Background: A pool of the nuclear transcription factor Stat3 in the mitochondria (mitoStat3) controls respiration and Ras transformation.
Results: Serine phosphorylation of mitoStat3 controls accumulation of reactive oxygen species and growth of breast cancer in mice.
Conclusion: These results provide the first evidence for a mechanism by which mitoStat3 contributes to tumorigenesis.
Significance: The data suggest new therapeutic approaches to treatment of breast cancer.
Keywords: Cancer, Mitochondria, Phosphorylation, Reactive Oxygen Species (ROS), Serine, STAT3, STAT3 Breast Cancer
Abstract
Signal transducer and activator of transcription 3 (Stat3) is a key mediator in the development of many cancers. For 20 years, it has been assumed that Stat3 mediates its biological activities as a nuclear localized transcription factor activated by many cytokines. However, recent studies from this laboratory and others indicate that Stat3 has an independent function in the mitochondria (mitoStat3) where it controls the activity of the electron transport chain (ETC) and mediates Ras-induced transformation of mouse embryo fibroblasts. The actions of mitoStat3 in controlling respiration and Ras transformation are mediated by the phosphorylation state of serine 727. To address the role of mitoStat3 in the pathogenesis of cells that are transformed, we used 4T1 breast cancer cells, which form tumors that metastasize in immunocompetent mice. Substitution of Ser-727 for an alanine or aspartate in Stat3 that has a mitochondrial localization sequence, MLS-Stat3, has profound effects on tumor growth, complex I activity of the ETC, and accumulation of reactive oxygen species (ROS). Cells expressing MLS-Stat3(S727A) display slower tumor growth, decreased complex I activity of the ETC, and increased ROS accumulation under hypoxia compared with cells expressing MLS-Stat3. In contrast, cells expressing MLS-Stat3(S727D) show enhanced tumor growth and complex I activity and decreased production of ROS. These results highlight the importance of serine 727 of mitoStat3 in breast cancer and suggest a novel role for mitoStat3 in regulation of ROS concentrations through its action on the ETC.
Introduction
Stats and the Jak tyrosine kinases that activate them have emerged as critical regulators of numerous fundamental biological processes important for health and disease, particularly inflammation, obesity, cardiovascular disease, and cancer. It is difficult to find an area of cell biology and physiology in which Stats do not have important if not key roles. In addition to its well described actions in the nucleus, our published studies have identified a pool of Stat3 that resides within the mitochondria (mitoSTAT3) where it regulates the activity of complexes of the electron transport chain (ETC)4 (1, 2). Furthermore, Ras transformation of mouse embryo fibroblasts (MEFs) requires expression of mitoStat3. In contrast to its actions in the nucleus, mitoStat3-mediated regulation of respiration and Ras transformation requires serine 727 but not tyrosine 705 or the SH2 or DNA binding domains of Stat3. These studies suggest that there is a completely novel function of Stat3 governed by its presence in the mitochondria where it controls energy balance. Subsequent studies have confirmed and extended these initial observations in a variety of non-transformed cells (3–8) and in the growth of pancreatic cancer cells in mice (9).
Numerous reports have indicated that overexpression of Stat3 in cells promotes cell growth and transformation (10). Approximately 75% of primary human breast and prostate cancers show increased levels of nuclear localized, tyrosine phosphorylated Stat3 (10). Although constitutive tyrosine phosphorylation of Stat3 has been implicated in the pathogenesis of many malignancies, there are a number of reports suggesting that phosphorylation of serine 727 (Ser-727) can contribute to, or be sufficient for, the growth and/or transformation of malignancies, including chronic lymphocytic leukemia, prostate, and breast cancer (11–13). It has been assumed that the many vital aspects of Stat3 in transformation are due to its actions as a nuclear transcription factor. Although it has been demonstrated that serine 727 of mitoStat3 is needed for Ras-mediated transformation of MEFs, the ability of mitoStat3 to influence tumor growth and metastasis of established tumor cells has not been explored. In this report, we describe a role for mitoStat3 in the regulation of growth and metastasis of breast cancer cells. The actions of Ser-727 on soft agar colony formation are also observed in Lewis lung carcinoma cells, suggesting that the phosphorylation state of Ser-727 mediates the behavior of tumors originating from a variety of tissues. Our results also are consistent with the hypothesis that mitoStat3 mediates its actions through affecting cellular levels of ROS, which may be mediated by changes in complex I activity of the ETC.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
6-Thioguanine (A-4882), methylene blue (M-4159), and hyaluronidase (H3884) were purchased from Sigma; elastase (2279), collagenase type 4 (4188), and collagenase type 1 (4196) were purchased from Worthington. Mouse monoclonal Stat3 antibodies were purchased from BD Transduction Laboratories. Complex II subunit 30 kDa (MS203) was purchased from Mito Sciences. Mouse monoclonal anti-α-tubulin was purchased from Sigma. Anti-nitro-tyrosine antibody (06-284) was purchased from Millipore. Anti-actin antibody (sc-56459) was purchased from Santa Cruz Biotechnology.
Cells
The 4T1 mouse mammary carcinoma cell line was purchased from the American Type Culture Collection (Rockville, MD). 4T cells were further authenticated in that they formed tumors in BALB/cJ mice and are 6-thiguanine resistant. Cells were grown in DMEM supplemented with 10% fetal bovine serum. 4T1 and Lewis lung cell lines were generated expressing various forms of Stat3 that contain a mitochondrial targeting sequence of human cytochrome c oxidase subunit VIII (MLS) fused to the N terminus of the protein (2). Pools of GFP-positive cells were selected by FACS sorting. Cells expressing various forms of Stat3, which contain mutations in the domains required to activate nuclear encoded genes, were used for these studies. These mutations include: tyrosine 705 to phenylalanine, the DNA binding domain (DBD) (14), the SH2 domain (15), and the nuclear localization sequence of Stat3 (16). We also mutated serine 727 to alanine or aspartic acid (2) alone or in combination with other mutations. mutMLS-Stat3 has mutations in the DBD and SH2 domains, tyrosine 705 to phenylalanine, and the nuclear localization sequence. Mutations in MLS-Stat3(Y705F/S727A) and MLS-Stat3(Y705F/S727D) constructs were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The following primers were used to generate arginine (Arg-609) to glutamine (Glu) mutation: forward primer, 5′-c ccg ggt acc ttc cta ctg cag ttc agc gag agc agc aaa g-3′ and reverse primer, 5′-t gaa ctg cag tag gaa ggt acc cgg ggg ctt tgt gc-3′. Other mutants have been described previously (2, 17). All mutations were sequenced for verification.
Transfection and Infection
The empty vector containing internal ribosome entry site (IRES) (MSCV-IRES-GFP), and vectors encoding wild type Stat3 and different MLS mutants of Stat3 were transfected into 293 T cells with Phoenix helper vector using FuGENE transfection reagent (Roche Diagnostics) (2). Cells expressing GFP were sorted by FACS (>95% GFP+) and used for experiments.
RT-PCR Assays
The following primers were used to analyze β-actin: 5′-GGT CAT CAC TAT TGG CAA CG-3′ (forward) and 5′-ACG GAT GTC AAC GTC ACA CT-3′ (reverse); C/EBPδ, 5′-CTCCCGCACACAACATACTG-3′ (forward) and 5′-CTTCGGCAACCACCTAAAAG-3′ (reverse).
Isolation of Mitochondria and Preparation of Cytosolic and Mitochondrial Lysates
Intact mitochondria were isolated from 4T1 cells as described (2).
NADH-Cytochrome c Reductase (NCR, Complex I/III)
Activities were determined spectrophotometrically. Incubations contained 2 μg of mitochondrial protein, 0.1 mm EDTA, 0.2% bovine serum albumin (w/v), 0.015% asolectin, 0.15 mm cytochrome c, 2 mm sodium azide, and 50 mm potassium phosphate, pH 7.4, in a final volume of 1 ml. After an equilibration period of 3 min, reactions were started by the addition of 0.5 mm NADH, and the increase in absorbance at 550 nm was monitored. Rotenone-sensitive activities were obtained by subtraction of the rate determined in incubations containing 7.5 μm rotenone from the rate in parallel incubations without rotenone. Activities were calculated using an extinction coefficient of 19.1 mm−1 cm−1 for cytochrome c.
Succinate-Cytochrome c Reductase (SCR, Complex II/III)
Assays were performed under similar conditions as NCR except that reactions contained 20 mm succinate, and antimycin A-sensitive activities were obtained by subtraction of the rate determined in incubations containing 10 μg of antimycin A from the rate in parallel incubations without antimycin A.
Measurement of Levels of Tyrosine-nitrated Proteins
4T1 cells expressing versions of MLS-Stat3 were incubated in 1% or 20% oxygen. Cell extracts were immunoprecipitated and then immunoblotted with anti-nitro-tyrosine antibodies. The amount of tyrosine-nitrated proteins was quantitated using a fluorescent-labeled secondary antibody (18).
Mitochondrial Superoxide Production
Production of mitochondrial superoxide was measured using Flow Cytometry as described (19, 20). Briefly, cells were grown to confluence and subjected to hypoxia or normoxia for 24 h. Cells were then incubated with MitoSOX Red superoxide indicator (Invitrogen) for 30 min. The labeled cells were washed twice with Hanks' balanced salt solution and then suspended in warm Hanks' balanced salt solution for analysis by flow cytometry. MitoSOX Red was excited by laser at 488 nm, and the data were collected at the forward scatter (FSC), side scatter (SSC) 585/42 nm (FL2) channel. In this work, the data were presented in the FL2 channel. Cell debris as represented by distinct low forward and side scatter were gated out for analysis. The data are presented as fold change of mean intensity of MitoSOX fluorescence compared with MSCV control with MitoSOX present.
In Vivo Tumorigenesis
Female mice were subcutaneously injected in the mammary gland with 7.0 × 103 4T1 cells or variants expressing different forms of Stat3. Tumor growth was assessed morphometrically using calipers, and tumor volumes were calculated according to the formula V (mm3) = L (length) × W2 (width)/2 (4). For experiments with MnTBAP, mice were injected subcutaneously with 300 μg/day MnTBAP or PBS as a control. This research was conducted under a protocol approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee.
Metastatic Tumor Cell Assay
4T1 tumor cells are 6-thioguanine-resistant. Metastatic cells were quantified by removing organs, plating dissociated cells in medium supplemented with 6-thioguanine, and counting the number of 6-thioguanine-resistant colonies as described (21). Clonogenic metastases were calculated on a per-organ basis.
Statistical Analysis
Statistical analysis was performed using ANOVA for multiple groups as indicated in figure legends and two-tailed Student t tests. All results are presented as means ± S.E. p < 0.05 was considered statistically significant.
RESULTS
Serine 727 Affects the Ability of MLS-Stat3 to Regulate Transformation in Vitro
Although it has been reported that mitoStat3 is required for transformation of MEFs, nothing is known about the role of mitoStat3 in the tumorigenesis of transformed cancer cell lines. To address this issue, we used 4T1 murine breast cancer cells. These cells mimic the pathology of human breast carcinomas in that they form large primary tumors that metastasize to many distant organs. The growth and metastasis of these cells is observed in immunocompetent BALB/c mice and Stat3 expression is required for the in vivo growth of 4T1 cells as well as their invasion in vitro and in vivo (22). We initially examined whether mitochondria-targeted Stat3 influenced the ability of 4T1 cells to grow and metastasize in vitro.
4T1 cell lines were generated that express various forms of Stat3 that contain a mitochondrial targeting sequence of human cytochrome c oxidase subunit VIII (MLS) fused to the N terminus of the protein (2). The targeting sequence places the protein of interest in the inner mitochondrial membrane. The vector also contains IRES, which drives the expression of GFP. GFP-positive cells were selected by FACS sorting. Pooled cells expressing various forms of Stat3, which contain mutations in the domains required to activate the expression of nuclear encoded early response genes, were used for these studies. These include the nuclear localization sequence, tyrosine 705, the DNA binding domain of Stat3, and the SH2 domain. We also mutated serine 727 alone or in combination with other mutations. Serine 727 is required for both optimal activity of the ETC and Ras transformation of MEFs (1, 2). Subcellular fractionation of these cell lines demonstrates that the MLS-Stat3 mutants are present primarily or only in the mitochondria and not in the cytoplasm (Fig. 1).
FIGURE 1.
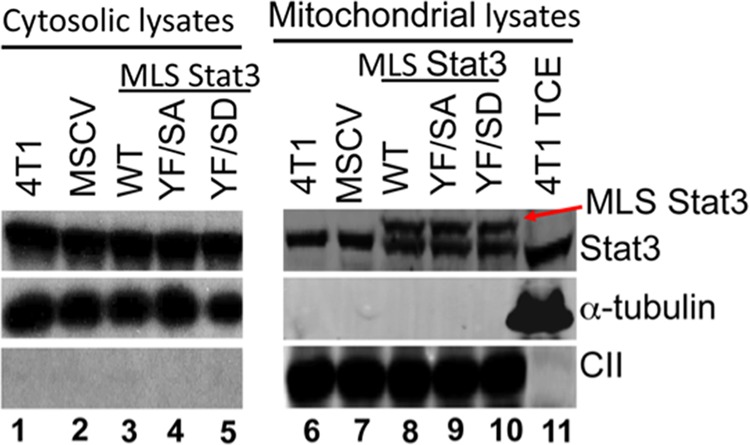
Expression of various mutants of Stat3 with a mitochondrial localization sequence. Mitochondria (30 μg), cytoplasm (10 μg), and total cell extract (TCE) (20 μg) were isolated from 4T1 cells and immunoblotted for Stat3 expression. MLS-Stat3 and its various mutant forms were visualized as the upper bands (right panel, indicated by the red arrow) in the mitochondrial fractions, whereas the endogenous Stat3 migrated as the lower band. The blot was also probed for tubulin (a cytosolic marker) and the 30-kDA subunit of complex II (a mitochondrial marker) (CII). A representative immunoblot from three independent experiments is shown.
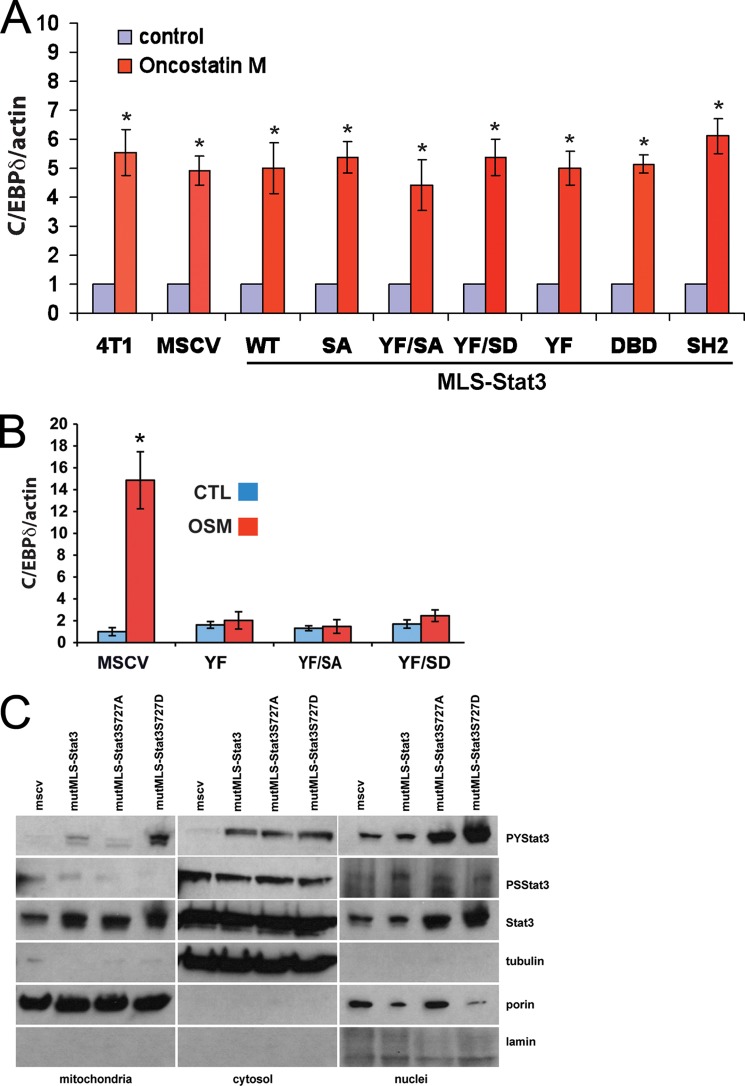
To determine whether expression of these various forms of MLS-Stat3 alters Stat3-dependent expression of cytokine-activated nuclear-encoded response genes, we examined oncostatin M (OSM) induced expression of C/EBPδ, a well described Stat3-dependent early response gene (23). OSM-induced expression of C/EBPδ is not altered in 4T1 cells expressing various forms of MLS-Stat3 (Fig. 2A), indicating that expression of these MLS-Stat3 proteins is not impinging on the actions of endogenous Stat3 that is activated through the Jak/Stat pathway. Mutation of tyrosine 705 to phenylalanine (Y705F) functions as a dominant negative Stat3 for cytokine signaling. The fact that the Y705F forms of Stat3 with a MLS did not alter the induction of endogenous Stat3 is consistent with a MLS preventing the protein from functioning in the nucleus. To confirm this idea, we made the same Y705F/S727A and Y705F/S727D mutants without an MLS. As expected, these forms of Stat3 inhibited OSM activation of CEBP/δ (Fig. 2B). We also expressed MLS-Stat3 that has multiple mutations in its DBD, its SH2 domain, Y705F, a deleted nuclear localization sequence (termed mutMLS-Stat3) and S727A or S727D in Stat3−/− MEFs. None of these constructs were able to support OSM induced expression of C/EBPδ (data not shown). Stat3, which is not tyrosine-phosphorylated, has also been reported to activate nuclear genes (24, 25). We examined the levels of transcripts of some of these genes (CCL5, IL-8, and IFNβ) in cells expressing different forms of MLS-Stat3. MLS-Stat3 variants did not alter expression of these RNAs.
FIGURE 2.
A, expression of various forms of MLS-Stat3 in 4T1 cells does not alter OSM-stimulated accumulation of C/EBPδ mRNA. 4T1 cells were incubated with OSM for 4 h. RNA was collected and analyzed for the expression of C/EBPδ by quantitative PCR. Samples were normalized to the expression of actin mRNA. *, p < 0.05 versus corresponding control; two-tailed Student's t test. B, expression of Stat3 (Y705F) without an MLS in 4T1 cells inhibits OSM-stimulated accumulation of C/EBPδ RNA. 4T1 cells were incubated with OSM for 4 h. RNA was collected and analyzed for the expression of C/EBPδ by quantitative PCR. Samples were normalized to the expression of actin mRNA. *, p < 0.05 versus MSCV control (CTL) group (two-way ANOVA). C, tyrosine phosphorylation of endogenous Stat3 is altered by the expression of mutMLS-Stat3(S727D). Cytosol, mitochondria, and nuclear extracts were prepared from 4T1 cells expressing mutMLS-Stat3 with Ser-727, S727A, or S727D. Western blots were probed for tyrosine-phosphorylated Stat3 (upper panel), serine-phosphorylated Stat3 (second panel), and total Stat3 (third panel). The blots were also probed for tubulin (cytosolic marker), porin (mitochondrial marker), and lamin (nuclear marker).
We also examined the levels of tyrosine phosphorylated endogenous Stat3 in 4T1 cells expressing mutMLS-Stat3 with S727A or S727D mutations. These constructs have Y705F so the exogenous protein cannot be tyrosine phosphorylated. Interestingly, expression of mutMLS-Stat3 (S727D) increases tyrosine but not serine phosphorylation of endogenous Stat3 even though there are no changes in basal or OSM induced expression of C/EBPδ. The reason for this result is not clear. One interpretation of is that there is a communication network between Stat3 in the nucleus and that in the mitochondria (Fig. 2C).
Under normal growth conditions there were no differences in rates of growth of 4T1 cells expressing various forms of MLS-Stat3. Because Ras transformation of MEFs required Ser-727, we examined the ability of 4T1 cell lines expressing various forms of MLS-Stat3 to form colonies in soft agar (Fig. 3A). Cells expressing mutMLS-Stat3 showed no differences in numbers of soft agar colonies compared with cells infected with empty MSCV or MLS-Stat3 (Fig. 3A, lanes 1–3). However, there were fewer colonies in cells expressing mutMLS-Stat3 where Ser-727 was mutated to an alanine, mutMLS-Stat3(S727A), compared with cells expressing MSCV (lanes 1 and 4). In contrast, cells expressing the phosphomimetic serine to aspartic acid mutMLS-Stat3(S727D) displayed increased numbers of colonies in soft agar (lanes 3 and 4 versus 5). We also expressed mutMLS-Stat3, mutMLS-Stat3(S727A), and mutMLS-Stat3(S727D) in Lewis lung carcinoma cells, and measured formation of colonies in soft agar. Similar to 4T1 cells, cells expressing mutMLS-Stat3(S727D) formed greater numbers of colonies (data not shown). These results indicate that the effects of the Ser-727 in the mitochondria are not limited to breast cancer cells.
FIGURE 3.
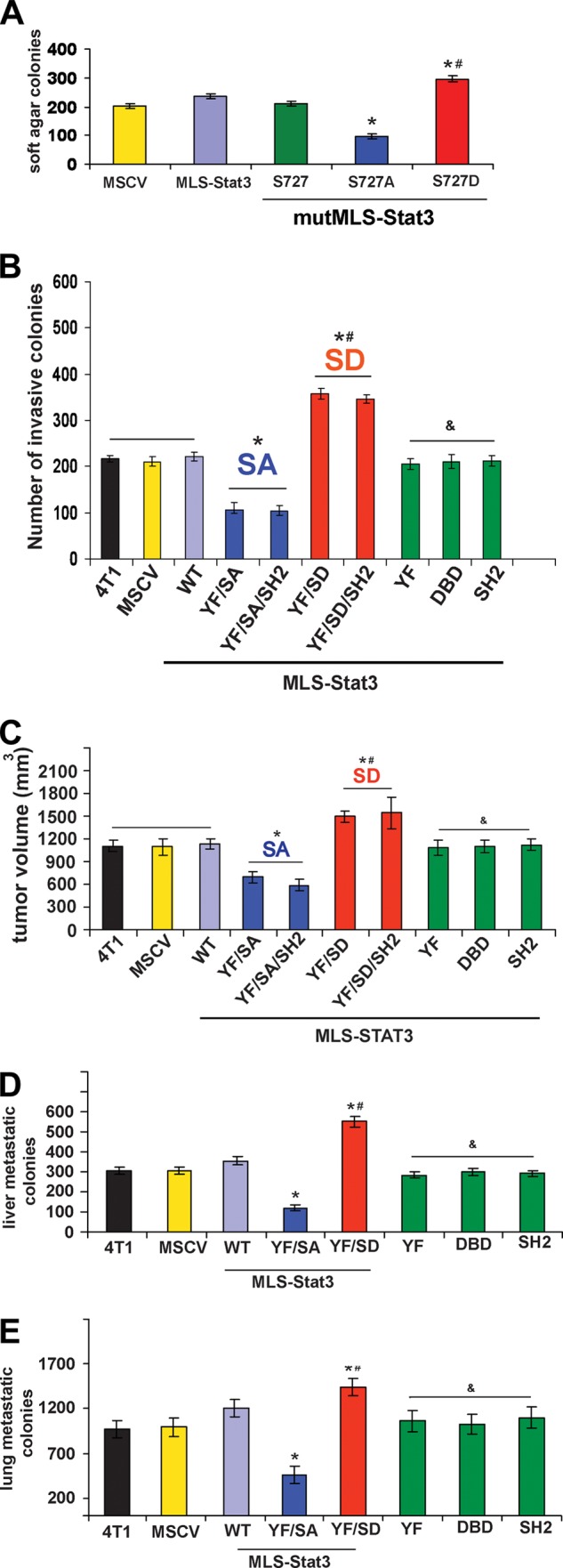
Ser-727 of MLS-Stat3 affects colony formation in soft agar, cell invasion, and tumor growth in mice. A, wild type Stat3 or Stat3 containing an MLS with mutations in the nuclear localization sequence, DBD, SH2, and Tyr-705 (mutMLS-Stat3) were expressed in 4T1 cells and assayed for their ability to form colonies in soft agar. mutMLS-Stat3 cells with Ser-727 mutated to alanine formed fewer colonies than cells with Ser-727 not mutated or mutated to aspartic acid. *, p < 0.05 versus Ser-727; #, p < 0.05 versus S727A (one-way ANOVA). B, Matrigel invasion assays were performed using 4T1 cells expressing various forms of Stat3. Single mutations in Y705F (YF), the DNA binding domain, and the SH2 domain (SH2) are the green bars. *, p < 0.05 versus MSCV; #, p < 0.05 versus S727A (SA); &, p < 0.05 versus S.D. (one-way ANOVA). C, tumor volumes at the site of injection of BALB/c mice with cell lines expressing various forms of MLS-Stat3. Cell lines that express Stat3 with an MLS in addition to the indicated mutations were injected into mice. Volumes were calculated using calipers. The animals were sacrificed after 27 days. *, p < 0.05 versus MSCV; #, p < 0.05 versus SA; &, p < 0.05 versus S.D. (one-way ANOVA). D and E, numbers of metastatic colonies in the livers and lungs formed in mice injected with cells expressing various forms of Stat3. *, p < 0.05 versus MSCV; #, p < 0.05 versus S727A (SA); &, p < 0.05 versus S.D. (one-way ANOVA).
Cell lines were also assayed for their ability to invade Matrigel (Fig. 3B). Similar to growth in soft agar, cells expressing MLS-Stat3(Y705F/S727A) invaded Matrigel less effectively than cells expressing MLS Stat3(Y705F/S727D). The presence of an intact SH2 and DNA binding domains as well as Tyr-705 is also not required for these effects of MLS-Stat3.
Serine 727 Plays an Important Role in the in Vivo Growth of 4T1 Cells
Cells expressing various forms of MLS-Stat3 were also analyzed for their ability to form tumors in BALB/c mice (Fig. 3C). Mice injected with 4T1 cells expressing MLS-Stat3(Y705F/S727A), (lane 4), MLS-Stat3(Y705F/S727A/SH2) (lane 5) formed smaller tumors than mice injected with MLS-Stat3(Y705F/S727D) (lane 6) or MLS-Stat3(Y705F/S727D/SH2) (lane 7). Single mutations, which inactivate the transcriptional actions of Stat3 (Y705F, SH2 domain, and DNA binding domain), did not influence the rates of tumor growth as measured by volume (data not shown).
The number of liver and lung metastases was also increased in mice injected with 4T1 cells expressing the S727D and decreased in mice expressing S727A (Fig. 3, D and E). This is not surprising in that larger 4T1 tumors have higher numbers of metastatic lesions. These observations demonstrate an important role for MLS-Stat3 Ser-727 in the transformative properties of 4T1 cells and the lack of a requirement for a functional DBD, SH2 domain, and Tyr-705, which are all essential for the actions of Stat3 as a nuclear transcription factor.
Activity of Complex I of the Electron Transport Chain Is Regulated by Stat3 Ser-727
Our previous studies have highlighted the role of serine 727 of mitoStat3 in regulation of the activity of the ETC (1, 2, 7). Because Ser-727 of mitoStat3 affected 4T1 cell growth and metastasis, we wanted to determine whether changes in complex I and II activity occurred in MLS-Stat3 cell lines. Activities of complexes I and III NADH cytochrome c reductase (NCR) or II and III succinate cytochrome c reductase (SCR) were measured in isolated mitochondria extracts (26). Under normoxia, we observed no differences in activities of either SCR or NCR in cells expressing different forms of MLS-Stat3. However, cells grown under conditions of hypoxia displayed clear differences in NCR (Fig. 4A) but not SCR (Fig. 4B). The fact that the activities of complexes II and III were not altered indicates the differences in NCR activity are due to changes in complex I and not complex III activity. These results are consistent with previous findings that overexpression of mitoStat3 can protect complex I activity and attenuate ROS production in hearts subjected to ischemia (1, 7).
FIGURE 4.
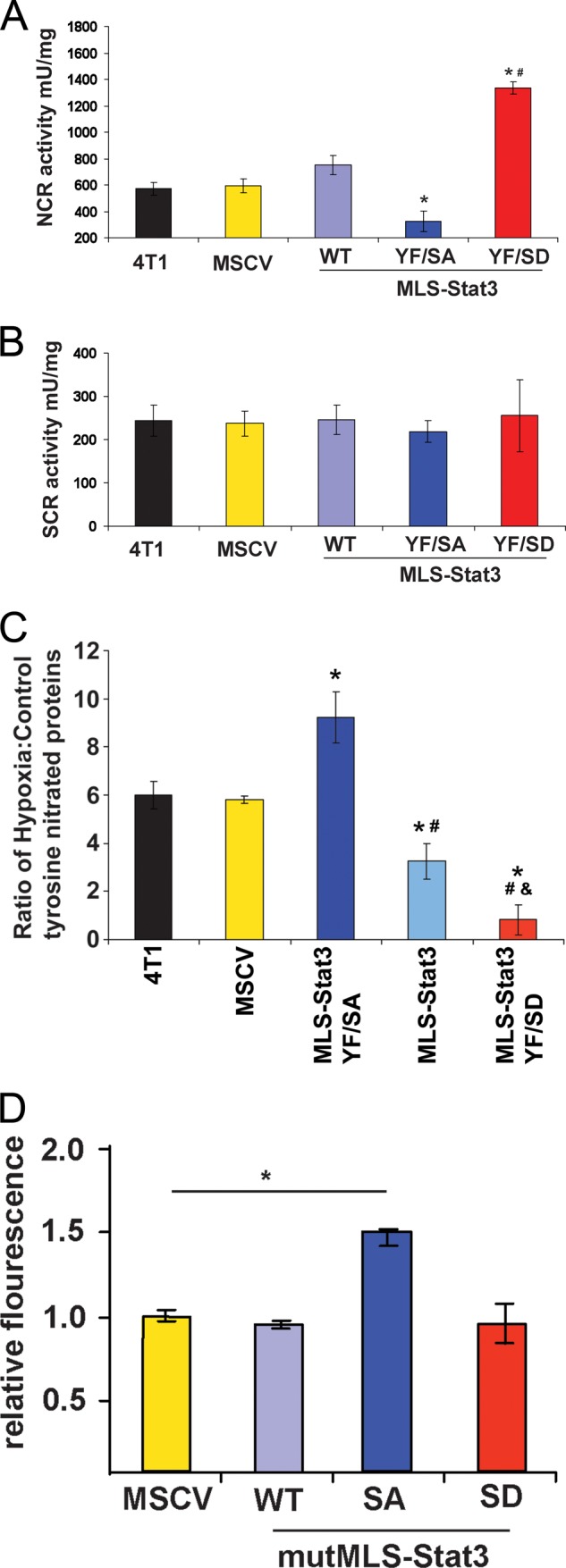
NCR activity and ROS levels are regulated by Ser-727. 4T1 cells expressing the indicated versions of MLS-Stat3 were cultured in 1% oxygen. Mitochondrial extracts were assayed for NCR (A), and SCR activities were as described previously (B) (17). *, p < 0.05 versus WT; #, p < 0.05 versus YF/SA (one-way ANOVA). C, levels of tyrosine-nitrated proteins are regulated by Ser-727. 4T1 cells expressing the indicated versions of MLS-Stat3 were incubated in 1% oxygen or 20%. Cell extracts were immunoprecipitated and then immunoblotted with anti-nitro-tyrosine antibodies. The amount of tyrosine-nitrated proteins was quantitated using a fluorescent-labeled secondary antibody. *, p < 0.05 versus MSCV; #, p < 0.05 versus MLS-Stat3 YF/SA; &, p < 0.05 versus MLS-Stat3 (one-way ANOVA). D, relative fluorescence of mitoSOX red in 4T1 cells expressing MSCV, mutMLS-Stat3, mutMLS-Stat3(S727A), and mutMLS-Stat3(S727D). *, p < 0.05 by two-tailed Student t test. mU, milliunits.
Hypoxia-induced ROS Correlates with 4T1 Tumorigenicity
Increased cellular levels of ROS have been reported to activate Stat3 (27). It is also known that if Stat3 expression is diminished or disrupted in the heart and several other tissues, levels of ROS are elevated (27, 28). Female mice that do not express Stat3 in the heart have increased incidence of postpartum mortality, and treatment of these mice with pharmacologic suppressors of ROS prevents postpartum mortality (29). It is known that tumor growth is influenced by levels of intracellular ROS (30–32). We therefore examined whether ROS production is changed in 4T1 cells expressing MLS-Stat3. As an indirect assay for ROS, we measured the relative amounts of protein tyrosine nitration in the various MLS-Stat3-expressing 4T1 cell lines subjected to hypoxia. Tyrosine can be nitrated by reactive nitric oxide formed from the reaction of superoxide anion or H2O2 and nitric oxide. Tyrosine-nitrated proteins immunoprecipitated from cell lysates were quantitated by densitometry (33). Results are expressed as a ratio of tyrosine-nitrated proteins in cells maintained in 1% O2 to those in 20% O2 (Fig. 4C). 4T1 cells and 4T1 cells expressing the MSCV vector (lanes 1 and 2) displayed similar ratios of tyrosine-nitrated proteins, whereas those expressing MLS-Stat3 (lane 4) or MLS-Stat3(Y705F/S727D) (lane 5) had significantly lower ratios of tyrosine-nitrated proteins compared with vector controls. Cells expressing MLS-Stat3(Y705F/S727A) (lane 3), which form less colonies in soft agar and smaller tumors in vivo expressed elevated levels of tyrosine-nitrated proteins compared with vector controls or cells expressing MLS-Stat3(Y705F/S727D).
We also measured the relative levels of superoxide produced in the mitochondria using mitoSOX dye (Fig. 4D). Similar to levels of nitrated proteins, cells expressing mutMLS-Stat3(S727A) contained more superoxide in the mitochondria compared with cells expressing MSCV or mutMLS-Stat3. We did not observe changes in superoxide in cells expressing mutMLS-Stat3(S727D). These results are discrepant with the decrease in tyrosine nitrated proteins observed in cells expressing mutMLS-Stat3(S727D) observed in Fig. 4C. One explanation for these differences are tyrosine-nitrated proteins are produced from H2O2 and superoxide, whereas mitoSOX red only detects superoxide. Another explanation is that the mitoSOX dye emission spectra overlaps GFP and decreases its sensitivity.
These results are consistent with the hypothesis that serine 727 controls the activity of complex I, which is a major source of intracellular ROS (34). To determine whether ROS production alters the ability of MLS-Stat3 expressing cells to transform, we examined soft agar colonies in the presence of the ROS scavenger N-acetyl cysteine. Incubation of all cell lines with N-acetyl cysteine increased their ability to form colonies compared with untreated cells. However, the ratio of number of colonies in N-acetyl cysteine:control cells was statistically increased in cells expressing MLS-Stat3(Y705F/S727A) or MLS-Stat3(Y705F/S727A/SH2) compared with the ratio N-acetyl cysteine:control in cells expressing MLS-Stat3(Y705F/S727D) tor MLS-Stat3(Y705F/S727D/SH2) (data not shown).
We also examined the ability of antioxidants to influence tumor growth of 4T1 cells using a pharmacological suppressor of ROS, MnTBAP. Twelve days after inoculation of mice with 4T1 cell lines expressing various forms of MLS-Stat3, animals were injected daily with MnTBAP. Tumor volumes were measured every 4 days until the mice were sacrificed (Fig. 5A). MnTBAP increased the rate of tumor growth only in mice inoculated with 4T1 cells expressing MLS-Stat3(Y705F/S727A). There were no significant changes in rates of tumor growth in cells expressing MSCV (the empty vector), MLS-Stat3(Y705F/S727D), or MLS-Stat3. Increases in tumor growth in cells expressing MLS-Stat3(Y705F/S727A) and treated with MnTBAP also resulted in increased liver metastasis (Fig. 5B). These experiments indicate that the ability of MLS-Stat3 to regulate tumor growth in vitro and in vivo is mediated by production of ROS. The fact that treatment of mice with MnTBAP selectively increased the growth of cells expressing MLS-Stat3(YF/SA) argues that the actions of this antioxidant are not unspecific.
FIGURE 5.
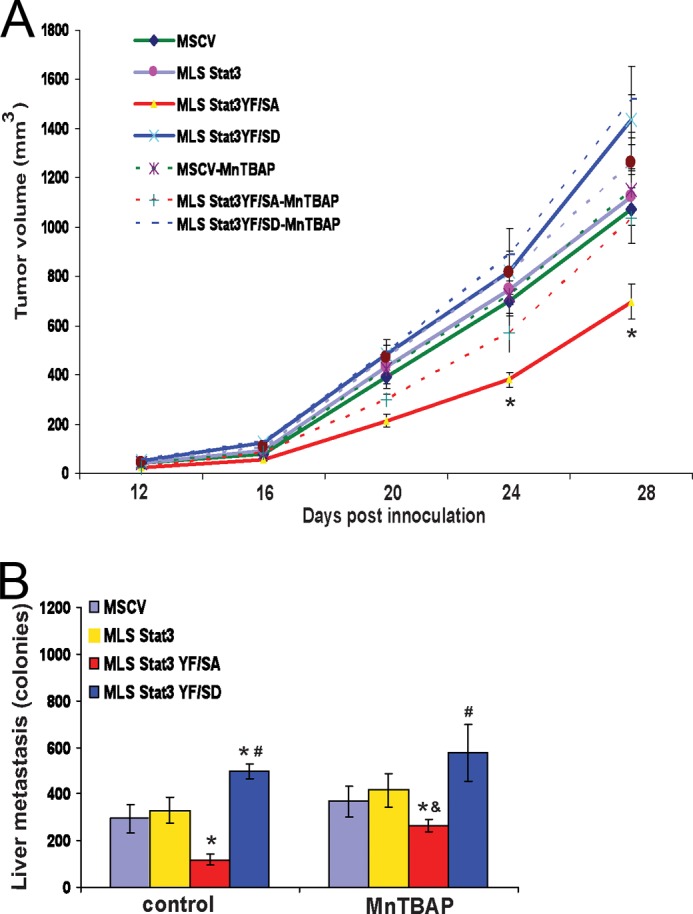
Treatment of mice with the antioxidant MnTBAP selectively increases tumorigenesis of cells expressing MLS-Stat3Y705F/S727A. Mice were inoculated with cells expressing various forms of MLS-Stat3. 12 days post inoculation, the mice were given daily injections of MnTBAP. A, tumor volume was measured by calipers. *, p < 0.05 versus control MLS-Stat3YF/SA-MnTBAP (two-way ANOVA on any given day of the analysis). B, after 28 days, mice were sacrificed, and the livers were assayed for metastatic colonies. *, p < 0.05 versus MLS-Stat3; #, p < 0.05 versus MLS-Stat3 YF/SA; &, p < 0.05 versus control MLS-Stat3 YF/SA (two-way ANOVA).
DISCUSSION
These studies were undertaken to examine the role of mitochondrial localized Stat3 in the pathogenesis of breast cancer. Previous reports have indicated that Stat3 in the mitochondria controls respiration and can restore the ability of Ras to transform Stat3−/− MEFs (1, 2). Serine 727 is required for both the actions of Stat3 on the ETC as well as Ras transformation. These studies address the role of mitoStat3 on the growth of cells that are already transformed. Constitutive serine phosphorylation of Stat3 has been observed in many human tumors and can be present in the absence of the protein being tyrosine-phosphorylated (11). Results from the laboratory of Farrar et al. (12) implicated serine 727 as being important for prostate cancer growth and recent studies implicate mitoStat3 in the growth of pancreatic cancer (9). We chose to examine the effects of mitoStat3 in 4T1 cells because expression of Stat3 is required for these cells to form tumors in immunocompetent mice (22). 4T1 cells mimic the effects of human disease in that morbidity is due to micrometastatic tumor cells that migrate to liver, brain, lung, etc. early during the growth of the primary tumor. However, human disease is multifaceted, and 4T1 cells by no means reflect the pathogenesis of all human breast tumors. Our studies indicate that mitoStat3 influences the ability of 4T1 cells to grow and invade both in vitro and in vivo. The observation that cells expressing various forms of MLS-Stat3 show similar activation of C/EBPδ by OSM indicate that mitoStat3 promotion of tumor growth and metastasis is independent of its actions as a nuclear transcription factor. The fact that the nuclear localization sequence, the DNA binding domain, the SH2 domain, and tyrosine 705 are not required for the actions of MLS-Stat3 is consistent with the hypothesis that MLS-Stat3 can influence cell growth independent of its functions in the nucleus. The mechanisms by which serine 727 in Stat3 enhances its oncogenic properties are speculative, but our results suggest that there is a strong connection between ROS accumulation and the ability of cells to grow in vitro and in vivo. ROS can play a dual in response to changes in homeostasis and cancer progression. However, it can function as a signaling molecule stimulating cell proliferation. Alternatively, when ROS concentrations reach a certain threshold they can cause release of cytochrome c and cell death. A major determinant of ROS production is the redox state of the ETC. Phosphorylation of Ser-727 of Stat3 enhances coupling of the complex I, resulting in decreased production of ROS. The actions of mitoStat3 are seen in 4T1 cells under conditions of hypoxia, not under conditions of normoxia. These results are consistent with our studies using mice that express an MLS-Stat3 transgene in the heart. Although base-line levels of ROS were the same in wild type and transgenic hearts, the increases in ROS were observed in wild type but not transgenic hearts subjected to ischemia (7). We speculate that mitoStat3 functions as a rheostat to optimize ROS concentrations to levels that enhance proliferation as opposed to increased apoptosis.
Furthermore, MnTBAP increases the growth of MLS-Stat3(Y705F/S727A) 4T1 cells in mice (Fig. 5). These studies are consistent with a mechanism of tumorigenesis where the amount of ROS released from complex I is minimized by serine phosphorylated mitoStat3. Decreased production of ROS leads to decreased apoptosis and increased tumor cell growth.
Although these studies indicate that mitoStat3 influences the tumorigenic potential of 4T1 cells, they do not address the role of the effects of Stat3 as a nuclear transcription factor on tumor growth and metastasis. It is likely that the actions of Stat3 in the nucleus are essential for 4T1 cell growth and the effects of mitoStat3 work in concert with nuclear Stat3.
Previous studies in heart suggest that mitoStat3 may be exerting its effect by interaction with the iron sulfur clusters in the distal region of complex I (7). It has been reported that Stat3 can be oxidized by peroxide to form multimers (35, 36). This has led to speculation that when ROS is elevated in the mitochondria secondary to decreased coupling efficiency of complex I, oxidation of cysteines in Stat3 could serve as an electron sink resulting in diminished ROS production and enhanced complex I and complex II activities. Reduction of oxidized Stat3 might either occur in the mitochondria or multimerized Stat3 might be exported from the mitochondria to be reduced in the cytosol by thioredoxins. Consistent with this latter possibility, when hearts are subject to ischemia, or 4T1 cells are incubated with H2O2, there is increased accumulation of Stat3 in the mitochondria, which could serve as a mechanism to reduce ROS accumulation, protecting cells from apoptosis and favoring cell survival (7). In the case of cancer, decreased ROS would favor cell proliferation.
It remains to be determined what the Stat3 effectors are that allow it to influence ROS production. One possibility is GRIM-19, which has been shown to bind Stat3 (37, 38). We speculate that phosphorylated Ser-727 of mitoStat3 favors optimal activity of complex I, resulting in less production of ROS, decreased release of cytochrome c from the inner membrane and/or decreased apoptosis, and a net increase in cell growth. Expression of GRIM-19 potentiates ROS production (39). Phosphorylation of Ser-727 facilitates the binding of mitoStat3 to GRIM-19, which serves to dampen the ability of GRIM-19 to produce ROS, resulting in the positive growth actions of mitoStat3. If the effects of dampening of GRIM-19 are diminished either by naturally occurring mutations in GRIM-19, which have been documented in thyroid cancers (40), or by other unknown mechanisms, then the growth promoting actions of serine phosphorylated mitoStat3 become more dominant and result in enhanced tumorigenesis. Future studies will address these possibilities.
This work was supported in part by National Institutes of Health Grants R01 1GM101677 and R21 AI088487 (to A. C. L.).
- ETC
- electron transport chain
- MLS
- mitochondrial localization sequence
- ROS
- reactive oxygen species
- DBD
- DNA binding domain
- MnTBAP
- Mn(III)tetrakis(4-benzoic acid)porphyrin chloride
- ANOVA
- analysis of variance
- OSM
- oncostatin M
- NCR
- NADH cytochrome c reductase
- SCR
- succinate cytochrome c reductase
- MEF
- mouse embryo fibroblast
- MSCV
- murine stem cell virus.
REFERENCES
- 1. Gough D. J., Corlett A., Schlessinger K., Wegrzyn J., Larner A. C., Levy D. E. (2009) Mitochondrial Stat3 supports Ras-dependent cellular transformation. Science 324, 1713–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., Moh A., Moghaddas S., Chen Q., Bobbili S., Cichy J., Dulak J., Baker D. P., Wolfman A., Stuehr D., Hassan M. O., Fu X. Y., Avadhani N., Drake J. I., Fawcett P., Lesnefsky E. J., Larner A. C. (2009) Function of mitochondrial Stat3 in cellular respiration. Science 323, 793–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Szczepanek K., Lesnefsky E. J., Larner A. C. (2012) Multi-tasking: Nuclear transcription factors with novel roles in the mitochondria. Trends Cell Biol. 22, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haghikia A., Stapel B., Hoch M., Hilfiker-Kleiner D. (2011) STAT3 and cardiac remodeling. Heart Fail. Rev. 16, 35–47 [DOI] [PubMed] [Google Scholar]
- 5. Heusch G., Musiolik J., Gedik N., Skyschally A. (2011) Mitochondrial STAT3 activation and cardioprotection by ischemic postconditioning in pigs with regional myocardial ischemia/reperfusion. Circ. Res. 109, 1302–1308 [DOI] [PubMed] [Google Scholar]
- 6. Sarafian T. A., Montes C., Imura T., Qi J., Coppola G., Geschwind D. H., Sofroniew M. V. (2010) Disruption of astrocyte STAT3 signaling decreases mitochondrial function and increases oxidative stress in vitro. PLoS One 5, e9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Szczepanek K., Chen Q., Derecka M., Salloum F. N., Zhang Q., Szelag M., Cichy J., Kukreja R. C., Dulak J., Lesnefsky E. J., Larner A. C. (2011) Mitochondrial-targeted signal transducer and activator of transcription 3 (STAT3) protects against ischemia-induced changes in the electron transport chain and the generation of reactive oxygen species. J. Biol. Chem. 286, 29610–29620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou L., Too H. P. (2011) Mitochondrial localized STAT3 is involved in NGF induced neurite outgrowth. PLoS One 6, e21680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackenzie G. G., Huang L., Alston N., Ouyang N., Vrankova K., Mattheolabakis G., Constantinides P. P., Rigas B. (2013) Targeting mitochondrial STAT3 with the novel phospho-valproic acid (MDC-1112) inhibits pancreatic cancer growth in mice. PLoS One 8, e61532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frank D. A. (2007) STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 251, 199–210 [DOI] [PubMed] [Google Scholar]
- 11. Frank D. A., Mahajan S., Ritz J. (1997) B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J. Clin. Invest. 100, 3140–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qin H. R., Kim H. J., Kim J. Y., Hurt E. M., Klarmann G. J., Kawasaki B. T., Duhagon Serrat M. A., Farrar W. L. (2008) Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation. Cancer Res. 68, 7736–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yeh Y. T., Ou-Yang F., Chen I. F., Yang S. F., Wang Y. Y., Chuang H. Y., Su J. H., Hou M. F., Yuan S. S. (2006) STAT3 ser727 phosphorylation and its association with negative estrogen receptor status in breast infiltrating ductal carcinoma. Int. J. Cancer 118, 2943–2947 [DOI] [PubMed] [Google Scholar]
- 14. Horvath C. M., Wen Z., Darnell J. E., Jr. (1995) A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding Domain. Genes Dev. 9, 984–994 [DOI] [PubMed] [Google Scholar]
- 15. Fu X. Y., Zhang J. J. (1993) Transcription factor p91 interacts with the epidermal growth factor receptor and mediates activation of the c-fos gene promoter. Cell 74, 1135–1145 [DOI] [PubMed] [Google Scholar]
- 16. Liu L., McBride K. M., Reich N. C. (2005) STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-α3. Proc. Natl. Acad. Sci. U.S.A. 102, 8150–8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McBride K. M., Banninger G., McDonald C., Reich N. C. (2002) Regulated nuclear import of STAT1 transcription factor by direct binding of importin-α. EMBO J. 21, 1754–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yakovlev V. A., Barani I. J., Rabender C. S., Black S. M., Leach J. K., Graves P. R., Kellogg G. E., Mikkelsen R. B. (2007) Tyrosine nitration of IκBα: a novel mechanism for NF-κB activation. Biochemistry 46, 11671–11683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bulua A. C., Simon A., Maddipati R., Pelletier M., Park H., Kim K. Y., Sack M. N., Kastner D. L., Siegel R. M. (2011) Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J. Exp. Med. 208, 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mukhopadhyay P., Rajesh M., Yoshihiro K., Haskó G., Pacher P. (2007) Simple quantitative detection of mitochondrial superoxide production in live cells. Biochem. Biophys. Res. Commun. 358, 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pulaski B. A., Ostrand-Rosenberg S. (2001) Mouse 4T1 breast tumor model. Curr. Protoc. Immunol., 10.1002/0471142735.im2002s39 [DOI] [PubMed] [Google Scholar]
- 22. Ling X., Arlinghaus R. B. (2005) Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 65, 2532–2536 [DOI] [PubMed] [Google Scholar]
- 23. Zhang F., Li C., Halfter H., Liu J. (2003) Delineating an oncostatin M-activated STAT3 signaling pathway that coordinates the expression of genes involved in cell cycle regulation and extracellular matrix deposition of MCF-7 cells. Oncogene 22, 894–905 [DOI] [PubMed] [Google Scholar]
- 24. Yang J., Liao X., Agarwal M. K., Barnes L., Auron P. E., Stark G. R. (2007) Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFκB. Genes Dev. 21, 1396–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J., Stark G. R. (2008) Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 26. Chen Q., Camara A. K., Stowe D. F., Hoppel C. L., Lesnefsky E. J. (2007) Modulation of electron transport protects mitochondria and decreases myocardial injury during ischemia and reperfusion. Am. J. Physiol. Cell Physiol. 292, C137–147 [DOI] [PubMed] [Google Scholar]
- 27. Simon A. R., Rai U., Fanburg B. L., Cochran B. H. (1998) Activation of the JAK-STAT pathway by reactive oxygen species. Am. J. Physiol. Cell Physiol. 275, C1640–1652 [DOI] [PubMed] [Google Scholar]
- 28. Waris G., Huh K. W., Siddiqui A. (2001) Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-κB via oxidative stress. Mol. Cell Biol. 21, 7721–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hilfiker-Kleiner D., Kaminski K., Podewski E., Bonda T., Schaefer A., Sliwa K., Forster O., Quint A., Landmesser U., Doerries C., Luchtefeld M., Poli V., Schneider M. D., Balligand J. L., Desjardins F., Ansari A., Struman I., Nguyen N. Q., Zschemisch N. H., Klein G., Heusch G., Schulz R., Hilfiker A., Drexler H. (2007) A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell 128, 589–600 [DOI] [PubMed] [Google Scholar]
- 30. Sharma L. K., Fang H., Liu J., Vartak R., Deng J., Bai Y. (2011) Mitochondrial respiratory complex I dysfunction promotes tumorigenesis through ROS alteration and AKT activation. Hum. Mol. Genet. 20, 4605–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sharma L. K., Lu J., Bai Y. (2009) Mitochondrial respiratory complex I: structure, function and implication in human diseases. Curr. Med. Chem. 16, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pizzimenti S., Toaldo C., Pettazzoni P., Dianzani M. U., Barrera G. (2010) The “two-faced” effects of reactive oxygen species and the lipid peroxidation product 4-hydroxynonenal in the hallmarks of cancer. Cancers 2, 338–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yakovlev V. A., Bayden A. S., Graves P. R., Kellogg G. E., Mikkelsen R. B. (2010) Nitration of the tumor suppressor protein p53 at tyrosine 327 promotes p53 oligomerization and activation. Biochemistry 49, 5331–5339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kushnareva Y., Murphy A. N., Andreyev A. (2002) Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 368, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L., Cheung S. H., Evans E. L., Shaw P. E. (2010) Modulation of gene expression and tumor cell growth by redox modification of STAT3. Cancer Res. 70, 8222–8232 [DOI] [PubMed] [Google Scholar]
- 36. Shaw P. E. (2010) Could STAT3 provide a link between respiration and cell cycle progression? Cell Cycle 9, 4294–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lufei C., Ma J., Huang G., Zhang T., Novotny-Diermayr V., Ong C. T., Cao X. (2003) GRIM-19, a death-regulatory gene product, suppresses Stat3 activity via functional interaction. EMBO J. 22, 1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J., Yang J., Roy S. K., Tininini S., Hu J., Bromberg J. F., Poli V., Stark G. R., Kalvakolanu D. V. (2003) The cell death regulator GRIM-19 is an inhibitor of signal transducer and activator of transcription 3. Proc. Natl. Acad. Sci. U.S.A. 100, 9342–9347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang G., Chen Y., Lu H., Cao X. (2007) Coupling mitochondrial respiratory chain to cell death: an essential role of mitochondrial complex I in the interferon-β and retinoic acid-induced cancer cell death. Cell Death Differ. 14, 327–337 [DOI] [PubMed] [Google Scholar]
- 40. Máximo V., Botelho T., Capela J., Soares P., Lima J., Taveira A., Amaro T., Barbosa A. P., Preto A., Harach H. R., Williams D., Sobrinho-Simões M. (2005) Somatic and germline mutation in GRIM-19, a dual function gene involved in mitochondrial metabolism and cell death, is linked to mitochondrion-rich (Hurthle cell) tumours of the thyroid. Br. J. Cancer 92, 1892–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]