Background: SNF1/AMP-activated protein kinases (AMPKs) are central energy regulators that are inactivated by dephosphorylation of the activation loop.
Results: Mutation of Ptc1 protein phosphatase 2C (PP2C) affects dephosphorylation of SNF1/AMPK in Saccharomyces cerevisiae.
Conclusion: Ptc1 and Sit4 type 2A-related phosphatase contribute to glucose-regulated dephosphorylation, whereas Reg1-Glc7 PP1 plays the major role.
Significance: At least three phosphatases regulate yeast SNF1/AMPK in vivo.
Keywords: AMP-activated Kinase (AMPK), Glucose, Phosphorylation, Protein Phosphatase, Yeast Genetics
Abstract
The SNF1/AMP-activated protein kinases (AMPKs) function in energy regulation in eukaryotic cells. SNF1/AMPKs are αβγ heterotrimers that are activated by phosphorylation of the activation loop Thr on the catalytic subunit. Protein kinases that activate SNF1/AMPK have been identified, but the protein phosphatases responsible for dephosphorylation of the activation loop are less well defined. For Saccharomyces cerevisiae SNF1/AMPK, Reg1-Glc7 protein phosphatase 1 and Sit4 type 2A-related phosphatase function together to dephosphorylate Thr-210 on the Snf1 catalytic subunit during growth on high concentrations of glucose; reg1Δ and sit4Δ single mutations do not impair dephosphorylation when inappropriate glycogen synthesis, also caused by these mutations, is blocked. We here present evidence that Ptc1 protein phosphatase 2C also has a role in dephosphorylation of Snf1 Thr-210 in vivo. The sit4Δ ptc1Δ mutant exhibited partial defects in regulation of the phosphorylation state of Snf1. The reg1Δ ptc1Δ mutant was viable only when expressing mutant Snf1 proteins with reduced kinase activity, and Thr-210 phosphorylation of the mutant SNF1 heterotrimers was substantially elevated during growth on high glucose. This evidence, together with findings on the reg1Δ sit4Δ mutant, indicates that although Reg1-Glc7 plays the major role, all three phosphatases contribute to maintenance of the Snf1 activation loop in the dephosphorylated state during growth on high glucose. Ptc1 has overlapping functions with Reg1-Glc7 and Sit4 in glucose regulation of SNF1/AMPK and cell viability.
Introduction
The SNF1/AMPK2 family has roles in energy regulation and stress responses, ranging from control of metabolic enzymes to transcription, at both the cellular and organismal levels (for review, see Ref. 1). The SNF1/AMPK enzymes are αβγ heterotrimers and are activated by phosphorylation of the activation loop Thr on the catalytic Snf1/α subunit. Protein kinases that phosphorylate the activation loop have been identified in Saccharomyces cerevisiae and mammals, but the set of protein phosphatases responsible for dephosphorylation is less well defined.
In the yeast S. cerevisiae, SNF1 is required for adaptation to environmental stresses, notably reduced availability of carbon source, the major energy supply for the cell (for review, see Ref. 2). During growth of S. cerevisiae cells on high concentrations of glucose, SNF1 is largely inactive, and Thr-210 on the activation loop is predominantly in the dephosphorylated state. Reg1-Glc7 PP1, comprising the regulatory subunit Reg1 and the catalytic subunit Glc7, was implicated in regulating the phosphorylation state of Thr-210 and SNF1 function by analysis of reg1Δ mutants (3–8); moreover, ADP protected SNF1 from dephosphorylation by purified Glc7 in vitro (9, 10). However, during growth on high glucose, the reg1Δ mutant exhibited not only elevated Thr-210 phosphorylation but also elevated glycogen accumulation, and mutations that blocked this inappropriate glycogen synthesis also restored Thr-210 dephosphorylation (11). It is likely that elevated glycogen synthesis promotes SNF1 activation by reducing glucose signaling to the SNF1 pathway.
A search for another phosphatase that acts on Thr-210 identified the Sit4 type 2A-related protein phosphatase (11), which also affects the mitotic G1/S transition (12), TOR (target of rapamycin) pathway (13), and eIF2α phosphorylation (14). The sit4Δ mutation similarly caused both a defect in Thr-210 dephosphorylation and inappropriate glycogen synthesis during growth on glucose, and blocking the latter restored dephosphorylation (11). The reg1Δ sit4Δ double mutation was lethal in cells expressing SNF1, but a truncated Snf1 kinase domain with reduced activity (residues 1–309; lacking the C-terminal regulatory region) could be expressed in glycogen-deficient reg1Δ sit4Δ snf1Δ cells, and phosphorylation of Thr-210 was highly elevated during growth on glucose (15). These findings indicated that both Reg1-Glc7 and Sit4 function in glucose-regulated dephosphorylation of Thr-210 and do so independently of the intact SNF1 heterotrimer. Evidence for physical interaction of Reg1 and Sit4 with Snf1 (4, 5, 11) supported the idea that these phosphatases directly dephosphorylate Thr-210.
For mammalian AMPK, the relevant phosphatases for the activation loop in vivo are not yet firmly established. Type 2A and 2Cα protein phosphatases dephosphorylate the activation loop Thr in vitro (16, 17); however, studies of mammalian cells implicated Ppm1E, and probably Ppm1F (18), and PP1-R6, a member of the PP1 family (19). The S. cerevisiae homolog of Ppm1E and Ppm1F is Ptc1, which is named for its identity as a PP2C family member. We have here used the yeast genetic system to address the role of Ptc1 in regulating activation-loop phosphorylation of SNF1/AMPK.
S. cerevisiae encodes a family of seven PP2C catalytic subunits, Ptc1 through Ptc7; the catalytic subunits of the PP2C family are not usually associated with regulatory subunits. Ptc1 is the best characterized, and it is structurally and functionally distinct from the others, although it may share some functions (for review, see ref. 20). Ptc1 is implicated in MAPK pathways such as the high-osmolarity glycerol and cell wall integrity pathways, the TOR (target of rapamycin) pathway, cation homeostasis, inheritance of cellular organelles, and other functions (20). Analysis of the ptc1Δ mutant using DNA microarrays showed no significant effects on expression of glucose-regulated genes (21, 22). Ptc1 binds to the adaptor protein Nbp2, which mediates its association with multiple protein kinases, but SNF1 was not identified as an interaction partner (23). We here examine the effects of the ptc1Δ mutation and Ptc1 overexpression, in combination with reg1Δ and/or sit4Δ mutations, on Thr-210 phosphorylation of wild-type and mutant forms of SNF1.
EXPERIMENTAL PROCEDURES
Strains
S. cerevisiae strains had the W303 (ade2 can1 ura3 leu2 his3 trp1) genetic background. The alleles SNF1–8xmyc::TRP1, snf1Δ::LEU2, snf4Δ::kanMX4, sit4Δ::nat1, reg1Δ::HIS3 and glc3Δ::kanMX4 were used previously (11, 24). The SSD1-v1 allele, which is present in many common strain backgrounds, is essential for viability of sit4Δ mutants (25) and does not affect phosphorylation of Thr-210 (11). ptc1Δ::nat1 and ptc1Δ::hphMX4 were constructed by replacing the 0.7-kb SnaBI/BamHI fragment, and ptc2Δ::TRP1 by replacing the 0.9-kb XbaI/NruI fragment. ptc3Δ::LEU2 (26) and ptc6Δ::kanMX4 (27) have been described. ptc4Δ::URA3, ptc5Δ::HIS3, ptc7Δ::hphMX4, and glc3Δ::URA3 were constructed using marker swap plasmids (28) to replace kanMX4 in the cognate alleles (Open Biosystems). Standard methods for genetic analysis and transformation were used.
Plasmids
Snf1–8xmyc and Snf1(1–309)-8xmyc were expressed from the SNF1 promoter on centromeric plasmids pYL199, pYL411, and pYL414 (15, 24). pXX7 and pYL200 are derivatives of pYL411 and pYL199 in vectors pRS316 (29) and YCp50 (30), respectively. Mutated versions of Snf1, expressed from the native promoter on a centromeric plasmid, have been described (31); catalytic activity of Snf1EA toward a synthetic peptide substrate was reduced 5-fold relative to WT (32). Ptc1 and Ptc1D58N coding sequences were amplified from YEp195-PTC1 (33) and YEp195-Ptc1(D58N) (34) by PCR and used to transform yeast for gap repair of the centromeric vector pMK547 (35); the recovered plasmids, pAR12 and pAR32, express 3xHA-Ptc1 and 3xHA-Ptc1D58N, respectively, from the ADH1 promoter and terminator. pAR48 was generated by gap repair of centromeric vector pRS314 and expressed 3xHA-Ptc1 from its own promoter and terminator. Recovered plasmids were sequenced.
Growth of Cultures
Cells were grown to mid-log phase in selective synthetic complete medium (SC) containing 2% (high) glucose, and an aliquot of the culture was harvested by rapid filtration to preserve the phosphorylation state of Thr-210 and was frozen in liquid nitrogen. Another aliquot was collected by rapid filtration, resuspended in SC containing 0.05% (low) glucose for 10 min, collected by filtration, and frozen.
Immunoblot Analysis
For analysis of Snf1 phosphorylation, whole cell extracts were prepared (11), and proteins (10 μg) were separated by SDS-PAGE on 7.5% polyacrylamide and analyzed by immunoblotting with anti-Thr(P)-172-AMPK (Cell Signaling Technologies). Membranes were incubated in 0.2 m glycine, pH 2, for 10 min and reprobed with anti-Myc (9E10, Santa Cruz Biotechnology) or anti-polyhistidine (Sigma; Snf1 has a stretch of His residues) to detect Snf1 polypeptides. ECL Plus (GE Healthcare) was used for visualization. Intensity of bands was quantified on appropriate exposures using ImageJ software (36), and phosphorylated Thr-210 was normalized to Snf1 protein. Analysis of HA-Ptc1 was carried out using 10% polyacrylamide and anti-HA (12CA5, Roche Applied Science) and anti-α-tubulin (Sigma).
RESULTS
Ptc1 Is Dispensable for Snf1 Thr-210 Dephosphorylation in Cells Containing Reg1-Glc7 and Sit4
To determine whether glucose regulation of the phosphorylation state of Snf1 Thr-210 depends on Ptc1, we first examined Myc-tagged Snf1, expressed from a centromeric plasmid in ptc1Δ snf1Δ and ptc1Δ snf1Δ glc3Δ cells. The glc3Δ mutation deletes the gene encoding glycogen branching enzyme, thereby avoiding the possibility of inappropriate glycogen synthesis, which results in elevated Snf1 phosphorylation (11). Cells were grown to mid-log phase on a high (2%) concentration of glucose, and an aliquot was shifted to low (0.05%) glucose for 10 min. Cell extracts were prepared, and proteins were analyzed by immunoblotting with antibodies that recognize phospho-Thr-210 and Myc. No defect in Thr-210 dephosphorylation was observed in cells grown on high glucose, and phosphorylation increased appropriately in response to glucose limitation (Fig. 1A). Glucose replenishment of glucose-depleted ptc1Δ cultures resulted in rapid dephosphorylation of Thr-210 (Fig. 1B). Mutants lacking all seven Ptc family members showed normal glucose-regulated dephosphorylation of both Snf1 and the truncated Snf1 kinase domain, Snf1(1–309)-myc (Fig. 1C).
FIGURE 1.
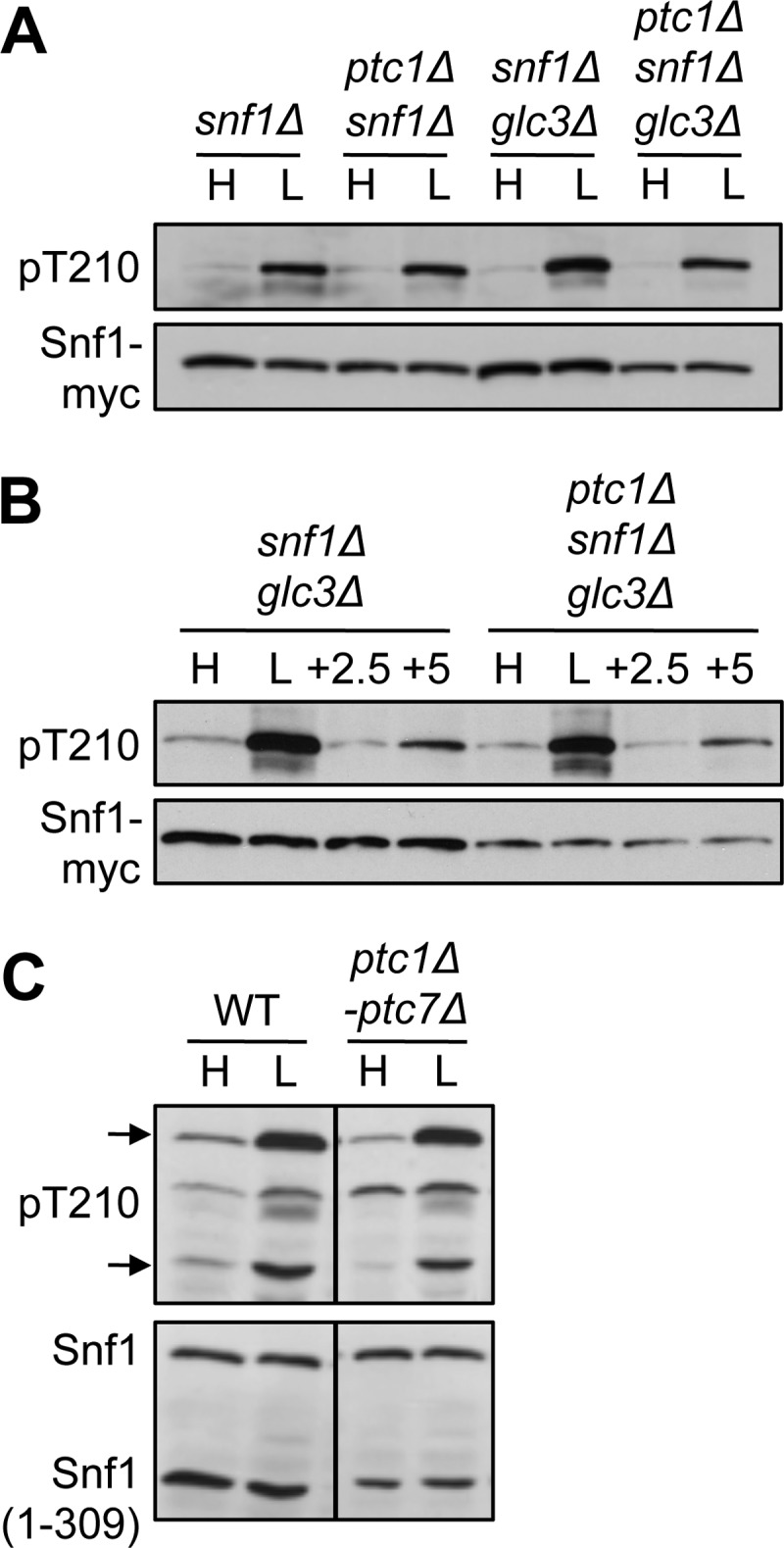
Snf1 Thr-210 phosphorylation in mutants lacking Ptc1 and other PP2C phosphatases. Cells of the indicated relevant genotype were collected after growth on 2% (H, high) glucose or after a shift to 0.05% (L, low) glucose for 10 min. For B, aliquots of cultures shifted to low glucose were replenished with 2% glucose for 2.5 or 5 min, as indicated. Protein extracts were separated by SDS-PAGE and subjected to immunoblot analysis with anti-Thr(P)-172-AMPK to detect phosphorylated Thr-210 (pT210) and anti-Myc (A and B) or anti-polyhistidine (C) to detect Snf1 proteins. A and B, independent transformants expressed Snf1-myc from the SNF1 promoter on a centromeric plasmid. B, analysis of snf1Δ and ptc1Δ snf1Δ cells (with wild-type GLC3) gave results similar to those shown. C, cells carrying mutations ptc1Δ through ptc7Δ expressed Snf1 from the genomic locus and Snf1(1–309)-myc from pYL411. All lanes are from the same blot. Arrows mark the positions of Snf1 and Snf1(1–309).
Ptc1 Affects the Phosphorylation State of Thr-210 in sit4Δ Cells
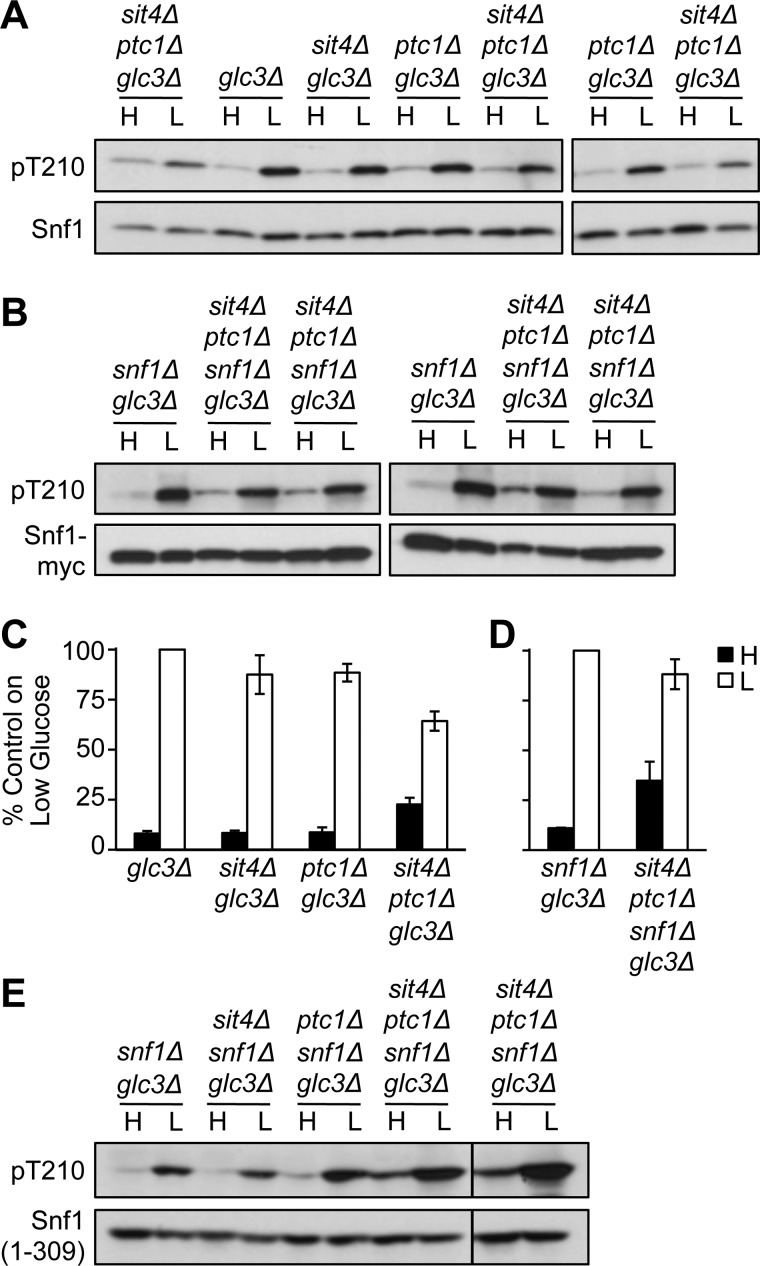
To assess the possibility that a role of Ptc1 was masked by the presence of other phosphatases that dephosphorylate Thr-210, we first constructed mutants lacking Sit4 and Ptc1. We examined sit4Δ ptc1Δ glc3Δ mutants to avoid the indirect effects of inappropriate glycogen accumulation caused by sit4Δ, which results in elevated Snf1 phosphorylation (11); all strains used in subsequent experiments carried glc3Δ, which will henceforth be omitted from the relevant genotypes. During growth on high glucose, Thr-210 phosphorylation was elevated to a small extent relative to Snf1 protein levels, and Thr-210 phosphorylation increased somewhat less robustly in response to glucose limitation than in control cells (Fig. 2A), perhaps because cells growing on high glucose are already somewhat preadapted for growth on low glucose. Similar results were obtained in sit4Δ ptc1Δ snf1Δ cells expressing Snf1-myc from a centromeric plasmid (Fig. 2B), although one of the four transformants showed somewhat greater phosphorylation than the others. Quantification of these results is shown in Fig. 2, C and D. An elevation of Thr-210 phosphorylation during growth on glucose was easily evident in sit4Δ ptc1Δ snf1Δ cells expressing Snf1(1–309)-myc (Fig. 2E); in this case, phosphorylation increased robustly in response to glucose limitation, perhaps reflecting the fact that Snf1(1–309) has little activity (15), and thus, cells on high glucose are not pre-adapted for growth on low glucose. These results support a modest role for Ptc1 in regulating the phosphorylation state of Snf1 Thr-210 in sit4Δ cells. Nonetheless, Thr-210 phosphorylation was largely glucose-regulated in cells lacking both Sit4 and Ptc1 but containing Reg1-Glc7.
FIGURE 2.
Snf1 Thr-210 phosphorylation in sit4Δ ptc1Δ cells. Cells of the indicated genotype were grown, extracts were prepared, and Snf1 phosphorylation was assayed as in Fig. 1. A, cells expressed Snf1 from the genomic locus. Three independent sit4Δ ptc1Δ glc3Δ strains are shown. B, four independent transformants expressed Snf1-myc from centromeric plasmid pYL200. C, quantification of results shown in panel A and obtained on other blots. Intensity of the bands was quantified, and phosphorylated Thr-210 was normalized to Snf1 protein. Values for high glucose (dark bars) and low glucose (open bars) were plotted as a percentage of the value obtained on the same blot for control glc3Δ samples on low glucose (n = 6, except n = 11 for sit4Δ ptc1Δ glc3Δ strains). D, quantification of results, relative to control snf1Δ glc3Δ samples, shown in B. E, cells expressed Snf1(1–309)-myc from pYL411. Transformants of two independent sit4Δ ptc1Δ glc3Δ snf1Δ strains were assayed on the same blot. H, high; L, low.
Ptc1 Is Required for Viability of reg1Δ Cells and for Thr-210 Dephosphorylation of Mutant SNF1
To assess the role of Ptc1 in the absence of Reg1-Glc7, we attempted to construct reg1Δ ptc1Δ mutant cells, but they were inviable. This finding, together with the inviability of reg1Δ sit4Δ cells (11), indicates that the three phosphatases have overlapping function(s) that are essential for cell viability.
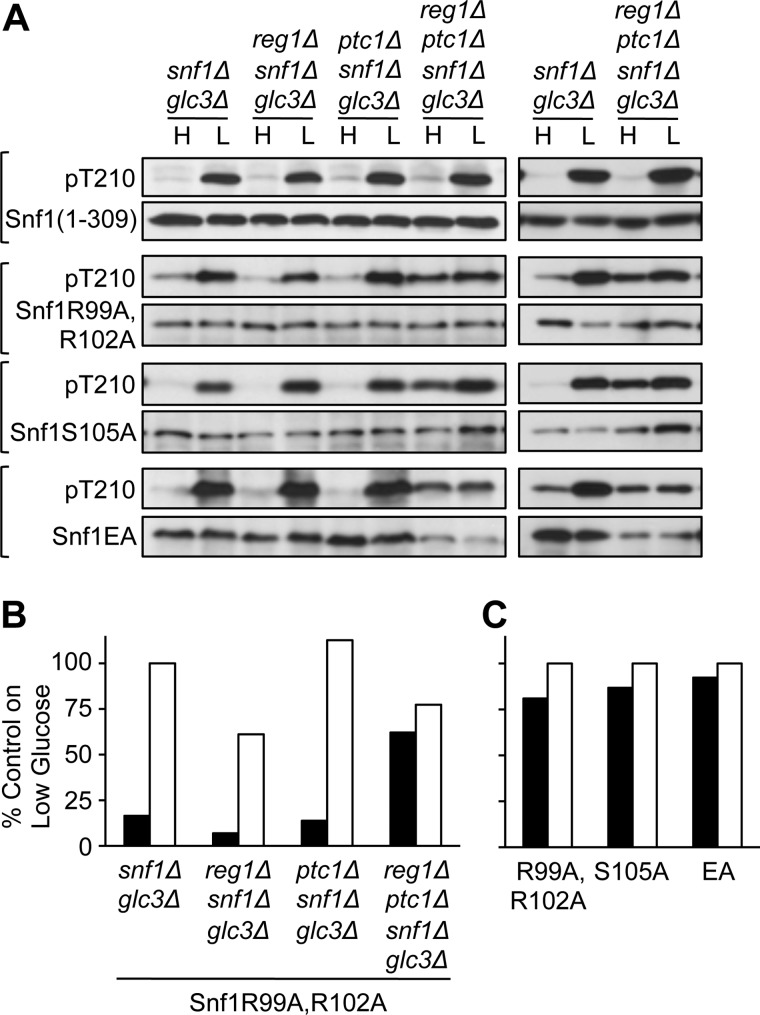
The snf1Δ mutation suppressed lethality, and reg1Δ ptc1Δ snf1Δ cells expressing Snf1(1–309), which has reduced catalytic activity (15), remained viable. These cells manifested no defect in Thr-210 dephosphorylation (Fig. 3A). However, Thr-210 may be more readily accessible to Sit4 in the context of the truncated kinase domain, which does not associate with β or Snf4/γ subunits (15), than in the context of the intact SNF1 heterotrimer.
FIGURE 3.
Thr-210 phosphorylation in reg1Δ ptc1Δ cells expressing mutant Snf1 proteins. A, cells of the indicated genotype expressed each mutant Snf1 protein from the SNF1 promoter on a centromeric plasmid. Snf1(1–309)-myc was expressed from pXX7. Other mutant Snf1 proteins carried the indicated Ala substitutions. The left and right blots show independent transformants. Immunoblot analysis of Snf1 Thr-210 phosphorylation was as described in Fig. 1. B, quantification of phosphorylated Thr-210, normalized to Snf1 protein, as in Fig. 2C, for the transformants expressing Snf1 R99A,R102A shown on the left blot of A. Values for high (H) glucose (dark bars) and low (L) glucose (open bars) are shown as percentage of the value obtained for the snf1Δ glc3Δ sample on low glucose. C, quantification of phosphorylated Thr-210, normalized to Snf1 protein, for reg1Δ ptc1Δ snf1Δ glc3Δ transformants expressing each mutant Snf1 protein. Values are shown as percentage of the value obtained for the same transformant on low glucose and are averages for the two transformants shown.
To address the possibility that Ptc1 is required for dephosphorylation of the SNF1 heterotrimer in a reg1Δ background, we expressed three mutant Snf1 proteins that have reduced catalytic activity in reg1Δ ptc1Δ snf1Δ cells. Snf1R99A,R102A and Snf1S105A have Ala substitutions of the indicated residues located on the αC helix of the kinase domain, and Snf1EA has Ala substitutions of four Glu residues (324–326 and 328) located immediately C-terminal to the kinase domain (31, 32). All three mutant kinases showed normal glucose regulation of Thr-210 phosphorylation in wild-type cells (31, 32). In cells lacking only Reg1-Glc7 or Ptc1, all three exhibited glucose-regulated phosphorylation (Fig. 3A). However, in reg1Δ ptc1Δ snf1Δ cells, Thr-210 phosphorylation of these mutant kinases was elevated during growth on high glucose and showed little increase upon glucose limitation (Fig. 3A). Quantification of these results is shown in Fig. 3, B and C. With the caveat that mutant SNF1 forms were examined, these results support a role of Ptc1 in dephosphorylation of Snf1 Thr-210 within the context of the SNF1 heterotrimer and suggest that Sit4 does not suffice for dephosphorylation in the absence of Reg1-Glc7 and Ptc1.
Snf1 Ubiquitin-associated Domain Does Not Mediate Effects of Ptc1 on Thr-210 Phosphorylation
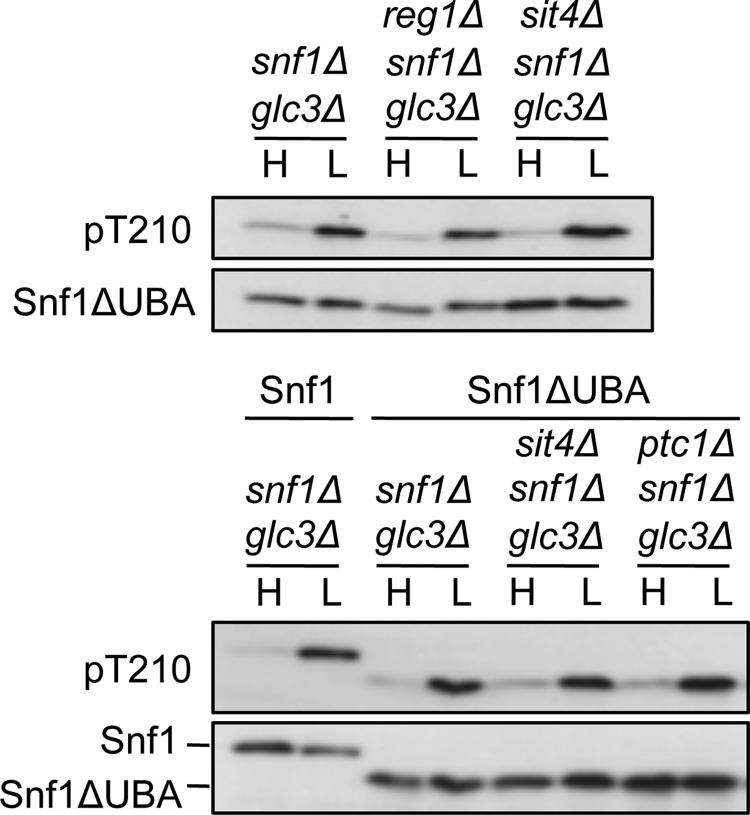
We next explored a possible mechanism for interaction of Ptc1 with SNF1 suggested by studies of Ppm1F and AMPK. The effects of the deubiquitylase inhibitor N-ethylmaleimide and Ppm1F depletion on AMPK phosphorylation in mammalian cell lysates led Voss et al. (18) to propose that polyubiquitylation and a proposed ubiquitin-associated domain (UBA) on the AMPK catalytic subunit mediate interaction with Ppm1F. Studies of AMPK-related kinases, however, indicated that the UBA mediates their phosphorylation and activation by LKB1 (37). In S. cerevisiae, the absence of Ubp8-mediated deubiquitylation reduced Snf1 Thr-210 phosphorylation ∼2-fold (38); ubiquitylation sites on Snf1 have not yet been identified. Deletion of the UBA (residues 347–398), located immediately C-terminal to the kinase domain, increased phosphorylation and SNF1 activity ∼2-fold during growth on high glucose (31). We reasoned that if the UBA mediates interaction with Ptc1, then Thr-210 phosphorylation of Snf1ΔUBA would be substantially elevated in reg1Δ snf1Δ cells on high glucose. However, this was not the case in reg1Δ snf1Δ cells or in sit4Δ snf1Δ or ptc1Δ snf1Δ cells (Fig. 4).
FIGURE 4.
Thr-210 phosphorylation of Snf1 deleted for its UBA. Cells of the indicated genotype expressed wild-type Snf1 or Snf1ΔUBA, which lacks residues 347–398, from the SNF1 promoter on centromeric plasmids (31). Immunoblot analysis of Snf1 Thr-210 phosphorylation was as described in Fig. 1. A duplicate experiment with independent transformants yielded similar results. H, high glucose; L, low glucose.
Reg1-Glc7 or Sit4 But Not Ptc1 Suffices for Thr-210 Dephosphorylation Independent of the SNF1 Heterotrimer
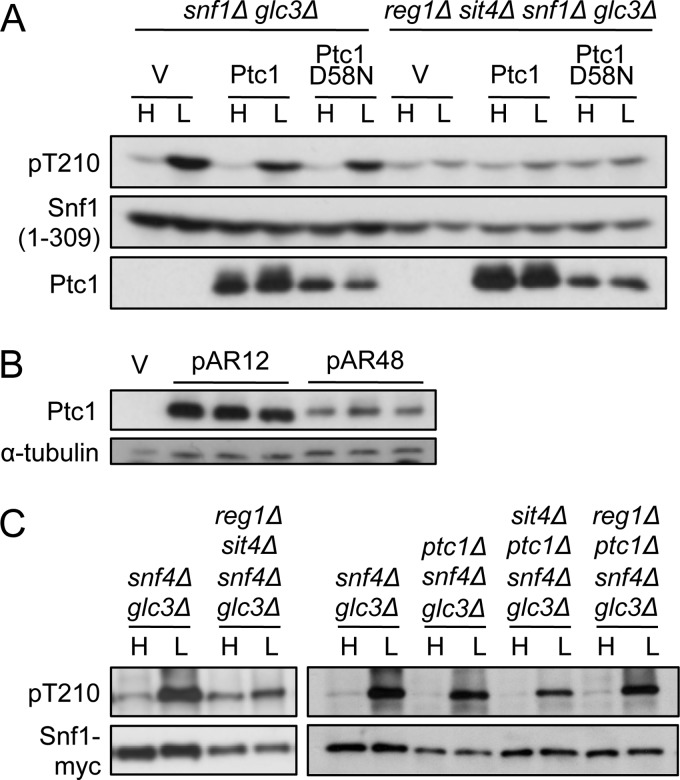
Whereas Thr-210 phosphorylation of Snf1(1–309) was substantially glucose-regulated in reg1Δ ptc1Δ cells and sit4Δ ptc1Δ cells (Figs. 2B and 3A), previous studies showed that in reg1Δ sit4Δ cells, phosphorylation of Snf1(1–309) was greatly elevated during growth on high glucose and increased <2-fold in response to glucose limitation (15). In an effort to detect a role of Ptc1 in regulating Snf1(1–309), we overexpressed HA-Ptc1 from the ADH1 promoter on plasmid pAR12 in reg1Δ sit4Δ snf1Δ cells. Overexpression did not reduce phosphorylation relative to the control, catalytically inactive HA-Ptc1D58N (Fig. 5A) (34). We confirmed that HA-Ptc1 was expressed at higher levels from pAR12 than from its native promoter on pAR48 (Fig. 5B).
FIGURE 5.
Effects of Ptc1 on Thr-210 phosphorylation in the absence of the SNF1 heterotrimer. A, cells of the indicated genotype expressed Snf1(1–309)-myc from pXX7 and carried empty vector (V) or overexpressed HA-Ptc1 or HA-Ptc1D58N from pAR12 or pAR32, respectively. Snf1 Thr-210 phosphorylation was assayed as described in Fig. 1. Similar results were observed in snf1Δ and reg1Δ sit4Δ snf1Δ cells. B, ptc1Δ cells carried vector, overexpressed HA-Ptc1 from pAR12, or expressed HA-Ptc1 from its own promoter from pAR48. Protein extracts were prepared from three independent transformants of each type and subjected to immunoblot analysis with anti-HA. The membrane was reprobed with anti-α-tubulin to control for loading. C, cells of the indicated genotype also carried genomic SNF1–8xmyc, and Thr-210 phosphorylation was assayed as described above. Snf1 protein levels are reduced in the reg1Δ sit4Δ snf4Δ glc3Δ samples, relative to snf4Δ glc3Δ controls. In additional controls, reg1Δ snf4Δ glc3Δ SNF1–8xmyc and sit4Δ snf4Δ glc3Δ SNF1–8xmyc cells exhibited glucose-regulated Thr-210 phosphorylation. H, high glucose; L, low glucose.
To further assess the role of Ptc1, we examined reg1Δ sit4Δ snf4Δ cells, which lack the Snf4/γ subunit of SNF1. These cells were expected to be viable because Snf4 is required for catalytic activity (39, 40) but not for glucose regulation of Thr-210 phosphorylation (15). Phosphorylation was elevated, relative to Snf1 protein levels, during growth on high glucose and showed only a small further increase in response to glucose limitation (Fig. 5C, left panel). In contrast, reg1Δ ptc1Δ snf4Δ and sit4Δ ptc1Δ snf4Δ cells showed no defect in dephosphorylation of Thr-210 on high glucose (Fig. 5C, right panel). These findings indicate that when the heterotrimer was disrupted by the absence of the Snf4 subunit, either Reg1-Glc7 or Sit4 sufficed for efficient dephosphorylation, whereas Ptc1 did not. These results, together with the defect of reg1Δ ptc1Δ snf1Δ cells expressing mutant SNF1 heterotrimers, suggest that the effects of Ptc1 on the phosphorylation state of Thr-210 require the intact SNF1 heterotrimer. Because cells normally express intact SNF1 heterotrimers, these findings are consistent with a physiological role for Ptc1.
Overexpression of Ptc1 Rescues Viability of reg1Δ sit4Δ Cells Expressing SNF1
To further examine the role of Ptc1 in regulating the heterotrimer, we first attempted to express Ala-substituted Snf1 subunits in reg1Δ sit4Δ snf1Δ cells. We did not recover the desired strains by transformation or genetic crossing, presumably because these mutant SNF1 heterotrimers are more highly functional than Snf1(1–309) (15, 31).
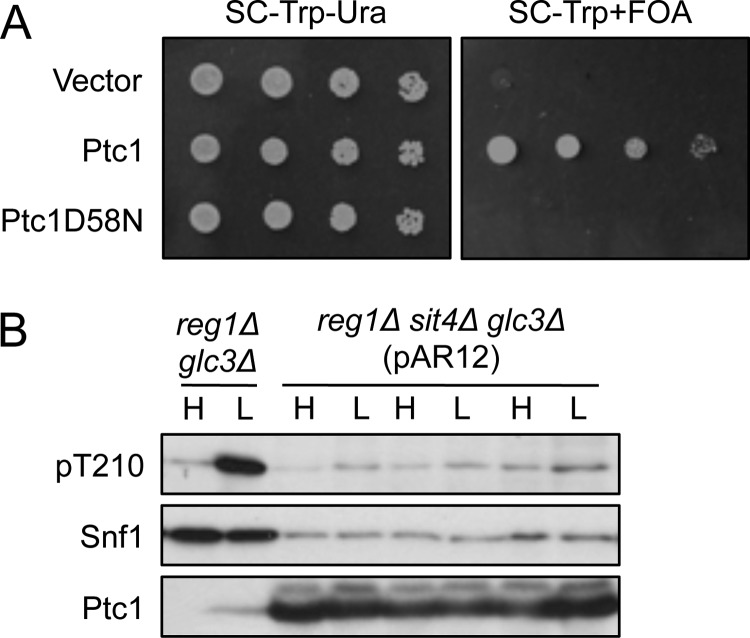
We then reasoned that overexpression of Ptc1 might reduce levels of activated SNF1 heterotrimer and thereby restore viability to the reg1Δ sit4Δ double mutant. We used pAR12 to transform reg1Δ sit4Δ cells carrying REG1 on a plasmid and explored whether overexpression of HA-Ptc1 allowed loss of the REG1 plasmid. Transformants were spotted on medium containing 5-fluorouracil (FOA) to select for loss of the URA3-marked REG1 plasmid. Colonies were recovered from cultures overexpressing HA-Ptc1 but not from HA-Ptc1D58N controls (Fig. 6A).
FIGURE 6.
Rescue of viability of reg1Δ sit4Δ mutant cells by overexpression of Ptc1. A, the TRP1 marked plasmid pAR12, which overexpresses HA-Ptc1, was used to transform reg1Δ sit4Δ glc3Δ trp1Δ ura3Δ cells carrying REG1 on the URA3-marked multicopy plasmid pSB16 (5). Control transformants expressed HA-Ptc1D58N from pAR32 or carried the empty vector pMK547. Overnight cultures of transformants were grown in selective SC-Trp-Ura. Cultures were spotted with 5-fold dilutions on SC-Trp-Ura solid medium or SC-Trp medium containing FOA (1 mg/ml) to select for loss of the URA3-marked REG1 plasmid, and plates were photographed after 2 or 3 days, respectively. Four independent transformants of each type were tested, with similar results to those shown. Similar results were also obtained when cultures were plated for single colonies on solid SC-Trp+FOA medium. B, cells from six FOA-resistant reg1Δ sit4Δ glc3Δ colonies overexpressing HA-Ptc1, derived from four independent transformants, were assayed for Thr-210 phosphorylation; three are shown. For comparison, reg1Δ glc3Δ cells are shown because the appropriate Ptc1D58N control was inviable. Genotypes of FOA-resistant colonies (presence of reg1Δ, absence of REG1) were confirmed using PCR. H, high glucose; L, low glucose.
To address the mechanism by which Ptc1 overexpression rescued viability, we examined six FOA-resistant colonies for phosphorylation of Thr-210 (Fig. 6B). The inviability of the Ptc1D58N control precluded any quantitative assessment, but Ptc1 overexpression clearly did not confer glucose regulation of phosphorylation, although it may have reduced phosphorylation during growth on both high and low glucose. However, because Snf1 protein levels were very low, and the phosphorylation state results from the opposing actions of activating kinases and phosphatases, these findings are not incompatible with other evidence that, in wild-type cells, Ptc1 participates in glucose regulation of Thr-210 phosphorylation. The mechanism by which Ptc1 overexpression rescued viability remains unclear and may reflect effects of Ptc1 on the levels or activity of SNF1 and/or other aspects of cellular regulation. Regardless of mechanism, rescue of reg1Δ sit4Δ cell viability by either overexpression of Ptc1 or deletion of SNF1 provides another line of genetic evidence linking Ptc1 to the SNF1 pathway.
DISCUSSION
We present genetic evidence that Ptc1, a member of the PP2C family, has a role in regulating the dephosphorylation of Thr-210 on the activation loop of Snf1 and hence the inactivation of SNF1. Previous studies implicated both Reg1-Glc7 and Sit4, members of the PP1 and type 2A-related phosphatase families, respectively, in glucose regulation of the phosphorylation state of Thr-210. Thus, at least three protein phosphatases have overlapping functions leading to robust regulation of yeast SNF1/AMPK in vivo.
We show here that the ptc1Δ mutation, in combination with reg1Δ, strongly impaired dephosphorylation of Thr-210 of the SNF1 heterotrimer during growth of cells on high glucose. Mutant forms of SNF1 with reduced catalytic activity were examined because the reg1Δ ptc1Δ double mutation caused inviability of cells containing wild-type SNF1. The reg1Δ sit4Δ double mutation similarly resulted in greatly elevated Thr-210 phosphorylation of mutant SNF1 on high glucose and inviability of cells with wild-type SNF1 (11, 15). In contrast, Thr-210 phosphorylation was largely glucose-regulated in sit4Δ ptc1Δ cells. Together, these findings indicate that, although Reg1-Glc7 plays a major role, all three protein phosphatases, Reg1-Glc7, Sit4, and Ptc1, contribute to glucose regulation of the phosphorylation state of Thr-210.
The effects of ptc1Δ on the phosphorylation state of SNF1 most simply suggest that Ptc1 directly dephosphorylates Thr-210. Physical interaction of Ptc1 and Snf1 has not been reported, and, in preliminary experiments, overexpressed Snf1 did not co-purify with tandem affinity purification-tagged Ptc1 expressed from the genomic locus; however, dephosphorylation need not depend on stable association. It is possible that Ptc1 acts indirectly to enhance the efficacy of dephosphorylation of SNF1 by Sit4, thereby accounting for the defect of reg1Δ ptc1Δ cells; for example, Ptc1 could affect SNF1 such that Thr-210 is more accessible or could affect Sit4 function, as suggested for the TOR pathway (34). However, the partial defect of sit4Δ ptc1Δ mutants indicates that Ptc1 regulates SNF1 by at least one mechanism that is independent of Sit4. The functional interaction of Ptc1 with SNF1 differs from that of the other two phosphatases, based on evidence that the effects of Ptc1 on the phosphorylation state of Thr-210 require the intact SNF1 heterotrimer, whereas Reg1-Glc7 and Sit4 affected dephosphorylation of the truncated kinase domain and of Snf1 in the absence of the Snf4 subunit.
The inviability of reg1Δ sit4Δ and reg1Δ ptc1Δ cells indicates that Reg1-Glc7, Sit4, and Ptc1 have overlapping functions with respect to cell viability, which speaks to the physiological importance of all three phosphatases. This inviability, together with rescue by the snf1Δ mutation, indicates that inappropriately elevated SNF1 activity during growth on high glucose is lethal when both Reg1-Glc7 and one of the other two phosphatases are absent. The phosphatases may counteract deleterious effects of elevated SNF1 activity; perhaps they have overlapping functions in the dephosphorylation of key substrates of SNF1.
The evidence presented here for a role of Ptc1 in regulating activation loop phosphorylation of SNF1 in yeast is in accord with a report implicating Ppm1E, and probably Ppm1F, in dephosphoryation of AMPK in mammalian cells (18). We note that Ptc1 is a less major player than Reg1-Glc7 PP1 in yeast and that other studies in mammalian cells implicated a member of the PP1 family, PP1-R6, in dephosphorylation of AMPK (19). Different phosphatases may play critical roles under different environmental conditions, or, in mammals, in different cell types. Here we have shown that during growth of S. cerevisiae on high glucose, Ptc1 functions together with at least two other phosphatases to confer robust regulation of the phosphorylation status of SNF1 and that this regulation of SNF1 is essential for cell viability.
This work was supported, in whole or in part, by National Institutes of Health Grant GM34095 (to M. C.).
- AMPK
- AMP-activated protein kinase
- PP
- protein phosphatase
- PP1
- protein phosphatase 1
- SC
- selective synthetic complete medium
- UBA
- ubiquitin-associated domain
- FOA
- 5-fluorouracil.
REFERENCES
- 1. Hardie D. G. (2011) AMP-activated protein kinase: an energy sensor that regulates all aspects of cell function. Genes Dev. 25, 1895–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hedbacker K., Carlson M. (2008) SNF1/AMPK pathways in yeast. Front Biosci. 13, 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tu J., Carlson M. (1995) REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14, 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ludin K., Jiang R., Carlson M. (1998) Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 95, 6245–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanz P., Alms G. R., Haystead T. A., Carlson M. (2000) Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell Biol. 20, 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCartney R. R., Schmidt M. C. (2001) Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276, 36460–36466 [DOI] [PubMed] [Google Scholar]
- 7. Hong S. P., Momcilovic M., Carlson M. (2005) Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase α as Snf1-activating kinases in yeast. J. Biol. Chem. 280, 21804–21809 [DOI] [PubMed] [Google Scholar]
- 8. Rubenstein E. M., McCartney R. R., Zhang C., Shokat K. M., Shirra M. K., Arndt K. M., Schmidt M. C. (2008) Access denied: Snf1 activation loop phosphorylation is controlled by availability of the phosphorylated threonine 210 to the PP1 phosphatase. J. Biol. Chem. 283, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mayer F. V., Heath R., Underwood E., Sanders M. J., Carmena D., McCartney R. R., Leiper F. C., Xiao B., Jing C., Walker P. A., Haire L. F., Ogrodowicz R., Martin S. R., Schmidt M. C., Gamblin S. J., Carling D. (2011) ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 14, 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chandrashekarappa D. G., McCartney R. R., Schmidt M. C. (2011) Subunit and domain requirements for adenylate-mediated protection of Snf1 kinase activation loop from dephosphorylation. J. Biol. Chem. 286, 44532–44541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruiz A., Xu X., Carlson M. (2011) Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc. Natl. Acad. Sci. U.S.A. 108, 6349–6354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sutton A., Immanuel D., Arndt K. T. (1991) The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell Biol. 11, 2133–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Como C. J., Arndt K. T. (1996) Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10, 1904–1916 [DOI] [PubMed] [Google Scholar]
- 14. Cherkasova V., Qiu H., Hinnebusch A. G. (2010) Snf1 promotes phosphorylation of the α subunit of eukaryotic translation initiation factor 2 by activating Gcn2 and inhibiting phosphatases Glc7 and Sit4. Mol. Cell Biol. 30, 2862–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ruiz A., Liu Y., Xu X., Carlson M. (2012) Heterotrimer-independent regulation of activation-loop phosphorylation of Snf1 protein kinase involves two protein phosphatases. Proc. Natl. Acad. Sci. U.S.A. 109, 8652–8657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanders M. J., Grondin P. O., Hegarty B. D., Snowden M. A., Carling D. (2007) Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 403, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suter M., Riek U., Tuerk R., Schlattner U., Wallimann T., Neumann D. (2006) Dissecting the role of 5′-AMP for allosteric stimulation, activation, and deactivation of AMP-activated protein kinase. J. Biol. Chem. 281, 32207–32216 [DOI] [PubMed] [Google Scholar]
- 18. Voss M., Paterson J., Kelsall I. R., Martín-Granados C., Hastie C. J., Peggie M. W., Cohen P. T. (2011) Ppm1E is an in cellulo AMP-activated protein kinase phosphatase. Cell Signal. 23, 114–124 [DOI] [PubMed] [Google Scholar]
- 19. Garcia-Haro L., Garcia-Gimeno M. A., Neumann D., Beullens M., Bollen M., Sanz P. (2010) The PP1-R6 protein phosphatase holoenzyme is involved in the glucose-induced dephosphorylation and inactivation of AMP-activated protein kinase, a key regulator of insulin secretion, in MIN6 β cells. FASEB J. 24, 5080–5091 [DOI] [PubMed] [Google Scholar]
- 20. Ariño J., Casamayor A., González A. (2011) Type 2C protein phosphatases in fungi. Eukaryot. Cell 10, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. González A., Ruiz A., Serrano R., Ariño J., Casamayor A. (2006) Transcriptional profiling of the protein phosphatase 2C family in yeast provides insights into the unique functional roles of Ptc1. J. Biol. Chem. 281, 35057–35069 [DOI] [PubMed] [Google Scholar]
- 22. van Wageningen S., Kemmeren P., Lijnzaad P., Margaritis T., Benschop J. J., de Castro I. J., van Leenen D., Groot Koerkamp M. J., Ko C. W., Miles A. J., Brabers N., Brok M. O., Lenstra T. L., Fiedler D., Fokkens L., Aldecoa R., Apweiler E., Taliadouros V., Sameith K., van de Pasch L. A., van Hooff S. R., Bakker L. V., Krogan N. J., Snel B., Holstege F. C. (2010) Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143, 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hruby A., Zapatka M., Heucke S., Rieger L., Wu Y., Nussbaumer U., Timmermann S., Dünkler A., Johnsson N. (2011) A constraint network of interactions: protein-protein interaction analysis of the yeast type II phosphatase Ptc1p and its adaptor protein Nbp2p. J. Cell Sci. 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 24. Liu Y., Xu X., Carlson M. (2011) Interaction of SNF1 protein kinase with its activating kinase Sak1. Eukaryot. Cell 10, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luke M. M., Della Seta F., Di Como C. J., Sugimoto H., Kobayashi R., Arndt K. T. (1996) The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell Biol. 16, 2744–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ruiz A., González A., García-Salcedo R., Ramos J., Ariño J. (2006) Role of protein phosphatases 2C on tolerance to lithium toxicity in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 62, 263–277 [DOI] [PubMed] [Google Scholar]
- 27. Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., Chu A. M., Connelly C., Davis K., Dietrich F., Dow S. W., El Bakkoury M., Foury F., Friend S. H., Gentalen E., Giaever G., Hegemann J. H., Jones T., Laub M., Liao H., Liebundguth N., Lockhart D. J., Lucau-Danila A., Lussier M., M'Rabet N., Menard P., Mittmann M., Pai C., Rebischung C., Revuelta J. L., Riles L., Roberts C. J., Ross-MacDonald P., Scherens B., Snyder M., Sookhai-Mahadeo S., Storms R. K., Véronneau S., Voet M., Volckaert G., Ward T. R., Wysocki R., Yen G. S., Yu K., Zimmermann K., Philippsen P., Johnston M., Davis R. W. (1999) Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 [DOI] [PubMed] [Google Scholar]
- 28. Voth W. P., Jiang Y. W., Stillman D. J. (2003) New 'marker swap' plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20, 985–993 [DOI] [PubMed] [Google Scholar]
- 29. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. (1987) A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60, 237–243 [DOI] [PubMed] [Google Scholar]
- 31. Momcilovic M., Carlson M. (2011) Alterations at dispersed sites cause phosphorylation and activation of SNF1 protein kinase during growth on high glucose. J. Biol. Chem. 286, 23544–23551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Momcilovic M. (2010) Regulation of the SNF1 complex. Ph.D. thesis, Columbia University [Google Scholar]
- 33. Muñoz I., Simón E., Casals N., Clotet J., Ariño J. (2003) Identification of multicopy suppressors of cell cycle arrest at the G1-S transition in Saccharomyces cerevisiae. Yeast 20, 157–169 [DOI] [PubMed] [Google Scholar]
- 34. González A., Ruiz A., Casamayor A., Ariño J. (2009) Normal function of the yeast TOR pathway requires the type 2C protein phosphatase Ptc1. Mol. Cell Biol. 29, 2876–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y., Xu X., Kuo M. H. (2010) Snf1p regulates Gcn5p transcriptional activity by antagonizing Spt3p. Genetics 184, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider C. A., Rasband W. S., Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jaleel M., Villa F., Deak M., Toth R., Prescott A. R., Van Aalten D. M., Alessi D. R. (2006) The ubiquitin-associated domain of AMPK-related kinases regulates conformation and LKB1-mediated phosphorylation and activation. Biochem. J. 394, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wilson M. A., Koutelou E., Hirsch C., Akdemir K., Schibler A., Barton M. C., Dent S. Y. (2011) Ubp8 and SAGA regulate Snf1 AMP kinase activity. Mol. Cell Biol. 31, 3126–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woods A., Munday M. R., Scott J., Yang X., Carlson M., Carling D. (1994) Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269, 19509–19515 [PubMed] [Google Scholar]
- 40. Celenza J. L., Carlson M. (1989) Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and evidence for functional interaction with the SNF4 protein. Mol. Cell Biol. 9, 5034–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]