Background: p87 and p101 represent non-catalytic subunits of class IB PI3Kγ.
Results: Expression and activity of PI3Kγ is modified differently by p87 and p101 in vitro and in living cells.
Conclusion: Non-catalytic subunits of PI3Kγ represent two different regulators in the absence of Gβγ or Ras.
Significance: p87 and p101 determine diversity within class IB PI3Kγ and allow integration in distinct PI3Kγ signaling pathways.
Keywords: G-proteins, Phosphatidylinositol 3-Kinase, Phosphatidylinositol Signaling, Phospholipid, Signal Transduction, Gβγ p101, p87, Phosphoinositide 3-Kinase γ (PI3Kγ)
Abstract
Class IB phosphoinositide 3-kinase γ (PI3Kγ) comprises a single catalytic p110γ subunit, which binds to two non-catalytic subunits, p87 or p101, and controls a plethora of fundamental cellular responses. The non-catalytic subunits are assumed to be redundant adaptors for Gβγ enabling G-protein-coupled receptor-mediated regulation of PI3Kγ. Growing experimental data provide contradictory evidence. To elucidate the roles of the non-catalytic subunits in determining the specificity of PI3Kγ, we tested the impact of p87 and p101 in heterodimeric p87-p110γ and p101-p110γ complexes on the modulation of PI3Kγ activity in vitro and in living cells. RT-PCR, biochemical, and imaging data provide four lines of evidence: (i) specific expression patterns of p87 and p101, (ii) up-regulation of p101, providing the basis to consider p87 as a protein forming a constitutively and p101 as a protein forming an inducibly expressed PI3Kγ, (iii) differences in basal and stimulated enzymatic activities, and (iv) differences in complex stability, all indicating apparent diversity within class IB PI3Kγ. In conclusion, expression and activities of PI3Kγ are modified differently by p87 and p101 in vitro and in living cells, arguing for specific regulatory roles of the non-catalytic subunits in the differentiation of PI3Kγ signaling pathways.
Introduction
Class I phosphoinositide 3-kinases (PI3K) are heterodimeric lipid kinases generating phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3),2 a principal second-messenger of the plasma membrane. PtdIns(3,4,5)P3 plays an essential role in the regulation of various cellular processes such as differentiation, proliferation, growth, and motility of cells (1–4). Class I PI3Ks harbor non-catalytic and catalytic subunits and are assigned to two groups. Enzymes containing non-catalytic p85-related subunits belong to class IA, whereas enzymes containing p87 (also known as p84) or p101 subunits belong to class IB.
In the case of class IA PI3Ks, p85 is responsible for the stabilization and inhibition of catalytic p110α, p110β, or p110δ subunits (5–8). Upon activation, p85 serves as an adaptor protein that interacts with phosphorylated tyrosine residues of membrane-localized receptor-tyrosine kinases, mediating translocation of PI3Ks from the cytosol to the plasma membrane (9, 10). Association with the receptor induces conformational alterations within the PI3K, relieving the p85-mediated inhibition of p110 (11, 12). Although p85 possesses adaptor and regulatory functions essential for appropriate spatial distribution and internal modulation in heterodimers, catalytic subunits of class IA determine the specificity and selectivity of PI3K signaling (13–17). This is reflected by the fact that an additional regulator, such as Ras or Gβγ, interacts directly with the catalytic subunits of PI3Kα and PI3Kδ or PI3Kβ, respectively (18–25).
In contrast, there is only one catalytic subunit, p110γ, representing class IB. It forms two heterodimeric PI3Kγ variants, p87–p110γ and p101–p110γ. Both variants can be regulated by G-protein-coupled receptors via interaction with Gβγ (21, 26–34). Based on initial data, it was proposed that p87 and p101 are functionally similar, acting as Gβγ adapters anchoring PI3Kγ to the plasma membrane (31–33). The adapter function of the class IB non-catalytic subunits fairly resembles the recruitment process involved in the activation of class IA PI3K. However, in contrast to the well characterized inhibitory function of class IA p85, the role of p87 and p101 in the modulation of p110γ activity remains obscure. More stringent examinations of PI3Kγ revealed that p87 and p101 do not function equivalently as Gβγ adapters (34, 35). Furthermore, we and others have demonstrated differential regulation of p87–p110γ and p101–p110γ and their integration in separate signaling cascades in vitro and in vivo (34, 36–39). These findings in combination with the fact that only a single catalytic subunit of PI3Kγ is known led us to hypothesize additional non-redundant functions of p87 and p101 apart from the Gβγ adapter function that should contribute to the specificity and selectivity of PI3Kγ signaling. Consistent with these assumptions, we have recently demonstrated that p101 but not p87 was able to rescue the stimulatory activity of Gβ1 mutants incapable of activating p110γ (35).
In the present study we show that PI3Kγ functions as an obligate heterodimeric enzyme, p87–p110γ or p101–p110γ. Based on the expression pattern in human tissues, every tissue tested expresses both non-catalytic and catalytic subunits of PI3Kγ, where p87 appears to be widely distributed and represents the dominant non-catalytic subunit of PI3Kγ. Hence, p87–110γ may be defined as a constitutively expressed enzyme, whereas p101–p110γ acquires more selective roles in an inducibly expressed manner. We demonstrate that differences in PI3Kγ activities are determined and defined by the non-catalytic subunits, p87 and p101, in vitro and in living cells. In particular, aside from their impact on the differential regulation by Gβγ and Ras, they exhibit distinct regulatory roles even in the absence of upstream stimulators, arguing for specific regulatory roles for the constitutively expressed p87–110γ and the inducibly expressed p101–p110γ.
EXPERIMENTAL PROCEDURES
Real-time PCR Analysis for Expression of PI3Kγ Subunits in Human Tissues
TissueScanTM human major tissue quantitative PCR arrays (OriGene: catalogue number HMRT102) were used as cDNA templates for RNA expression analysis in various normal (non-cancerous) human tissues. Real-time PCR amplifications were performed using RT2 quantitative PCR primer assays (Qiagen: #PPH15199A, #PPH15703A, #PPH02226A, and #PPH00150F for p87, p101, p110γ, and GAPDH, respectively) and RT2 SYBR Green quantitative PCR mastermix (Qiagen) according to the manufacturer's specifications. The real-time PCR cycling was carried out in a LightCycler 480 (Roche Diagnostics), and the data were analyzed using LightCycler 480 (1.5.0) software. The data were normalized by relative quantification of the target gene to a reference gene, GAPDH, based on crossing point (Cp) values.
Cell Culture and Expression Plasmids
HEK 293 cells (German Resource Centre for Biological Material) were cultured and transfected as described previously (32, 34). Expression plasmids encoding CFP-p85α, p87, p101, p110γCAAX, FLAG-neurofibromin 1, and GFP-Grp1PH were described previously (30, 34). A plasmid encoding β-adrenergic receptor kinase-CFP was a generous gift of Michael Schaefer.
Isolation of Human Peripheral Blood Mononuclear Cells
Human peripheral blood mononuclear cells were isolated from buffy coats obtained from the blood donation center of the university hospital of Tübingen using Ficoll-Paque (GE Healthcare) density gradient centrifugation. After lysis of any remaining red blood cells in ACK buffer (0.155 m NH4Cl, 0.01 m KHCO3, 0.1 mm EDTA), the cells were washed in PBS and adjusted to a density of 2 × 106 cells/ml cell culture medium (RPMI 1640 supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine) with or without 10% FCS (all cell culture products from PAA). After 0, 4, and 24 h, the cells were harvested and washed in PBS.
Confocal Microscope Imaging
Cell imaging was performed using a Zeiss Axiovert 100M confocal laser scanning microscope as described previously (30, 33). The confocal images were analyzed using Zeiss LSM Image Examiner (3.2.0.70) software. The subcellular distribution of GFP-Grp1PH was evaluated as detailed earlier (34, 40).
Gel Electrophoresis, Immunoblotting, and Antibodies
Generation and characterization of the antisera against the Gβ1 subunits and p110γ are detailed elsewhere (28, 41). A specific antibody against p101 was a generous gift from Len Stephens. In the current study two preparations of anti-p87 antibodies were used. The antibody against murine p87 was a generous gift from Michael Schaefer. For generation of an antibody against human p87, rabbits were immunized with peptide conjugates corresponding to the N-terminal sequence of p87 (amino acids ESSDVELDLQRSVQAVLREC). In the actual experiment, affinity-purified antibodies were used for visualization of p87.
Anti-GFP and anti-FLAG antibodies were purchased from Cell Signaling (#29565) and from Sigma (#F3165), respectively. Total protein lysates of human adult tissues, i.e. brain, lung, rectum, thymus, were purchased from BioChain (Newark). Proteins were fractionated by SDS/PAGE (10% acrylamide) and transferred onto nitrocellulose membranes (HybondTM-C Extra, GE Healthcare). Visualization of specific antisera was performed using the ECL chemiluminescence system (GE Healthcare) or the SuperSignal® West Pico Chemiluminescent Substrate (Pierce) according to the manufacturers' instructions. Chemiluminescence signals were estimated using the VersaDocTM 4000 MP imaging system (Bio-Rad).
Expression and Purification of Recombinant Proteins
Sf9 (Fall Armyworm Ovary; Invitrogen) cells were cultured and infected as described previously (35). Recombinant baculoviruses for expression of Gβ1γ2 and PI3Kγ subunits as well as their expression in Sf9 cells and purification of recombinant Gβ1(His)6γ2, (His)6p110γ, p87-(His)6p110γ, and p101-(His)6p110γ have been described elsewhere (34, 35, 42). Non-catalytic His6-fused p85α, p87, and p101 subunits of PI3K were expressed in Sf9 cells and purified using the same purification protocol as for heterodimeric PI3Kγ. Purified proteins were quantified by Coomassie Brilliant Blue staining after SDS/PAGE (10% acrylamide) with BSA as the standard. The proteins were stored at −80 °C.
To analyze the exchange of non-catalytic subunits with heterodimeric PI3Kγ variants, Sf9 cells (100 ml) were infected with baculoviruses encoding tag-free p87 or p101. After 48 h, cells were harvested and resuspended in lysis buffer containing 25 mm HEPES/NaOH, pH 7.5, 300 mm NaCl, 10 mm β-mercaptoethanol, 45 mm imidazole, and EDTA-free protease inhibitor mixture tablets (1 tablet/50-ml solution, Roche Diagnostics). Cells were lysed by forcing them through a 22-gauge needle 5 times and subsequently through a 26-gauge needle 7 times in 8 ml of lysis buffer. The cytosolic fraction of Sf9 cells (2 ml) was incubated with purified (His)6p110γ, p87-(His)6p110γ, or p101-(His)6p110γ (10 μg of catalytic p110γ subunit in the assay) for 1 h at 4 °C. Thereafter, 50 μl of Ni2+-SepharoseTM 6 Fast Flow beads (GE Healthcare) was added to the mixture and incubated for a further 30 min at 4 °C. After extensive washing with lysis buffer, the proteins were eluted by adding 150 μl of 1× sample buffer according to Laemmli (50). Alternatively, after incubation, the mixture was ultrafiltered using Amicon® Ultra-4 MWCO 100-kDa (Millipore) centrifugal filter devices according to manufacturer's specifications. The proteins in the Amicon filtrate were then analyzed by immunoblotting using specific antibodies.
Immunoprecipitation of PI3Kγ
Purified recombinant p87-(His)6p110γ or p101-(His)6p110γ (1 μg of catalytic p110γ subunit in the assay) were mixed with 2.2 μg of anti-p110γ antibody (Cell Signaling #5405) in precipitation buffer containing 20 mm Tris/HCl, pH 7.7, 150 mm NaCl, 1 mm β-mercaptoethanol, and 0.033% polyoxyethylene-10-lauryl ether (C12E10). The assays were conducted in a final volume of 200 μl. After an incubation period of 3 h at 4 °C, 20 μl of Protein A-Sepharose CL-4B beads (GE Healthcare) preincubated in blocking buffer (20 mm Tris/HCl pH 7.7, 150 mm NaCl, and 1% BSA) were added, and the mixture was incubated overnight at 4 °C. The beads were isolated using Micro Bio-SpinTM columns (Bio-Rad) and washed using precipitation buffer. Bound proteins were eluted by adding 1× Laemmli sample buffer (50).
Preparation of Phospholipid Vesicles
Phospholipid vesicles were prepared as described previously with some modifications (35). A 30-μl phospholipid mixture containing 320 μm phosphatidylethanolamine, 140 μm phosphatidylcholine, 30 μm sphingomyelin, and 40 μm phosphatidylinositol 4,5-diphosphate with or without supplementation with different concentrations of phosphatidylserine (see below) was dried using N2 gas and sonicated in buffer containing 40 mm Tris/HCl, pH 7.7, 0.1% BSA, 1 mm EGTA, 7 mm MgCl2, 120 mm NaCl, 1 mm DTT, and 1 mm β-glycerophosphate. To achieve equal association of each PI3Kγ variant with phospholipid vesicles in the absence of Gβ1γ2, the phospholipid vesicles containing 300 μm phosphatidylserine were incubated with 32 nm PI3Kγ variant for 10 min at 4 °C. To achieve equal association of each PI3Kγ variant in the presence of Gβ1γ2, p101–p110γ (32 nm) was incubated with phospholipid vesicles lacking phosphatidylserine, whereas p110γ or p87–p110γ (32 nm concentrations each variant) were incubated with vesicles containing 180 μm phosphatidylserine in the presence of 100 nm Gβ1γ2 in every experimental setup for 10 min at 4 °C.
Analysis of PI3Kγ Enzymatic Activity
The lipid kinase activity, autophosphorylation of PI3Kγ, and determination of Gβ1γ2 and PI3Kγ association with phospholipid vesicles were performed as described previously (21, 29, 35, 43).
Statistical Analysis
Results (mean ± S.E.) were analyzed using Student's t test (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.005).
RESULTS AND DISCUSSION
Expression of PI3Kγ Subunits in Human Tissues
Recent evidence suggests that PI3Kγ variants integrate into different signaling pathways (37–39). In particular, it was reported that p87–p110γ and p101–p110γ generate distinct cellular pools of PtdIns(3,4,5)P3 (36). These data suggest either a cell- or tissue-specific nature of PI3Kγ variants or differential modulation of PI3Kγ depending on the non-catalytic subunit associated. To examine a specific distribution of PI3Kγ subunits, we analyzed their expression levels in human non-cancerous tissues by real-time PCR and protein translation in some selected tissues by immunoblot analyses (Fig. 1). All human tissues expressing p110γ also expressed either p87 or p101 or both (Table 1). We did not observe any tissue that contained p110γ without non-catalytic subunits, suggesting that PI3Kγ exists as an obligate heterodimer, either p87–p110γ or p101–p110γ. As reported earlier (31–33, 36), the highest levels of PI3Kγ expression were found in tissues involved in immune responses, such as bone marrow, lymph nodes, blood leukocytes, spleen, and thymus (Table 1). However, we found significant differences in the expression of p87 and p101 in various tissues. Whereas p87 was expressed in almost all p110γ-positive tissues tested, distribution of p101 was more restricted. These unequal levels of expression and differential distribution of non-catalytic subunits in human tissues point to different cellular functions of both enzymes. Based on the broad expression of the heterodimeric p87–p110γ, it may be considered as a constitutively expressed enzyme, whereas the p101–p110γ heterodimer fulfills selective roles as an inducible enzyme. To further support this hypothesis, we studied the protein expression of p87 and p101 in an easily available human cell model, i.e. primary cultured peripheral blood mononuclear cells, by immunoblots (Fig. 2). For semiquantitative assessment, the intensities of the p87 and p101 signals were normalized to Hsp90 expression. Under these conditions p87 protein levels remained unchanged, whereas p101 protein signals increased in a time-dependent manner (Fig. 2A). Statistical analysis of at least 12 independent experiments using blood samples from 12 different donors revealed that the increase in the p101 signal was statistically significant (Fig. 2B).
FIGURE 1.

Expression of PI3Kγ regulatory subunits in human tissues. Human adult normal tissue total protein lysates obtained from BioChain were analyzed for expression of p87 and p101. A, representative blots showing expression of p87 and p101 in various human tissues. 50 μg of protein was loaded per lane for brain, lung, and rectum, and 20 μg of protein was loaded for thymus. B, p87 antibody raised against a human p87 peptide in rabbit was characterized by peptide blocking against human p87 overexpressed in HEK cells.
TABLE 1.
Real-time PCR analysis of mRNA expression of PI3Kγ subunits in human tissues
Real-time PCR amplification was performed using the TissueScanTM human non-cancerous qPCR array (OriGene) as described under “Experimental Procedures.” Shown are human tissues with p110γ mRNA expression. The target (p87 or p101) mRNA to GAPDH mRNA ratio is indicated as follows: not detected (ND), <0.001; +, 0.001 to 0.005; ++, 0.005 to 0.01; +++, >0.01. Human tissues expressing p110γ always express either p87 or p101 or both, indicating that PI3Kγ exists as a heterodimeric enzyme in vivo, p87–p110γ or p101–p110γ.
| Human tissues | p87 | p101 |
|---|---|---|
| Adrenal gland | + | + |
| Bone marrow | +++ | +++ |
| Brain | + | + |
| Cervix | ++ | ND |
| Colon | + | + |
| Duodenum (descending part) | + | + |
| Epididymis | + | ND |
| Esophagus | + | ND |
| Fat | ++ | + |
| Heart | + | ND |
| Intestine (small) | ++ | + |
| Intracranial artery | + | ND |
| Liver | ND | + |
| Lung | ++ | +++ |
| Lymph node | +++ | +++ |
| Mammary gland | + | ND |
| Optic nerve | + | ND |
| Ovary | + | ND |
| Oviduct | + | ND |
| Pancreas | + | ND |
| Pituitary gland | + | + |
| Placenta | + | + |
| Plasma blood leukocytes | +++ | +++ |
| Prostate | ++ | + |
| Rectum | +++ | ND |
| Retina | + | + |
| Seminal vesicles | + | ND |
| Skin | ++ | + |
| Spinal cord | + | + |
| Spleen | +++ | +++ |
| Stomach | ++ | ++ |
| Testis | + | ND |
| Thymus | ++ | +++ |
| Tonsil | ++ | + |
| Trachea | + | + |
| Urethra | ++ | ND |
| Urinary bladder | + | ND |
| Uterus | + | + |
| Uvula | + | ND |
| Vagina | + | ND |
| Vena cava | + | ND |
FIGURE 2.
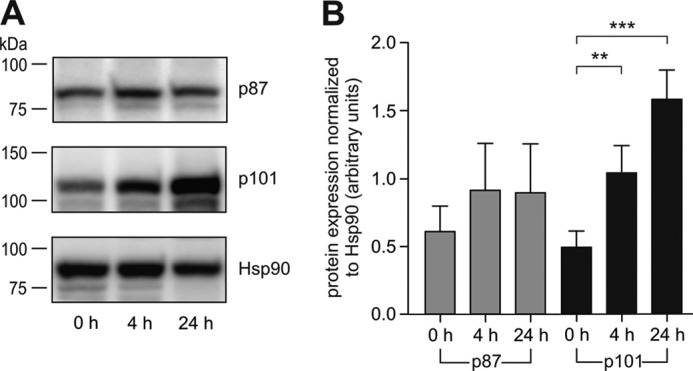
Expression of PI3Kγ regulatory subunits upon serum stimulation in human peripheral blood mononuclear cells. The cells were isolated from human blood samples and cultured in the presence of fetal calf serum. At different time points the cells were harvested and analyzed by Western blotting. A, representative blots showing expression of p87, p101, and Hsp90. B, the histogram represents statistical evaluation of the expression of p87 and p101, normalized to Hsp90 (n = 12).
These results are reminiscent of mouse data published earlier by Perino et al. (37). The authors showed increased p101 levels in mouse heart upon transverse aortic constriction, whereas p87 protein levels remained unchanged. The elevated protein levels of p101 in two different systems support not only the idea of different roles for the non-catalytic subunits of PI3Kγ but also the concept of p87–p110γ as constitutive and p101–p110γ as inducible heterodimers. Having found evidence for an isoform-specific regulation of PI3Kγ expression, we next looked for differences in their biochemical properties.
Biochemical Characterization of p87–p110γ and p101–p110γ
The activation mechanism of lipid kinases may be dissected into two steps: translocation of the enzyme to its substrate at the plasma membrane and stimulation of its catalytic activity. In a previous study we showed that p87 and p101 differ with respect to membrane recruitment of PI3Kγ by upstream regulators (34). To study the impact of the non-catalytic subunits for stimulation of p110γ, we had to eliminate the impact of membrane recruitment in our biochemical approach. In doing so, we took advantage of the finding described by Kirsch et al. (44), i.e. the binding of anionic phosphatidylserine (PS) to p110γ. We first determined the optimal concentration of PS for the different experimental conditions to achieve similar amounts of p110γ associated with phospholipid vesicles in the presence or absence of Gβ1γ2 (Fig. 3A). In addition, we ensured that the selected PS concentrations did not interfere with Gβ1γ2 association (Table 2). Under these optimized conditions, equal amounts of PI3Kγ variants could be detected in the vesicular fractions of each experimental setup (Fig. 3, B and C, upper panels, top rows, Table 3). In parallel setups, we determined lipid kinase activities (Fig. 3, B and C, upper panels, bottom rows) that were subsequently quantified (Fig. 3, B and C, graphs). In the absence of Gβ1γ2, the data show that the basal activity of p110γ is slightly reduced in the presence of p87 but drastically enhanced in the presence of p101. These data indicate that the two non-catalytic subunits represent distinct regulators independent of their adaptor function and Gβγ.
FIGURE 3.
Lipid kinase activities of phospholipid vesicle-associated class IB PI3Ks. A, an experimental model of phospholipid vesicles recruiting equal amounts of PI3Kγ variants. To obtain equal recruitment of PI3Kγ variants, we designed phospholipid vesicles where p110γ and p87–p110γ were recruited via interaction with anionic phosphatidylserine (PS) and p101–p110γ via interaction with Gβ1γ2. B and C, lipid kinase activities of PI3Kγ variants equally associated with phospholipid vesicles in the presence and absence of 100 nm Gβ1γ2 were examined as described previously (35). To achieve equal association of PI3Kγ variants, phospholipid vesicles were prepared and incubated with 32 nm concentrations of enzyme as detailed under “Experimental Procedures.” The upper panels show representative immunoblots of sedimented phospholipid vesicles probed with specific anti-p110γ antiserum (top rows) and autoradiographs demonstrating formation of 32P-labeled PtdIns(3,4,5)P3 under identical experimental conditions (bottom rows). The histograms represent statistical evaluations (mean values ± S.E.) of four (A) or three (B) independent experiments. p101–p110γ displays higher basal and Gβ1γ2-induced lipid kinase activities as compared with p110γ and p87–p110γ, whereas the association of each PI3Kγ variants with phospholipid vesicles was comparable.
TABLE 2.
Association of Gβ1γ2 dimer with phospholipid vesicles
Recombinant purified Gβ1γ2 dimers were incubated with phospholipid vesicles containing different amounts of anionic PS in the presence and absence of PI3Kγ variants. Phospholipid vesicles were prepared as detailed under “Experimental Procedures.” Aliquots of sedimented phospholipid vesicles and their supernatants were subjected to SDS/PAGE followed by immunoblotting using Gβ1–4-specific antiserum. Chemiluminescence signals were estimated with a VersaDocTM 4000 MP imaging system (Bio-Rad). For calculation of vesicle-associated Gβ1γ2, signal intensities in the sedimented phospholipid vesicles and its supernatant were added and considered as 100%. Shown here are the mean values ± S.E. of four separate experiments. PI3Kγ variants do not affect the association of Gβ1γ2 with phospholipid vesicles.
| Phospholipid vesicles | Coincubation of Gβ1γ2 with PI3Kγ variants | Phospholipid vesicle-associated Gβ1γ2 |
|---|---|---|
| % | ||
| +0 mm PS | p110γ | 57.2 ± 9.3 |
| p87–p110γ | 56.1 ± 8.6 | |
| p101–p110γ | 51.4 ± 9.9 | |
| +0.18 mm PS | p110γ | 59.5 ± 8.5 |
| p87–p110γ | 57.7 ± 10.4 | |
| p101–p110γ | 58.6 ± 9.2 | |
| +0.3 mm PS | p110γ | 57.8 ± 7.2 |
| p87–p110γ | 60.2 ± 9.7 | |
| p101–p110γ | 61.8 ± 7.7 |
TABLE 3.
Association of PI3Kγ variants with phospholipid vesicles
To achieve equal association of PI3Kγ variants in the presence or absence of 100 nm Gβ1γ2, phospholipid vesicles were prepared and incubated with 32 nm enzyme as detailed under “Experimental Procedures.” Aliquots of sedimented phospholipid vesicles and their supernatants were subjected to SDS/PAGE followed by immunoblotting using anti-p110γ antiserum. Chemiluminescence signals were estimated with a VersaDocTM 4000 MP imaging system (Bio-Rad). For calculation of vesicle-associated PI3Kγ, signal intensities in the sedimented phospholipid vesicles and its supernatant were added and considered as 100%. Shown here are the mean values ± S.E. of four (−Gβ1γ2) or three (+Gβ1γ2) independent experiments.
| P13Kγ variants | Phospholipid vesicle-associated P13Kγ |
|
|---|---|---|
| −Gβ1γ2 | +Gβ1γ2 | |
| % | ||
| p110γ | 8.8 ± 3.1 | 37.6 ± 16.9 |
| p87–p110γ | 8.7 ± 3.5 | 23.4 ± 14.4 |
| p101–p110γ | 9.1 ± 2.8 | 32.8 ± 17.4 |
Adding Gβ1γ2 to the PI3Kγ variants resulted in increased activity of all enzymes studied (Fig. 3C, see change in y axis scale). Both non-catalytic subunits promoted stimulatory modulation of p110γ in the Gβ1γ2-activated heterodimers. The effect of p101 was significantly higher as compared with p87 (Fig. 3C, graphs). This underlines that p87 and p101 have distinct profiles with respect to their role as independent regulatory subunits and as non-catalytic adaptors determining specificity toward upstream regulators. Moreover, in light of its tissue distribution, p87–p110γ can be considered as a widely expressed enzyme that exhibits only low activity upon Gβγ stimulation, whereas the p101–p110γ variant may represent a selectively expressed PI3Kγ with a high rate of PtdIns(3,4,5)P3 production. The low activity of p87–p110γ upon stimulation by G-proteins can be interpreted as a possible way to maintain membrane homeostasis and/or serve as a coincidence detector integrating upstream signals from different pathways. The latter assumption is supported by our previous finding that the p87–p110γ variant co-requires Ras-stimulation to gain full enzymatic activity (34). In this scenario the p87–p110γ variant may be reminiscent of a characteristic feature of class IA PI3Kβ, which was found by us and others to integrate signals from receptor-tyrosine kinases and G-protein-coupled receptors (19, 21, 29, 45). The fact that some tissues and cells express both p87 and p101 prompted us to examine whether non-catalytic subunits of PI3Kγ can be exchanged.
Reconstitution of Heterodimeric PI3Kγ
To address the question of the interchangeability of the non-catalytic subunits, we studied the enzymatic activity of both heterodimers in coexpression and reconstitution approaches. Therefore, we purified p85α (a non-catalytic subunit of class IA PI3Ks) as a control, p87, p101, p110γ monomers, and p87–p110γ and p101–p110γ heterodimers to apparent homogeneity (Fig. 4A). p85α and p87 appeared to be stable in the absence of their catalytic subunits, whereas the yield of purified p101 was about 20–40 times lower compared with expression of the other subunits. This confirms and extends our previous observations regarding the in vivo instability of p101 after expression in HEK 293 cells (30, 33). Individually expressed and purified p87 or p101 were incubated with p110γ in the presence of substrate-containing liposomes followed by assessment of the lipid kinase activity. Both p87 and p101 together with p110γ were able to almost fully reconstitute Gβ1γ2-stimulated enzymatic activity compared with the activity determined in parallel experiments using the coexpressed heterodimers (Fig. 4B). The activity of PI3Kγ was selectively reconstituted in the presence of p87 or p101, whereas p85α, a subunit of PI3Kα, -β, and -δ, failed to reconstitute a functional enzyme (Fig. 4B). For concentration-response studies, we chose p101–p110γ because it possessed the best signal-to-noise ratio. Coincubation of p101 with p110γ enhanced Gβ1γ2-induced translocation of p110γ to phospholipid vesicles, which was indistinguishable from the translocation of coexpressed and preformed heterodimer (Fig. 4C). Stimulation of lipid kinase activity by Gβ1γ2 revealed a similar concentration-response correlation of the reconstituted complexes as compared with coexpressed p101–p110γ preparations (Fig. 4D). Although the cellular function of p110γ autophosphorylation is not yet clear (43), we studied the autophosphorylation of p101–p110γ as an additional enzymatic feature of PI3Kγ. The high basal and autonomous activity of monomeric p110γ was transformed into a Gβγ-dependent autophosphorylation of p101–p110γ, which was similar between coexpressed and reconstituted dimers (Fig. 4E). The reconstitution of fully active heterodimeric enzymes from individually purified PI3Kγ subunits prompted us to further validate the stability of the heterodimers and their ability to exchange non-catalytic subunits.
FIGURE 4.
Reconstitution of heterodimeric PI3Kγ. A, recombinant class IA p85α subunit, individual subunits of PI3Kγ, and heterodimeric variants were expressed in and purified from Sf9 cells. Proteins were subjected to SDS/PAGE (10% acrylamide) and analyzed by Coomassie Brilliant Blue staining. Molecular mass is given in kDa. B, lipid kinase activity of monomeric p110γ subunit in the presence of 200 nm Gβ1γ2 was significantly increased after coincubation with either p87 or p101 and reached the level of intensity of coexpressed p87–p110γ or p101–p110γ, respectively. The assays were performed as described previously (35). For the reconstitution of heterodimeric PI3Kγ variants, p110γ was incubated with a 10-fold molar excess of p87 or p101. Coincubation of p110γ with class IA p85α served as a negative control. The data shown here are mean values ± S.E. (n = 3). C, Gβ1γ2-mediated recruitment of p110γ, p101–p110γ, and p110γ coincubated with p101 to phospholipid vesicles. Pulldown assays were performed in the presence of 32 nm p110γ or p110γ/p101 and 32 nm p101 subunit as detailed previously (35). Sedimented phospholipid vesicles were subjected to SDS/PAGE (10% acrylamide) followed by immunoblotting using specific anti-p110γ antiserum. D and E, stimulation of lipid kinase activity and autophosphorylation of p110γ (○), p101–p110γ (●), and p110γ coincubated with p101 (▾) in response to increasing concentrations of Gβ1γ2 were studied. The assays were conducted as detailed previously (35) with some modifications. The lipid kinase assays (D) were performed in the presence of 1.6 nm p110γ or p101–p110γ and 16 nm p101, whereas autophosphorylation (E) was studied in the presence of 6.4 nm p110γ or p101–p110γ and 32 nm p101. Gβ1γ2-induced activation of PI3Kγ is illustrated as a percentage of the maximal stimulation of coexpressed heterodimeric p101–p110γ. The lipid kinase activity of monomeric p110γ subunit is known to be sensitive to Gβ1γ2-induced stimulation in vitro (26, 34, 35). The almost complete loss of p110γ sensitivity to Gβ1γ2 was due to the higher concentrations of non-ionic detergent, polyoxyethylene-10-lauryl ether, in the assay. The data shown here represent the averages of two independent experiments.
Interaction of p87 and p101 with p110γ in Heterodimeric Complexes
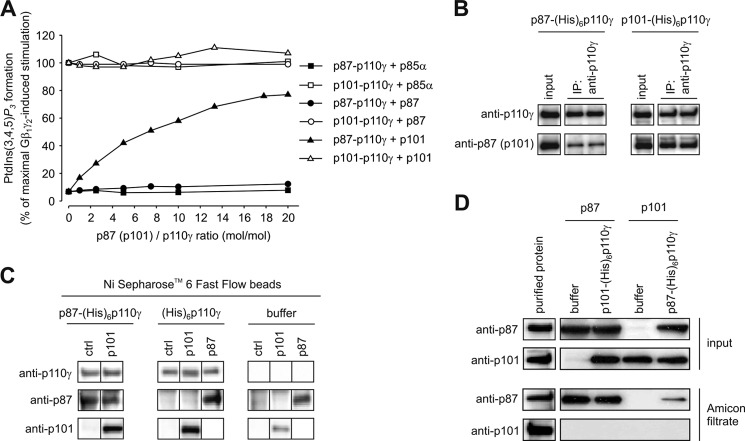
We incubated heterodimeric p87–p110γ and p101–p110γ complexes with increasing concentrations of p85α, p87, or p101 in the presence of constant concentrations of Gβ1γ2. Subsequently, their lipid kinase activities were tested (Fig. 5A). Class IA p85α, unable to form a complex with p110γ, did not alter the activities of p87–p110γ and p101–p110γ heterodimers. Class IB p87 and p101 also did not affect the activities of p87–p110γ and p101–p110γ, respectively. These results demonstrate the homogeneous nature of the purified heterodimeric complexes, as increases in lipid kinase activity in these experimental setups would be indicative of concomitant purified monomeric p110γ. The situation changed drastically in configurations where the opposing non-catalytic subunits were applied in a concentration-dependent manner. Incubation of p87–p110γ with increasing concentrations of p101 significantly enhanced Gβ1γ2-stimulated lipid kinase activity in a concentration-dependent manner (Fig. 5A). In contrast, the application of increasing concentrations of p87 to p101–110γ had no impact on lipid kinase activity. This was surprising, as one would expect a significant decrease in activity due to the formation of new p87–p110γ complexes, which are less sensitive to Gβ1γ2.
FIGURE 5.
Different stabilities of heterodimeric PI3Kγ complexes. A, heterodimeric p87–p110γ and p101–p110γ complexes were incubated with increasing concentrations of individually purified p87 or p101 as indicated above for 30 min at 4 °C, and the lipid kinase activities were estimated in the presence of 200 nm Gβ1γ2 as described previously (35). The assays are represented as the percentage of p101–p110γ stimulation by 200 nm Gβ1γ2. Coincubation of PI3Kγ with class IA p85α subunit served as a negative control. The data shown here are the averages of three independent experiments. B, purified heterodimeric p87–p110γ and p101–p110γ complexes were immunoprecipitated (IP) using specific anti-p110γ antibody as detailed under “Experimental Procedures.” Interaction of the antibody with catalytic p110γ subunit leads to significant release of p87 from the heterodimeric p87–p110γ complex, whereas the heterodimeric state of p101–p110γ is unaltered. C, interaction of p87 and p101 with recombinant purified PI3Kγ variants. Incubation of proteins and copurification using Ni2+-SepharoseTM 6 Fast Flow beads (GE Healthcare) was performed as detailed under “Experimental Procedures.” Aliquots of the eluates were separated by SDS/PAGE (10% acrylamide) and analyzed by immunoblotting using antibodies raised against p87, p101, and p110γ. p101 associated with p110γ in heterodimeric p87–p110γ complexes. D, p87–p110γ releasing the non-catalytic p87 subunit. Incubation of PI3Kγ variants with non-catalytic subunits and filtration through Amicon® Ultra-4 MWCO 100 kDa (Millipore) centrifugal filtration devices was done as detailed under “Experimental Procedures.” Aliquots of inputs (5 μl) and filtrates (15 μl) were subjected to SDS/PAGE (10% acrylamide) followed by immunoblotting using specific antibodies against p87 and p101. p87 was observed in the filtrate of the centrifugal filtration device indicating dissociation of the heterodimeric p87–p110γ complex.
In light of the instability of solitary purified p101, the inability of p87 to change the activity of p101–p110γ argues for strong binding of p101 to p110γ and hence a high stability for heterodimeric p101–p110γ complexes. Further support for this feature comes from an immunoprecipitation approach (Fig. 5B). A commercially obtained anti–p110γ antibody was used to precipitate heterodimeric PI3Kγ variants. In the case of the p101–p110γ dimers, both subunits showed comparable signals in immunoblots of the precipitated samples and also of the unprecipitated sample. However, when the p87–p110γ dimers were immunoprecipitated, the signals of p87, but not p110γ, were reduced compared with their starting products. This argues for a reduced recovery of p87, suggesting the complex stability of p87 with p110γ is weaker compared with complexes with p101.
The fact that p101 and p110γ can be purified independently and reconstituted into a stable heterodimeric complex can be described as a “click-in” mechanism in which p101 finds its final stable conformation resistant to protein degradation and protein replacement by p87. On the other hand, the concentration-dependent increase in activity after application of p101 to the heterodimeric p87–p110γ complex raised the question of whether the increase in activity resulted from a newly formed heterodimeric or just from a heterotrimeric PI3Kγ complex. To address this question, we chose two independent experimental approaches, affinity copurification (Fig. 5C) and ultrafiltration (Fig. 5D).
The affinity copurification approach was based on immobilized p87–p110γ complexes using hexahistidine-tagged p110γ. The immobilized complexes bound to Ni2+-SepharoseTM beads were coincubated with tag-free p101 and subsequently washed, eluted, and analyzed by immunoblot (Fig. 5C). Parallel experiments in the presence and absence of monomeric p110γ subunits incubated with non-catalytic subunits were used as controls. Incubation of p87 or p101 with p110γ resulted in reconstitution of the heterodimers, supporting the functional data (Fig. 5C, center panel). A strong nonspecific binding of p87 to the Ni2+-SepharoseTM beads was obvious in the control experiments (Fig. 5C, right panel), making it difficult to conclude whether a heterodimeric or a heterotrimeric complex was formed in the experiments testing the association of monomeric p101 with p87–110γ. Nevertheless, we saw a tight association of p101 with p110γ (Fig. 5C, left panel), which corresponds to the functional data shown in Fig. 5A.
The ultrafiltration approach was chosen to study the dissociation of p87 or p101 from p87–p110γ or p101–p110γ, respectively. The protein mixtures were separated using a 100-kDa molecular weight cutoff centrifugal filter device and analyzed by immunoblot (Fig. 5D). In mixtures testing p87 in the presence and absence of p101–p110γ, p87 was present in the filtrate as well as in mixtures testing p101 in the presence of p87–p110γ, indicating release of p87 from the heterodimeric complex (Fig. 5D). This finding suggests that p87–p110γ represents a less stable PI3Kγ variant than p101–p110γ and argues against the occurrence of a heterotrimeric complex.
Our data expand the differences between the two PI3Kγ variants with respect to several new aspects; that is, an inducible, selectively expressed, highly stable, and highly active p101–p110γ and a constitutive, ubiquitously expressed, and less stable p87–p110γ. Their physiological roles can be discussed in different ways. Up-regulation of p101 would enable the cell to switch PI3Kγ activity from a low to a high G-protein-sensitive activity state. Alternatively, the differences in activity and stability argue for the integration of PI3Kγ into independent pathways, e.g. p101–p110γ in a pathway dominated by Gβγ and a second p87–p110γ preferred pathway with yet unidentified players that may use Ras proteins and/or protein kinases (34, 46).
Characterization of p87–p110γ and p101–p110γ in Living Cells
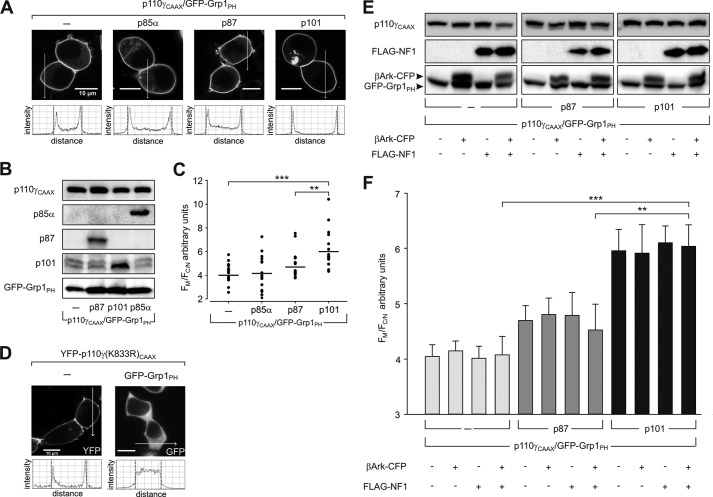
To extend and validate the in vitro data, the ability of p87 and p101 to affect the activity of p110γ was studied in living cells. HEK 293 cells were cotransfected with the plasmids encoding different PI3Kγ variants and the PtdIns(3,4,5)P3 sensor, GFP-Grp1PH (Fig. 6A). Stimulus-dependent translocation of PI3Kγ to the membrane and, therefore, to the substrate was eliminated by using p110γ C-terminally fused to a CAAX-box motif (p110γCAAX) (30). Expression of each protein was verified by immunoblotting (Fig. 6B). The amount of catalytically active p110γCAAX was not affected by the coexpression of additional proteins. Accumulation of p110γCAAX at the plasma membrane correlated with enhanced generation of PtdIns(3,4,5)P3 (Fig. 6, A and C). To rule out kinase-independent effects due to the presence of a superfluous protein at the plasma membrane, we analyzed PtdIns(3,4,5)P3-induced translocation of GFP-Grp1PH in the presence of the catalytically inactive p110γ(K833R)CAAX mutant (30, 33, 43, 47, 48). Although the K833R mutation did not affect localization of p110γCAAX at the plasma membrane, the PtdIns(3,4,5)P3 sensor GFP-Grp1PH was not translocated from the cytosol due to the blunted catalytic activity of the enzyme (Fig. 6D). Statistical analysis revealed a significant increase in p110γCAAX activity in the presence of p101 (Fig. 6C). In contrast, p87 displayed a weak and insignificant stimulatory effect on p110γCAAX. The inability of class IA p85α to modulate the activity of p110γCAAX validated the specificity of class IB non-catalytic subunits in these experiments.
FIGURE 6.
Lipid kinase activities of constitutively membrane-associated class IB PI3Ks in living cells. A–C, HEK 293 cells were transfected with plasmids encoding PI3Kγ (p110γCAAX, p110γCAAX with CFP-p85α, p87–p110γCAAX, and p101–p110γCAAX) and GFP-Grp1PH. After starvation for 18 h the cells were imaged (confocal laser-scanning microscope slices of 1 μm) and then lysed. A, cellular distribution of GFP-Grp1PH in PI3K-expressing cells. Shown are representative cells (confocal laser-scanning microscope slices of 1 μm) from three independent experiments (scale bar, 10 μm). B, protein expression in HEK 293 cells evaluated by immunoblotting using anti-p110γ, anti-p87, anti-p101, and anti-GFP (CFP) antibodies. Anti-GFP (CFP) was used to detect CFP-p85α and GFP-Grp1PH. C, the scatter plot represents the statistical evaluation of the membrane translocation of the PtdIns(3,4,5)P3 sensor, GFP-Grp1PH, in the corresponding experiments. The data shown here are mean values ± S.E. of three independent experiments comprising a total of 15–18 cells per condition. D, generation of PtdIns(3,4,5)P3 requires catalytic activity of the constitutively membrane-associated p110γCAAX. HEK 293 cells were transfected with plasmids encoding kinase-defective YFP-p110γ(K833R)CAAX mutant and GFP-Grp1PH. Shown here are representative images of cells starved for 18 h (confocal laser-scanning microscope slices of 1 μm) from three independent experiments (scale bar, 10 μm). Although YFP-p110γ(K833R)CAAX is localized at the plasma membrane, loss of its catalytic activity impairs PtdIns(3,4,5)P3 synthesis and, hence, translocation of its sensor, GFP-Grp1PH, to the plasma membrane. E and F, activity of constitutively membrane-associated p110γCAAX is not affected by endogenous Gβ1γ2 or Ras. E, HEK 293 cells were transfected with plasmids encoding PI3Kγ (p110γCAAX, p87–p110γCAAX and p101–p110γCAAX), GFP-Grp1PH, β-adrenergic receptor kinase (βArk)-CFP and FLAG-neurofibromin 1 (NF1). After starvation for 18 h the cells were imaged (confocal laser-scanning microscope slices of 1 μm) and then lysed. The cell lysates were analyzed by immunoblotting for the expression of the plasmids using anti-p110γ, anti-FLAG, and anti-GFP (CFP) antibodies. F, the histogram represents statistical evaluation of the membrane translocation of GFP-Grp1PH. The data represent the mean values ± S.E. of three independent experiments comprising a total of 15–18 cells per condition. Although p87 displays some stimulation of membrane-associated p110γCAAX, this effect was not statistically significant. The activity of p110γCAAX was significantly enhanced by p101.
To evaluate the impact of the known activation of PI3Kγ by endogenous Gβγ and Ras, we studied the effect by coexpressing either with β-adrenergic receptor kinase, a scavenger of Gβγ, or with neurofibromin 1, a RasGAP protein, or both (Fig. 6E) (34, 49). Neither β-adrenergic receptor kinase nor neurofibromin 1 altered the activities of the PI3Kγ variants, arguing for PtdIns(3,4,5)P3 production by heterologously expressed PI3Kγ variants independent of endogenous Gβγ and Ras activation (Fig. 6F). In summary, the findings in living cells support our in vitro data of differential regulation of p110γ by p87 and p101.
Conclusion
Our study provides strong evidence for several new isoform-specific features of the non-catalytic PI3Kγ subunits, i.e. (i) direct but divergent regulation of p110γ by p87 and p101, (ii) different complex stabilities of the two subunits with p110γ, (iii) diverse spatial and temporal distribution of the PI3Kγ variants in human tissues, considering p87 as a protein forming a constitutively and p101 as a protein forming an inducibly expressed PI3Kγ. Together with the previously detected distinct Gβγ adapter functions of p87 and p101 and their different sensitivities toward upstream activators, these differences establish the basis for a specific, multifaceted, and finely tuned PI3K-dependent signaling network. We, therefore, conclude that p87–p110γ and p101–p110γ represent different and non-redundant variants of PI3Kγ assigned to independent signaling cascades.
Acknowledgments
The expert technical assistance of Renate Riehle, Inken Dillmann, and Claudia Müller is greatly appreciated. We thank all members of the Nürnberg laboratory previously located in Düsseldorf and in Tübingen.
This work was supported in part by the Deutsche Forschungsgemeinschaft.
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- CFP
- cyan fluorescent protein
- GFP
- green fluorescent protein
- Grp
- general receptor for phosphoinositides
- PS
- phosphatidylserine.
REFERENCES
- 1. Fruman D. A., Bismuth G. (2009) Fine-tuning the immune response with PI3K. Immunol. Rev. 228, 253–272 [DOI] [PubMed] [Google Scholar]
- 2. Bunney T. D., Katan M. (2010) Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat. Rev. Cancer 10, 342–352 [DOI] [PubMed] [Google Scholar]
- 3. Damilano F., Perino A., Hirsch E. (2010) PI3K kinase and scaffold functions in heart. Ann. N.Y. Acad. Sci. 1188, 39–45 [DOI] [PubMed] [Google Scholar]
- 4. Vanhaesebroeck B., Stephens L., Hawkins P. (2012) PI3K signaling: the path to discovery and understanding. Nat. Rev. Mol. Cell Biol. 13, 195–203 [DOI] [PubMed] [Google Scholar]
- 5. Yu J., Zhang Y., McIlroy J., Rordorf-Nikolic T., Orr G. A., Backer J. M. (1998) Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110α catalytic subunit by the p85 regulatory subunit. Mol. Cell Biol. 18, 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geering B., Cutillas P. R., Nock G., Gharbi S. I., Vanhaesebroeck B. (2007) Class IA phosphoinositide 3-kinases are obligate p85–p110 heterodimers. Proc. Natl. Acad. Sci. U.S.A. 104, 7809–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 [DOI] [PubMed] [Google Scholar]
- 8. Zhang X., Vadas O., Perisic O., Anderson K. E., Clark J., Hawkins P. T., Stephens L. R., Williams R. L. (2011) Structure of lipid kinase p110β/p85β elucidates an unusual SH2-domain-mediated inhibitory mechanism. Mol. Cell 41, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu H., Yan Y., Backer J. M. (2007) Regulation of class IA PI3Ks. Biochem. Soc. Trans. 35, 242–244 [DOI] [PubMed] [Google Scholar]
- 10. Williams R., Berndt A., Miller S., Hon W. C., Zhang X. (2009) Form and flexibility in phosphoinositide 3-kinases. Biochem. Soc. Trans. 37, 615–626 [DOI] [PubMed] [Google Scholar]
- 11. Dbouk H. A., Pang H., Fiser A., Backer J. M. (2010) A biochemical mechanism for the oncogenic potential of the p110β catalytic subunit of phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 107, 19897–19902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vadas O., Burke J. E., Zhang X., Berndt A., Williams R. L. (2011) Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 4, re2. [DOI] [PubMed] [Google Scholar]
- 13. Okkenhaug K., Ali K., Vanhaesebroeck B. (2007) Antigen receptor signaling: a distinctive role for the p110δ isoform of PI3K. Trends Immunol. 28, 80–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ciraolo E., Iezzi M., Marone R., Marengo S., Curcio C., Costa C., Azzolino O., Gonella C., Rubinetto C., Wu H., Dastrù W., Martin E. L., Silengo L., Altruda F., Turco E., Lanzetti L., Musiani P., Rückle T., Rommel C., Backer J. M., Forni G., Wymann M. P., Hirsch E. (2008) Phosphoinositide 3-kinase p110β activity: key role in metabolism and mammary gland cancer but not development. Sci. Signal. 1, ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guillermet-Guibert J., Bjorklof K., Salpekar A., Gonella C., Ramadani F., Bilancio A., Meek S., Smith A. J., Okkenhaug K., Vanhaesebroeck B. (2008) The p110β isoform of phosphoinositide 3-kinase signals downstream of G protein-coupled receptors and is functionally redundant with p110γ. Proc. Natl. Acad. Sci. U.S.A. 105, 8292–8297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia S., Liu Z., Zhang S., Liu P., Zhang L., Lee S. H., Zhang J., Signoretti S., Loda M., Roberts T. M., Zhao J. J. (2008) Essential roles of PI3K-p110β in cell growth, metabolism, and tumorigenesis. Nature 454, 776–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luk S. K., Piekorz R. P., Nürnberg B., Tony To S. S. (2012) The catalytic phosphoinositol 3-kinase isoform p110δ is required for glioma cell migration and invasion. Eur. J. Cancer 48, 149–157 [DOI] [PubMed] [Google Scholar]
- 18. Rodriguez-Viciana P., Warne P. H., Vanhaesebroeck B., Waterfield M. D., Downward J. (1996) Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 15, 2442–2451 [PMC free article] [PubMed] [Google Scholar]
- 19. Kurosu H., Maehama T., Okada T., Yamamoto T., Hoshino S., Fukui Y., Ui M., Hazeki O., Katada T. (1997) Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110β is synergistically activated by the βγ subunits of G proteins and phosphotyrosyl peptide. J. Biol. Chem. 272, 24252–24256 [DOI] [PubMed] [Google Scholar]
- 20. Vanhaesebroeck B., Welham M. J., Kotani K., Stein R., Warne P. H., Zvelebil M. J., Higashi K., Volinia S., Downward J., Waterfield M. D. (1997) p110δ, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U.S.A. 94, 4330–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maier U., Babich A., Nürnberg B. (1999) Roles of non-catalytic subunits in Gβγ-induced activation of class I phosphoinositide 3-kinase isoforms β and γ. J. Biol. Chem. 274, 29311–29317 [DOI] [PubMed] [Google Scholar]
- 22. Gupta S., Ramjaun A. R., Haiko P., Wang Y., Warne P. H., Nicke B., Nye E., Stamp G., Alitalo K., Downward J. (2007) Binding of Ras to phosphoinositide 3-kinase p110α is required for Ras-driven tumorigenesis in mice. Cell 129, 957–968 [DOI] [PubMed] [Google Scholar]
- 23. Zhao L., Vogt P. K. (2010) Hot-spot mutations in p110α of phosphatidylinositol 3-kinase (PI3K). Differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 9, 596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castellano E., Downward J. (2011) RAS interaction with PI3K: more than just another effector pathway. Genes Cancer 2, 261–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dbouk H. A., Vadas O., Shymanets A., Burke J. E., Salamon R. S., Khalil B. D., Barrett M. O., Waldo G. L., Surve C., Hsueh C., Perisic O., Harteneck C., Shepherd P. R., Harden T. K., Smrcka A. V., Taussig R., Bresnick A. R., Nürnberg B., Williams R. L., Backer J. M. (2012) G protein-coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci. Signal. 5, ra89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stoyanov B., Volinia S., Hanck T., Rubio I., Loubtchenkov M., Malek D., Stoyanova S., Vanhaesebroeck B., Dhand R., Nürnberg B., Gierschik P., Seedorf K., Hsuan J. J., Waterfield M. D., Wetzker R. (1995) Cloning and characterization of a G protein-activated human phosphoinositide 3-kinase. Science 269, 690–693 [DOI] [PubMed] [Google Scholar]
- 27. Stephens L. R., Eguinoa A., Erdjument-Bromage H., Lui M., Cooke F., Coadwell J., Smrcka A. S., Thelen M., Cadwallader K., Tempst P., Hawkins P. T. (1997) The Gβγ sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell 89, 105–114 [DOI] [PubMed] [Google Scholar]
- 28. Leopoldt D., Hanck T., Exner T., Maier U., Wetzker R., Nürnberg B. (1998) Gβγ stimulates phosphoinositide 3-kinase-γ by direct interaction with two domains of the catalytic p110 subunit. J. Biol. Chem. 273, 7024–7029 [DOI] [PubMed] [Google Scholar]
- 29. Maier U., Babich A., Macrez N., Leopoldt D., Gierschik P., Illenberger D., Nürnberg B. (2000) Gβ5γ2 is a highly selective activator of phospholipid-dependent enzymes. J. Biol. Chem. 275, 13746–13754 [DOI] [PubMed] [Google Scholar]
- 30. Brock C., Schaefer M., Reusch H. P., Czupalla C., Michalke M., Spicher K., Schultz G., Nürnberg B. (2003) Roles of Gβγ in membrane recruitment and activation of p110γ/p101 phosphoinositide 3-kinase γ. J. Cell Biol. 160, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Suire S., Coadwell J., Ferguson G. J., Davidson K., Hawkins P., Stephens L. (2005) p84, a new Gβγ-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110γ. Curr. Biol. 15, 566–570 [DOI] [PubMed] [Google Scholar]
- 32. Voigt P., Brock C., Nürnberg B., Schaefer M. (2005) Assigning functional domains within the p101 regulatory subunit of phosphoinositide 3-kinase γ. J. Biol. Chem. 280, 5121–5127 [DOI] [PubMed] [Google Scholar]
- 33. Voigt P., Dorner M. B., Schaefer M. (2006) Characterization of p87PIKAP, a novel regulatory subunit of phosphoinositide 3-kinase γ that is highly expressed in heart and interacts with PDE3B. J. Biol. Chem. 281, 9977–9986 [DOI] [PubMed] [Google Scholar]
- 34. Kurig B., Shymanets A., Bohnacker T., Prajwal, Brock C., Ahmadian M. R., Schaefer M., Gohla A., Harteneck C., Wymann M. P., Jeanclos E., Nürnberg B. (2009) Ras is an indispensable coregulator of the class IB phosphoinositide 3-kinase p87–p110γ. Proc. Natl. Acad. Sci. U.S.A. 106, 20312–20317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shymanets A., Ahmadian M. R., Kössmeier K. T., Wetzker R., Harteneck C., Nürnberg B. (2012) The p101 subunit of PI3Kγ restores activation by Gβ mutants deficient in stimulating p110γ. Biochem. J. 441, 851–858 [DOI] [PubMed] [Google Scholar]
- 36. Bohnacker T., Marone R., Collmann E., Calvez R., Hirsch E., Wymann M. P. (2009) PI3Kγ adaptor subunits define coupling to degranulation and cell motility by distinct PtdIns(3,4,5)P3 pools in mast cells. Sci. Signal. 2, ra27. [DOI] [PubMed] [Google Scholar]
- 37. Perino A., Ghigo A., Ferrero E., Morello F., Santulli G., Baillie G. S., Damilano F., Dunlop A. J., Pawson C., Walser R., Levi R., Altruda F., Silengo L., Langeberg L. K., Neubauer G., Heymans S., Lembo G., Wymann M. P., Wetzker R., Houslay M. D., Iaccarino G., Scott J. D., Hirsch E. (2011) Integrating cardiac PIP3 and cAMP signaling through a PKA anchoring function of p110γ. Mol. Cell 42, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmid M. C., Avraamides C. J., Dippold H. C., Franco I., Foubert P., Ellies L. G., Acevedo L. M., Manglicmot J. R., Song X., Wrasidlo W., Blair S. L., Ginsberg M. H., Cheresh D. A., Hirsch E., Field S. J., Varner J. A. (2011) Receptor tyrosine kinases and TLR/IL1Rs unexpectedly activate myeloid cell PI3Kγ, a single convergent point promoting tumor inflammation and progression. Cancer Cell 19, 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brazzatti J. A., Klingler-Hoffmann M., Haylock-Jacobs S., Harata-Lee Y., Niu M., Higgins M. D., Kochetkova M., Hoffmann P., McColl S. R. (2012) Differential roles for the p101 and p84 regulatory subunits of PI3Kγ in tumor growth and metastasis. Oncogene 31, 2350–2361 [DOI] [PubMed] [Google Scholar]
- 40. Preuss I., Kurig B., Nürnberg B., Orth J. H., Aktories K. (2009) Pasteurella multocida toxin activates Gβγ dimers of heterotrimeric G proteins. Cell. Signal. 21, 551–558 [DOI] [PubMed] [Google Scholar]
- 41. Leopoldt D., Harteneck C., Nürnberg B. (1997) G proteins endogenously expressed in Sf 9 cells: interactions with mammalian histamine receptors. Naunyn. Schmiedebergs Arch. Pharmacol. 356, 216–224 [DOI] [PubMed] [Google Scholar]
- 42. Shymanets A., Ahmadian M. R., Nürnberg B. (2009) Gβγ-copurified lipid kinase impurity from Sf9 cells. Protein Pept. Lett. 16, 1053–1056 [DOI] [PubMed] [Google Scholar]
- 43. Czupalla C., Culo M., Müller E. C., Brock C., Reusch H. P., Spicher K., Krause E., Nürnberg B. (2003) Identification and characterization of the autophosphorylation sites of phosphoinositide 3-kinase isoforms β and γ. J. Biol. Chem. 278, 11536–11545 [DOI] [PubMed] [Google Scholar]
- 44. Kirsch C., Wetzker R., Klinger R. (2001) Anionic phospholipids are involved in membrane targeting of PI 3-kinase γ. Biochem. Biophys. Res. Commun. 282, 691–696 [DOI] [PubMed] [Google Scholar]
- 45. Murga C., Fukuhara S., Gutkind J. S. (2000) A novel role for phosphatidylinositol 3-kinase β in signaling from G protein-coupled receptors to Akt. J. Biol. Chem. 275, 12069–12073 [DOI] [PubMed] [Google Scholar]
- 46. Walser R., Burke J. E., Gogvadze E., Bohnacker T., Zhang X., Hess D., Küenzi P., Leitges M., Hirsch E., Williams R. L., Laffargue M., Wymann M. P. (2013) PKCβ phosphorylates PI3Kγ to activate it and release it from GPCR control. PLoS Biol. 11, e1001587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Walker E. H., Pacold M. E., Perisic O., Stephens L., Hawkins P. T., Wymann M. P., Williams R. L. (2000) Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol. Cell 6, 909–919 [DOI] [PubMed] [Google Scholar]
- 48. Patrucco E., Notte A., Barberis L., Selvetella G., Maffei A., Brancaccio M., Marengo S., Russo G., Azzolino O., Rybalkin S. D., Silengo L., Altruda F., Wetzker R., Wymann M. P., Lembo G., Hirsch E. (2004) PI3Kγ modulates the cardiac response to chronic pressure overload by distinct kinase-dependent and -independent effects. Cell 118, 375–387 [DOI] [PubMed] [Google Scholar]
- 49. Goubaeva F., Ghosh M., Malik S., Yang J., Hinkle P. M., Griendling K. K., Neubig R. R., Smrcka A. V. (2003) Stimulation of cellular signaling and G protein subunit dissociation by G protein βγ subunit-binding peptides. J. Biol. Chem. 278, 19634–19641 [DOI] [PubMed] [Google Scholar]
- 50. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]