Background: The NS5 protein from dengue virus comprises a methyltransferase and a polymerase domain connected by a linker region.
Results: Linker residues enhance polymerase activity and thermostability.
Conclusion: A crystal structure of the dengue virus polymerase reveals that linker residues contribute to protein stability.
Significance: These results should accelerate the development of antivirals against dengue virus, a major human pathogen.
Keywords: Enzyme Kinetics, Flaviviruses, Protein Stability, Viral Polymerase, X-ray Crystallography, NS5 Protein, RNA-dependent RNA Polymerase, Dengue Virus, Flavivirus, Linker Region
Abstract
The dengue virus (DENV) non-structural protein 5 (NS5) comprises an N-terminal methyltransferase and a C-terminal RNA-dependent RNA polymerase (RdRp) domain. Both enzymatic activities form attractive targets for antiviral development. Available crystal structures of NS5 fragments indicate that residues 263–271 (using the DENV serotype 3 numbering) located between the two globular domains of NS5 could be flexible. We observed that the addition of linker residues to the N-terminal end of the DENV RdRp core domain stabilizes DENV1–4 proteins and improves their de novo polymerase initiation activities by enhancing the turnover of the RNA and NTP substrates. Mutation studies of linker residues also indicate their importance for viral replication. We report the structure at 2.6-Å resolution of an RdRp fragment from DENV3 spanning residues 265–900 that has enhanced catalytic properties compared with the RdRp fragment (residues 272–900) reported previously. This new orthorhombic crystal form (space group P21212) comprises two polymerases molecules arranged as a dimer around a non-crystallographic dyad. The enzyme adopts a closed “preinitiation” conformation similar to the one that was captured previously in space group C2221 with one molecule per asymmetric unit. The structure reveals that residues 269–271 interact with the RdRp domain and suggests that residues 263–268 of the NS5 protein from DENV3 are the major contributors to the flexibility between its methyltransferase and RdRp domains. Together, these results should inform the screening and development of antiviral inhibitors directed against the DENV RdRp.
Introduction
Several flaviviruses such as yellow fever virus, West Nile virus, Japanese encephalitis virus, and dengue virus are significant human pathogens. No specific antiviral therapy is available for treating flavivirus infections that together affect a large number of the world population. The flavivirus non-structural protein 5 (NS5)3 is about 900 amino acids long and comprises a methyltransferase domain at its N terminus and an RNA-dependent RNA polymerase (RdRp) domain at its C-terminal end. Both enzymatic activities form attractive targets for antiviral development (1–4). Based on small angle x-ray scattering studies in solution as well as limited proteolysis studies of NS5, the two globular protein domains appear to be connected by a flexible linker and could adopt a range of orientations with respect to each other (5). Crystal structures of the separate methyltransferase (6, 7) and polymerase domains of NS5 (8, 9) as well as limited proteolysis studies done on the full-length NS5 protein (NS5FL) (8) suggest that residues 263HVNAEPETP271 (using the DENV3 NS5 sequence numbering scheme; see Fig. 1a) could form a flexible linker region that connects the two globular enzymatic domains. However, a precise mapping of residues belonging to the linker region and conferring interdomain flexibility to the NS5 protein has not been conducted. In addition, limited information is available on the influence of residues from the linker region in modulating protein stability, viral replication, and NS5 enzymatic activities. We previously reported the characterization of the enzymatic activity and a crystal structure for the DENV3 RdRp catalytic domain spanning residues 272–900 of the NS5 protein (hereafter named RdRp272–900) (8). Well diffracting crystals of RdRp272–900 were initially obtained at a temperature of 4 °C (10) and subsequently at 18 °C following improvements in the protein purification and crystallization procedures (11). However, RdRp272–900 possesses a low de novo initiation activity in vitro and is prone to inactivation upon prolonged incubation at room temperature, which renders inhibitor screening via enzyme inhibition assay impractical. To obtain a robust and enzymatically active protein that can be used for both inhibition studies and structure-based drug discovery, two main strategies are possible: variants of the target protein such as RdRp domains from other DENV serotypes or from related flaviviruses can be used; alternatively, truncations of the target protein at its N- or C-terminal end may be engineered. Here we report the enzymatic characterization and a crystal structure at 2.6-Å resolution of a fragment bearing an N-terminal extension comprising residues 265–900 of the NS5 protein from DENV3. This protein, which incorporates residues from the putative linker region, shows a significantly improved thermostability and de novo RNA polymerization activity compared with a shorter fragment spanning residues 272–900. We also found that proteins bearing N-terminal extensions can be readily crystallized at 20 °C in several conditions. The crystal asymmetric unit contains two polymerase molecules that assemble as a tight head-to-tail dimer with each molecule adopting a closed conformation like that in the previously reported structure obtained in space group C2221 (8, 11). The structure also points to residues 263–268 as the main determinants for the observed flexibility between the MTase and RdRp domains of the isolated NS5 protein from DENV3. We also report a mutation study of a linker residue that leads to reversion to the wild-type residue, indicating the importance of linker residues for viral replication.
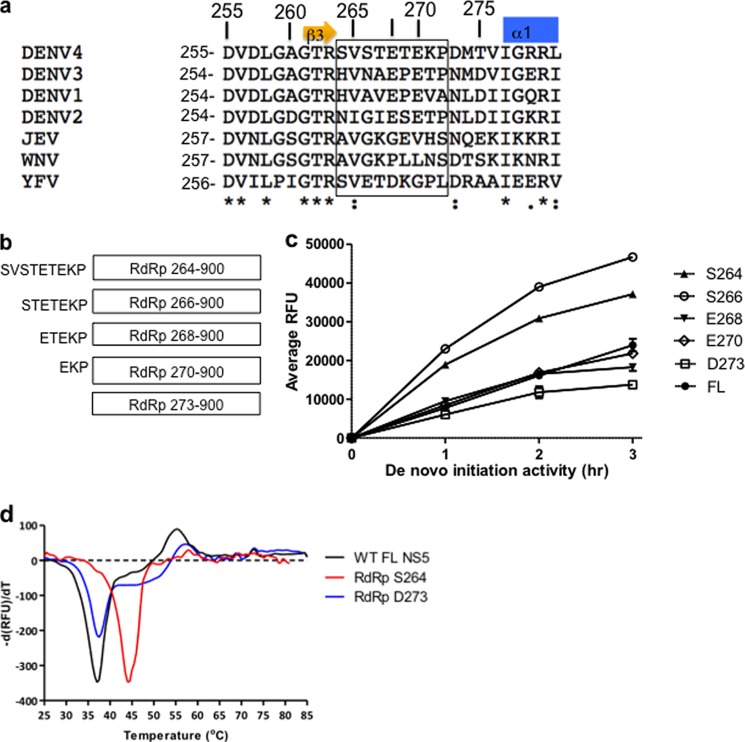
FIGURE 1.
a, alignment of amino acid residues spanning flavivirus NS5 interdomain linker sequences from dengue serotypes 1–4 (DENV1–4) (GenBank accession numbers ABG75766.1, AY037116.1, EU081206, and ABR13879.1), yellow fever virus (YFV) (GenBank accession number AFH35044), JEV (sequence obtained from Protein Data Bank code 4K6M (24)), and West Nile virus (GenBank accession number AAF20092.2) using the ClustalW program (28). The C-terminal β3 strand of the MTase domain (23) and the N-terminal α-helix α1 of the RdRp domain of NS5 (8, 11) are indicated above the sequence. Linker residues as inferred from known crystal structures of NS5 fragments (6–8, 11, 23) and the full-length NS5 protein from JEV (23) are boxed. Strictly conserved amino acids are marked with an asterisk below the sequences, conserved residues are marked with a colon, and non-conserved residues are marked with a dot. Residues that were individually mutated to alanine via site-directed mutagenesis in DENV4 NS5FL can be found using numbers above the sequences that correspond to the DENV4 RdRp numbering scheme. b, schematic diagram of the N-terminal extended DENV4 RdRp domain proteins that were expressed and tested for activity (MY01-22713 strain). The longest RdRp construct (264–900) contains all 9 amino acid residues from the linker region. The other deletion constructs tested have 7, 5, and 3 amino acids of the linker sequence (RdRp266–900, RdRp268–900, and RdRp270–900) respectively. RdRp273–900 denotes the RdRp domain devoid of residues from the linker region. c, graphical representation of results obtained from a typical activity assay obtained for DENV4 NS5FL and RdRp proteins in the de novo initiation FAPA assay measured over a period of 3 h. The data shown are the average RFU obtained from triplicate wells. d, representative thermofluor experiments showing the increased thermostability for DENV4 RdRp264–900 (red curve) compared with DENV4 NS5FL (black curve) and DENV4 RdRp273–900 (blue curve) proteins. Error bars, S.D. from the mean.
EXPERIMENTAL PROCEDURES
Cloning of DENV NS5FL and RdRp Fragments
cDNAs encoding the NS5FL proteins from DENV1–4 (GenBankTM accession number EU848545.1, GenBank accession number AF038403.1, MY00-22366 strain, and MY01-22713 strain) were cloned in pET28a. cDNAs encoding the RdRp domains from DENV3 and DENV4 were cloned into a modified pET28a vector comprising a PreScission protease cleavage site inserted downstream of the vector N-terminal His tag sequence. Site-directed alanine mutations of DENV4 NS5FL were performed using pET28-D4-MY01-22713 NS5FL (12) as a template according to the manufacturer's protocol (Stratagene). A list of primers used for cloning and mutagenesis are available in supplemental Table 1.
Reverse Genetic Analysis of the DENV4 T269A Linker Mutation
A cDNA fragment containing the DENV4 (GenBank accession number AF326825) T269A mutation (ACA to GCC) was isolated from a synthetic gene construct (containing DENV4 nucleotides 8121–9340; GenScript) by PstI/SacII digest and substituted with the corresponding wild-type sequence in the DENV4 infectious cDNA clone p4 (a kind gift from Stephen Whitehead, National Institutes of Health). The wild-type and mutant plasmids were used to produce in vitro RNA transcripts as described previously (13). The in vitro transcripts were transfected into BHK-21 cells to recover infectious virus, and the virus was analyzed for the presence of the introduced mutation as described previously (14).
Protein Purification of DENV RdRp Constructs
Expression plasmids for the RdRp domains from DENV1–4 and NS5FL proteins were transformed into BL21 cells and expressed as described previously (8, 10, 13). The cell pellet was lysed by sonication in buffer A (20 mm Hepes at pH 7.0, 300 mm NaCl, 5 mm imidazole, and EDTA-free Complete protease inhibitors (Roche Applied Science)). The lysate was clarified by centrifugation at 20,000 rpm for 1 h at 4 °C. The supernatant was purified by nickel-nitrilotriacetic acid affinity chromatography by washing unbound protein with buffer A supplemented with 40 mm imidazole. The RdRp was eluted in a linear imidazole gradient ranging from 40 to 500 mm. For removing the N-terminal His tag from RdRp proteins, 500 units of PreScission protease (GE Healthcare) was added to the pooled fractions containing the RdRp; the mixture was dialyzed overnight against buffer A. The RdRp or NS5FL protein was further purified by size exclusion chromatography using buffer A with 5 mm tris(2-carboxyethyl)phosphine. SDS-PAGE analysis of the resulting RdRp indicated a purity of >95% (data not shown).
DENV Polymerase de Novo Initiation Assays
A DENV polymerase de novo initiation fluorescence-based alkaline phosphatase-coupled polymerase (FAPA) assay was adapted from the DENV elongation FAPA assay (12). Briefly, the reaction comprised 100 nm enzyme, 100 nm in vitro translated DENV 5′-UTR–3′-UTR RNA, 20 μm ATP, 20 μm GTP, 20 μm UTP, and 5 μm Atto-CTP (TriLink BioTechnologies) in a total volume of 30 μl in assay buffer comprising 50 mm Tris/HCl, pH 7.5, 10 mm KCl, 1 mm MgCl2, 0.3 mm (DENV2 and -4) or 1 mm (DENV1 and -3) MnCl2, 0.001% Triton X-100, and 10 μm cysteine.4 The reactions were allowed to proceed for up to 3 h at room temperature. At the indicated time points, 30 μl of 2.5× STOP buffer (200 mm NaCl, 25 mm MgCl2, 1.5 m diethanolamine, pH 10; Promega) with 25 nm calf intestinal alkaline phosphatase (New England Biolabs) was added to the wells to terminate the reactions. The plate was shaken and centrifuged briefly at 1200 rpm followed by incubation at room temperature for 60 min and then read on a Tecan Saffire II microplate reader at excitationmax and emissionmax wavelengths of 422 and 566 nm, respectively. All data points were collected in triplicate wells in 96-well black opaque plates (Corning), and the experiments were performed at least three times.
Measurement of Steady-state Kinetic Parameters
To determine the Michaelis-Menten constant (Km) and Vmax of the NTP and RNA substrates in the de novo FAPA assay, NTPs or RNA was serially diluted 2-fold for up to 10 times to provide a range of substrate concentrations. The NTP concentrations tested ranged from 30 to 0.5 μm (with a fixed RNA concentration of 250 nm), whereas RNA concentrations tested ranged from 250 to 0.5 nm (at fixed NTP concentrations of 10 μm each). To measure the Km for NTP, 10 μl of 250 nm RNA was added to the well followed by 10 μl of the different serially diluted NTP to their respective wells. For the Km measurement of RNA, 10 μl of 10 μm NTPs was added to the wells followed by 10 μl of the different serially diluted RNA to their respective wells. The reaction was started with the addition of 10 μl of enzyme at a concentration of 100 nm. After incubation at room temperature for 2 h, 2.5× STOP buffer containing calf intestinal alkaline phosphatase was added to terminate the reactions, and the plate was processed as described above. All data points were performed in triplicate wells in 96-well black opaque plates (Corning), and the experiments were performed three times. Amounts of Atto-Phos (2′-[2-benzothiazoyl]-6′-hydroxybenzothiazole phosphate; nm) released from Atto-CTP incorporation were calculated from a standard curve comprising 10 points of 2-fold serially diluted Atto-Phos reagent (500–0.98 nm; Promega). Km values were obtained by plotting the observed 2′-[2-benzothiazoyl]-6′-hydroxybenzothiazole production (nm/min or velocity, v) as a function of nucleotide or RNA concentrations, and the data were fitted to the equation v = Vmax × [S]/(Km + [S]) or v = Vmax × [S]/(Km + [S] × (1 + [S]/Ki)) where [S] is the substrate concentration for RNA and NTP, respectively, using GraphPad® Prism software.
Thermofluorescence Assay
Each reaction mixture comprised 2.5 μm protein and 62.5× SYPRO Orange dye (Invitrogen) in 1× assay buffer (50 mm Tris, pH 7.5, 100 mm KCl, 0.001% Triton X-100, 0.1 mm MnCl2, 0.1 mm MgCl2) in a total volume of 20 μl in a 96-well PCR white opaque plate (Bio-Rad). The plate was sealed and heated from 25 to 85 °C with increments of 0.5 °C using an iQTM5 Multicolor Real-Time PCR Detection System (Bio-Rad). Fluorescence was detected at an excitationmax of 485 nm and an emissionmax of 625 nm. Signals were recorded as relative fluorescence units (RFU) with respect to temperature, and their derivatives (−dRFU/dT) were plotted using GraphPad Prism software. Melting temperatures for each protein were measured in triplicate wells.
Crystallization and Structure Determination
Crystallization trials were set up at 20 °C using a Phoenix crystallization robot using sitting drop vapor diffusion. RdRp265–900 (or RdRp263–900) from DENV3 (GenBank accession number AY662691.1) was concentrated to 7–10 mg/ml in 20 mm Hepes, pH 7.0, 300 mm NaCl, 5 mm tris(2-carboxyethyl)phosphine and screened against 96 unique conditions from the Hampton Research index screen and crystal screen II. Crystals were obtained in numerous conditions including conditions 33 and 34 (Index HT Screen HR2-134, Hampton Research) and in condition 30 (crystal screen II, Hampton Research). Larger crystals of size 100 × 100 × 300 μm3 were obtained over 2–4 days by mixing a volume of 1 μl of RdRp265–900 (or RdRp263–900) with 1 μl of condition 34 (1.0 m succinic acid, 0.1 m Hepes, pH 7, 1% (w/v) PEG monomethyl ether 2000). For cryoprotection, crystals were transferred to the crystallization solution supplemented with 20% glycerol. Diffraction data, collected on an R-Axis IV++ mounted on a rotating anode (Rigaku FRE), were integrated using MOSFLM (15) and scaled and truncated to structure factor amplitudes using SCALA/TRUNCATE from the CCP4 suite (16). The structure was determined by molecular replacement using the previously determined structure of RdRp272–900 (Protein Data Bank code 2J7U (8) as a search probe. Residues from the linker region were built using the computer graphics program Coot (17), and the structure was refined using REFMAC5 (16).
RESULTS
DENV4 NS5 Interdomain Linker Residues Enhance Protein Thermostability and the Polymerase de Novo Initiation and Elongation Activities
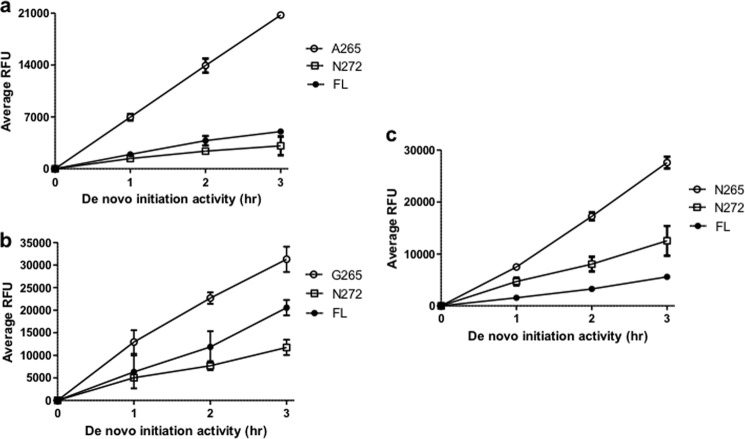
To investigate the impact of residues from the putative NS5 interdomain linker on protein stability and polymerase activities, we cloned and expressed a series of recombinant DENV4 RdRp domains with N-terminal amino acid residues starting respectively at Ser264, Ser266, Glu268, or Glu270 of the linker sequence (Fig. 1, a and b). We compared the in vitro polymerase activities of these RdRp domains bearing N-terminal extensions against the NS5FL protein comprising amino acids 1–900 and with the RdRp domain devoid of the linker sequence spanning residues 273–900 (Fig. 1c and Table 1). De novo initiation activities of all four extended RdRp proteins were higher than that of RdRp273–900. In addition, RdRp264–900 and RdRp266–900 were also more active than NS5FL. Both proteins exhibited >2- and >3-fold higher activities than NS5FL and RdRp273–900, respectively (Fig. 1c and Table 1). All RdRp proteins bearing N-terminal extensions were more stable against thermal denaturation than NS5FL and RdRp273–900 as demonstrated by their higher melting temperatures with increments ranging from 4 to 6 °C compared with the latter protein (Fig. 1d and Table 1). Overall, these results indicate that residues from the linker region, especially residues 264–267 (DENV4 numbering), affect protein stability and increase the de novo initiation activity of the polymerase from DENV4. Similar observations were made when these experiments were repeated with proteins from the other three DENV serotypes (Fig. 2): RdRp265–900 proteins from DENV1–3 were consistently 2–5-fold more active than their respective RdRp272–900 and NS5FL proteins.
TABLE 1.
Thermostability and time course activity data of DENV4 FL and RdRp proteins
Protein melting temperatures (Tm; °C) were determined by thermofluorescence as described under “Experimental Procedures.” Each protein was measured in triplicate wells. Protein activity was measured in a de novo initiation FAPA assay. Results shown are the average percentage of activity compared with DENV4 FL WT NS5 protein derived from average RFU obtained for each protein from at least three independent experiments. All data points were performed in triplicate wells.
|
De novo initiation activity |
Thermofluorescence, Tm | |||
|---|---|---|---|---|
| 1 h | 2 h | 3 h | ||
| % | °C | |||
| FL WT | 100 | 100 | 100 | 37.5 |
| RdRp264–900 | 242.5 ± 21.7 | 216.7 ± 16.5 | 174.9 ± 9.6 | 44 |
| RdRp266–900 | 293.4 ± 42.2 | 267.4 ± 20.0 | 244.8 ± 22.0 | 42 |
| RdRp268–900 | 115.4 ± 11.1 | 109.2 ± 8.7 | 87.2 ± 14.6 | 43 |
| RdRp270–900 | 108.9 ± 15.3 | 117.3 ± 9.4 | 109.8 ± 22.5 | 42 |
| RdRp273–900 | 82.0 ± 13.0 | 80.2 ± 18.0 | 77.2 ± 21.5 | 38 |
FIGURE 2.
Graphical representation of typical activity data obtained for DENV1 (GenBank accession number EU848545.1) (a), DENV2 (GenBank accession number AF038403.1) (b), and DENV3 (MY00-22366 strain) (c) FL NS5, RdRp265–900, and RdRp272–900 proteins in de novo initiation FAPA assay measured over a period of 3 h. The data shown are the average RFU obtained from triplicate wells. Error bars, S.D. from the mean.
Linker Residues Differentially Affect DENV4 Polymerase de Novo Initiation Activity
Alignment of flavivirus NS5 interdomain linker sequences reveals that only amino acid residues Glu268 and Glu270 (DENV4 numbering) are conserved across the four DENV1–4 serotypes but not across other flaviviruses (Fig. 1a). To investigate the roles of linker residues in DENV polymerase activities, we performed individual alanine mutations of residues Glu268 and Glu270 as well as Ser264, Val265, Thr269, Lys271, and Pro272 in the context of the NS5FL protein from DENV4 and compared their activities against the wild-type DENV4 NS5FL protein (Table 2). Alanine substitutions provoked mostly minor changes (less than 2-fold changes) in de novo initiation activities. The E270A mutation exhibited approximately a 2-fold reduction in de novo initiation. By contrast, the mutation T269A induced an ∼40% increase in de novo initiation activity. Interestingly, mutation of residues Ser264 to Thr269 led to an increased activity (with the exception of V265A that has an activity equivalent to the NS5FL WT protein), whereas mutations E270A, K271A, and P272A decreased polymerase activity. This is in line with the observation that in the crystal structure of RdRp265–900 amino acids 270–272 form an integral part of the RdRp globular domain, whereas residues 265–267 are flexible (see below).
TABLE 2.
Time course activity data of DENV4 FL WT and mutant NS5 proteins measured in de novo initiation FAPA assay
Results shown are the average percentage of activity compared with DENV4 FL WT NS5 protein derived from average RFU obtained for each protein from at least three independent experiments. All data points were performed in triplicate wells.
|
De novo initiation activity |
|||
|---|---|---|---|
| 1 h | 2 h | 3 h | |
| % | |||
| FL NS5 WT | 100 | 100 | 100 |
| S264A | 140.8 ± 10.9 | 137.1 ± 13.6 | 131.9 ± 11.8 |
| V265A | 102.0 ± 1.7 | 99.9 ± 9.9 | 97.5 ± 5.8 |
| E268A | 132.8 ± 12.1 | 123.9 ± 1.3 | 146.4 ± 1.7 |
| T269A | 164.0 ± 13.8 | 141.4 ± 14.3 | 140.2 ± 14.6 |
| E270A | 47.2 ± 6.4 | 47.4 ± 5.6 | 54.6 ± 10.2 |
| K271A | 82.9 ± 4.2 | 84.5 ± 6.0 | 69.4 ± 13.3 |
| P272A | 66.4 ± 11.6 | 58.7 ± 9.3 | 55.6 ± 10.4 |
DENV NS5 Interdomain Linker Is Involved in Turnover of NTP and RNA Substrates in Polymerase de Novo Initiation
To better understand the mechanism for the enhanced activity of DENV4 RdRp266–900, we performed steady-state kinetic measurements with this protein as well as with NS5FL mutants T269A and E270A that show the greatest changes in de novo initiation activity and compared them against the WT NS5FL protein and RdRp273–900 (Fig. 3 and Table 3). RdRp273–900 exhibited a slightly higher Km for NTP and lower catalysis (kcat) for both substrates than the WT NS5FL protein, which could account for its lower de novo initiation activity as seen in the time course studies (Fig. 1c). RdRp266–900 displayed about 2-fold faster turnover of both substrates than the former two proteins, which is consistent with its enhanced activity observed in the time course experiments (Fig. 1c). In the case of the NS5FL mutants, Km values for both NTP and RNA substrates are slightly improved in T269A compared with the WT protein, whereas the lower activity of mutant E270A could be explained by its ∼2-fold poorer turnover of NTP and RNA. Overall, these experiments suggest that residues in the linker sequence primarily affect the turnover of the NTP and RNA substrates during enzyme catalysis and have less influence on substrate binding to the proteins. We hypothesize that during de novo initiation events the linker residues facilitate conformational changes in the protein-RNA complex as well as the release of PPi.
FIGURE 3.
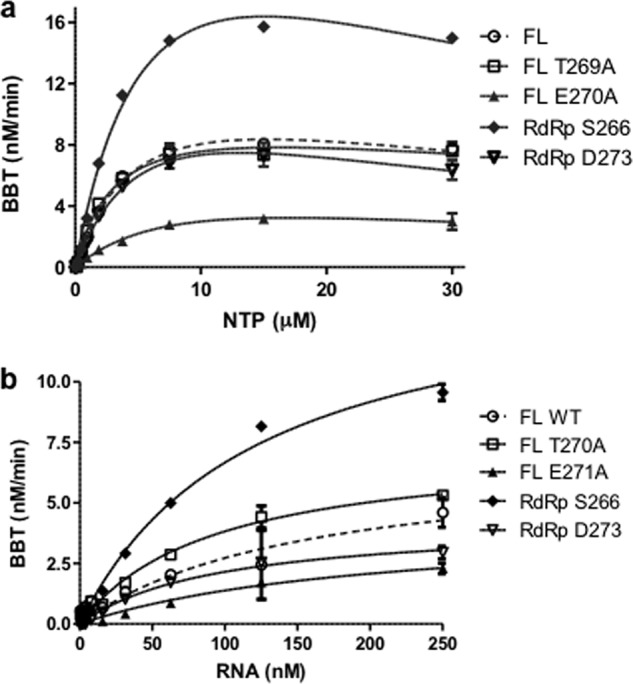
Graphical representation of steady-state kinetics parameters determined for substrates NTP (a) and RNA (b) in DENV de novo initiation FAPA assay using DENV4 FL WT, FL mutant T269A and E270A proteins, and DENV4 RdRp266–900 and RdRp273–900. The reactions were performed at room temperature for 2 h and comprised decreasing concentrations of 2-fold serially diluted NTP ranging from 30–0.05 μm with constant 250 nm RNA (a) or RNA concentrations ranging from 250–0.5 nm with constant 10 μm NTPs (b). For each data point, the average RFU from triplicate wells was measured. Enzyme kinetics parameters were determined using GraphPad Prism software as described under “Experimental Procedures.” BBT, 2′-[2-benzothiazoyl]-6′-hydroxybenzothiazole. Error bars, S.D. from the mean.
TABLE 3.
Kinetic constants for NS5FL WT and mutant proteins
Vmax, kcat, and Km values were obtained using GraphPad Prism software as described under “Experimental Procedures.” Results shown are the average values obtained for each protein from three independent experiments. All data points were performed in triplicate wells.
| Average | NTP |
RNA |
||||
|---|---|---|---|---|---|---|
| Vmaxapp | Kmapp | kcatapp | Vmaxapp | Kmapp | kcatapp | |
| nm/min | μm | min−1 | nm/min | nm | min−1 | |
| NS5FL WT | 11. 66 ± 3.94 | 3.52 ± 0.95 | 0.12 ± 0.04 | 8.71 ± 2.56 | 185.55 ± 45.89 | 0.09 ± 0.03 |
| NS5FL T269A | 9.94 ± 1.57 | 2.93 ± 0.59 | 0.10 ± 0.02 | 8.32 ± 1.37 | 121.96 ± 42.19 | 0.08 ± 0.01 |
| NS5FL E270A | 5.58 ± 0.36 | 5.94 ± 1.69 | 0.06 ± 0.01 | 4.39 ± 0.27 | 252.35 ± 68.94 | 0.04 ± 0.01 |
| RdRp266–900 | 25.86 ± 8.04 | 5.24 ± 2.45 | 0.26 ± 0.08 | 21.54 ± 6.54 | 166.77 ± 52.88 | 0.22 ± 0.07 |
| RdRp273–900 | 12.71 ± 3.85 | 5.69 ± 1.79 | 0.13 ± 0.04 | 7.71 ± 3.03 | 149.19 ± 52.68 | 0.08 ± 0.03 |
The DENV Linker Domain Is Critical for Virus Replication in Cells
In a previous report using a DENV4 infectious cDNA clone, paired mutation of Glu270/Lys272 abolished virus replication in cells (13). Our finding that mutating Glu270 to alanine also negatively impacts in vitro de novo polymerase activity (Table 2) corroborates these data. To assess whether Thr269 is also important for virus replication, the mutation T269A was introduced into the DENV4 genome by reverse genetics, and its effect on viral replication was examined (Fig. 4). However, it was found during the virus recovery process that the threonine to alanine mutation (ACA to GCC) reverted to threonine (ACC) via a single nucleotide substitution. This result suggests the presence of selective pressure to retain a threonine residue at position 269 of the linker region of DENV4 RdRp (Fig. 1a).
FIGURE 4.
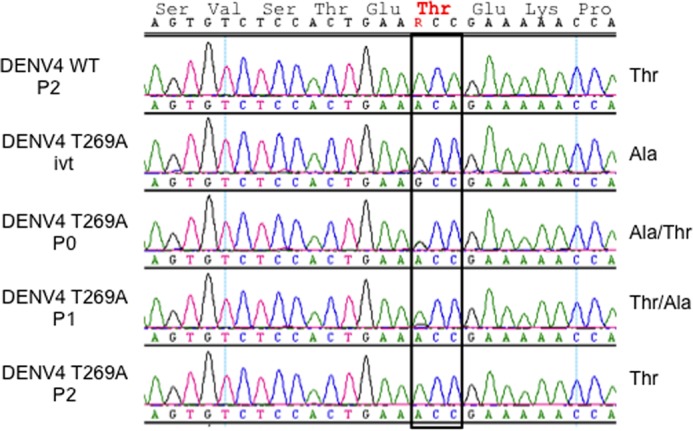
Effects of the NS5 mutation T269A on virus replication. In vitro RNA transcripts corresponding to the DENV4 WT virus or containing nucleotide substitutions encoding an NS5 T269A mutation were transfected into BHK-21 cells. After 2 days, the culture supernatants were harvested (P0) and either further passaged on BHK-21 cells or used for viral RNA extraction. The process was repeated to isolate first (P1) and second (P2) round BHK-21 culture supernatants. RNA extracted from the culture supernatants or prepared in vitro (ivt) was used for RT-PCR and sequencing to confirm the presence of the WT and mutant codons at amino acid residue 269.
DENV3 RdRp265–900 Crystallizes as a Compact Dimer
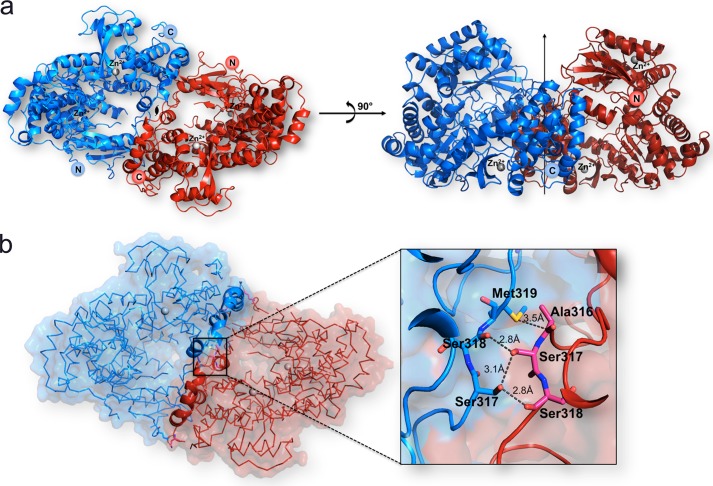
To provide a structural explanation for the enhanced thermostability and enzymatic activity of RdRp fragments bearing N-terminal extensions, we attempted to crystallize the RdRp263–900 and RdRp265–900 proteins from DENV3 as well as RdRp266–900 from DENV4. We obtained crystals for both DENV3 RdRp fragments in several different conditions and optimized the most promising condition obtained for RdRp263–900 and RdRp265–900. The structure of DENV3 RdRp265–900 was refined using data extending to 2.6-Å resolution (Tables 4 and 5) and is essentially identical to the structure obtained using DENV3 RdRp263–900. Residues 268–888 were included in the refined model, whereas residues 265–267 and the C-terminal residues 889–900 are disordered. Like the structure of RdRp272–900 reported earlier at 1.79-Å resolution (Protein Data Bank code 4HHJ) (11), residues 407–419 (loop 3) and 455–469 from the finger domain are not visible in the electron density map, confirming their intrinsic mobility in the absence of an RNA template regardless of specific crystal packing contacts. The asymmetric unit comprises two RdRp265–900 molecules assembled as a head-to-tail dimer related by a non-crystallographic dyad (Fig. 5). The interface area between both polymerase molecules extends over 1,604 Å2 and is stabilized via seven hydrogen bonds involving residues 296, 316–319, and 584 from the finger domain as well as residues 747 and 876 from the thumb domain (Fig. 5). Although RdRp265–900 appears to be predominantly monomeric in solution, dimerization modes have been put forward for the RdRp from poliovirus and from hepatitis C virus (18, 19). Moreover, the poliovirus RdRp can assemble into an ordered lattice involving two sets of polymerase-polymerase interactions (20). Thus, the extensive interface observed here for RdRp265–900 from DENV3 could participate in the formation of an enzymatic lattice whose function could be to cluster and orient viral enzymes and their substrates at the cytoplasmic face of intracellular membranes to facilitate RNA replication in the infected cells (20).
TABLE 4.
Data collection statistics
| Free RdRp265–900 DENV-3 | |
|---|---|
| Space group | P21212 |
| Cell parameters (a, b, c; Å) | 122.99, 136.05, 103.24 |
| Mosaicity (°) | 0.55 |
| Wavelength (Å) | 1.5418 |
| Resolution rangea (Å) | 20.00–2.60 (2.69–2.60) |
| No. of observed reflections | 520,768 (38,136) |
| No. of unique reflections | 53,052 (4,658) |
| Completeness (%) | 98.5 (87.9) |
| Multiplicity | 9.8 (8.2) |
| Rmergeb | 0.121 (0.629) |
| I/σ(I) | 11.4 (2.6) |
| Solvent content (%) | 52 |
a The numbers in parentheses refer to the last (highest) resolution shell.
b Rmerge = ΣhΣi|Ihi − Ih /Σh,iIhi where Ihi is the ith observation of reflection h and Ih is its mean intensity.
TABLE 5.
Refinement statistics
r.m.s., root mean square.
| No. of reflections | |
| Used for refinementa | 52,736 (3,384) |
| Used for Rfree calculation | 2,676 (196) |
| No. of non-hydrogen atoms | 9,706 |
| No. of water molecules | 433 |
| No. of Zinc ions/molecule | 2 |
| Average B factors (Å2) | |
| Wilson | 70.1 |
| Protein atoms (monomer A, B) | (60.6, 70.2) |
| Water molecules | 61.2 |
| Zinc atoms (monomer A, B) | 50.8, 73.4 |
| Rfactorb (%) | 19.21 (22.3) |
| Rfreec (%) | 24.51 (26.5) |
| r.m.s. deviations from ideality | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.15 |
| Residues in Ramachandran plot | |
| Residues in favored regions (%) | 99.2 |
| Ramachandran outliers (%) | 0.6 |
| Protein Data Bank code | 4C11 |
a The numbers in parentheses refer to the last (highest) resolution shell.
b Rfactor = Σ‖Fobs| − |Fcalc‖/Σ|Fobs|.
c Rfree was calculated with 5% of reflections excluded from the refinement.
FIGURE 5.
The DENV3 RdRp265–900 dimer. a, “front view” along the non-crystallographic dyad of the DENV-3 RdRp265–900 dimer (Protein Data Bank code 4C11; this work). Protein monomers are displayed as ribbons colored red and blue, respectively. The two zinc ions bound per monomer are depicted as gray spheres and labeled as are the N- and C-terminal ends of the polypeptide chains. The right panel shows the polymerase dimer following a 90° rotation along a horizontal axis, bringing the dyad axis vertical (depicted as an arrow). b, surface view of the RdRp265–900 dimer showing their complementarity. The inset shows a magnified view of residues involved in forming the dimer interface through hydrogen bond contacts (see text). Residues, displayed as sticks, are labeled and colored in blue or red according to the molecule to which they belong.
RdRp265–900 adopts a closed preinitiation conformation similar to the one captured for RdRp272–900 that was crystallized in space group C2221 with one molecule per asymmetric unit (8, 11). A comparison of each RdRp265–900 molecule with the structure of RdRp272–900 returns root mean square deviations of 0.67 and 0.89 Å, respectively, after superposition of main-chain atoms of residues 272–883. When the two molecules forming the dimer in the asymmetric unit are compared, a value of 0.66 Å is obtained, showing that the closed conformation of the polymerase is preserved despite different packing forces. The largest conformational change observed between the two monomers occurs in the loop region that connects helices α21 and α22, corresponding to residues 742–746 of RdRp265–900 of monomer B. This loop is displaced by up to 6 Å due to crystal packing contacts with a neighboring molecule.
Residues That Form the Linker Region between the Methyltransferase and Polymerase Domain
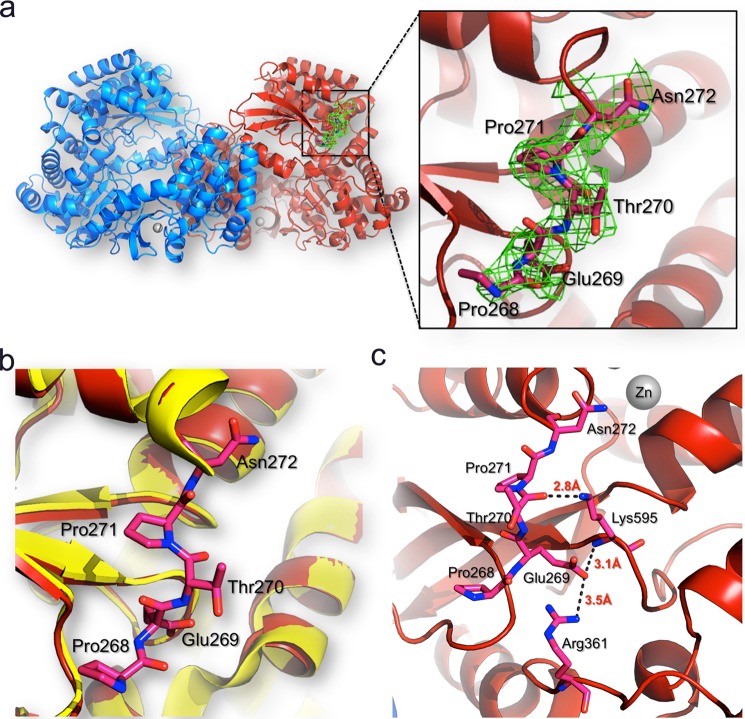
The structure reveals that residues 268–271 (using the numbering scheme from the DENV3 RdRp) form a short β-strand and establish several interactions with the RdRp protein core domain (Fig. 6, a and b). The interactions observed include hydrogen bonds between the main-chain carbonyl oxygen of Thr270 with the ϵ-amino group of the side chain of Lys595. In addition, the carboxylate group of Glu269 forms a hydrogen bond with the main-chain nitrogen amide of Lys595 and is also at the right distance to make a salt bridge with the side chain of Arg361 (Fig. 6c). Interestingly, in DENV1–4, the residue at position 595 is either a lysine or an arginine, and residues Glu269 and Arg361 are strictly conserved across DENV1–4 serotypes (corresponding, respectively, to Glu270 and Arg362 in DENV4 RdRp). Thus, these interactions are likely to be conserved across the four serotypes and could restrict the mobility of the linker from residue 268 to 272. This is also consistent with the observation that mutating Glu270 to alanine in the DENV4 polymerase reduced the turnover for the NTP and RNA substrates by ∼2-fold (Table 3). However, we observed that temperature factors for residues 268 and 269 are higher than for the rest of the protein; thus, we cannot completely rule out the possibility that these residues could adopt alternative conformations in the context of the full-length NS5 protein. Moreover, the conformation of the linker segment of NS5 might also be affected by the RNA substrate and through interactions with other proteins of the replication complex, such as the NS2B-NS3 protease-helicase with which the polymerase domain interacts with a Kd of 1 μm (21, 22).
FIGURE 6.
Interactions of residues 268–271 with the DENV3 RdRp protein core. a, location of residues 268–271 (represented as sticks and colored in pink) in the RdRp dimer structure. The two protein monomers are represented as blue and red ribbons, respectively. For clarity, the linker is shown only for one monomer but has the same conformation in the other monomer. Inset, residues 268–271 are visible in the refined DENV-3 RdRp265–900 structure (this work). The 2Fo − Fc electron density map (green) calculated with phases from the refined model and displayed at 1σ level is overlaid on the current model for linker residues. b, the RdRp272–900 structure (Protein Data Bank code 2J7U; in yellow) is overlaid on the RdRp265–900 structure (Protein Data Bank code 4C11; in red), and linker residues are displayed as red sticks and labeled. c, atomic interactions established between the linker residues with the core domain of the polymerase. Hydrogen bonds between linker residues and the core polymerase domain are represented by dashes with the respective distances indicated.
DISCUSSION
The crystal structure of the MTase domain from DENV2 reveals ordered residues up to amino acid 267 (Protein Data Bank code 1L9K) (23). The segment comprising amino acids 268–296 is disordered, but it is included in the protein fragment used for crystallization. Likewise, the structure of the MTase domain from DENV3 (comprising residues 1–272) determined at 1.7-Å resolution (Protein Data Bank code 3P8Z) (7) reveals the polypeptide chain up to residue Arg262, which has been strictly conserved during flavivirus evolution (Fig. 1a). Interestingly, in both structures, the guanidinium group of the arginine side chain establishes a strong hydrogen bond with the main-chain carbonyl oxygen of Val97. Thus, the C-terminal subdomain of the DENV3 MTase consisting of strand β3 spanning residues 259–262 belongs to the MTase core domain, whereas residues beyond 263 extend away from it. This is in agreement with the pattern of sequence conservation for NS5 across flaviviruses that is observed up to residue Arg262, especially the “GTR” sequence motif that forms strand β3 (Fig. 1a). This is also consistent with limited trypsin digestion studies of the full-length NS5 protein from DENV3 that produces a fragment starting at position 264 (8). Here we report a novel crystal form diffracting to 2.6-Å resolution of an RdRp fragment from DENV3 spanning residues 265–900. This fragment has significantly enhanced thermostability and catalytic properties compared with the RdRp fragment described previously and should therefore be of great use for compound screening using enzyme assay and structure-based drug discovery.
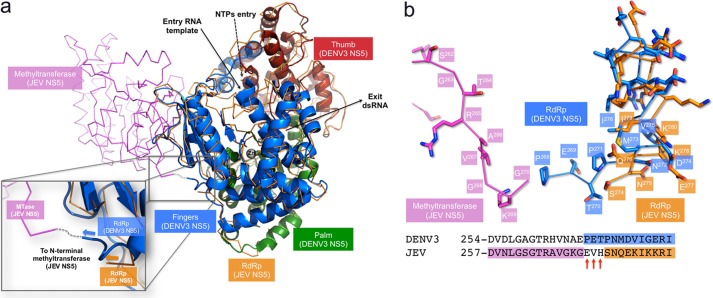
Based on interferences revealed through reverse genetic experiments combined with molecular docking, a model for a molecular interaction between the MTase and RdRp domains of NS5 from West Nile virus was put forward (9). In this model, Lys46/Arg47/Glu49 from the MTase domain are in the vicinity of residue 512, giving a rather compact shape to the NS5. However, a single stable conformation for the isolated NS5 protein is not fully consistent with the small angle x-ray scattering data, which suggested that NS5 can adopt a range of conformations from compact to fully extended (5). In this respect, the segment comprising amino acids 263–267 of the NS5 protein (DENV3 numbering; see Fig. 1a) appears to be the major determinant that imparts flexibility between the MTase and RdRp domains. This hypothesis is supported by a recent structure of the full-length NS5 protein from Japanese encephalitis virus (JEV) (24): a superposition of the DENV3 polymerase domain (Protein Data Bank code 4C11; this work) onto the full-length NS5 protein from JEV (Protein Data Bank code 4K6M) is shown in Fig. 7. Interestingly, residues Ser274-Asn275-Gln276-Glu277 from the well resolved segment of the NS5 linker region from JEV closely superimpose with residues Pro271-Asn272-Met273-Asp274 of DENV3 RdRp, respectively (Figs. 1a and 7). Linker residues of both the JEV and DENV3 NS5 proteins point toward the same direction with respect to the polymerase core domain. Moreover, the conformation adopted by linker residues 268–274 in the DENV3 RdRp structure reported here would enable the connection with the MTase domain as it is placed in the JEV NS5 full-length structure (Fig. 7). This suggests that the relative orientations observed for the MTase and RdRp domains in the crystal structure of the NS5 protein from JEV could also constitute one of the stable conformations adopted by the NS5 protein from DENV.
FIGURE 7.
Superposition of the DENV3 RdRp265–900 structure onto the full-length NS5 protein from JEV. a, the DENV3 RdRp265–900 structure (Protein Data Bank code 4C11; this work) that comprises the polymerase domain and residues from the linker region is represented as ribbons with its three domains colored in green (palm domain), blue (finger domain), and red (thumb domain); the full-length NS5 protein from JEV (Protein Data Bank code 4K6M (24)) is represented via its α-carbon chain trace in brown for the polymerase domain and in purple for the MTase domain. The paths taken by the RNA template, the double-stranded RNA (dsRNA) product, and NTP molecules are indicated by arrows. The inset shows a magnified view of the linker region that connects the MTase to the RdRp domain. The putative connection between the MTase and RdRp polypeptide chains is depicted by a dashed line. b, magnified view of the α-carbon chain trace of the linker region with amino acid side chains represented as sticks and labeled (MTase from JEV, purple; RdRp from JEV, orange; RdRp from DENV3, blue) showing the conformational similarity between both linker structures (see text). Other main-chain atoms were omitted for clarity. The conformation adopted by linker residues 268–274 in the DENV3 RdRp structure reported here would enable the connection with the MTase domain as it is placed in the JEV NS5 full-length structure with very minimal structural adaptation required as the distance between Gly270 (JEV; purple) and Pro268 (DENV3; blue) α-carbons is 4.1 Å. The alignment of amino acid sequences of linker regions from the NS5 proteins from JEV and DENV3 uses the same color code. The three non-resolved residues presumably disordered in the structure of NS5 from JEV are indicated by red arrows below the sequence.
The T269A mutation results in a 40% increase in de novo polymerase activity (Table 2); however, the same mutation introduced into an infectious clone system results in reversion to the wild-type amino acid (Fig. 4). How can we reconcile these seemingly conflicting results? Assuming a relative orientation between the RdRp and MTase domains similar to the one seen in the full-length NS5 structure from JEV, the side chain of Thr269 that points toward the solvent away from the polymerase core domain would not make contact with the MTase domain. Hence, the T269A mutation is unlikely to directly affect MTase activity but instead could have pleiotropic effects on the function of NS5. Thus, the observed increase in the de novo polymerase activity for this mutant may come at the expense of changes in the interaction of NS5 with other viral or cellular proteins, leading to the observed reversion during virus culture.
Interestingly, as was observed for the NS3 protease-helicase linker (25, 26), most residues from the interdomain linker of NS5 have been poorly conserved during flavivirus evolution (Fig. 1a), suggesting limited functional constraints on the precise amino acid sequence of this region. The relatively small impact of our alanine mutagenesis study on enzymatic activity also supports this observation. However, strict conservation of the length of this protein segment suggests evolutionary pressure to preserve the flexibility needed to maintain interactions with other proteins of the replication complex (27) such as the NS3 protease-helicase and to coordinate the distinct steps of RNA replication.
Acknowledgments
We thank colleagues at Novartis Institute for Tropical Diseases for technical help and scientific discussions during the course of this work. We thank Shamala Devi (University of Malaya) for providing RNA viruses for cloning the DENV3 and DENV4 NS5 proteins. We acknowledge beam-time allocation from the Swiss Light Source and expert scientific advice from the staff.
This work was supported by Biomedical Research Council Grant 0912219/599 and in part by Competitive Research Programme Grant CRP2008 from the National Research Foundation (to the J. L. laboratory).

This article contains supplemental Table 1.
The atomic coordinates and structure factors (code 4C11) have been deposited in the Protein Data Bank (http://wwpdb.org/).
P. Niyomrattanakit, Y.-L. Chen, S. P. Kim, and P.-Y. Shi, manuscript in preparation.
- NS5
- non-structural protein 5
- DENV
- dengue virus
- MTase
- methyltransferase
- RdRp
- RNA-dependent RNA polymerase
- FL
- full length
- FAPA
- fluorescence-based alkaline phosphatase-coupled polymerase
- RFU
- relative fluorescence units
- JEV
- Japanese encephalitis virus.
REFERENCES
- 1. Davidson A. D. (2009) New insights into flavivirus non-structural protein 5. Adv. Virus Res. 74, 41–101 [DOI] [PubMed] [Google Scholar]
- 2. Noble C. G., Shi P. Y. (2012) Structural biology of dengue virus enzymes: towards rational design of therapeutics. Antiviral Res. 96, 115–126 [DOI] [PubMed] [Google Scholar]
- 3. Sampath A., Padmanabhan R. (2009) Molecular targets for flavivirus drug discovery. Antiviral Res. 81, 6–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malet H., Massé N., Selisko B., Romette J.-L., Alvarez K., Guillemot J.-C., Tolou H., Yap T. L., Vasudevan S. G., Lescar J., Canard B. (2008) The flavivirus polymerase as a target for drug discovery. Antiviral Res. 80, 23–35 [DOI] [PubMed] [Google Scholar]
- 5. Bussetta C., Choi K. H. (2012) Dengue virus non-structural protein 5 adopts multiple conformations in solution. Biochemistry 51, 5921–5931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yap L. J., Luo D., Chung K. Y., Lim S. P., Bodenreider C., Noble C., Shi P.-Y., Lescar J. (2010) Crystal structure of dengue virus methyltransferase bound to a 5′-capped octameric RNA. PLoS One 5, e12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lim S. P., Sonntag L. S., Noble C., Nilar S. H., Ng R. H., Zou G., Monaghan P., Chung K. Y., Dong H., Liu B., Bodenreider C., Lee G., Ding M., Chan W. L., Wang G., Yap L. J., Chao A. T., Lescar J., Yin Z., Vedananda T. R., Keller T. H., Shi P.-Y. (2011) Small-molecule inhibitors that selectively block dengue virus methyltransferase. J. Biol. Chem. 286, 6233–6240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yap T. L., Xu T., Chen Y. L., Malet H., Egloff M.-P., Canard B., Vasudevan S. G., Lescar J. (2007) The crystal structure of the Dengue virus RNA-dependent RNA polymerase at 1.85 Å resolution. J. Virol. 81, 4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Malet H., Egloff M. P., Selisko B., Butcher R. E., Wright P. J., Roberts M., Gruez A., Sulzenbacher G., Vonrhein C., Bricogne G., Mackenzie J. M., Khromykh A. A., Davidson A. D., Canard B. (2007) Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 282, 10678–10689 [DOI] [PubMed] [Google Scholar]
- 10. Yap T. L., Chen Y. L., Xu T., Wen D., Vasudevan S. G., Lescar J. (2007) A Multi-step strategy to obtain crystals of the dengue virus RNA-dependent RNA polymerase that diffract to high resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Noble C. G., Lim S. P., Chen Y.-L., Liew C. W., Yap L., Lescar J., Shi P.-Y. (2013) Conformational flexibility of the dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 87, 5291–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niyomrattanakit P., Chen Y.-L., Dong H., Yin Z., Qing M., Glickman J. F., Lin K., Mueller D., Voshol H., Lim J. Y. H., Nilar S., Keller T. H., Shi P.-Y. (2010) Inhibition of dengue virus polymerase by blocking of the RNA tunnel. J. Virol. 84, 5678–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanley K. A., Lee J. J., Blaney J. E., Jr., Murphy B. R., Whitehead S. S. (2002) Paired charge-to-alanine mutagenesis of dengue virus type 4 NS5 generates mutants with temperature-sensitive, host range, and mouse attenuation phenotypes. J. Virol. 76, 525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kroschewski H., Lim S. P., Butcher R. E., Yap T. L., Lescar J., Wright P. J., Vasudevan S. G., Davidson A. D. (2008) Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J. Biol. Chem. 283, 19410–19421 [DOI] [PubMed] [Google Scholar]
- 15. Leslie A. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 16. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 17. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 18. Pata J. D., Schultz S. C., Kirkegaard K. (1995) Functional oligomerization of poliovirus RNA-dependent RNA polymerase. RNA 1, 466–477 [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Q. M., Hockman M. A., Staschke K., Johnson R. B., Case K. A., Lu J., Parsons S., Zhang F., Rathnachalam R., Kirkegaard K., Colacino J. M. (2002) Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76, 3865–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tellez A. B., Wang J., Tanner E. J., Spagnolo J. F., Kirkegaard K., Bullitt E. (2011) Interstitial contacts in an RNA-dependent RNA polymerase lattice. J. Mol. Biol. 412, 737–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zou G., Chen Y.-L., Dong H., Lim C. C., Yap L. J., Yau Y. H., Shochat S. G., Lescar J., Shi P.-Y. (2011) Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 286, 14362–14372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kapoor M., Zhang L., Ramachandra M., Kusukawa J., Ebner K. E., Padmanabhan R. (1995) Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J. Biol. Chem. 270, 19100–19106 [DOI] [PubMed] [Google Scholar]
- 23. Egloff M. P., Benarroch D., Selisko B., Romette J. L., Canard B. (2002) An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21, 2757–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lu G., Gong P. (2013) Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 9, e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo D., Wei N., Doan D. N., Paradkar P. N., Chong Y., Davidson A. D., Kotaka M., Lescar J., Vasudevan S. G. (2010) Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 285, 18817–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Assenberg R., Mastrangelo E., Walter T. S., Verma A., Milani M., Owens R. J., Stuart D. I., Grimes J. M., Mancini E. J. (2009) Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. J. Virol. 83, 12895–12906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C. K., Walther P., Fuller S. D., Antony C., Krijnse-Locker J., Bartenschlager R. (2009) Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]