Background: The appropriate protein levels of ECF (extra-cytoplasmic function) σ factors are essential for bacteria functions.
Results: SigT degradation is dependent on ClpP protease and accelerated by secondary metabolites.
Conclusion: SigT degradation is regulated in a dual positive feedback manner.
Significance: This novel mechanism expands our understanding of ingenious cooperation of intracellular molecules for proper physiological functions of bacteria.
Keywords: Actinobacteria, Antibiotics, Protein Degradation, Secondary Metabolism, Transcription Regulation
Abstract
Here we report that in Streptomyces coelicolor, the protein stability of an ECF σ factor SigT, which is involved in the negative regulation of cell differentiation, was completely dependent on its cognate anti-σ factor RstA. The degradation of RstA caused a ClpP/SsrA-dependent degradation of SigT during cell differentiation. This was consistent with the delayed morphological development or secondary metabolism in the ΔclpP background after rstA deletion or sigT overexpression. Meanwhile, SigT negatively regulated clpP/ssrA expression by directly binding to the clpP promoter (clpPp). The SigT-clpPp interaction could be disrupted by secondary metabolites, giving rise to the stabilized SigT protein and retarded morphological development in a non-antibiotic-producing mutant. Thus a novel regulatory mechanism was revealed that the protein degradation of the ECF σ factor was initiated by the degradation of its anti-σ factor, and was accelerated in a dual positive feedback manner, through regulation by secondary metabolites, to promote rapid and irreversible development of the secondary metabolism. This ingenious cooperation of intracellular components can ensure economical and exquisite control of the ECF σ factor protein level for the proper cell differentiation in Streptomyces.
Introduction
Throughout evolutionary history, organisms have adopted various regulatory measures for rapid accommodation to diverse environments. Extra-cytoplasmic function (ECF)2 σ factors, the largest group of σ70 family in bacteria, have been extensively studied. They are seen to alternatively recruit RNA polymerase core enzyme to modulate the gene expression of their regulons to facilitate a rapid response and adaptation to environmental stresses (1–3).
Meanwhile, to meet the physiological requirements of bacteria, the intracellular protein levels and transcriptional regulatory activities of ECF σ factors are also elaborately modulated at multiple levels. These include transcription, translation, antagonism from cognate anti-σ factors, and protein degradation. The gene expression of most ECF σ factor genes is rapidly induced by extracellular environmental signals and is frequently autoregulated to confer a prompt resistance to damages (4–8). BldN, an ECF σ factor from Streptomyces coelicolor, can be initially translated as a pro-protein, and becomes matured during cell differentiation (9). Anti-σ factors traditionally function as antagonists to inhibit the transcriptional activities of cognate ECF σ factors, and ECF σ factors are activated after conformation changes (10) or sequential proteolysis of anti-σ factors by intra-membrane and intracellular proteases (11–16). The intracellular ECF σ factors are also maintained at an appropriate level by protein degradation. In bacteria, protein degradation is executed mainly by the ATP-dependent proteases ClpP, Lon, HslUV, and FstH (17, 18). Under conditions of oxidative stress, the degradation of SigR′, an isoform of the ECF σ factor SigR in S. coelicolor, is known to be partially dependent on the ClpP protease (19).
We previously described that loss of the ECF σ factor SigT or its anti-σ factor RstA in S. coelicolor resulted in retarded production of the blue pigment actinorhodin (Act) on nitrogen-limited media. We also found that RstA had a positive role on autoregulation of sigT expression (20). Meanwhile, although the locations of sigT and rstA are close to each other (21), they are not co-transcribed (20). This is another distinctive characteristic of SigT from other classical ECF σ factors. Moreover, on rich media, both ΔsigT and ΔrstA mutants show accelerated morphological development and secondary metabolism. This unusual phenomena triggered us to discover that the SigT protein had disappeared in the ΔrstA mutant (21). This had not previously been reported and suggested a potentially unique regulatory mechanism of RstA on SigT, beyond the putative traditional antagonistic effects.
Here we present further evidence that RstA is required for protein stability of SigT, and that degradation of RstA causes a rapid ClpP/SsrA pathway-dependent degradation of SigT in a dual positive feedback manner. We also suggest that secondary metabolites could act as regulators on SigT degradation for the timely and irreversible progress of cell differentiation. This novel mechanism enriches our understanding of how cellular components work cooperatively to keep the appropriate protein levels of transcriptional regulatory factors for proper cell development and environmental adaptation.
EXPERIMENTAL PROCEDURES
Media
Escherichia coli strains were routinely cultured in a LB medium. Liquid Tryptic Soy Broth (TSB) plus 5% PEG6000 was used for mycelium preparation in the primary metabolism. Solid R2YE and liquid R5 media were used for cell differentiation of S. coelicolor, and MS medium for spore preparation (22, 23).
Plasmid Construction
The plasmids and primers used in this study are listed in Tables 1 and 2, respectively. E. coli TG1 was used for routine plasmid construction. Plasmid pLM29 (21) was digested with BamHI/HindIII, and the purified sigT fragment was inserted into the BamHI/HindIII site of pET28a to create pL85. The 3flag-sigT fragment was amplified with primers 1 and 2, ligated to pTA2 after dA addition, and digested with NdeI/NotI. After purification, the 3flag-sigT fragment was inserted into the NdeI/NotI site of pLM26 (21) to generate plasmid pL86. Primers 3, 4, and primers 5, 6 were used to amplify clpPp and sigTp, respectively, and each was ligated into pTA2 for plasmids pL87 and pL88, respectively. About 200 bp of the clpP1 coding region and 300 bp of the sigT coding region were obtained with primers 7, 8 and primers 9, 10, respectively, and each was inserted into pTA2 for plasmids pL89 and pL90, respectively. sigTp was digested with BamHI from pL88 and replaced ermEp* in pLM26 (21) to produce plasmid pL149. The sigT-linker was digested with NdeI from pLM34 (21) and fused to egfp in pL149 to create plasmid pL150. The streptomycin-resistant gene aadA was amplified from pIJ779 with primers 21 and 22, and ligated to the pTA2 vector to generate plasmid pL146. Then the EcoRI fragment aadA was ligated to EcoRI site of pIJ8660 (24) to create pL147. The rstA-3flag fragment was digested from pLM36 (21) with NdeI/NotI, and inserted into the NdeI/NotI site of pL147 for plasmid pL148.
TABLE 1.
Plasmids and cosmids
| Plasmid or cosmid | Description | References |
|---|---|---|
| pLM29 | sigT in pPROEX-HTb | 21 |
| pET28a | Expression vector in E. coli | Novagen |
| pL85 | BamHI/HindIII sigT fragment from pLM29 into pET28a | This study |
| pET32a | Expression vector in E. coli | Novagen |
| pLM26 | Kanamycin resistant gene and ermEp* in pIJ8630 | 21 |
| pL86 | NdeI/NotI 3flag-sigT into pLM26 | This study |
| pLM36 | NdeI/NotI rstA-3flag into pLM26 | 21 |
| pTA2 | T vector, self-ligation | Toyobo, Japan |
| pL87 | clpP1/P2 promoter (clpPp) in pTA2 | This study |
| pL88 | sigT promoter (sigTp) in pTA2 | This study |
| pL89 | 200 bp of clpP1 in pTA2 | This study |
| pL90 | 300 bp of sigT in pTA2 | This study |
| Cosmid StC49 | Cosmid containing clpP1/P2 | John Innes Centre |
| pL92 | ΔclpP1/P2::oriT-aac (3)IV in cosmid StC49 | This study |
| Cosmid N6–68 | Cosmid containing ssrA | This study |
| pL94 | ΔssrA::aadA in cosmid N6–68 | This study |
| pL95 | ΔssrA::FRT in cosmid N6–68 | This study |
| pIJ779 | Plasmid with FRT-aadA-FRT for disruption cassette amplification | 25 |
| pIJ773 | Plasmid with FRT-oriT-aac (3)IV-FRT for disruption cassette amplification | 25 |
| pIJ8660 | Promoter-probing plasmid | 24 |
| pL146 | aadA in pTA2 | This study |
| pL147 | aadA into EcoRI site of pIJ8660 | This study |
| pL148 | rstA-3flag in NdeI/NotI site of pL147 | This study |
| pL149 | sigTp replaces ermEp* in pLM26 | This study |
| pL150 | sigT-linker fused to egfp in pL149 | This study |
TABLE 2.
Oligonucleotides used in this study
| Primer no. | Sequence |
|---|---|
| 1 | CATATGGACTACAAGGACCACGACGGCGATTACAAGGACCACGACATCGACTACAAGGACGACGACGACAAGGCGGGCGGCGCGAGTCATGAC |
| 2 | TCACGCTCGTCCACCTCCGCCCTTC |
| 3 | GGATCCCCTCGCGCCGTCCCCGAC |
| 4 | GGATCCCCGTCTCACGTTTCCCTGCG |
| 5 | GGATCCGCACAACCATCGTCCGCCGCTG |
| 6 | GGATCCCGTCGTTCCTGCCTCCCCATTC |
| 7 | CGTCGTAAGACGAGCAGGTGG |
| 8 | CGGGTCGGAGGCAAGGAGCAG |
| 9 | CGCTGGGGGACCGTGAGGAG |
| 10 | TCGCTGGTCGGCGGGCAAGG |
| 11 | GAGTCCCGGAAAGCGGCTCCAGCCCCGCAGGGAAACGTGATTCCGGGGATCCGTCGACC |
| 12 | CCGTGCCAAGGAGCGGCAGACGATACAGCCTCTTGCCTATGTAGGCTGGAGCTGCTTC |
| 13 | GCGAGCCCCTCTTCGGAGGACTTGAAAAATCAACATGGGATTCCGGGGATCCGTCGACC |
| 14 | TCGCGCTTACGCGAGCACACGCCGCCCTCGACCTCGTGGTGTAGGCTGGAGCTGCTTC |
| 15 | TACAAGCTTATCAAGCGCAAGCAGCGGGC |
| 16 | ATCGAATTCGGGGGTGCGGGTGGCAGAGC |
| 17 | GCCAGGGTTTTCCCAGTCACGA |
| 18 | Biotin-GCCAGGGTTTTCCCAGTCACGA |
| 19 | GAGCGGATAACAATTTCACACAGG |
| 20 | 6FAM-GTTGTAAAACGACGGC |
| 21 | CTGCAGTTCGAAGTTCCTATTC |
| 22 | GCTTGATGTGTAGGCTGGAG |
Construction of S. coelicolor Strains
S. coelicolor strains used in this study are listed in Table 3. The clpP1/P2 and ssrA genes were disrupted by the PCR-targeting strategy (25). clpP1/P2::FRT-oriT-aac(3)IV-FRT disruption cassette was amplified with primers 11, 12 from EcoRI/HindIII fragments of pIJ773, and electrotransformed into E. coli BW25113/pIJ790 containing cosmid StC49 to generate disruption cosmid pL92. The ssrA::FRT-aadA-FRT disruption cassette was amplified with primers 13, 14 from EcoRI/HindIII fragments of pIJ779, and electrotransformed into E. coli BW25113/pIJ790 containing cosmid N6–68 to generate disruption cosmid pL94. The in-frame deletion cosmid pL95 was created by passage of cosmid pL94 through E. coli BT340 at 42 °C. Disruption cosmids pL92 and pL94 were conjugated through E. coli ET12567/pUZ8002 into LM22 (ΔrstA) to knock out clpP1/P2 and ssrA, respectively, to give rise to strains L43 and L44, respectively. Cosmid pL95 was introduced into M145 for strain L46 by in-frame deletion of ssrA (25).
TABLE 3.
S. coelicolor strains used in this study
Protein Techniques
E. coli BL21(DE3) containing pL85 was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C to express the soluble His-SigT protein. The protein was purified with Ni2+-NTA according to the manufacturer's instructions (Qiagen). S. coelicolor mycelia were collected after inoculation of spores in liquid medium or on cellophane for the time indicated, and lysed for protein preparation as previously described (21). About 20 μg of total protein was subject to Coomassie Blue staining or Western blot with mouse α-FLAG antibody (M2, Sigma) or rabbit α-GFP antibody (Proteintech Group). Purified E. coli RNA polymerase core enzyme (Epicenter), purified SigT, and E. coli cell lysate with expressed SigT were separated on a 6% SDS-PAGE for Western blot with antibodies against E. coli RNA polymerase subunit β and β′ (Santa Cruz Biotechnology), or on 12% SDS-PAGE for Coomassie Blue staining.
RNA Analysis
Total RNA was extracted from mycelia cultured in TSB or on cellophane overlaid on R2YE medium and treated with RNase-free DNase I as described (21). About 10 μg of RNA was subject to formaldehyde gel-based Northern blot analysis (26). The RNA on the nylon membrane was hybridized with an internal biotin-labeled ssrA DNA probe, and detected with streptavidin-HRP and BeyoECL plus, as described by the manufacturer (Beyotime, China). Ribonulease protection assays were demonstrated to analyze sigT and clpP1 mRNA in a low abundance. Briefly, about 10 μg of total RNA was co-precipitated with 0.1 ng of biotin-labeled antisense RNA probe. Protected RNA fragments were prepared as described by the manufacturer (Ambion), and separated on a 7 m urea, 5% polyacrylamide gel after heat-denaturing for clpP1, or subject to Northern blot for sigT. After electrophoresis, the RNA fragments were blotted to a positive charged nylon membrane, UV-fixed, and then detected with streptavidin-HRP and North2South Chemiluminescent Substrate (Thermo Scientific).
Electrophoretic Mobility Shift Assay (EMSA)
About 1 ng of the probe was incubated with 0, 0.4, 1.0, 2.0, and 4.0 μm purified His-SigT, respectively, at 25 °C in buffer (10 mm Tris, 100 mm Na2HPO4, pH 8.0, 50 μg/ml of sheared sperm DNA) for 30 min and loaded on a 5% native polyacrylamide gel for separation in 0.5× TBE running buffer. The probe was then electroblotted to the nylon membrane, UV-fixed, and detected with streptavidin-HRP and BeyoECL plus (Beyotime, China). For inhibitory assays by secondary metabolites, the crude extract of secondary metabolites, HPLC-purified γ-actinorhodin or undecylprodigiosin with a decreased concentration as indicated, were incubated with His-SigT at 25 °C for 30 min, and 1 ng of 5′-labeled clpPp probe was then added for a further 30-min incubation. For the E. coli RNAP core enzyme binding assay, an increased amount of purified SigT protein was incubated without or with 0.4 μg of core enzyme for about 30 min at room temperature, and then 1 ng of biotin-labeled clpPp probe was added for a further 30-min incubation.
Preparation of Secondary Metabolites
Preparation of crude extract of undecylprodiginine (Red), Act, and their intermediates has been described previously (27, 28). Briefly, Red and its intermediates were extracted with acidified methanol (0.5 m HCl) from wild type mycelia cultured in a R5-medium for 1 day. The supernatant was collected by a brief centrifugation, filtered, vacuum evaporated, and solubilized in methanol as 100 μg/μl. For the preparation of extracellular Act and its intermediates, the 6-day wild type culture supernatant in the R5-medium was acidified to pH 3.0, extracted with 2 volumes of ethyl acetate, and vacuum evaporated. The 6-day wild type cells in the R5-medium were lysed with ultrasonication, and the supernatant was extracted with 2 volumes of ethyl acetate and vacuum evaporated for preparation of intracellular Act and its intermediates. The residues were resuspended in DMSO as 100 μg/μl.
The extracellular γ-actinorhodin was purified by HPLC (29) and resuspended in DMSO as 10 μm. For purification of undecylprodigiosin, the total prodiginine was prepared from the mycelia of a S. coelicolor Δact mutant M1141 cultured in R5-medium for 6 days as described above. The concentrated extract was resolved by HPLC in a C18 column with solution A (CH3OH:CH3CN:H2O = 40%:10%:50%) and solution B (CH3OH in 0.1% trifluoroacetic acid) and with detection at 530 nm. Undecylprodigiosin was eluted in a linear A:B gradient from 100:0% to 20:80% in 10 min, followed by 20:80% to 0:100% in 20 min, and collected at about 26.5 min. The purity and molecular weight were determined by HPLC and mass spectrometry.
DNase I Footprinting Assay
The FAM-labeled non-radioactive DNase I footprinting assay was carried out as previously described with minor modifications (30–32). Briefly, the purified SigT protein was ultrafiltered with YM-10 (Millipore) for a 10-kDa cut-off and eluted in the 20 mm Tris buffer, pH 7.5. A 5′-FAM-labeled clpPp probe was amplified with universal primers 19 and 20 from plasmid pL87 and then gel purified. About 50 ng of clpPp probe was incubated with SigT protein at room temperature for 30 min, and 0.03 units of DNase I (Promega) was added with 10 mm MgCl2 and 1 mm CaCl2. After exact incubation at room temperature for 1 min, reactions were stopped with an equal volume of 100 mm EDTA, pH 8.0, immediately followed by phenol/chloroform extraction and precipitation with 0.75 m NH4Ac, 40 μg of glycogen and ethanol. DNA mixed with Liz-500 DNA marker (MCLAB) was loaded on ABI 3130 DNA sequencer and electropherograms were analyzed with Genemapper version 4.0 software (Applied Biosystems) to align and determine the protected region. A DNA sequencing ladder was prepared with the 5′-FAM-labeled primer 20 according to Thermo Sequenase Dye Primer Manual Cycle Sequencing Kit (U.S. Biochemical Corp.).
Non-radioactive Labeling of DNA and RNA Probes
The ssrA DNA probe for Northern blot was internally labeled by PCR. Primers 15 and 16 were used to amplify ssrA from the M145 genome with Taq polymerase. Then the gel-purified ssrA fragment was used to amplify the biotin-labeled probe with biotin-11-dUTP (Fermentas) with universal primers 17 and 19.
clpP1 and sigT antisense RNA were internally labeled with biotin by in vitro transcription. The template for in vitro transcription was amplified with KOD plus from plasmids pL89 and pL90, respectively, with universal primers 17 and 19, then gel purified and eluted in diethyl pyrocarbonate-treated water. The in vitro transcription was carried out with T3 RNA polymerase (Fermentas) and the Biotin RNA Labeling Mix (Roche) at 37 °C for at least 3 h. The DNA template was removed by digestion with RNase-free DNase I (Takara) and the RNA probe was purified with the RNeasy Mini Kit (Qiagen) and eluted in diethyl pyrocarbonate-treated water.
For preparation of EMSA probes, the 5′-biotin-labeled universal primers 18 and 19 were used to amplify the 5′-biotin clpPp and sigTp probes from plasmid pL87 and pL88, respectively. The unlabeled universal primers 17 and 19 were used to amplify the unlabeled clpPp probe for competition assays.
RESULTS
ClpP/SsrA Is Required for Degradation of SigT without RstA
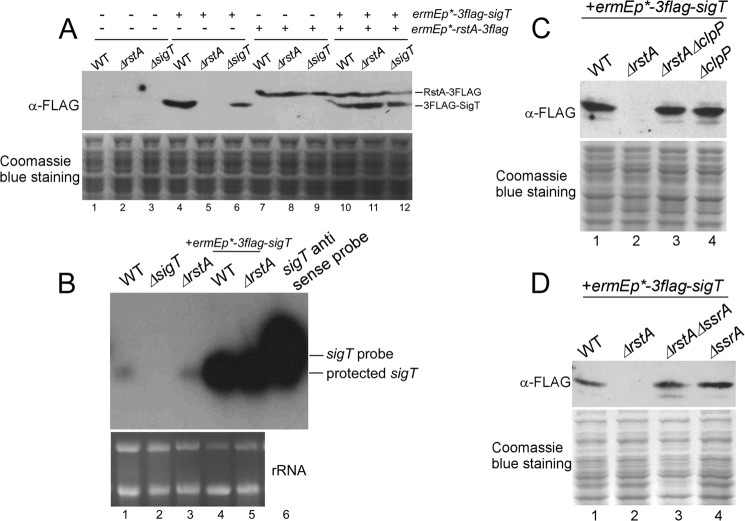
Traditionally, the antagonistic effects of anti-σ factors often result in opposite phenotypes between σ factor and anti-σ factor mutants (7, 33, 34). We have previously reported an exception that deletion of the ECF σ factor SigT or its cognate anti-σ factor RstA in S. coelicolor causes accelerated morphological development and increased antibiotic production. This prompted us to discover the disappearance of SigT protein in the ΔrstA mutant at the post-transcription level (21). Here we further demonstrate that complementation of rstA in the ΔrstA mutant could restore the SigT protein (Fig. 1A, lane 11). In contrast, deletion of sigT did not affect the RstA protein (Fig. 1A, lane 9). These results further suggest the requirement of RstA for the presence of SigT. We then checked whether the absence of the SigT protein in the ΔrstA mutant resulted from a repressed expression of sigT or a decay of sigT mRNA. It was observed that sigT mRNA in the ΔrstA mutant was stable and present in comparable levels to those of the wild type, both under sigT operon promoter (Fig. 1B, lanes 1 and 3) and under the strong constitutive promoter ermEp* (Fig. 1B, lanes 4 and 5). sigT mRNA expressed from ermEp* was about 30 times higher than that from sigT native promoter. This suggested that absence of the SigT protein in the ΔrstA mutant could not have resulted from the possible disappearance of sigT mRNA.
FIGURE 1.
ClpP/SsrA is involved in degradation of SigT without RstA. A, complementation of RstA for SigT protein stability. Total protein was prepared from strains M145 (wild type, WT), LM22 (ΔrstA), LM21 (ΔsigT) containing either plasmid pL86 (3flag-sigT) or plasmid pL148 (rstA-3flag) or both, in TSB liquid medium for 2 days. SigT and RstA proteins were analyzed by Western blot with α-FLAG antibody. Coomassie Blue staining of total protein served as a loading control. B, sigT expression profile in wild type M145 (WT), LM21 (ΔsigT), LM22 (ΔrstA), M145/pL86 (WT + 3flag-sigT), and LM22/pL86 (ΔrstA + 3flag-sigT) as demonstrated by a ribonuclease protection assay. rRNA was shown as a loading control in a formaldehyde gel. C and D, ClpP/SsrA is required for SigT degradation without RstA. Total protein was prepared from strains M145/pL86 (wild type + 3flag-sigT), LM22/pL86 (ΔrstA + 3flag-sigT), L43/pL86 (ΔrstAΔclpP + 3flag-sigT), and L45/pL86 (ΔclpP + 3flag-sigT) (C), strains M145/pL86 (wild type + 3flag-sigT), LM22/pL86 (ΔrstA + 3flag-sigT), L44/pL86 (ΔrstAΔssrA + 3flag-sigT), and L46/pL86 (ΔssrA + 3flag-sigT) (D) in TSB liquid medium for 2 days and analyzed as in A.
Given the direct protein interaction between SigT and RstA (21), our results suggest that RstA is probably required for SigT protein stability. In bacteria, ClpP/SsrA is one of the major ATP-dependent protein degradation systems that includes ClpP1/P2 (ClpP) protease encoded by clpP1/P2 (clpP) and the tmRNA-mediated recognition tag encoded by ssrA (35). After further deletion of clpP1/P2 in a ΔrstA mutant (ΔrstAΔclpP) or ssrA in a ΔrstA mutant (ΔrstAΔssrA), reappearance of the SigT protein was observed, with protein levels that were comparable with those in the wild type, ΔclpP1/P2 mutant (ΔclpP) and ΔssrA mutant, respectively (Fig. 1, C and D). Thus, our results strongly suggest that RstA can stabilize and protect SigT from degradation, and that ClpP/SsrA is required for SigT degradation after rstA deletion.
SigT Degradation Is Dependent on ClpP Protease during Cell Differentiation
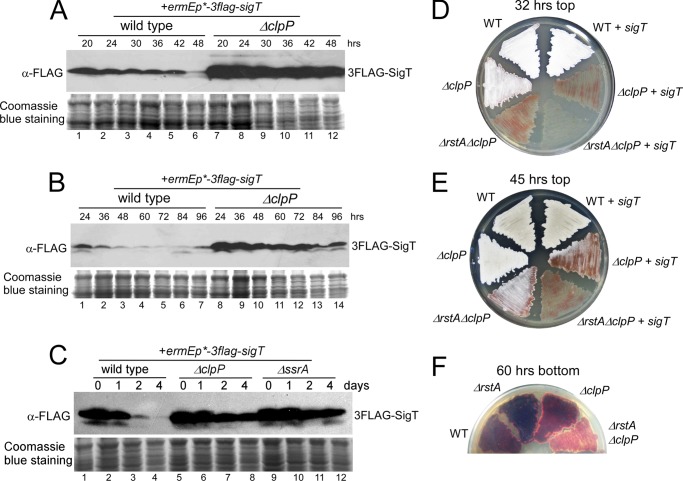
We then checked the physiological conditions under which SigT was degraded. On a solid R2YE medium, the SigT protein in the wild type was degraded gradually when cells were developing from substrate mycelia to aerial mycelia (Fig. 2A, lanes 1-5), and the protein level dropped sharply after sporulation (Fig. 2A, lane 6). The constitutively low level of SigT protein in the prolonged spore phase (48 h later) was repeatedly observed (Fig. 2B, lanes 1-7). However, the SigT protein was much more abundant in the ΔclpP mutant under all developmental phases than that in the wild type, especially after sporulation (Fig. 2, A and B). It should be noted that the prominent morphological development and secondary metabolism of S. coelicolor concurred on the solid R2YE medium. SigT protein levels were then checked in the liquid R5-medium, where S. coelicolor cells grow only as mycelia but produce abundant red and blue secondary metabolites, undecylprodiginine (Red) and actinorhodin (Act), respectively (22). When the cells cultured in the TSB medium at the logarithmic phase were transferred to R5-medium, SigT protein levels in the wild type decreased slightly in the first day (Fig. 2C, lanes 1 and 2). This corresponded to the time when cells mainly produce Red (22). Beyond this, however, the protein level fell dramatically after 2 days, and was almost undetectable 4 days later (Fig. 2C, lanes 3 and 4). This corresponded to the time when Act is also produced (22). These phenomena were consistent with the observations on the R2YE medium, where cells mainly produce Red in the aerial phase (36 h) and then both antibiotics in the spore phase (48 h and later). In contrast, almost no degradation of the SigT protein was observed in the ΔclpP mutant even after 4 days in R5-medium (Fig. 2C, lanes 5-8). This suggests, on a genetic level, that ClpP protease is required for developmental phase-dependent SigT degradation.
FIGURE 2.
SigT is degraded dependent on ClpP/SsrA during cell differentiation. A–C, ClpP protease is required for SigT degradation during cell differentiation. SigT protein dynamics of strains M145/pL86 (wild type + 3flag-sigT) and L45/pL86 (ΔclpP + 3flag-sigT) on R2YE medium covered with cellophane (A and B) or in a liquid R5-medium (C) for the times indicated as demonstrated by Western blot. Coomassie Blue staining of total protein served as a loading control. Spores of strains M145 (wild type, WT), M145/pL86 (WT + sigT) L45 (ΔclpP), L43 (ΔrstAΔclpP), L45/pL86 (ΔclpP + sigT), and L43/pL86 (ΔrstAΔclpP + sigT) were streaked on R2YE medium for 32 (D) or 45 h (E), respectively, and photographed. F, spores of strains M145 (wild type, WT), LM22 (ΔrstA), L45 (ΔclpP), and L43 (ΔrstAΔclpP) were streaked on R2YE medium for 60 h and photographed.
In the wild type, loss of the ECF σ factor SigT or anti-σ factor RstA results in accelerated morphological development and secondary metabolism (21). This could be at least partially explained by the observations that SigT was unstable in the ΔrstA mutant, whereas RstA remained stable in the ΔsigT mutant, as shown above. Meanwhile, overexpression of sigT in the wild type did not have a significant impact on morphological development (Fig. 2, D and E). This is possibly because SigT would be degraded in the wild type during cell differentiation. However, when the SigT protein was stabilized in the ΔclpP mutant (Fig. 2, A–C), further deletion of rstA caused a delay in aerial hyphae formation (compare ΔclpP and ΔrstAΔclpP in Fig. 2, D and E). Meanwhile, overexpression of sigT in the ΔclpP or ΔclpPΔrstA mutant also led to a retarded aerial hyphae development, as compared with their parental strains, respectively (Fig. 2, D and E). S. coelicolor can produce a red antibiotic undecylprodiginine (Red) and a blue antibiotic actinorhodin (Act) and mixture of both makes the lawn appear purple. The wild type and ΔclpP mutant was more purple than the ΔclpPΔrstA mutant on a R2YE medium, and the ΔclpPΔrstA mutant appeared much redder (Fig. 2F). This suggests Act production was dramatically lessened after rstA deletion in the ΔclpP mutant, a situation that contrasted to that of the ΔrstA mutant in the wild type background (Fig. 2F) (21). This suggests a positive role of RstA on antibiotic production if SigT is stabilized. All of these phenotypes were consistent with the hypothesis that SigT negatively regulates cell differentiation and RstA was an anti-σ factor not only protecting SigT from degradation but also antagonizing SigT activity when the SigT protein was stabilized.
SigT-independent Degradation of RstA during Cell Differentiation
Next we examined how RstA regulated SigT stability. It should be reasonable that RstA conformation alteration or protein degradation should result in dissociation of the SigT-RstA complex to mimic rstA deletion and leave SigT liable to degradation. RstA has an HXXXCXXC (where X is any amino acid) motif for possible disulfide formation (21), as does another anti-σ factor RsrA, which can release the ECF σ factor SigR under oxidation after conformation changes in S. coelicolor (10). However, we did not observe RstA mobility shift or SigT degradation under oxidative stresses from diamide or H2O2 (data not shown). It was then speculated that RstA should also be degraded during cell differentiation. Indeed, we found RstA degradation in wild type cells both on the solid R2YE medium (Fig. 3A, lanes 1-6) and in the liquid R5-medium (Fig. 3B, lanes 1-5) as observed for the SigT protein. Furthermore, we observed that deletion of sigT did not significantly affect RstA protein kinetics either on the R2YE medium (Fig. 3A, lanes 7-12) or in liquid R5-medium (Fig. 3B, lanes 6-10). This suggests that SigT is not essential for RstA stability. These data also strongly suggest that RstA has protective effects on SigT, and that RstA must be degraded before SigT degradation to leave SigT isolated and susceptible to ClpP/SsrA pathway-dependent degradation. Furthermore, we found RstA was not stabilized in the ΔclpP mutant either on R2YE or in R5-liquid medium (Fig. 3, C and D), suggesting that the RstA and SigT degradations are dependent upon different pathways, and that stabilized SigT in the ΔclpP mutant during cell differentiation does not result from a stabilized RstA, also supporting ClpP-dependent SigT degradation.
FIGURE 3.
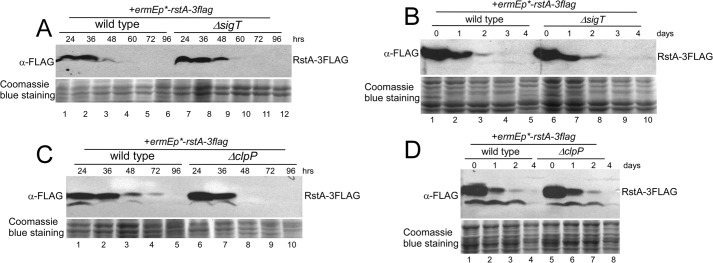
A and B, RstA degradation independent of SigT during cell differentiation. RstA protein dynamics in strains M145/pLM36 (wild type + rstA-3flag) and LM21/pLM36 (ΔsigT + rstA-3flag) on R2YE medium (A) or in liquid R5-medium (B) for the times indicated as demonstrated by Western blot with α-FLAG antibody. C and D, RstA degradation independent of ClpP protease. RstA protein dynamics in strains M145/pL36 (wild type + rstA-3flag) and L45/pL36 (ΔclpP + rstA-3flag) on R2YE medium (C) or in liquid R5-medium (D) for the times indicated as demonstrated by Western blot. Coomassie Blue staining of total protein served as a loading control.
Endogenous Kinetics of the SigT Protein
We have shown that SigT expressed from a strong constitutive promoter ermEp* is protected by RstA and its degradation is dependent on ClpP. Next we checked the endogenous SigT protein level during cell differentiation by monitoring the SigT-GFP fusion protein expressed under the sigT promoter (sigTp). It was observed that the SigT-GFP protein was rapidly degraded after the cells developed into its spore phase in wild type, and the ΔclpP mutant once again showed more abundant SigT-GFP fusion protein at all developmental phases (Fig. 4A), although a slightly higher SigT-GFP protein level was observed at the aerial hyphae phase (Fig. 4A, lanes 2 and 7). With the GFP protein as a reporter for the sigTp activity assay during cell differentiation, both wild type and the ΔclpP mutant exhibited a similar kinetics of the GFP protein, which gradually increased and then decreased when cells were developing into the aerial mycelium and spore phases, respectively (Fig. 4B). This expression pattern excluded the possibility that the SigT protein difference in wild type and in the ΔclpP mutant might result from different sigTp activities. We suspected that increased sigTp activity, occurring before the aerial hyphae, could buffer the fast degradation of the endogenous SigT protein, although decreased sigTp activity in the spore phase might speed up the disappearance of the SigT protein, thus leading to an appropriate intracellular SigT protein level and timely cell differentiation. Therefore, our results suggest that developmental phase-dependent SigT degradation requires ClpP protease during cell differentiation.
FIGURE 4.
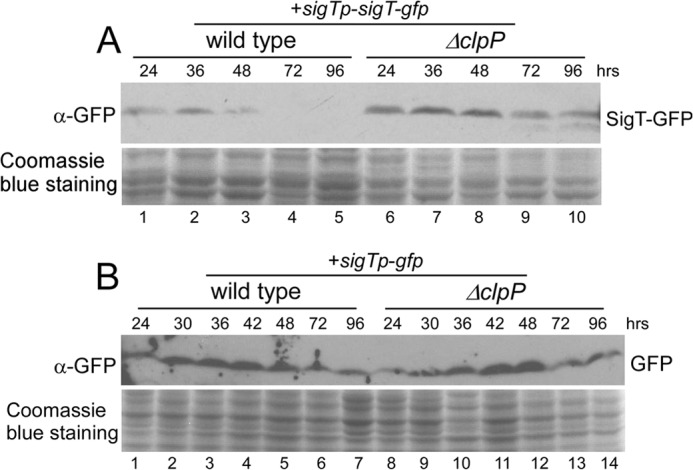
Endogenous SigT protein dynamics. Protein dynamics of SigT-GFP fusion protein (A) and GFP expressed under sigTp (B) in strains M145/pL97 (wild type + sigTp-sigT-gfp) and L45/pL97 (ΔclpP + sigTp-sigT-gfp) (A), in strains M145/pL96 (wild type + sigTp-gfp) and L45/pL96 (ΔclpP + sigTp-gfp) (B). Cells were collected on R2YE medium overlaid with cellophane after the indicated times and lysed for Western blot with α-GFP antibody. Coomassie Blue staining of total protein was used as a loading control.
Negative Regulation of clpP and ssrA Expression by SigT and RstA
clpP1, clpP2 are co-transcribed in one operon, and ClpP1 and ClpP2 are the subunits of the ClpP protease (19). Our gene expression analysis showed that clpP1 expression was undetectable before 36 h, but increased after 48 h, when cells developed into spores (Fig. 5A). Wild type cells showed similar expression levels of ssrA at 24- and 36-h time points, when cells were in substrate mycelia and aerial mycelia, respectively, but 2 times higher expression was observed after sporulation, or 48 h later (Fig. 5B). This was consistent with the sharp drop of the SigT protein level after sporulation (Fig. 2, A and B). Deletion of sigT or rstA could enhance the expression of clpP1 at all developmental phases and a more dramatic expression augment was observed in the ΔrstA mutant (Fig. 5A), which might also be responsible for degradation of the SigT protein in the ΔrstA mutant. Meanwhile, ΔsigT and ΔrstA mutants showed about 5 times higher ssrA expression than wild type at 24 and 36 h, and about 3 times higher at 48 h, but no significant difference after 72 h (Fig. 5B). Furthermore, the clpP1 expression profile was verified by a GFP reporter assay, in which GFP was expressed under the control of the clpP promoter (clpPp). It was found that the GFP protein level gradually increased with cell differentiation in the wild type on a R2YE medium, and we also observed more GFP protein in both the ΔsigT mutant and in the ΔrstA mutant at all developmental phases (data not shown). Thus, our results indicate that both SigT and RstA negatively regulated the expression of clpP1/P2 and ssrA.
FIGURE 5.
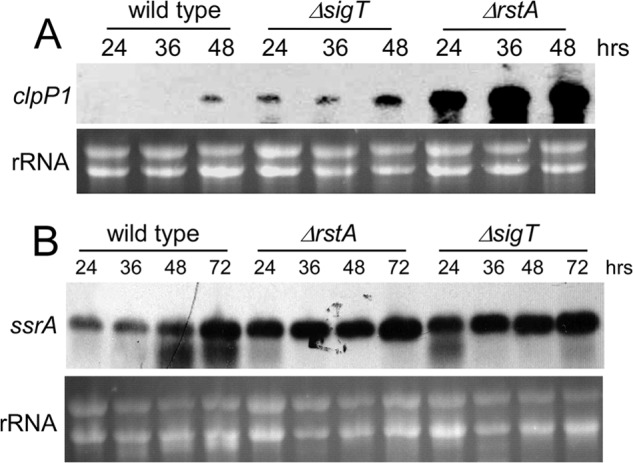
A and B, SigT negatively regulates clpP1/P2 and ssrA expression. Ribonuclease protection assay of clpP1 expression (A) and Northern blot analysis of ssrA expression (B) in M145 (wild type), LM21 (ΔsigT) and LM22 (ΔrstA) grown on R2YE medium overlaid with cellophane at various time points. About 10 μg of total RNA run in a formaldehyde gel was for a loading control.
Interaction between SigT and the clpP Promoter
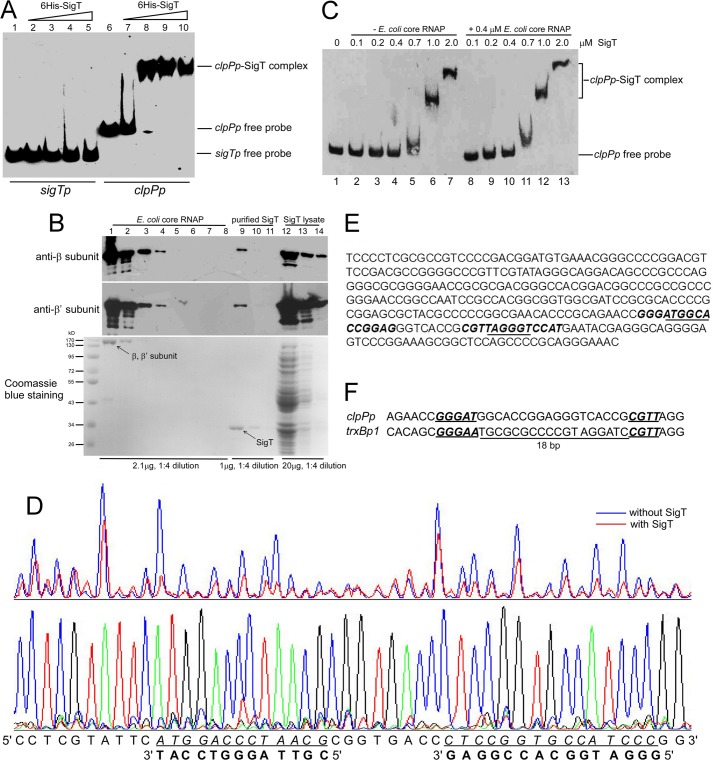
We then investigated whether SigT could directly regulate the expression of clpP1/P2 and/or ssrA. The SigT protein was expressed and purified from E. coli. EMSA showed that SigT could bind to the clpP1/P2 promoter (clpPp) (Fig. 6A, lanes 6-10) but not to the sigT promoter (sigTp) (Fig. 6A, lanes 1-5). Another σ factor, SigN (36) from S. coelicolor, was also purified from E. coli, but no binding activity of SigN to clpPp was observed (data not shown). Furthermore, the addition of an excessive unlabeled clpPp probe could restore the shifted band to the free probe position (data not shown). These results suggest a specific interaction between SigT and clpPp. Traditionally, the promoter recognition and DNA-binding activity of σ factors is exerted after formation of holoenzyme together with the core enzyme of RNA polymerase (RNAP) (37). We then checked whether SigT bound to clpPp with the E. coli RNAP core enzyme. About 2.1 μg of purified E. coli core enzyme was co-separated with about 1 μg of purified SigT and 20 μg of E. coli lysate with expressed SigT. Subunit β and β′ co-migrated almost to the same position in a 12% polyacrylamide gel, and showed about 2 times the signal strength of purified SigT by Coomassie Blue staining. This was consistent with loading of about 1 μg of β, β′ subunit and SigT, respectively (Fig. 6B, lanes 1 and 9). Immuno-blots with anti-β and β′ subunits showed that a weak signal (about 0.02 μg) was also observed in the purified SigT (Fig. 6B, lane 9), which suggested SigT was present to a level of about a 200-fold molar excess of RNAP. Meanwhile, about 2 μg of SigT and 1 μg of β, β′ subunit were observed in total lysate (Fig. 6B, lanes 1, 9, and 12). This suggested that most of the RNAP was not co-purified with SigT, and that RNAP might not be involved in formation of the SigT-core enzyme complex for SigT-clpPp binding. Moreover, supplementation of another additional RNAP core enzyme did not significantly increase the binding affinity of SigT to clpPp (Fig. 6C), further supporting the idea that SigT can solely bind to clpPp. The binding of σ factors to promoters without core enzymes has also been in evidence in studies of several other σ factors, such as CorE in Myxococcus xanthus (38), SigA from Bacillus subtilis (39), and Tt-RpoE1 from Thermoanaerobacter tengcongensis (40). Furthermore, two binding sites of SigT on clpPp were determined by a DNase I footprinting assay (Fig. 6D). These two sites comprised the −10 and −35 regions of clpPp, respectively (Fig. 6E) (41), consistent with SigT as an ECF σ factor. Recently, SigT was classified as an ECF27 with SigM from Corynebacterium glutamicum, and proposed to bind to a conserved element GGGAAN18-CGTT on trxBp1 (3), which was also recognized by another ECF σ factor SigR from S. coelicolor (7, 10). The SigT recognition sites on clpPp had a GGGATN18-CGTT element, which matched that of trxBp1 (Fig. 6F). Thus all our results indicate that SigT can regulate clpP1/P2 expression by directly binding to its promoter.
FIGURE 6.
SigT recognizes clpP promoter. A, EMSA for SigT-clpPp binding. About 1 ng of 5′-biotin-labeled sigTp and clpPp probes were incubated with an increasing amount of purified SigT protein, subjected to electrophoresis, and detected by ECL. B, evaluation of RNAP in purified SigT. About 2.1 μg of purified E. coli RNAP core enzyme (lanes 1-8), 1.0 μg of purified SigT (lanes 9-11), and 20 μg of E. coli lysate containing SigT (lanes 12-14) were loaded on a 6% SDS-PAGE for Western blot with antibodies against β and β′ subunits of E. coli RNAP, respectively, or on 12% SDS-PAGE for Coomassie Blue staining. All samples were diluted by 1:4. C, E. coli RNAP core enzyme was not required for SigT-clpPp binding. EMSA of SigT-clpPp binding with an increased amount of purified SigT as indicated without core enzyme (lanes 1-7) or with core enzyme (lanes 8-14). D, DNase I footprinting assay to determine the SigT recognition site on clpPp. 5′-FAM-labeled clpPp probe amplified from pL87 was used in the DNase I footprinting assay with purified SigT. The bottom strand of clpPp was analyzed. The protected region is underlined and in italics. The complementary strand is also shown. The sequencing ladder was generated with the same 5′-FAM-labeled primer. E, the promoter region of clpPp. The SigT binding site deduced from the DNase I footprinting assay is in italics and bold, and the −10 and −35 regions are underlined. F, alignment of recognition sites of SigT on clpPp with trxBp1. The conserved element is bold italic and underlined. The 18-bp interval region between two recognition sites is also shown.
Secondary Metabolites Prevent SigT-clpPp Binding
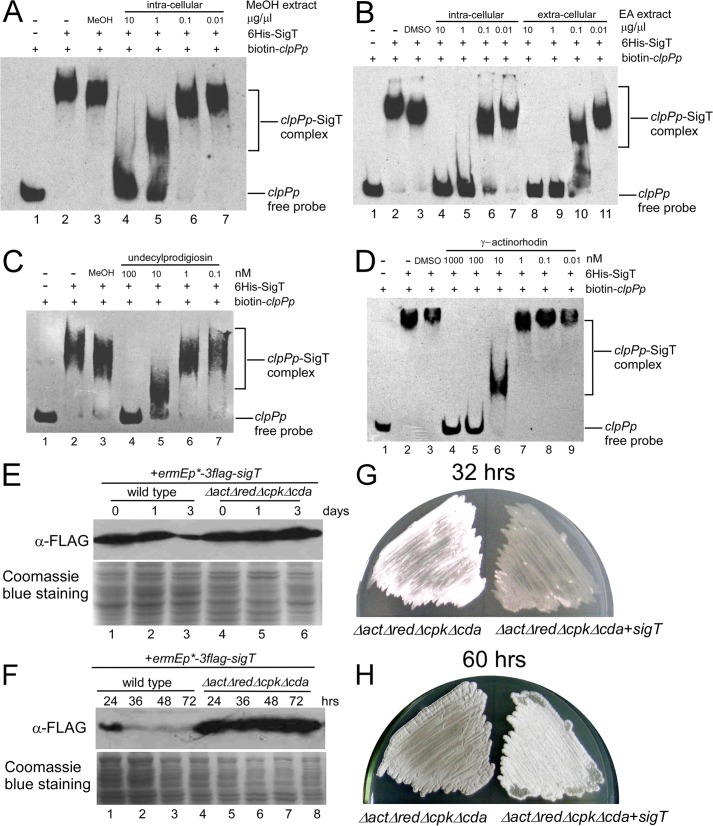
Next we examined whether a cell differentiation program could promote SigT degradation, probably by interfering with the SigT-clpPp interaction, to remove its negative effects. It was found that intracellular methanol (MeOH) extracts with undecylprodiginine (Red) and its intermediates (27), prepared from wild type cells cultured in R5-medium for 1 day, could inhibit SigT-clpPp binding (Fig. 7A). Meanwhile, γ-Act and its intermediates, extracted with ethyl acetate (31) from intracellular or extracellular parts of wild type cells cultured in R5-medium for 6 days, could also destroy SigT-clpPp binding (Fig. 7B). These results suggest that these are intracellular and secreted secondary metabolites that can damage the SigT-clpPp interaction. Furthermore, HPLC-purified undecylprodigiosin (data not shown) and γ-actinorhodin (29) from S. coelicolor could impede SigT-clpPp binding under a low concentration (10−7 m), respectively (Fig. 7, C and D). These results strongly suggest that during cell differentiation, a small amount of secondary metabolites can damage SigT-clpPp binding, and that continuously increased production of secondary metabolites can cause a gradually increased expression of clpP1/P2 during cell differentiation as described in Fig. 5.
FIGURE 7.
A–D, inhibition of SigT-clpPp binding by secondary metabolites. Intracellular crude extracts with methanol (MeOH) from wild type cells (A), intracellular or extracellular crude extracts with ethyl acetate (EA) from wild type cells (B), HPLC-purified undecylprodigiosin (C) or γ-actinorhodin (D) was incubated with SigT in a concentration gradient before addition of biotin-labeled clpPp probe, respectively, and subjected to EMSA. Methanol or DMSO were used as the solution control. E–H, SigT is stabilized in non-antibiotic-producing cells. SigT protein dynamics in strains M145/pLM86 (wild type + 3flag-sigT) and M1146/pLM86 (ΔactΔredΔcpkΔcda + 3flag-sigT) grown in liquid R5- (E) or on R2YE (F) medium for the times indicated as demonstrated by Western blot with α-FLAG antibody. Coomassie Blue staining of total protein was for a loading control. Spores of strains LM1146 (ΔactΔredΔcpkΔcda) and M1146/pLM86 (ΔactΔredΔcpkΔcda + sigT) were streaked on R2YE medium for 32 (G) and 60 h (H), respectively, and photographed.
Stabilized SigT Protein in a Non-antibiotic-producing Mutant
Based on our observations, it was speculated that the removal of secondary metabolites could result in a tight binding of SigT to clpPp to suppress clpP expression, and lead to the stabilization of the SigT protein. Indeed, in R5-liquid medium, where cells grew as mycelia and underwent secondary metabolism, we did not observe SigT protein degradation in a mutant (ΔactΔredΔcpkΔcda) (Fig. 7E), whose production of four secondary metabolites in S. coelicolor is abolished (42). Moreover, SigT protein was present at a higher and steadier level in the ΔactΔredΔcpkΔcda mutant than in the wild type on R2YE solid medium, where both morphological development and secondary metabolism were observed (Fig. 7F). Meanwhile, overexpression of sigT in ΔactΔredΔcpkΔcda caused delayed aerial hyphae development (Fig. 7G) and sporulation (Fig. 7H). These results further support our hypothesis that SigT, as a negative regulator for cell differentiation, is stabilized in vivo with reduced antibiotic production.
DISCUSSION
After fulfilling their mission to selectively regulate gene expression after stress responses, ECF σ factors are then rapidly degraded, as coordinated by the positive feedback autoregulation of gene expression (7, 18). In bacteria, this process remains essential for proper physiological adaptation and cell development in that it prevents the constitutive hyper-expression of their regulons. However, exactly how the cellular components, including the ECF σ factors themselves, work collaboratively to promote degradation of ECF σ factors, thus enabling proper physiological adaptation, remains to be fully elucidated.
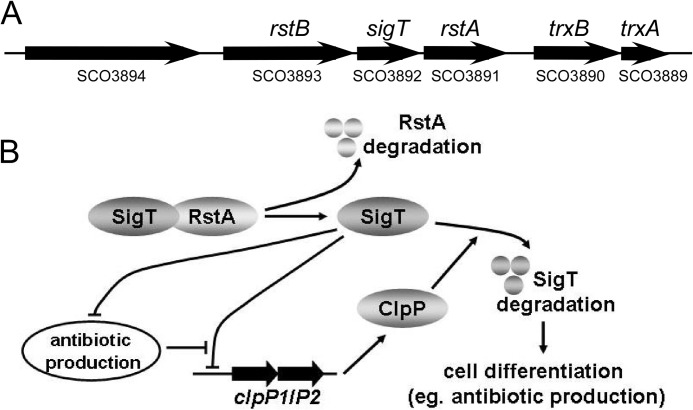
SigT negatively regulates cell differentiation in S. coelicolor (21). In this report, we show that ClpP protease is required for SigT protein degradation to remove its inhibitory effects on cell differentiation. This is consistent with the phenotypes of the ΔclpP and ΔssrA mutants that display delayed morphological development and secondary metabolism (41, 43). Furthermore, SigT negatively regulates the clpP1/P2 gene expression by binding to its promoter. So the initiation of SigT degradation results in up-regulation of clpP1/P2 expression and enhancement of ClpP protease activity in vivo, which in turn causes more SigT to degrade. Furthermore, SigT negatively regulates the secondary metabolism, whereas antibiotics can disturb the SigT-clpPp binding. Thus initiation of SigT degradation also gives rise to increased production of antibiotics, whose interference with SigT-clpPp binding, in turn causes increased expression of clpP1/P2 and accelerated degradation of SigT, for more rapid production of secondary metabolites (Fig. 8).
FIGURE 8.
A proposed regulatory model for protein degradation of ECF σ factor SigT during cell differentiation. A, genomic organization of sigT, rstA, and their adjacent genes. B, a proposed model of dual positive feedback regulatory mechanism of SigT degradation dependent on ClpP protease and secondary metabolites.
Much evidence has supported the classical hypothesis that the transcription activity of a σ factor is antagonized by its cognate anti-σ factor. Consequently, the σ factor and its anti-σ factor mutants display opposite phenotypes (37). The antagonism of anti-σ factor on its cognate σ factor is a trans-regulation, which includes repression after binding but then derepression after disassociation, and during this whole process the matured σ factor is stable (19). However, as we have reported, the protein stability of the ECF σ factor SigT is entirely dependent on its anti-σ factor RstA, which protects SigT from degradation, and RstA degradation causes the instability of SigT. It should be noted that the wild type and the ΔclpP mutant have similar SigT protein levels at the logarithmic phase in the TSB medium (Figs. 1C and 2C). This suggests that SigT is stable during primary metabolism. However, when cells are entering the secondary metabolism, RstA degradation-triggered SigT degradation occurs, and the dual positive feedback regulation takes place for a rapid SigT degradation and accelerated cell differentiation (Fig. 8).
Our results also suggest that secondary metabolites, whose production is under the negative control of SigT, can act as regulators to promote SigT degradation, thus leading to rapid occurrence of the secondary metabolism as an irreversible biological process. Meanwhile, because γ-actinorhodin and undecylprodigiosin are structurally unrelated compounds, how they play a similar role on inhibiting SigT binding to clpPp remains for further investigations. They might interfere with the σ2 and/or σ4 regions of SigT for DNA binding. However, we observed that many antibiotics produced from other microbial cannot destroy SigT-clpPp interaction (data not shown), suggesting that only the antibiotics from S. coelicolor can have the specific inhibitory effects on SigT-clpPp binding. One of the approaches is to obtain the crystal structures of the SigT-compound complex or SigT-DNA complex to elucidate the interactive residues of SigT to DNA and antibiotics, just as actinorhodin and its pathway-specific regulator ActR in S. coelicolor (44).
Here we first report that an ECF σ factor is directly involved in a feedback regulation by secondary metabolites in Streptomyces. Given the common observations that the regulatory activities of “pseudo” γ-butyrolactone receptors (29), pathway-specific regulators (45, 46), and a LysR-type regulator (31) were also regulated by the secondary metabolites for antibiotic production, we believe that the secondary metabolite-mediated feedback regulation on the transcriptional regulator activities in Streptomyces is a general physiological strategy in bacteria, and that this evolutionary-based, economical, fine-tuned and cooperative regulation is essential for harmonic environmental adaptation and proper cell development.
Acknowledgments
We gratefully thank Prof. Jung-Hye Roe of Seoul National University, South Korea, for the clpP1/P2 mutant (ΔclpP), Prof. Mervy J. Bibb of the John Innes Centre, UK, for the Δact and ΔactΔredΔcpkΔcda mutants, Prof. Zhong-Jun Qin of the Institute of Plant Physiology and Ecology, Chinese Academic Sciences, for cosmid N6–68 to delete ssrA, Gawain Bennett of the John Innes Centre for cosmid StC49 to delete clpP1/P2, and Prof. Ke-Qian Yang of the Institute of Microbiology, Chinese Academic Sciences, for HPLC-purified Act, respectively. We also thank Chris Wood of the College of Life Sciences, Zhejiang University for his critical reading of this manuscript.
This work was supported by National Basic Research Program of China (973 Program) Grant 2012CB721005, National Science Foundation of China Grant 31070040, National High Technology Research and Development Program of China (863 Program) Grant 2012AA02A706, National Science and Technology Major Projects for “Major New Drugs Innovation and Development” Grant 2011ZX09202-101-11 and Specialized Research Fund for the Doctoral Program of Higher Education Grant 20120101110143. This work was also supported by fellowships from Chinese Scholarship Council (201208330062) and New Star Project from Zhejiang University.
- ECF
- extra-cytoplasmic function
- Red
- undecylprodiginine
- Act
- actinorhodin
- RNAP
- RNA polymerase.
REFERENCES
- 1. Paget M. S., Helmann J. D. (2003) The σ70 family of σ factors. Genome Biol. 4, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gruber T. M., Gross C. A. (2003) Multiple σ subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57, 441–466 [DOI] [PubMed] [Google Scholar]
- 3. Staroń A., Sofia H. J., Dietrich S., Ulrich L. E., Liesegang H., Mascher T. (2009) The third pillar of bacterial signal transduction. Classification of the extracytoplasmic function (ECF) σ factor protein family. Mol. Microbiol. 74, 557–581 [DOI] [PubMed] [Google Scholar]
- 4. Ho T. D., Hastie J. L., Intile P. J., Ellermeier C. D. (2011) The Bacillus subtilis extracytoplasmic function σ factor σ(V) is induced by lysozyme and provides resistance to lysozyme. J. Bacteriol. 193, 6215–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thackray P. D., Moir A. (2003) SigM, an extracytoplasmic function σ factor of Bacillus subtilis, is activated in response to cell wall antibiotics, ethanol, heat, acid, and superoxide stress. J. Bacteriol. 185, 3491–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hong H. J., Paget M. S., Buttner M. J. (2002) A signal transduction system in Streptomyces coelicolor that activates the expression of a putative cell wall glycan operon in response to vancomycin and other cell wall-specific antibiotics. Mol. Microbiol. 44, 1199–1211 [DOI] [PubMed] [Google Scholar]
- 7. Paget M. S., Kang J. G., Roe J. H., Buttner M. J. (1998) σR, an RNA polymerase σ factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2). EMBO J. 17, 5776–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bibb M. J., Domonkos A., Chandra G., Buttner M. J. (2012) Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σ(BldN) and a cognate anti-σ factor, RsbN. Mol. Microbiol. 84, 1033–1049 [DOI] [PubMed] [Google Scholar]
- 9. Bibb M. J., Buttner M. J. (2003) The Streptomyces coelicolor developmental transcription factor σBldN is synthesized as a proprotein. J. Bacteriol. 185, 2338–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kang J. G., Paget M. S., Seok Y. J., Hahn M. Y., Bae J. B., Hahn J. S., Kleanthous C., Buttner M. J., Roe J. H. (1999) RsrA, an anti-σ factor regulated by redox change. EMBO J. 18, 4292–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Damron F. H., Yu H. D. (2011) Pseudomonas aeruginosa MucD regulates the alginate pathway through activation of MucA degradation via MucP proteolytic activity. J. Bacteriol. 193, 286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho T. D., Ellermeier C. D. (2012) Extra cytoplasmic function σ factor activation. Curr. Opin. Microbiol. 15, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barik S., Sureka K., Mukherjee P., Basu J., Kundu M. (2010) RseA, the SigE specific anti-σ factor of Mycobacterium tuberculosis, is inactivated by phosphorylation-dependent ClpC1P2 proteolysis. Mol. Microbiol. 75, 592–606 [DOI] [PubMed] [Google Scholar]
- 14. Sklar J. G., Makinoshima H., Schneider J. S., Glickman M. S. (2010) M. tuberculosis intramembrane protease Rip1 controls transcription through three anti-σ factor substrates. Mol. Microbiol. 77, 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heinrich J., Hein K., Wiegert T. (2009) Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. Microbiol. 74, 1412–1426 [DOI] [PubMed] [Google Scholar]
- 16. Heinrich J., Wiegert T. (2006) YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-σ factor of Bacillus subtilis. Mol. Microbiol. 62, 566–579 [DOI] [PubMed] [Google Scholar]
- 17. Jenal U., Hengge-Aronis R. (2003) Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6, 163–172 [DOI] [PubMed] [Google Scholar]
- 18. Gur E., Biran D., Ron E. Z. (2011) Regulated proteolysis in Gram-negative bacteria. How and when? Nat. Rev. Microbiol. 9, 839–848 [DOI] [PubMed] [Google Scholar]
- 19. Kim M. S., Hahn M. Y., Cho Y., Cho S. N., Roe J. H. (2009) Positive and negative feedback regulatory loops of thiol-oxidative stress response mediated by an unstable isoform of σR in actinomycetes. Mol. Microbiol. 73, 815–825 [DOI] [PubMed] [Google Scholar]
- 20. Feng W. H., Mao X. M., Liu Z. H., Li Y. Q. (2011) The ECF σ factor SigT regulates actinorhodin production in response to nitrogen stress in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 92, 1009–1021 [DOI] [PubMed] [Google Scholar]
- 21. Mao X. M., Zhou Z., Cheng L. Y., Hou X. P., Guan W. J., Li Y. Q. (2009) Involvement of SigT and RstA in the differentiation of Streptomyces coelicolor. FEBS Lett. 583, 3145–3150 [DOI] [PubMed] [Google Scholar]
- 22. Huang J., Lih C. J., Pan K. H., Cohen S. N. (2001) Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15, 3183–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kieser T., Bibb M. J., Butter M. J., Chater K. F., Hopwood D. A. (2000) Practical Streptomyces Genetics, The John Innes Foundation, Norwich, CT [Google Scholar]
- 24. Sun J., Kelemen G. H., Fernández-Abalos J. M., Bibb M. J. (1999) Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145, 2221–2227 [DOI] [PubMed] [Google Scholar]
- 25. Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. (2003) PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U.S.A. 100, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sambrook J., MacCallum P., Russell D. (2000) Molecular Cloning: A Laboratory Manual, Third Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Mo S., Sydor P. K., Corre C., Alhamadsheh M. M., Stanley A. E., Haynes S. W., Song L., Reynolds K. A., Challis G. L. (2008) Elucidation of the Streptomyces coelicolor pathway to 2-undecylpyrrole, a key intermediate in undecylprodiginine and streptorubin B biosynthesis. Chem. Biol. 15, 137–148 [DOI] [PubMed] [Google Scholar]
- 28. Tahlan K., Ahn S. K., Sing A., Bodnaruk T. D., Willems A. R., Davidson A. R., Nodwell J. R. (2007) Initiation of actinorhodin export in Streptomyces coelicolor. Mol. Microbiol. 63, 951–961 [DOI] [PubMed] [Google Scholar]
- 29. Xu G., Wang J., Wang L., Tian X., Yang H., Fan K., Yang K., Tan H. (2010) “Pseudo” γ-butyrolactone receptors respond to antibiotic signals to coordinate antibiotic biosynthesis. J. Biol. Chem. 285, 27440–27448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zianni M., Tessanne K., Merighi M., Laguna R., Tabita F. R. (2006) Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17, 103–113 [PMC free article] [PubMed] [Google Scholar]
- 31. Mao X. M., Sun Z. H., Liang B. R., Wang Z. B., Feng W. H., Huang F. L., Li Y. Q. (2013) Positive feedback regulation of stgR expression for secondary metabolism in Streptomyces coelicolor. J. Bacteriol. 195, 2072–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sola-Landa A., Rodríguez-García A., Amin R., Wohlleben W., Martín J. F. (2013) Competition between the GlnR and PhoP regulators for the glnA and amtB promoters in Streptomyces coelicolor. Nucleic Acids Res. 41, 1767–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paget M. S., Bae J. B., Hahn M. Y., Li W., Kleanthous C., Roe J. H., Buttner M. J. (2001) Mutational analysis of RsrA, a zinc-binding anti-σ factor with a thiol-disulphide redox switch. Mol. Microbiol. 39, 1036–1047 [DOI] [PubMed] [Google Scholar]
- 34. Gehring A. M., Yoo N. J., Losick R. (2001) RNA polymerase sigma factor that blocks morphological differentiation by Streptomyces coelicolor. J Bacteriol 183, 5991–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore S. D., Sauer R. T. (2007) The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124 [DOI] [PubMed] [Google Scholar]
- 36. Dalton K. A., Thibessard A., Hunter J. I., Kelemen G. H. (2007) A novel compartment, the “subapical stem” of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 64, 719–737 [DOI] [PubMed] [Google Scholar]
- 37. Helmann J. D. (2002) The extracytoplasmic function (ECF) σ factors. Adv. Microb. Physiol. 46, 47–110 [DOI] [PubMed] [Google Scholar]
- 38. Gómez-Santos N., Pérez J., Sánchez-Sutil M. C., Moraleda-Muñoz A., Muñoz-Dorado J. (2011) CorE from Myxococcus xanthus is a copper-dependent RNA polymerase σ factor. PLoS Genet. 7, e1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yeh H. Y., Chen T. C., Liou K. M., Hsu H. T., Chung K. M., Hsu L. L., Chang B. Y. (2011) The core-independent promoter-specific interaction of primary σ factor. Nucleic Acids Res. 39, 913–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu J., Li J., Wu Z., Pei H., Zhou J., Xiang H. (2012) Identification and characterization of the cognate anti-σ factor and specific promoter elements of a T. tengcongensis ECF σ factor. PloS One 7, e40885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Crécy-Lagard V., Servant-Moisson P., Viala J., Grandvalet C., Mazodier P. (1999) Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32, 505–517 [DOI] [PubMed] [Google Scholar]
- 42. Gomez-Escribano J. P., Bibb M. J. (2011) Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 4, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang C., Glover J. R. (2009) The SmpB-tmRNA tagging system plays important roles in Streptomyces coelicolor growth and development. PLoS One 4, e4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Willems A. R., Tahlan K., Taguchi T., Zhang K., Lee Z. Z., Ichinose K., Junop M. S., Nodwell J. R. (2008) Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA. J. Mol. Biol. 376, 1377–1387 [DOI] [PubMed] [Google Scholar]
- 45. Li Q., Wang L., Xie Y., Wang S., Chen R., Hong B. (2013) SsaA, a novel class of transcriptional regulator, controls sansanmycin production in Streptomyces sp. SS involving a feedback mechanism. J. Bacteriol. 195, 2232–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., Yang K., Tan H. (2009) Autoregulation of antibiotic biosynthesis by binding of the end product to an atypical response regulator. Proc. Natl. Acad. Sci. U.S.A. 106, 8617–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bentley S. D., Chater K. F., Cerdeño-Tárraga A. M., Challis G. L., Thomson N. R., James K. D., Harris D. E., Quail M. A., Kieser H., Harper D., Bateman A., Brown S., Chandra G., Chen C. W., Collins M., Cronin A., Fraser A., Goble A., Hidalgo J., Hornsby T., Howarth S., Huang C. H., Kieser T., Larke L., Murphy L., Oliver K., O'Neil S., Rabbinowitsch E., Rajandream M. A., Rutherford K., Rutter S., Seeger K., Saunders D., Sharp S., Squares R., Squares S., Taylor K., Warren T., Wietzorrek A., Woodward J., Barrell B. G., Parkhill J., Hopwood D. A. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147 [DOI] [PubMed] [Google Scholar]