Background: Gene regulation in apicomplexan parasites is poorly understood.
Results: The plant-like nuclear factor TgAP2XI-5 is targeted at gene promoters, including those required for parasite virulence.
Conclusion: TgAP2XI-5 is a novel DNA sequence-specific transcription factor of T. gondii.
Significance: Identifying master regulators of virulence gene expression is crucial for understanding of pathogenicity of this pathogen.
Keywords: Gene Expression, Gene Regulation, Gene Transcription, Infectious Diseases, Promoters
Abstract
Gene regulation in apicomplexan parasites, a phylum containing important protozoan parasites such as Plasmodium and Toxoplasma, is poorly understood. The life cycle of Toxoplasma gondii is complex, with multiple proliferation and differentiation steps, of which tachyzoite proliferation is the most relevant to pathogenesis in humans and animals. Tachyzoites express invasion and virulence factors that are crucial for their survival and manipulation of host cell functions. The expression of those factors is tightly controlled during the tachyzoite cell cycle to permit their correct packaging in newly formed apical secretory organelles named micronemes and rhoptries in the daughter cells. However, little is known about the factors that control the expression of genes encoding the virulence factors present in these parasite-specific secretory organelles. We report that the plant-like nuclear factor TgAP2XI-5 targets more than 300 gene promoters and actively controls the transcription of these genes. Most of these target genes, including those that are essential for parasite virulence, showed a peak of expression in the S and M phases of the cell cycle. Furthermore, we identified the cis-regulatory element recognized by TgAP2XI-5 and demonstrated its ability to actively drive gene transcription. Our results demonstrated that TgAP2XI-5 is a novel DNA sequence-specific transcription factor associated with promoter activation. TgAP2XI-5 may regulate gene transcription of crucial virulence factors in T. gondii.
Introduction
Toxoplasma gondii is a unicellular eukaryote of the Apicomplexa phylum, which contains many deadly protozoan parasites such as Plasmodium (the cause of malaria) and Cryptosporidium (responsible for cryptosporidiosis). T. gondii is of critical importance to pregnant women, with first time infections having the potential to cause severe illness and even death in the developing fetus. In addition, the opportunistic nature of this obligate intracellular parasite can lead to the development of focal central nervous system infections in patients with AIDS/HIV. Paramount to the adaptability of T. gondii is its complex life cycle, which is characterized by multiple differentiation steps that are essential for its survival in both the human and definitive feline host. Humans can become infected by ingesting either oocysts shed by infected cats or cyst-contaminated meats. The oocysts and cysts contain sporozoites and bradyzoites, respectively, two developmental stages that differentiate into rapidly growing tachyzoites, the causative parasitic forms responsible for the clinical manifestations in humans. Bradyzoites typically cause chronic infection because of their ability to evade the immune system and resist common drug treatments, but they are also capable of reverting back to the more virulent tachyzoite stages in immunocompromised individuals.
Phenotypic differences are apparent between different T. gondii life cycle stages and also at various stages of the tachyzoite cell cycle. These changes are accompanied by the regulation of different gene populations. Transcriptome studies reveal that 18% of genes present in the T. gondii genome are co-regulated at specific stages of the life cycle (1), and more than 2,500 genes show regulated expression during the tachyzoite cell cycle (2). The distribution of these co-regulated genes throughout the T. gondii genome strongly implies that cis-acting elements and trans-acting factors regulate the transcription of individual genes. Indeed, a number of cis-acting elements (or sequence-specific DNA motifs) present in the promoters of developmentally regulated genes have been shown to be required for transcription (3). However, until recently trans-activating regulators such as DNA sequence-specific transcription factors were thought to be poorly represented in apicomplexan genomes, leading to speculation that other mechanisms such as epigenetics may play more active roles in T. gondii gene regulation. For example, the importance of chromatin structure in T. gondii gene regulation was highlighted by the discovery of specific chromatin marks at the promoters of transcriptionally active tachyzoite genes (4) and by the importance of the histone acetylase GCN5A in bradyzoite differentiation (5).
However, recent reanalysis of apicomplexan genomes uncovered a family of putative transcription factors that are defined by the possession of one or more plant-like AP2 domains (6). Intriguingly, the 68 putative AP2 transcription factors currently annotated in the T. gondii genome represent a significant expansion when compared with the 26 conserved AP2 proteins present in Plasmodium genomes. Importantly, like many tachyzoite genes, the steady-state transcript levels of 24 T. gondii AP2 genes appear to be cell cycle-dependent (2), implying that other members of the ApiAP2 (apicomplexan AP2) family may also participate in regulating the expression of stage-specific genes in Apicomplexa. Recently, we and others have demonstrated the involvement of a T. gondii AP2 transcription factor in cyst formation, indicating that ApiAP2 factors may play an important role in T. gondii gene regulation (7, 8). However, no genome-wide ChIP-chip data are available for T. gondii ApiAP2s. The ability to recognize and bind specific DNA motifs was confirmed for a number of Plasmodium and Cryptosporidium AP2s through protein-binding microarrays using recombinant proteins encompassing their respective AP2 DNA-binding domains (9). Furthermore, two putative AP2 transcription factors of Plasmodium appear to be essential for the development of mosquito invasive stages (10) and sporozoites (11, 12), and the disruption of these genes was accompanied by altered transcript profiles. In addition, genes of the ookinetes, which defines the fertilized form of the malarial parasite in a mosquito's body, formed by fertilization of a macrogamete by a microgamete and developing into an oocyst, contain a conserved TAGCTA motif in the promoters of several genes whose transcription was affected by disruption of AP2-O gene (10). This motif was shown to be essential for driving the expression of the ookinete protein, SOAP (secreted ookinete adhesive protein), and overlaps with the AGCTAGCT motif determined in silico for the AP2-O Plasmodium falciparum ortholog PF11_0442 (9).
In this study, we show that a putative AP2 transcription factor of T. gondii, TgAP2XI-5, binds to the promoters of hundreds of transcriptionally active genes. Furthermore, we identify the cis-regulatory element recognized by TgAP2XI-5 and demonstrate its ability to drive gene transcription. Thus, our results define TgAP2XI-5 as a novel sequence-specific transcription factor that regulates the promoters of T. gondii genes that are transcriptionally active during the S and M phases.
EXPERIMENTAL PROCEDURES
Parasite Tissue Culture and Manipulation
The virulent type I RH ΔKu80 strain of T. gondii tachyzoites (13) were propagated in vitro in human foreskin fibroblasts using DMEM supplemented with 10% FCS, 2 mm glutamine, and 1% penicillin-streptomycin. T. gondii tachyzoites were grown in ventilated tissue culture flasks at 37 °C and 5% CO2. Transgenes and promoter constructs were introduced by electroporation into tachyzoites of the T. gondii RH ΔKu80 strain (a gift from Vern Carruthers), and stable transformants were selected by culture in the presence of 2 μm pyrimethamine (for the DHFR selectable marker) or 25 μg/ml mycophenolic acid and 50 μg/ml xanthine (for the HXGPRT selectable cassette). Clonal lines were obtained by limiting dilution. Prior to protein purification and during ChIP, intracellular parasites were purified by sequential syringe passage with 17- and 26-gauge needles and filtration through a 3-μm polycarbonate membrane filter.
DNA Manipulation
TgAP2XI-5 (TGME49_016220) was amplified from genomic DNA of the parental Type I strain and cloned into either pLIC.HA9.DHFR or pLIC.TAP.HXGPRT vectors kindly provided by Vern Carruthers and Michael White (13). A region of the TgAP2XI-5 coding sequence corresponding to amino acids 540–867 was amplified by PCR and cloned into the pGex6P3 vector using EcoRI and BamHI sites. An entry vector containing 500 bp of the ROP18 (rhoptry protein 18) promoter (from T. gondii RH type I strain) was kindly provided by Jon Boyle (14). Two sites in the ROP18 promoter and one site in the histone H3 promoter were mutated using the QuikChange site-directed mutagenesis kit (Stratagene). Wild-type and mutated promoters were shuttled from the entry vector into a destination vector containing firefly luciferase and a 3′-untranslated region from the Toxoplasma DHFR gene (provided by Michael Behnke and Michael White, Department of Veterinary Molecular Biology, Montana State University) using the Gateway LR cloning strategy. Plasmids were selected using either ampicillin (100 μg/ml) or kanamycin (25 μg/ml). A list of primers used is provided in supplemental Table S1.
Dual Luciferase Assay
Equimolar quantities of each wild-type and mutated promoter construct (typically 50 μg) were co-transfected into T. gondii tachyzoites with 20 μg of Renilla luciferase driven by the T. gondii TUB1 promoter (4). Parasites were harvested 24 h post-transfection, and firefly and Renilla luciferase levels were determined using the dual-luciferase reporter assay system (Promega). Firefly luciferase measurements were standardized to those for Renilla luciferase, and the activity of mutated promoters was reported as a percentage of activity of the wild-type constructs.
Antibodies
Anti-HA mouse antibody (Invitrogen) was used at a dilution of 1:300 in immunofluorescence studies and 1:1,000 in Western blots. The anti-TgAP2XI-5 mouse antibodies were produced after immunization of mice with a truncated recombinant version of the protein and used at 1:500 dilutions in immunofluorescence studies and 1:1,000 in Western blots.
GST Fusion Protein Purification
The pTgAP2XI-5/Gex6P3 plasmid was used for expression of a truncated TgAP2XI-5 protein with the N-terminal GST tag. The plasmid was transformed into BL21 Escherichia coli, grown to A600 = 0.6, and induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside. After incubation for 12 h at 22 °C and 180 rpm, the bacteria were centrifuged, and the pellet was lysed by sonication in PBS supplemented with 1 mm PMSF. Following incubation with 1% Triton X-100 on ice for 30 min, the samples were centrifuged. The GST-tagged TgAP2XI-5 protein was purified from the soluble fraction by binding to glutathione-Sepharose 4B resin (GE Healthcare) and eluted with 20 mm reduced glutathione. The purified protein was washed with PBS using a 30-kDa cutoff centrifugal filter (Amicon Ultra) and quantified using the Bio-Rad kit for protein assay.
Chromatin Immunoprecipitation
ChIP was performed using a protocol described previously and parasites expressing a tandem affinity purification-tagged version of TgAP2XI-5 (4). Briefly, intracellular parasites were fixed for 10 min at room temperature using 1% formaldehyde and processed for DNA sonication using the Bioruptor device for 45 min at 4 °C with a 30-s on-off cycle. Protein-DNA complexes were then bound for 2 h with IgG beads (Amersham Biosciences), washed five times in ChIP wash buffer, washed once in ChIP equilibration buffer, and eluted in twice in 100 μl of ChIP elution buffer. ChIP DNA purification and amplification were carried out as previously described (4).
A tilling microarray was designed by Genotypic Technology (India) based on version 6 of the T. gondii ME49 genome (version 7.3) and printed by Agilent Technologies. The microarray encompasses more than 983,000 features representing the entire genome with an average coverage of one oligonucleotide every 63 bp (15). Purified ChIP material was processed according to the Agilent Mammalian ChIP-on-chip protocol version 10.11, and labeled DNA was hybridized to an Agilent T. gondii tiling array for 40 h at 65 °C (G4481-90010; Agilent Technologies). Microarrays were washed and scanned according to the manufacturer's protocol, and the results were processed with the Genomic workbench Standard edition. The Ringo R package was used with default values to identify peaks among the AP2XI-5 ChIP-chip experiments.
Quantitative PCR
All primers were designed online using Primer2 v.0.4.0 and are listed in supplemental Table S1. Quantitative PCR was carried out on an Mx3000P System (Agilent Technologies). Individual reactions were prepared with 0.5 μm of each primer, 5 ng of ChIP or input DNA, and SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) in a final volume of 20 μl. All experiments were performed twice with separate biological replicates. For each experiment, reactions were performed in triplicate with data being presented as percentages of input.
Calculation of Normalized Cell Cycle Transcript Profiles
Transcript microarray data generated from Behnke et al. (2) were downloaded from ToxoDB, the toxoplasma genomics resource. Robust multiarray average (RMA)3 Toxoplasma values represent the level of expression for each gene. Normalized expression profiles were produced for each gene by dividing the RMA value at each cell cycle time point by the RMA value in the asynchronous population and plotting the data for individual or groups of genes across the different time points. Charts and heat maps of different gene populations were generated using the log2 values of these relative RMA values.
DNA Motif Discovery
Genomic sequences corresponding to the 652 manually identified TgAP2XI-5 ChIP peaks (listed in supplemental Table S2) were analyzed using RSAT (Regulatory Sequence Analysis Tools) (16). Over-represented six-base motifs were identified using the “oligo-analysis (words)” tool under default settings, using Leishmania major, an unrelated protozoan parasite, as a background organism model. For a more specific frame of reference, the putative promoters of all 7,987 predicted T. gondii genes were downloaded from ToxoDB and subjected to the same analysis. The “position-analysis (words)” tool was used to identify six-base motifs over-represented at the center of TgAP2XI-5 peak sequences, using a class grouping interval of 50 bases.
Electromobility Shift Assays
A 40-bp region within the ROP18 promoter that contained the GCTAGC motif was labeled with biotin (supplemental Table S1). The annealing of sense and antisense single-stranded DNA probes was carried out by incubating complementary probes for 5 min at 95 °C in 10 mm Tris, 1 mm EDTA, 50 mm NaCl (pH 8.0) and then cooling overnight to room temperature. EMSA were performed using the Light Shift Chemiluminescent EMSA kit (Pierce) as follows. The provided 10× binding buffer was supplemented with 1 μg of poly(dI·dC) and 5 mm MgCl2, and preincubation of 10 pmol of recombinant protein or GST and different concentration of appropriate competitor (supplemental Table S1) was carried out for 10 min at room temperature. For the supershift assays, 0.5 μl of immune or preimmune serum were incubated with the recombinant protein. Next, 20 fmol of biotinylated probe was added, and the samples were incubated for a further 30 min at room temperature. The mixture was then resolved on a pre-run 5% acrylamide gel, prepared in 0.5× TBE. The DNA was then transferred onto a Nylon membrane and blotted with Streptavidin-HRP beads according to the manufacturer's instructions.
RESULTS
TgAP2XI-5 Is a Constitutively Expressed Nuclear Factor of T. gondii Tachyzoites
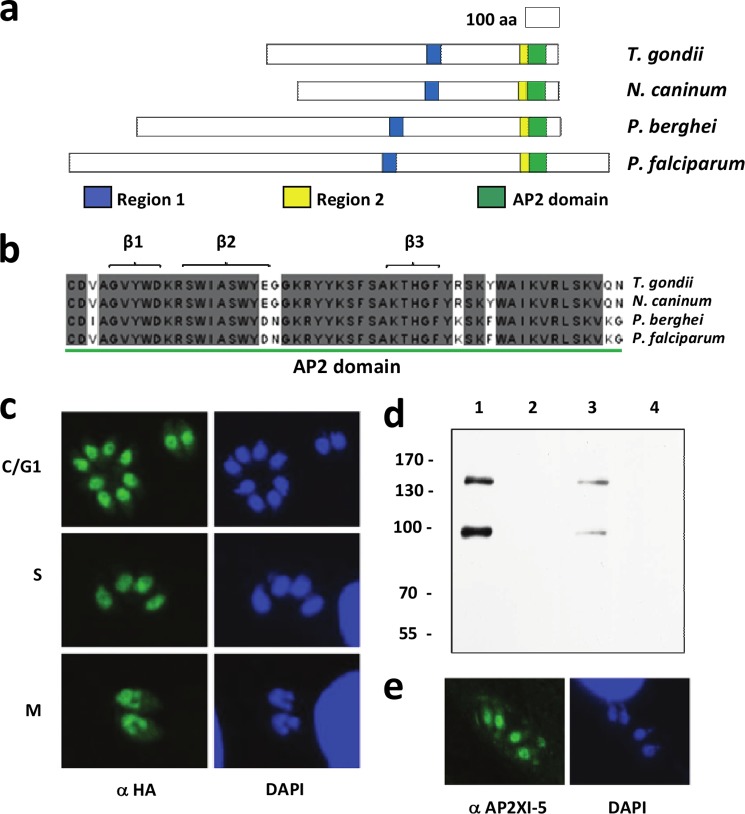
TgAP2XI-5 (TGME49_016220) is one of 68 T. gondii genes that encode a putative plant-like AP2 transcription factor. It is expressed at the highly virulent tachyzoite stage and was originally identified through an expression screen as a gene that is overexpressed in the slowly dividing and dormant bradyzoite forms isolated from brain tissue cysts of chronically infected mice (7). We decided to further characterize this novel putative plant-like transcription factor. Homologs of the AP2 domain of TgAP2XI-5 protein (TGME49_016220, 868 amino acids) were found in other apicomplexan parasites, including Neospora caninum (NCLIV_095590, 779 amino acids), Plasmodium berghei (PBANKA_090590, 1,258 amino acids), and P. falciparum (PF11_0442, 1,604 amino acids) with BLASTp E values of 7.8e−181, 6.4e−41, and 2.1e−40, respectively. Although there was no amino acid sequence variation in TgAP2XI-5 among the three lineages of T. gondii, sequence alignment of the protein with its more divergent apicomplexan homologs revealed conservation of only the C-terminal AP2 domain and two other domains, Regions 1 and 2 (Fig. 1a). Region 1 was located in the middle portion of each protein and was strongly conserved between T. gondii and N. caninum (97.5% identity) with moderate conservation between T. gondii and either P. falciparum or P. berghei (57.5 and 55% identity, respectively). Although region 1 had no clear matches to known Pfam domains, it was recently demonstrated to act as a trans-activator domain in Plasmodium (17). Region 2 was adjacent to and directly upstream of the AP2 domain and appeared to be highly conserved between T. gondii and N. caninum (100% identity), with moderate conservation between T. gondii and either P. falciparum or P. berghei (64% identity for both). In numerous apicomplexan AP2 transcription factors, the region directly upstream of the AP2 domain contains a small DNA-binding motif called the AT-hook (6, 18), but it is unclear whether region 2 of TgAP2XI-5 and its homologs is a true AT-hook, particularly given the lack of the characteristic glycine-arginine-proline (GRP) AT-hook tripeptide (19). Nevertheless, the TgAP2XI-5 AP2 domain itself was highly conserved between T. gondii and either N. caninum (100% identity), P. falciparum (88.5% identity), or P. berghei (86.5% identity) (Fig. 1b), particularly the three β-sheets involved in the direct binding of double-stranded DNA (18, 19). Interestingly, despite the apparent expansion in the number of AP2-containing proteins in T. gondii, there did not appear to be any paralogs of TgAP2XI-5, with the most closely related AP2 (TGME49_018960) showing only 40% identity over the DNA-binding domain.
FIGURE 1.
TgAP2XI-5 contains a conserved AP2 domain and is expressed throughout the tachyzoite cell cycle. a, schematic representation of conserved AP2-containing proteins from T. gondii (TgAP2XI-5 or TGME49_016220), N. caninum (NCLIV_095590), P. berghei (AP2-O or PBANKA_090590), and P. falciparum (PF11_0442) illustrating the position of region 1 (blue), region 2 (yellow), and the AP2 domain (green). b, multiple sequence alignment of the AP2 domains showing the stretches of amino acids (aa) corresponding to anti-parallel β-sheets (β1–3), which are responsible for DNA binding. Residues fully conserved between the four protein sequences are shaded gray. c, expression of TgAP2XI-5-HA was observed in the nuclei of T. gondii tachyzoites throughout the entire cell cycle. C, cytokinesis; G1, gap phase; S, DNA synthesis; M, mitosis. DAPI staining was used to indicate the location and morphology of the parasite and host nuclei. d, Western blot analyses of nuclear lysates of the TgAP2XI-5-HA mutant (lane 1), wild-type ΔKu80 Type I T. gondii (lanes 2 and 3), and lysate of uninfected host cell (lane 4). Monoclonal anti-HA antibody was used to detect the HA-tagged TgAP2XI-5 (lane 1), whereas polyclonal anti-TgAP2XI-5 sera were used to detect the native protein (lane 3). Molecular mass marker is indicated in kilodaltons. e, polyclonal anti-TgAP2XI-5 sera were used to confirm expression and nuclear localization of TgAP2XI-5 in T. gondii tachyzoites.
All of the 68 T. gondii AP2s were annotated as putative transcription factors solely because of their possession of at least one AP2 domain. Therefore, to partially validate the predicted biological function of TgAP2XI-5, localization experiments were carried out using parasites expressing a C-terminal HA-tagged endogenous version of TgAP2XI-5 that maintains the original TgAP2XI-5 endogenous promoter. As expected, expression of TgAP2XI-5-HA was clearly observed in the nucleus of T. gondii tachyzoites (Fig. 1c). Furthermore, TgAP2XI-5-HA was expressed at a strong level throughout the entire tachyzoite cell cycle, consistent with the steady-state transcript levels of AP2 previously determined by expression microarray (2). Western blot analysis of TgAP2XI-5-HA expression using monoclonal anti-HA antibody detected two clear bands of ∼100 and 150 kDa (Fig. 1d, lane 1), whereas the same antibody did not react against untagged RH ΔKu80 nuclear extracts (Fig. 1d, lane 2). In parallel, a recombinant protein encompassing both region 2 and the AP2 domain of TgAP2XI-5 with an N-terminal GST tag was expressed in E. coli. Polyclonal sera raised against purified TgAP2XI-5-GST were tested against untagged RH ΔKu80 nuclear extracts (Fig. 1d, lane 3) or lysate of uninfected host cell (Fig. 1d, lane 4). The presence of two proteins of 100 and 150 kDa was confirmed using polyclonal antibodies generated against the bacterial recombinant TgAP2XI-5-GST and nuclear extract from the wild-type parasites (Fig. 1d, lane 3) but not with cell lysate obtained from uninfected cells (Fig. 1d, lane 4). It is unclear why two bands were detected for TgAP2XI-5 (predicted molecular mass of 89 kDa), particularly given the lack of any clear paralogs within T. gondii. The polyclonal sera raised against purified bacterial recombinant TgAP2XI-5-GST were then used for localization studies in T. gondii tachyzoites, confirming constitutive expression of native TgAP2XI-5 in the parasite nucleus (Fig. 1e).
TgAP2XI-5 Is Enriched Exclusively at the Promoters of T. gondii Genes
ChIP experiments were performed to identify regions of the T. gondii genome that are potentially targeted and bound by TgAP2XI-5. A genome-wide analysis of the TgAP2XI-5 ChIP DNA was then carried out using a tandem affinity purification-tagged version of TgAP2XI-5 and a custom tiling microarray consisting of over 1 million oligonucleotides, with each oligonucleotide averaging 60 bp of the T. gondii genome. Manual screening of this ChIP-on-chip data using a cutoff log2[ChIP:INPUT] ratio of 0.9 for the maximum peak “height” resulted in the identification of 652 individual peaks. These peaks represent regions of the genome with localized TgAP2XI-5 enrichment and were distributed quite evenly over the 14 T. gondii chromosomes. The number of peaks identified was reduced to 384 and 69 by applying more stringent cutoff log2[ChIP:INPUT] ratios of 2.0 and 3.0, respectively. Information regarding the genomic location, width, and height of each peak, as well as the associated gene predicted for each peak, is listed in supplemental Tables S2–S4 for cutoff log2[ChIP:INPUT] ratios of 0.9, 2.0, and 3.0, respectively. As expected, a control ChIP-on-chip experiment using the wild-type nontagged ΔKu80 Type I strain showed very few peaks and was used to assess background fluctuations over the entire array.
We also used an automated peak finder to screen the ChIP-on-chip data. The Ringo peak finder (20) with increasing stringency cutoff scores of >30, >35, and >40 (supplemental Table S5) identified 237, 119, and 58 peaks, respectively. Among the Ringo >30 peak set, more than 60% were common to the manually identified peaks (with cutoff log2[ChIP:INPUT] ratios of 0.9 and 2.0). This number increased to 82% in the Ringo >40 peak set (supplemental Table S6). The good correlation between the manually and automated peak sets illustrates the quality of the peak predictions.
A clear majority of the 652 peaks were located within the putative promoters of protein coding genes. The observation that TgAP2XI-5 enrichment occurs exclusively in gene promoters was corroborated by the fact that virtually all of the 652 peaks (99.4%) co-localized genomic regions enriched with markers of active promoters, namely the modified histone markers H3K9ac and H3K4me3 (profiles of which are available at the ToxoDB website) (4). The four TgAP2XI-5 enrichment peaks with no correlating H3K9ac or H3K4me3 peaks were located in the putative promoters of genes encoding a putative microneme protein (TGME49_075790); two surface antigen-related proteins, SRS22I (TGME49_038850) and SRS53C (TGME49_115390); and ROP42 (TGME49_00980), with the latter displaying unreliable ChIP-on-chip data for the modified histones.
The 734 annotated genes showing TgAP2XI-5 enrichment at their promoter and therefore potentially targeted by TgAP2XI-5 included many important apicomplexan-specific genes. Those annotated as such include 24 rhoptry organelle proteins (ROPs), 21 AP2 transcription factors, 14 surface antigen-related proteins, 8 microneme proteins, 7 inner membrane complex proteins, and 6 rhoptry neck proteins. Interestingly, although the 734 genes represent 9.2% of the total 7,987 genes predicted for T. gondii (ME49 strain), the number of identified ROP and AP2 genes represents 52.2% (24 of 46) and 30.9% (21 of 68) of their respective families, suggesting preferential promoter binding by TgAP2XI-5. Clear enrichment of TgAP2XI-5 occupancy was observed as a single peak (Table 1 and Fig. 2a) at the bidirectional promoter of core histones H2A and H3 (TGME49_061250 and TGME49_061240, highlighted in yellow). Note that there was no corresponding peak for the negative wild-type control ChIP-on-chip experiment (in red). Further examples of TgAP2XI-5 binding were at the putative promoters of ROP18, TgAP2XII-2, ROP30, and TgAP2XI-5 itself (Fig. 2b), and these promoters were chosen for validation because they represent a range of highly and lowly enriched ChIP-on-chip peaks. The TgAP2XI-5 ChIP enrichment of these promoter regions was validated by quantitative PCR by targeting the promoters of each of these five genes. DNA pulled down by TgAP2XI-5 ChIP (Fig. 2c, black bars) was enriched for all five promoters compared with DNA pulled down in the wild-type (negative; Fig. 2c, white bars) ChIP experiments. The promoter region of a hypothetical gene (TGME49_106350) that displayed no TgAP2XI-5 enrichment in ChIP-on-chip analysis was included as a negative control (Neg 1), as was a region of chromosome Ia that was not located in a promoter (Neg 2). As expected, these regions were not significantly enriched in the TgAP2XI-5 ChIP DNA compared with the wild-type ChIP DNA (Fig. 2c).
TABLE 1.
Genomic loci with strongest TgAP2XI-5 enrichment
The top 20 peaks corresponding to TgAP2XI-5 ChIP enrichment are listed in descending order of peak height (represented as log2[ChIP:INPUT]). The genomic loci of these peaks are listed, including chromosome number and the start, end, and total size of each observed peak. The identities of genes located immediately downstream of these peaks are also given, with their putative annotations.
| Chromosome | Start | End | Size | Height | Gene | Annotation |
|---|---|---|---|---|---|---|
| bp | ||||||
| IX | 5,352,000 | 5,353,600 | 1,600 | 4.467 | TGME49_105160 | Histone H2B, putative |
| VI | 498,500 | 500,100 | 1,600 | 4.406 | TGME49_039260 | Histone H4, putative |
| VIIb | 1,798,400 | 1,800,100 | 1,700 | 4.278 | TGME49_061250/40 | Histone H2A (canonical)/H3 |
| IV | 885,400 | 887,500 | 2,100 | 4.194 | TGME49_119560 | Microneme protein MIC3 |
| IX | 1,165,700 | 1,167,500 | 1,800 | 4.094 | TGME49_066630 | Hypothetical |
| V | 359,400 | 360,900 | 1,500 | 4.037 | TGME49_020380 | Hypothetical |
| V | 1,523,400 | 1,525,500 | 2,100 | 3.95 | TGME49_087040 | Hypothetical |
| VIII | 2,072,100 | 2,074,100 | 2,000 | 3.909 | TGME49_032400 | PAN domain-containing protein |
| X | 5,952,300 | 5,954,000 | 1,700 | 3.881 | TGME49_014070/80 | Hypothetical/toxofilin |
| VIIb | 913,300 | 915,200 | 1,900 | 3.789 | TGME49_062920 | Trypsin, putative |
| Ib | 1,139,800 | 1,141,700 | 1,900 | 3.768 | TGME49_009240 | Hypothetical |
| IV | 1,277,300 | 1,279,200 | 1,900 | 3.701 | TGME49_118470 | AP2IV-4 |
| X | 411,500 | 414,200 | 2,700 | 3.691 | TGME49_028230 | Hypothetical |
| IX | 3,479,800 | 3,481,400 | 1,600 | 3.67 | TGME49_089710 | AP2IX-5 |
| IX | 562,900 | 565,500 | 2,600 | 3.667 | TGME49_067680 | Microneme protein, putative (MIC12) |
| V | 544,000 | 546,000 | 2,000 | 3.644 | TGME49_012770 | Hypothetical |
| IX | 3,330,700 | 3,332,700 | 2,000 | 3.629 | TGME49_089370 | Hypothetical |
| Ib | 1,095,300 | 1,098,200 | 2,900 | 3.599 | TGME49_009180 | PAN domain-containing protein, putative |
| XII | 541,600 | 542,900 | 1,300 | 3.525 | TGME49_108080 | Rhoptry kinase family protein ROP5 (incomplete catalytic triad) |
| VIIa | 1,402,100 | 1,403,800 | 1,700 | 3.487 | TGME49_005370 | Hypothetical |
FIGURE 2.
ChIP-on-chip analysis of TgAP2XI-5 occupancy demonstrates enrichment at specific gene promoters. a and b, linear representations of T. gondii chromosome regions encompassing the genes encoding H2A/H3 (a) and TgAP2XI-5 (b), ROP18, TgAP2XII-2, and ROP30. ChIP-on-chip data are presented as the log2 ratio of the hybridization signal given by DNA immunoprecipitated with TgAP2XI-5 over the signal given by the nonenriched input DNA. The log2 ratio of each oligonucleotide present on the tiled microarray has been plotted at the respective genomic position for the TgAP2XI-5 experiment (black) and the negative control wild-type experiment (red). Genes whose promoters appear to be targeted by TgAP2XI-5 are highlighted in yellow, and genes not targeted by TgAP2XI-5 are indicated in dark blue. Genes above the horizontal axis read (5′ to 3′) from left to right, whereas those below read right to left. The putative regions of TgAP2XI-5 enrichment are boxed. c, quantitative PCR was performed on ChIP, and input DNA was purified from the TgAP2XI-5 (black) and wild-type control (white) experiments. Amplification was carried out on regions within the promoter of each of the five genes listed, as well as the promoter for a hypothetical gene, TGME49_106350, displaying no TgAP2XI-5 enrichment (Neg1), and a non-promoter region of chromosome Ia (Neg2). The enrichment for corresponding ChIP DNA samples was presented as a percentage of INPUT for each target.
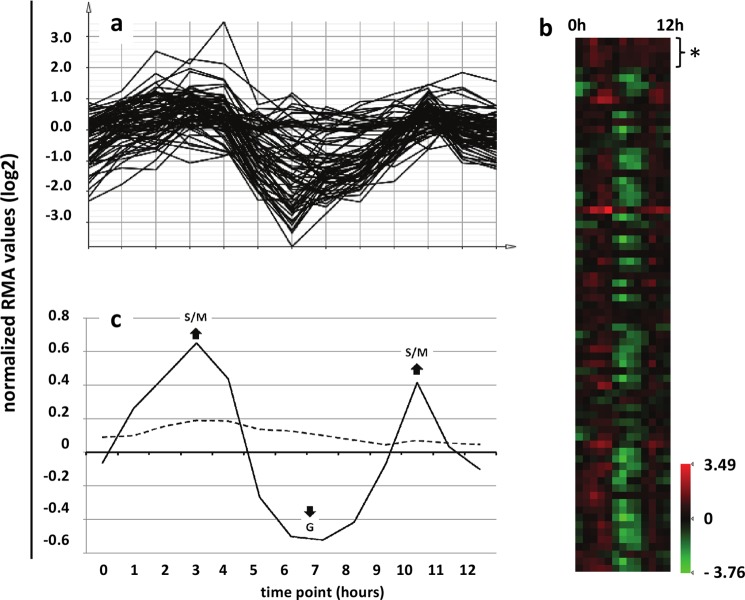
Many of the T. gondii AP2 transcription factors have previously been shown to display cell cycle-regulated transcription. Although TgAP2XI-5 itself appeared to be expressed consistently throughout the tachyzoite cell cycle, many of the genes targeted by this AP2 protein displayed fluctuating transcript profiles Indeed, the normalized transcript profiles of the 73 genes most clearly bound by TgAP2XI-5 (from supplemental Table S4) demonstrated a remarkably similar pattern of regulation, with peak transcripts being detected at ∼3 and 10 h (Fig. 3, a and b). Clear exceptions included genes encoding the four core histones (indicated in Fig. 3b), which demonstrate a similar steady-state expression to that of TgAP2XI-5 itself, although this may be due to limitations associated with microarray analysis of highly transcribed genes. Extending this analysis to all 734 genes potentially targeted by TgAP2XI-5 revealed that maximum transcript levels were detected at the 3- and 10-h time points, which corresponded to the S (DNA synthesis) and M (mitosis) phases of the tachyzoite cell cycle (Fig. 3c), even though the ChIP experiments were performed with an asynchronously growing parasite population (60% G1, 40% S/M). In contrast, averaging of the normalized transcript profiles of all T. gondii genes revealed very little net fluctuation throughout the tachyzoite cell cycle.
FIGURE 3.
TgAP2XI-5 targets a large group of genes whose peak transcript levels are observed during DNA synthesis and mitosis. Individual transcript profiles of T. gondii genes throughout the tachyzoite cell cycle generated by Behnke et al. (2) were modified to express normalized transcript levels (see “Experimental Procedures”) to facilitate comparison between different genes. a and b, the individual transcript profiles for the 73 genes whose promoters were most clearly targeted by TgAP2XI-5 were plotted as a chart where the x axis represents the tachyzoites cell cycle time points from the Behnke et al. (2) study (a) and a heat map (b). The heat map profiles of the four core histones, H2A, H2B, H3, and H4, are indicated by asterisks. c, the log2 average of normalized RMA values were plotted at each time point for all 687 genes targeted by TgAP2XI-5 with reported transcript profiles (solid line) and for all 7,138 genes from the study described by Behnke et al. (2) (dashed line). The tachyzoite cell cycle stages associated with the two peaks (S, DNA synthesis; M, mitosis) and single trough (G, gap phase) are indicated.
TgAP2XI-5 Enrichment Occurs Primarily at a GCTAGC Motif
A more comprehensive analysis of TgAP2XI-5 promoter binding was carried out using the RSAT on-line computational resource. Using this approach, the TgAP2XI-5-enriched peak sequences (ranging in size from 600–3100 bp) were scanned for over-represented six-base motifs. The palindromic motif GCTAGC was the highest ranked, with a total of 440 occurrences throughout the 652 peak sequences (Fig. 4). This DNA motif was also enriched in sequences extracted from the Ringo peaks set. This corresponded exactly with the motif uncovered for the P. falciparum ortholog, PFF11_0442, in protein binding microarrays (9) and overlapped with the motif identified in the promoters of genes potentially regulated by the P. berghei ortholog, PBANKA_090590, also known as AP2-O (10). Many of the other top ranked motifs uncovered in the present study overlap with the GCTAGC motif, including AGCTAG and CTAGCC (Fig. 4), and comparison of those motifs performed with the RSAT program showed that they could be amalgamated into two highly similar extended motifs (Fig. 4, row (i)), both conserving the GCTAGC sequence.
FIGURE 4.

A palindromic GCTAGC motif is over-represented in promoters targeted by TgAP2XI-5. DNA sequences corresponding to the 652 promoter regions targeted by TgAP2XI-5 were analyzed using the on-line Regulatory Sequence Analysis Tool. Over-represented six-base motifs are listed with the observed occurrence, expected occurrence, occurrence p value, and score.
The five motifs, GCTAGC, ACTAGC, CAAGAC, CAAGACAC, and GAGGAAAA, were analyzed more critically by comparing the frequency of their occurrence in both the populations of TgAP2XI-5 ChIP-on-chip peaks and in the putative promoters of all 7,987 T. gondii protein coding genes. The GCTAGC motif was observed in 54.6% of the 652 promoters enriched by TgAP2XI-5 ChIP compared with only 16.6% (i.e., background) of all T. gondii promoters (Fig. 5a), and a similar pattern was observed for the ACTAGC motif. In contrast, the GAGGAAAA motif had a background frequency of 23.1% and a frequency of only 21.3% in the enriched promoters and was therefore disregarded. Interestingly, although the frequency of the six-base CAAGAC motif showed no clear enhancement in the ChIP-enriched promoter sequences, the longer CAAGACAC motif appeared more frequently in promoter sequences than in the background sequences. Even clearer differences for the GCTAGC, ACTAGC, and CAAGACAC motifs were apparent when comparing the background frequencies with those of the 69 highly enriched promoters (i.e., using the log2[ChIP:INPUT] ratio cutoff of 3.0) (Fig. 5b).
FIGURE 5.

GCTAGC and ACTAGC motifs are enriched above background in TgAP2XI-5 targeted promoters. a, the frequency that gene promoters possessed an individual motif at least once was calculated for all 652 TgAP2XI-5 targeted promoters (white) and for the promoters all 7,987 predicted T. gondii ME49 genes (black). b, this comparison was also repeated for the 69 promoters (from supplemental Table S4) most clearly targeted by TgAP2XI-5.
Further evidence that GCTAGC represents the primary DNA motif recognized by TgAP2XI-5 was obtained by repeating the motif search using a positional bias, because it was expected that this motif would be located at the center of each peak sequence. The search was performed with RSAT using a positional bias for the center of each sequence with extending windows of 50 bp. GCTAGC was clearly identified as the highest ranked motif, and six of the remaining top seven motifs were overlapping derivatives of GCTAGC (Fig. 6a). Plotting the occurrences of each of these seven motifs across each of the 50-bp segments revealed a clear bias for each of these motifs at the very center of the peak sequences (Fig. 6b). In contrast, the remaining four motifs identified by the search did not demonstrate such a strong central bias (Fig. 6c). This shows that the regions of highest enrichment of TgAP2XI-5 on individual promoters coincide with the location of the GCTAGC motif, indicating that it may be the TgAP2XI-5 DNA-binding motif. A closer look at 9 of the top 20 TgAP2XI-5 ChIP-on-chip peaks (from Table 1) again demonstrated that the GCTAGC motif was located at the tip of each peak (Fig. 6d). Interestingly, the one exception shown here was for the putative promoter of the core histone H2B, which contained two almost tandem ACTAGC motifs that might substitute for the primary motif.
FIGURE 6.
The GCTAGC motif is located at the center of TgAP2XI-5 ChIP-on-chip peaks. a, the 652 promoter regions targeted by TgAP2XI-5 were reanalyzed using the “position-analysis (words)” tool on the on-line Regulatory Sequence Analysis Tool to identify six-base motifs over-represented at the center of peaks. Over-represented six-base motifs are listed with the observed occurrence, p value, E value, and score. Motifs observed to be overlapping with the GCTAGC motif are marked with asterisks. b and c, the occurrences of different six-base motifs were mapped against positions (using a 50-bp window) relative to the peak center. The relative peak distribution of motifs overlapping (b) and not overlapping (c) with the GCTAGC motif is shown. d, TgAP2XI-5 ChIP-on-chip peaks, corresponding to the putative promoters of nine genes listed in Table 1, have been plotted (as described in Fig. 2, a and b) along with the position of GCTAGC (green bar) and ACTAGC (red bar) motifs.
The GCTAGC Motif Is Involved in Promoter Activation
Given that TgAP2XI-5 AP2 clearly targets the GCTAGC motif in many active gene promoters, it is likely that this motif is responsible, at least in part, for activating gene transcription. To address this hypothesis, the putative promoter region of ROP18 was fused to firefly luciferase, and the single GCTAGC motif present in the promoter was mutated to GGTACC (Fig. 7a). In addition to a single GCTAGC motif, the putative promoter of ROP18 contained a GCTAGG motif, which was also mutated (CCATGG) for the following experiment. A dual luciferase assay was carried out following transient transfection with constructs containing wild-type or mutated promoters of the ROP18 gene in addition to vector encoding Renilla luciferase for standardization. Base pair mutations in the GCTAGG motif of the ROP18 promoter (pROP18m2) resulted in no apparent change in luciferase expression compared with the wild-type ROP18 promoter construct (pROP18). In contrast, a 2-bp mutation in the single GCTAGC motif (pROP18m1) resulted in a 70% decrease in luciferase expression compared with wild type. The combination of both mutations (pROP18m1m2) resulted in only a 55% reduction in luciferase expression compared with the wild type. In parallel, the level of the AP2XI-5 protein was verified for each sample (Fig. 7b) by Western blot. We showed that similar level of the AP2XI-5 protein was expressed in all samples, indicating that the mutation of the promoter sequence was responsible of the differences in the Rop18 promoter activity (Fig. 7a).
FIGURE 7.
The GCTAGC motif is required for efficient transcription of the rop18 gene. a, the putative promoter of rop18 (pROP18), corresponding to a 500-bp region upstream of the start codon, was subjected to site-directed mutagenesis, resulting in the disruption of a single GCTAGC motif (pROP18m1), a single GCTAGG motif (pROP18m2), or both motifs (pROP18m1m2). These three mutant promoters and the wild-type promoter were cloned upstream of a reporter luciferase construct and assayed for their ability to drive transcription. The transcriptional potential of mutated promoters, measured as normalized firefly luciferase activity, was reported as a percentage of the activity of the wild-type promoter construct. Statistical analysis was performed using a Student t test and showed nonsignificant (ns) or significant differences for the promoter activities (*, p < 0.05). b, Western blot analyses of the lysates prepared for the luciferase assay. The lysates corresponding to the parasites transfected with the WT ROP18 promoter (WT), the disruption of a single GCTAGC motif (m1), a single GCTAGG motif (m2), or both motifs (m1m2) were probed for the TgAP2XI-5 protein (top panel). The TgActin antibody was used as a loading control (bottom panel). Molecular mass marker is indicated in kilodaltons. c, an electrophoretic mobility shift assay was carried out using a recombinant protein spanning the TgAP2XI-5 AP2 domain and region 2. The shift caused by the binding of TgAP2XI-5 to the biotinylated probe is indicated by an arrow. TgAP2XI-5 binding was inhibited by 100- and 300-fold excesses of specific competitor but not by a nonspecific competitor. d, an electrophoretic mobility shift assay was carried out using the TgAP2XI-5 recombinant protein or GST. The shift caused by the binding of TgAP2XI-5 to the biotinylated probe is indicated by a solid arrow. GST was not able to form a shifted band. The binding of TgAP2XI-5 to the probe was supershifted as indicated by the dashed arrows after the addition of the immune serum specific to TgAP2XI-5 but not the preimmune serum.
TgAP2XI-5 Binds to the GCTAGC Motif
The GCTAGC motif is present in most of the TgAP2XI-5 enriched promoter regions. Because this motif appears to be crucial for the activation of the ROP18 promoter, we measured the ability of the TgAP2XI-5 recombinant protein to bind in vitro to the GCTAGC motif and its mutated version. For that, we performed an EMSA using labeled oligonucleotides encompassing the 40 base pairs surrounding the ROP18 promoter GCTAGC motif. As suggested by Fig. 7c, the recombinant TgAP2XI-5 protein is able to bind to this DNA sequence. This binding is specifically mediated by the GCTAGC motif because an excess of unlabeled oligonucleotide encompassing the motif is able to compete the interaction with the probe, whereas an excess of unlabeled oligonucleotide encompassing the mutated version of the motif (GGTACC) is not. We also verified that GST alone was not able to bind to this oligonucleotide (Fig. 7d). The addition of the polyclonal serum raised against TgAP2XI-5, induced the formation of a specific supershift, whereas the preimmune serum did not (Fig. 7d), indicating that the TgAP2XI-5 protein is indeed responsible for the formation of the retarded bands.
DISCUSSION
TgAP2XI-5, a putative AP2 transcription factor of T. gondii, is a homolog of two partially characterized AP2 transcription factors in P. falciparum and P. berghei. Typical of different apicomplexan AP2 family members (6), the conservation of TgAP2XI-5 with its putative apicomplexan orthologs is restricted primarily to its AP2 domain. This is important because proteins with highly conserved AP2 domains tend to bind to the same DNA motifs, even between organisms of different genera (18, 19). It was not surprising, therefore, to find preferential binding of TgAP2XI-5 to a GCTAGC motif within the promoters of T. gondii genes. This motif is identical to that recognized by the P. falciparum (PF11_0442) homolog and overlaps with the motif recognized by the P. berghei (PBANKA_090590 or AP2-O) homolog. In P. berghei, this motif was identified in the promoters of a number of ookinete-specific genes, an observation consistent with the fact that AP2-O is essential for the formation of invasive ookinetes (10). In T. gondii, promoters targeted by TgAP2XI-5 typically contained a single GCTAGC motif, contrary to observations in P. berghei where at least two copies of the cis-regulatory element are required for efficient AP2-O binding (10).
Although the ChIP experiment was restricted to asexual tachyzoite stages, the genome-wide ChIP-on-chip analysis demonstrated the binding of TgAP2XI-5 to hundreds of putative gene promoters. Putative promoters of genes coding for core histones, rhoptry organelle proteins, and other AP2 proteins were over-represented in the TgAP2XI-5 ChIP data. Perhaps more importantly, genes targeted by TgAP2XI-5 display elevated transcript abundance at the S phase and mitosis (M), particularly compared with gap phase (G). Indeed, enrichment of the GCTAGC motif that is recognized by TgAP2XI-5 has previously been observed in the promoters of genes displaying elevated S/M phase transcript abundance (2), and it would appear that TgAP2XI-5 plays a role in mediating this regulation. Attempted knockouts of TgAP2XI-5 were unsuccessful using both direct and inducible strategies, and it is tempting to conclude that this reflects the essential role of TgAP2XI-5 in the tachyzoite cell cycle.
Nonetheless, it is surprising that TgAP2XI-5 itself shows no clear fluctuations in transcript abundance during the tachyzoite cell cycle, particularly given its likely role in activating S/M genes. Some transcription factors require additional activation before they can begin to regulate the transcription of specific genes. For example, STAT transcription factors latently present in the cytoplasm undergo phosphorylation leading to dimer formation and translocation to the nucleus, where they regulate gene transcription (21). Analysis of the phosphoproteome of T. gondii revealed extensive phosphorylation of TgAP2XI-5 (22), indicating its potential for post-translational activation, possibly through homo- and heterodimer formation. Dimer formation is particularly important because it can alter the temporal nature of transcription factor activities (23), offering one explanation for the lack of correlation between the expression of TgAP2XI-5 and the timed transcription of its putative gene targets. Interestingly, the P. falciparum AP2 protein, PF14_0633, has been shown to form a homodimer upon binding to DNA (18), although in this case dimer formation is probably mediated by disulfide bonds rather than phosphorylation. Yeast two-hybrid screening has also highlighted the potential for heterodimer formation between other AP2s of P. falciparum (24). Western blot analysis of TgAP2XI-5 consistently detected two protein bands of 100 and 150 kDa, suggesting the possible presence of a heterodimer.
Among many of the important gene promoters targeted by TgAP2XI-5 is that of ROP18, an important virulence factor of T. gondii. Intriguingly, the expression of exogenous copies of types I and II (25) rop18 alleles in the avirulent type III strain restored virulence to the parasite. Further analysis of lineage-specific sequence variation in the rop18 promoter revealed a 2.1-kb insert in the type III strain, with virtually undetectable transcription of this gene compared with type I and II strains (25). Among other effects, including a 6-fold reduction of activity in promoter assays (14), this 2.1-kb insertion results in the absence of any GCTAGC motifs in the type III promoter, compared with the presence of a single motif in type I and three motifs in type II. We can now report that the single type I GCTAGC motif represents a cis-regulatory element required for efficient rop18 transcription. Furthermore, we demonstrated that the recombinant protein encompassing the TgAP2XI-5 AP2 domain is able to bind to this GCTAGC motif in the rop18 promoter sequence environment, indicating that TgAP2XI-5 may act as a crucial activator of the rop18 promoter activity. Mutation of this motif did not result in complete ablation of gene transcription, indicating that other cis-regulatory elements are present in the rop18 promoter or that this important virulence gene exhibits a basal level of transcription. Nevertheless, with the cautious assumption that TgAP2XI-5 represents the only transcription factor targeting this GCTAGC motif, these data highlight the requirement of AP2 binding for efficient transcriptional activation.
Previous studies of T. gondii chromatin complexes have highlighted the potential role of TgAP2s in regulating gene expression. Proteomic analysis of the T. gondii corepressor complex (TgCRC), co-purified with the histone deactelyase, TgHDAC3, led to the identification of TgAP2VIII-4 (TgCRC-350) and speculation that the AP2 protein is required for transcriptional repression via TgHDAC3 activation (26). Given that TgAP2XI-5 promoter occupancy was observed to co-localize almost exclusively with enriched regions of acetylated histone H3K9, a marker of transcriptionally active promoters (4), it may well associate with histone modification enzymes.
Motif analyses of TgAP2XI-5-bound gene promoter sequences indicated that the GCTAGC motif was not always required for binding of the AP2 protein, although a secondary ACTAGC motif was also identified. The potential for TgAP2XI-5 to form heterodimers with transcription factors that bind different cis-regulatory elements could offer one explanation for how the AP2 protein binds to promoters lacking either a GCTAGC or ACTAGC motif. Furthermore, Jun-ATF2 heterodimers have been shown to display binding affinities to DNA sequences different from those bound by either of their parental homodimers (27).
CONCLUSIONS
Our results demonstrate that TgAP2XI-5 is a novel DNA sequence-specific transcription factor that occupies promoter regions that are mostly active at the S/M phase of the cell cycle. We showed that TgAP2XI-5 bound to a GCTAGC motif and that this interaction was important for full rop18 promoter activity. Furthermore, TgAP2XI-5 was enriched at numerous active promoters including those for crucial virulence factors such as the rhoptry and microneme proteins. In summary, our data indicate that TgAP2XI-5 regulates gene transcription of crucial virulence factors in T. gondii.
Acknowledgments
We thank T. Mouveaux and J. Duflot for excellent technical assistance and other members of the laboratory for fruitful discussions
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI087625 and RC4AI092801. (to K. K.). This work was also supported by the CNRS, INSERM, French National Research Agency Grant ANR-09-MIEN-002-01 (BradyToxoFT), and Grant ANR-11-LABX-0024 from the National Agency for Research to the Laboratoire d'Excellence ParaFrap.

This article contains supplemental Tables S1–S6.
- RMA
- robust multiarray average
- ROP
- rhoptry organelle protein.
REFERENCES
- 1. Radke J. R., Behnke M. S., Mackey A. J., Radke J. B., Roos D. S., White M. W. (2005) The transcriptome of Toxoplasma gondii. BMC Biol. 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Behnke M. S., Wootton J. C., Lehmann M. M., Radke J. B., Lucas O., Nawas J., Sibley L. D., White M. W. (2010) Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS ONE 5, e12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gissot M., Kim K., Schaap D., Ajioka J. W. (2009) New eukaryotic systematics. A phylogenetic perspective of developmental gene expression in the Apicomplexa. Int. J. Parasitol. 39, 145–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gissot M., Kelly K. A., Ajioka J. W., Greally J. M., Kim K. (2007) Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 3, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naguleswaran A., Elias E. V., McClintick J., Edenberg H. J., Sullivan W. J., Jr (2010) Toxoplasma gondii lysine acetyltransferase GCN5-A functions in the cellular response to alkaline stress and expression of cyst genes. PLoS Pathog. 6, e1001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Balaji S., Babu M. M., Iyer L. M., Aravind L. (2005) Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 33, 3994–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walker R., Gissot M., Croken M. M., Huot L., Hot D., Kim K., Tomavo S. (2013) The Toxoplasma nuclear factor TgAP2XI-4 controls bradyzoite gene expression and cyst formation. Mol. Microbiol. 87, 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Radke J. B., Lucas O., De Silva E. K., Ma Y., Sullivan W. J., Jr., Weiss L. M., Llinas M., White M. W. (2013) ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proc. Natl. Acad. Sci. U.S.A. 110, 6871–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell T. L., De Silva E. K., Olszewski K. L., Elemento O., Llinás M. (2010) Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 6, e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuda M., Iwanaga S., Shigenobu S., Mair G. R., Janse C. J., Waters A. P., Kato T., Kaneko I. (2009) Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 71, 1402–1414 [DOI] [PubMed] [Google Scholar]
- 11. Yuda M., Iwanaga S., Shigenobu S., Kato T., Kaneko I. (2010) Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol. 75, 854–863 [DOI] [PubMed] [Google Scholar]
- 12. Iwanaga S., Kaneko I., Kato T., Yuda M. (2012) Identification of an AP2-family protein that is critical for malaria liver stage development. PLoS One 7, e47557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huynh M.-H., Carruthers V. B. (2009) Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyle J. P., Saeij J. P., Harada S. Y., Ajioka J. W., Boothroyd J. C. (2008) Expression quantitative trait locus mapping of Toxoplasma genes reveals multiple mechanisms for strain-specific differences in gene expression. Eukaryot. Cell 7, 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gissot M., Walker R., Delhaye S., Huot L., Hot D., Tomavo S. (2012) Toxoplasma gondii chromodomain protein 1 binds to heterochromatin and colocalises with centromeres and telomeres at the nuclear periphery. PLoS One 7, e32671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995) MatInd and MatInspector. New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pino P., Sebastian S., Kim E. A., Bush E., Brochet M., Volkmann K., Kozlowski E., Llinás M., Billker O., Soldati-Favre D. (2012) A tetracycline-repressible transactivator system to study essential genes in malaria parasites. Cell Host Microbe 12, 824–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindner S. E., De Silva E. K., Keck J. L., Llinás M. (2010) Structural determinants of DNA binding by a P. falciparum ApiAP2 transcriptional regulator. J. Mol. Biol. 395, 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Silva E. K., Gehrke A. R., Olszewski K., León I., Chahal J. S., Bulyk M. L., Llinás M. (2008) Specific DNA-binding by apicomplexan AP2 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 105, 8393–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toedling J., Huber W. (2008) Analyzing ChIP-chip data using bioconductor. PLoS Comput. Biol. 4, e1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Levy D. E., Darnell J. E., Jr. (2002) Stats. Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 22. Treeck M., Sanders J. L., Elias J. E., Boothroyd J. C. (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe 10, 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Amoutzias G. D., Robertson D. L., Van de Peer Y., Oliver S. G. (2008) Choose your partners. Dimerization in eukaryotic transcription factors. Trends Biochem. Sci. 33, 220–229 [DOI] [PubMed] [Google Scholar]
- 24. LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. (2005) A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–107 [DOI] [PubMed] [Google Scholar]
- 25. Saeij J. P., Boyle J. P., Coller S., Taylor S., Sibley L. D., Brooke-Powell E. T., Ajioka J. W., Boothroyd J. C. (2006) Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science 314, 1780–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saksouk N., Bhatti M. M., Kieffer S., Smith A. T., Musset K., Garin J., Sullivan W. J., Jr., Cesbron-Delauw M. F., Hakimi M. A. (2005) Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25, 10301–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hai T., Curran T. (1991) Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. U.S.A. 88, 3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]