblasts. Invest. Ophthalmol. Vis. Sci. 41, 404–410 [PubMed] [Google Scholar]
- 42. Kridel S. J., Chen E., Kotra L. P., Howard E. W., Mobashery S., Smith J. W. (2001) Substrate hydrolysis by matrix metalloproteinase-9. J. Biol. Chem. 276, 20572–20578 [DOI] [PubMed] [Google Scholar]
- 43. Kaijzel E. L., van Heijningen P. M., Wielopolski P. A., Vermeij M., Koning G. A., van Cappellen W. A., Que I., Chan A., Dijkstra J., Ramnath N. W., Hawinkels L. J., Bernsen M. R., Löwik C. W., Essers J. (2010) Multimodality imaging reveals a gradual increase in matrix metalloproteinase activity at aneurysmal lesions in live fibulin-4 mice. Circ. Cardiovasc. Imaging 3, 567–577 [DOI] [PubMed] [Google Scholar]
- 44. Jackson V., Olsson T., Kurtovic S., Folkersen L., Paloschi V., Wågsäter D., Franco-Cereceda A., Eriksson P. (2012) Matrix metalloproteinase 14 and 19 expression is associated with thoracic aortic aneurysms. J. Thorac. Cardiovasc. Surg. 144, 459–466 [DOI] [PubMed] [Google Scholar]
- 45. Yue W., Sun Q., Landreneau R., Wu C., Siegfried J. M., Yu J., Zhang L. (2009) Fibulin-5 suppresses lung cancer invasion by inhibiting matrix metalloproteinase-7 expression. Cancer Res. 69, 6339–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cardin A. D., Weintraub H. J. (1989) Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis 9, 21–32 [DOI] [PubMed] [Google Scholar]
- 47. Brune D. C. (1992) Alkylation of cysteine with acrylamide for protein sequence analysis. Anal. Biochem. 207, 285–290 [DOI] [PubMed] [Google Scholar]