Background: The NEDD8 protein and neddylation levels in cells are modulated by NUB1L.
Results: NUB1L down-regulated the NEDD8 levels by directly interacting with NEDD8 and P97.
Conclusion: NUB1L joins to the P97UFD1/NPL4 complex and promotes transfer of NEDD8 to proteasome for degradation.
Significance: This study provides a molecular mechanism for modulation of neddylation.
Keywords: Proteasome, Protein Degradation, Protein Motifs, Protein-Protein Interactions, Ubiquitination, NEDD8, NUB1L, P97/VCp Complex, Cullin 1, Neddylation
Abstract
The NEDD8 protein and neddylation levels in cells are modulated by NUB1L or NUB1 through proteasomal degradation, but the underlying molecular mechanism is not well understood. Here, we report that NUB1L down-regulated the protein levels of NEDD8 and neddylation through specifically recognizing NEDD8 and P97/VCP. NUB1L directly interacted with NEDD8, but not with ubiquitin, on the key residue Asn-51 of NEDD8 and with P97/VCP on its positively charged VCP binding motif. In coordination with the P97-UFD1-NPL4 complex (P97UFD1/NPL4), NUB1L promotes transfer of NEDD8 to proteasome for degradation. This mechanism is also exemplified by the canonical neddylation of cullin 1 for SCF (SKP1-cullin1-F-box) ubiquitin E3 ligases that is exquisitely regulated by the turnover of NEDD8.
Introduction
NEDD8 (neural precursor cell expressed developmentally down-regulated 8, Rub in Saccharomyces cerevisiae) is a ubiquitin (Ub)2-like protein sharing almost 60% identity and 80% homology with Ub (1, 2). Analogous to Ub, mature NEDD8 can attach to its substrates by an isopeptide linkage between its C-terminal glycine and a lysine of the substrate, as called neddylation. The pathway is catalyzed by APPBP1-UBA3 heterodimer (NEDD8-activating, E1) (3–5), UBC12, or UbE2F (NEDD8-conjugating, E2) (3, 6) and various NEDD8 E3 ligases. The neddylation of certain substrates are carried out by particular NEDD8 ligases. For example, Rbx1 and Dcn1 are responsible for the neddylation of cullin family proteins, which are the best characterized NEDD8 targets. Cullins are scaffold proteins of cullin-RING ligases that comprise the largest subfamily of Ub E3 ligases (7, 8). Neddylation of cullins stimulates the activities of cullin-RING ligases by introducing conformational changes and prohibiting their binding with the inhibitor CAND1 (9–11). By controlling the turnover of a large number of key regulators, cullin-RING ligases are contributable to many biological processes including cell cycle progression, cell signaling, transcription, and so on (7, 12, 13).
Similar to ubiquitination, protein neddylation can also be reversed by NEDD8 isopeptidases, called deneddylation, which is carried out by COP9 signalosome (CSN) and other deneddylating enzymes (14–18). Besides deneddylation, the protein levels of NEDD8 and neddylated substrates are also negatively regulated by some cellular factors. Interferon-inducible proteins NUB1L (NEDD8 ultimate buster-1 long) and its splicing variant NUB1 can interact with NEDD8 and promote the proteasomal degradation of NEDD8 and its protein conjugates (19, 20). Although NUB1L contains a 14-residue insertion in the NUB1 sequence, their biologically functional specificities are currently unknown. In addition, NUB1L can also facilitate the proteasomal degradation of FAT10, another Ub-like protein (21, 22). As NUB1L and NUB1 have been identified to interact with the Rpn10/S5a and Rpn1 subunits of proteasome, they are postulated to recruit NEDD8 for proteasomal degradation (23–25). Probably linking with the protein level of NEDD8 and the activity of SCF (SKP1-cullin1-F-box) protein (12, 26), NUB1 plays a role in modulation of anti-proliferation actions through affecting the levels of p27 and cyclin E (27). NUB1 also interacts with AIPL1 (aryl hydrocarbon receptor-interacting protein-like 1), a protein whose mutation causes a severe inherited blindness, Leber congenital amaurosis (28, 29). AIPL1 modulates the cellular localization of NUB1 and suppresses the inclusion formation by NUB1 fragments, indicating that NUB1 may be related to the pathogenesis of Leber congenital amaurosis (30, 31). Moreover, NUB1 can also reduce the transcriptional activity of p53 by inhibiting its neddylation and promoting cytoplasmic localization (32). NUB1 has also been found in the formation of inclusion bodies though regulating synphilin-1 and glycogen synthase kinase 3β (33, 34), suggesting that NUB1 is probably related to neurodegenerative diseases (35–38).
Although accumulating studies have shown that NUB1L/NUB1 can promote proteasomal degradation of NEDD8 and its conjugates (19, 20, 39), the underlying molecular mechanism is still obscure. In the Ub-proteasome system, a ternary complex comprised of P97/VCP, UFD1, and NPL4 (P97UFD1/NPL4) plays a key role in recruiting and delivering ubiquitinated substrates to the proteasome (40–43). P97/VCP (valosin-containing protein, Cdc48 in yeast) is a member of the AAA-ATPase family (44), implicated in a variety of cellular processes including Ub-proteasome related degradation, membrane fusion, autophagy, and so on (40, 43, 45, 46). Normally, NPL4 binds with Ub, and then P97UFD1/NPL4 delivers ubiquitinated proteins to the proteasome for degradation (47–52). During ATP hydrolysis, P97 undergoes significant conformational changes that may affect its interaction with cofactors and clients (53–55). As for NEDD8 and neddylation, whether P97UFD1/NPL4 is involved and how NUB1L/NUB1 functions in this pathway remain to be explored.
We investigated NUB1L/NUB1 in neddylation and elucidated the molecular mechanism by which NUB1L promotes transfer of NEDD8 for proteasomal degradation. NUB1L specifically recognizes NEDD8 and interacts with P97; it joins to the P97UFD1/NPL4 machinery and mediates NEDD8 delivery. We propose that P97UFD1/NPL4 recruits NEDD8 to NUB1L during ATP hydrolysis and ultimately transfers it to the proteasome for degradation.
EXPERIMENTAL PROCEDURES
Plasmids, Reagents, and Antibodies
All plasmids were constructed using standard molecular biology techniques. For HA-tagged proteins, the cDNAs coding for NEDD8, Ub, and NUB1(L) were subcloned into HA-pcDNA3.0 (Invitrogen). For FLAG-tagged proteins, the cDNAs coding for NUB1L and P97 were subcloned into pCMV-tag 2B (Agilent). The pEGFP-N1 vector was used to express enhanced GFP in HEK 293T cells. For GST-tagged proteins, the cDNAs coding for NEDD8, Ub, NUB1(L)-VBM-(414–443), NUB1-UBAs-(373–520), and NUB1L-UBAs-(373–534) were subcloned into pGEX-4T3 (GE Healthcare). The cDNAs for P97-ND1-(1–458) and NUB1L were subcloned into pET-22b (Novagen). For His-tagged full-length P97 and NPL4, their cDNAs were subcloned into pET-28a (Novagen). For Trx-tagged proteins, the cDNAs coding for UFD1 and the NUB1L fragments (including 36–615, 82–615, 148–615, 211–615, and 373–615) were subcloned into pET-32 M (Novagen). For GB1-tagged NUB1L-UBA2 (414–472) and NUB1L-PEST (545–615), the cDNAs were subcloned into pHGB (56), whereas for C-terminal GB1-tagged NUB1L-(1–372), the cDNA was subcloned into pET-GB1. Antibodies against HA and FLAG were from Sigma. Anti-GFP, anti-actin, and anti-NUB1L were from Santa Cruz. Antibodies against NEDD8 and P97 were from Cell Signaling. Anti-cullin1 was from Bioworld. MG132 was from Calbiochem, whereas ATP, ADP, and ATPγS were from Sigma.
Cell Culture, Transfection, Western Blotting, Immunoprecipitation, and Immunofluorescence Microscopy
HEK 293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and penicillin-streptomycin at 37 °C in 5% CO2. Transient transfection of cells with expression vectors were carried out using PolyJetTM reagent (SignaGen) according to the manufacturer's instructions. Cells were harvested and lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, mixture protease inhibitor (Roche Applied Science)). The lysates were subjected to SDS-PAGE with 8, 12, or 15% acrylamide gels and transferred onto PVDF membranes (PerkinElmer Life Sciences). The indicated proteins were probed with the respective primary and secondary antibodies and visualized by using an ECL detection kit (Thermoscientific). For immunoprecipitation, cell lysates were incubated with HA-antibody-conjugated agarose for 1 h at 4 °C. The beads were washed with radioimmune precipitation assay buffer three times and subjected to immunoblotting analysis. For immunofluorescence, cells were grown on glass coverslips for 24 h after transfection. Images were obtained on a Leica TCS SP2 confocal microscope (Leica Microsystems).
Protein Expression and Purification
Proteins were expressed in Escherichia coli BL21 (DE3). The His-tagged proteins were purified by Ni2+-nitrilotriacetic acid columns (Qiagen), and the GST-fused proteins were purified using glutathione Sepharose-4B columns (Amersham Biosciences). They were further purified by gel filtration chromatography with respective columns (GE Healthcare).
GST Pulldown Experiment
The pulldown experiments for GST-UFD1, NPL4, and P97 were carried out in a Tris-HCl buffer (50 mm Tris-HCl, pH 8.0, 5 mm MgCl2, 150 mm KCl, 1 mm DTT, 5% glycerol). The other pulldown experiments were carried out in a PBS buffer (10 mm Na2HPO4, 140 mm NaCl, 2.7 mm KCl, 1.8 mm KH2PO4, pH 7.3). GST and GST-fused proteins were incubated with glutathione Sepharose-4B beads at 4 °C for 0.5 h. Other proteins were then incubated with immobilized GST or GST fusion proteins at 4 °C for 1 h. The beads were collected by centrifugation and washed 3 times, then eluted by a GSH buffer (50 mm Tris-HCl, 10 mm GSH, pH 8.0).
siRNA Knockdown Experiment
The siRNA target sequence is 5′-CGAUGGUGCUUGAACUAAA-3′ for NUB1L, 5′-AAGUAGGGUAUGAUGACAUUG-3′ for P97, and 5′-UUCUCCGAACGUGUCACGU-3′ for the siRNA control sequence. Cells were transfected with the duplex siRNA using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
NMR Titration Experiment
The chemical-shift assignments for GB1-NUB1L-VBM (residues 414–443) were obtained by triple resonance NMR experiments with a 13C,15N-double-labeled sample. 15N-labeled GB1-NUB1L-VBM was expressed in M9 minimal medium containing 15NH4Cl as the sole nitrogen resource. A sample of GB1-NUB1L-VBM (100 μm) was dissolved in a buffer containing 20 mm phosphate, 50 mm NaCl, pH 6.5. All spectra were recorded at 25 °C on a 600-MHz NMR spectrometer (Bruker). P97-N213 (residues 1–213) was titrated to GB1-NUB1L-VBM at different molar ratios, and the 1H,15N HSQC spectra were acquired to monitor the chemical-shift changes upon titration. The NMR titration of 15N-labeled NEDD8 with GB1-UBA2 or GB1-PEST followed this procedure (57).
RESULTS
NUB1L Down-regulates the Protein Levels of NEDD8 and Neddylation
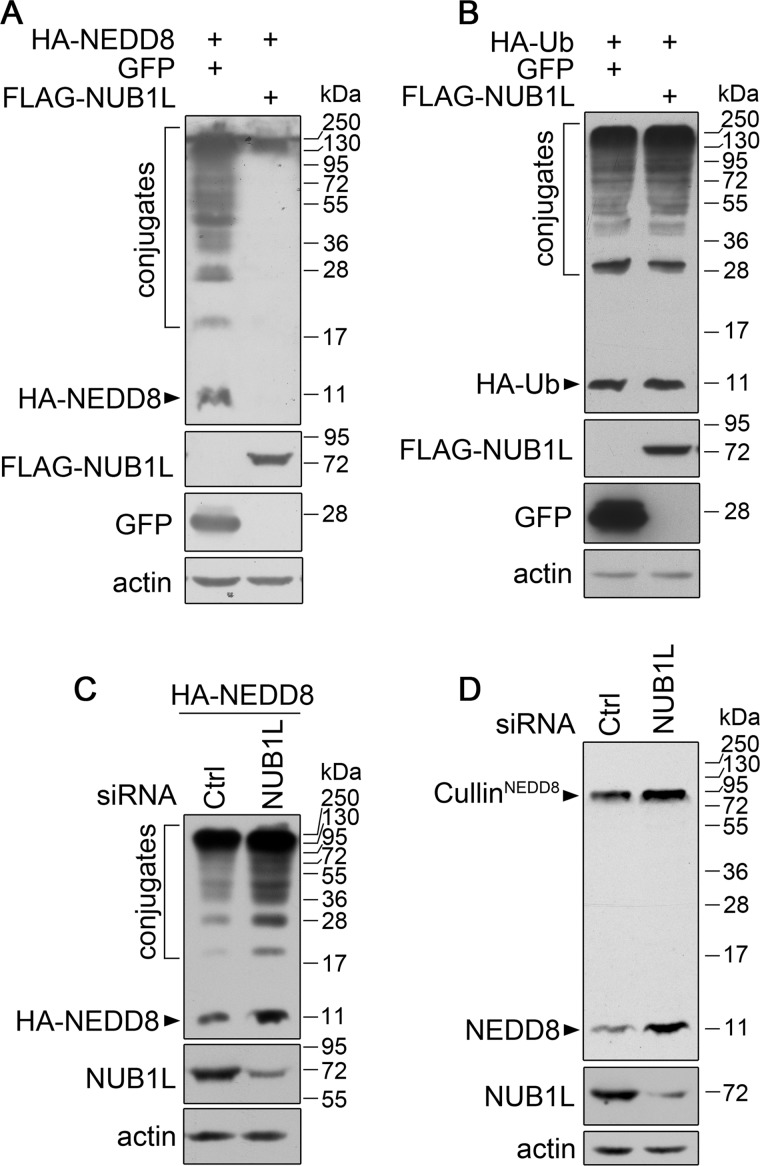
It was previously reported that NUB1L was a negative regulator of NEDD8 (39). In accordance with those previous studies, we found that, compared with the negative control GFP, NUB1L specifically down-regulated the protein levels of overexpressed NEDD8 and its protein conjugates (Fig. 1A). However it had no effect on Ub or ubiquitinated proteins (Fig. 1B). Knockdown of NUB1L (and NUB1) by siRNA caused increases of both overexpressed (Fig. 1C) and endogenous NEDD8 and its conjugates (Fig. 1D). Interestingly, the endogenous level of neddylated cullins was also significantly enhanced by reducing NUB1L. These results indicate that NUB1L negatively regulates the protein levels of NEDD8 and its conjugates.
FIGURE 1.
NUB1L down-regulates the protein levels of NEDD8 and its conjugates. A and B, effect of NUB1L overexpression on the protein level of NEDD8 (A) or Ub (B). FLAG-tagged NUB1L was co-transfected with HA-NEDD8 or HA-Ub into HEK 293T cells (GFP as a control). About 24 h after transfection, the cell lysates were subjected to immunoblotting with respective antibodies. C and D, effect of NUB1L knockdown on the protein level of NEDD8. HEK 293T cells were transfected with siRNA against NUB1L, then the protein level of overexpressed (C) or endogenous NEDD8 (D) was analyzed by Western blotting. The neddylated cullins are indicated in the gel as instructions from the manufacturer supplying this antibody against NEDD8.
NUB1L Specifically Interacts with NEDD8 for Regulating Its Protein Level
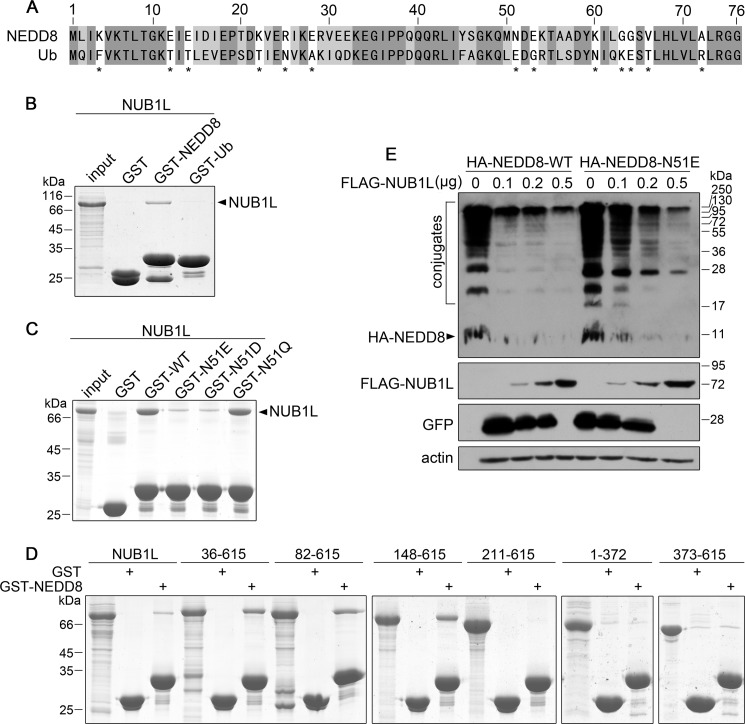
To understand how NUB1L distinguishes NEDD8 from Ub, we studied their interactions by GST pulldown. Although NEDD8 has the highest homology with Ub among the Ub-like proteins (Fig. 2A), NUB1L only interacted with NEDD8 but not with Ub (Fig. 2B). In comparison of the sequences, there are 13 residues significantly different between NEDD8 and Ub (Fig. 2A). We separately replaced these residues in NEDD8 with the corresponding ones in Ub and studied their interactions with NUB1L (Fig. 2C). The result showed that N51E mutation severely abolished the interaction, indicating that Asn-51 in NEDD8 played a critical role in binding with NUB1L. Because the N51E and N51D mutations, but not N51Q, could abolish this interaction (Fig. 2C), the negative charges might be involved in the breakdown of the interaction between NUB1L and NEDD8. However, the E51N mutant in Ub still could not obtain the ability to bind NUB1L (data not shown), possibly due to other residues responsible for this specific interaction. NUB1L is a multidomain protein and contains three UBA domains that potentially bind with Ub (21, 39). Previous studies suggested that NEDD8 bound NUB1L through the C-terminal PEST motif and the linker between UBA1 and UBA2 (39). We investigated whether these segments in NUB1L were responsible for NEDD8 binding by GST pulldown (Fig. 2D). It seems that a large portion of NUB1L (Ala-148–Asn-615) except for the N terminus contributes to binding with NEDD8, suggesting that the specific interaction requires the overall conformation of NUB1L.
FIGURE 2.
Asn-51 of NEDD8 determines its specific interaction with NUB1L. A, sequence alignment of NEDD8 and Ub. The 13 residues that are significantly different are marked with asterisks. B, GST pulldown showing NUB1L specifically interacts with NEDD8 but not with Ub. GST-tagged NEDD8 or Ub was applied to pull down NUB1L, whereas GST was used as a control. C, Asn-51 mutation influences its interactions with NUB1L. GST-tagged Asn-51 mutants of NEDD8 were applied to pull down NUB1L. D, GST pulldown examining the NEDD8 binding fragments of NUB1L. GST-fused NEDD8 was used to pull down different NUB1L fragments. The fragments include 36–615, 82–615, 148–615, 211–615, 373–615, and 1–372. Except for fragment 1–372 that has attached a GB1 tag in C terminus, all the fragments have an N-terminal Trx tag for pulldown experiments. E, effect of NUB1L on the protein level of wild-type NEDD8 or its N51E mutant in a dose-dependent manner. HA-tagged wild-type NEDD8 or its N51E mutant was co-transfected with different dose of NUB1L plasmid (GFP as a control), and then the protein amounts of NEDD8 or N51E mutant were detected by Western blotting.
We next performed co-transfection of FLAG-NUB1L and HA-NEDD8 or its N51E mutant in HEK 293T cells. The result showed that NUB1L significantly reduced the protein level of wild-type NEDD8 in a dose-dependent manner (Fig. 2E). However, NUB1L could not effectively down-regulate the N51E mutant of NEDD8. This demonstrates that the interaction between NUB1L and NEDD8 is responsible for the down-regulation of NEDD8 by NUB1L.
NUB1L and Its Splicing Variant NUB1 Interact with P97 through a VCP Binding Motif (VBM)
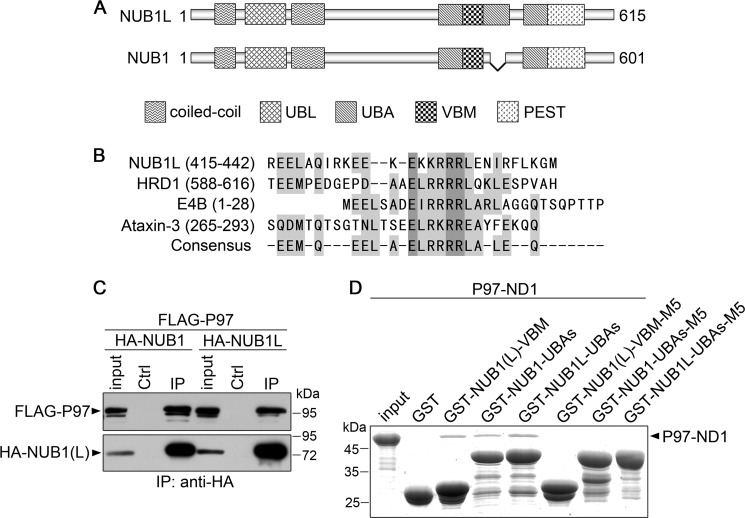
NUB1L as well as its splicing variant NUB1 is characterized by multidomains or motifs in its sequence, whereas NUB1 lacks a UBA domain (UBA2) due to 14 residues missing (Fig. 3A). From sequence analysis, a potential VBM resided in between two UBA domains was identified from both NUB1L and NUB1 (Fig. 3B). This motif shares high similarity with those known VBM motifs from HRD1 (58), E4B (59), and ataxin-3 (60). To get insights into the underlying mechanism that NUB1L down-regulates the protein levels of NEDD8 and its conjugates, we examined whether NUB1L or NUB1 interacted with P97. In the co-immunoprecipitation experiment, P97 could co-immunoprecipitate with NUB1L or NUB1 (Fig. 3C), indicating that P97 associated with NUB1L or NUB1. We then tested whether NUB1L/NUB1 directly interacted with P97 in vitro. The result showed that the GST-fused NUB1L/NUB1 fragments containing the VBM motif could pull down the ND1 domain of P97 (P97-ND1) (Fig. 3D, third through fifth lanes). NMR titration further confirmed that the VBM peptide of NUB1L physically interacted with the N-terminal domain of P97 (P97-N213) (Fig. 4). Because VBM is characterized by its core basic residues (Fig. 3B), we prepared three mutants of NUB1L/NUB1 (M5) by substituting the five positively charged residues (KKRRR) with non-charged residues (GGPGG) (see Fig. 6A). The result showed that all the three mutants appeared to have lost their binding abilities with P97-ND1 (Fig. 3D, sixth to eight lanes). It suggests that NUB1L/NUB1 interacts with P97 through its respective VBM motif and the core basic residues are important for this specific interaction.
FIGURE 3.
The VBM motif of NUB1L (or NUB1) is responsible for interacting with P97. A, domain architectures of NUB1L and NUB1. With insertion of 14 residues, NUB1L has three UBA domains, whereas NUB1 has only two; both of them contain a VBM motif in between UBA1 and UBA2. B, sequence alignment of some typical VBMs. The VBM motif of NUB1L is compared with those of three proteins, HRD1, E4B and ataxin-3. C, co-immunoprecipitation (IP) experiment for the interaction between NUB1L/NUB1 and P97. HA-tagged NUB1L or NUB1 and FLAG-tagged P97 were co-transfected into HEK 293T cells, and 48 h after transfection the cell lysates were subjected to immunoprecipitation by anti-HA antibody. D, GST pulldown showing the interactions between the VBM motif of various NUB1L/NUB1 and the ND1 domain of P97. GST-fused NUB1L/NUB1 fragments or their M5 mutants were applied to pull down P97-ND1. VBM, residues Arg-414–Gly-443; NUB1L-UBAs, residues Leu-373–Ser-532; NUB1-UBAs, residues Leu-373–Ser-518; M5, mutation of 428KKRRR432 to 428GGPGG432; P97-ND1, residues M1-Q458.
FIGURE 4.
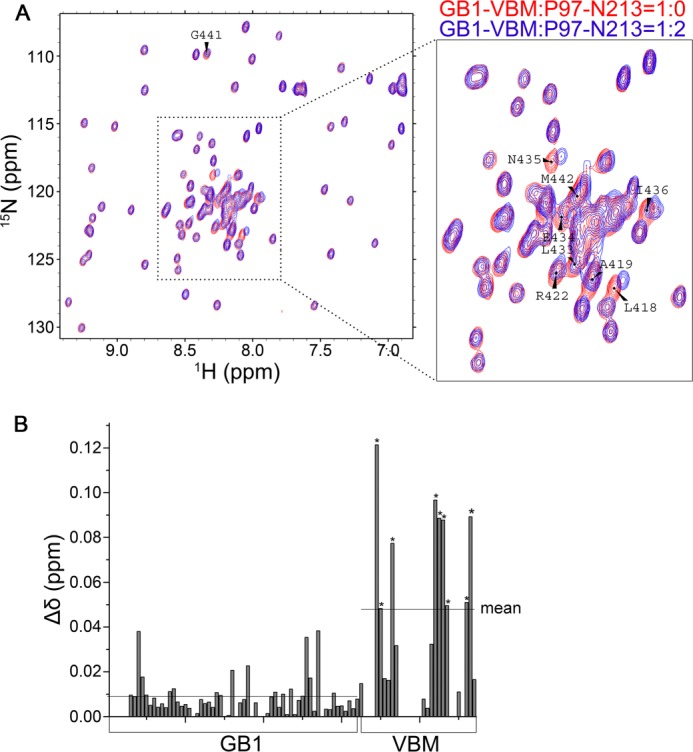
NMR titration showing the interaction between the VBM motif of NUB1L (or NUB1) and the N-terminal domain of P97. A, overlay of the HSQC spectra of 15N-labeled GB1-tagged NUB1L-VBM (100 μm) (red) and titration with P97-N213 at a molar ratio of 1: 2 (blue). VBM, residues 414–443 of NUB1L; P97-N213, residues 1–213 of P97. B, plot of the chemical-shift change against the residue number of GB1-tagged NUB1L-VBM. The mean chemical-shift change in GB1 or VBM is shown with a solid line. The asterisks stand for the residues (shown in A) with a significant chemical-shift change.
FIGURE 6.
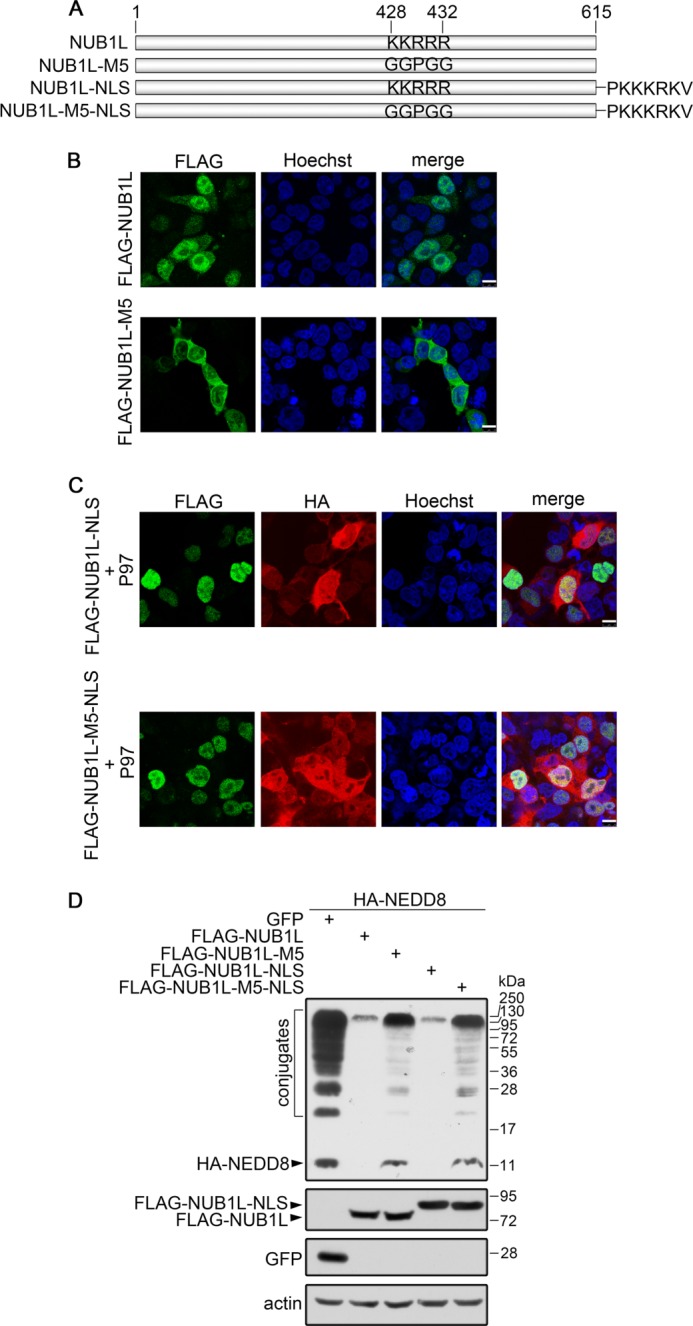
The cellular localization of NUB1L. A, diagram of the constructs for NUB1L and its mutants. NUB1L-NLS, attachment of the NLS sequence of SV40 large T-antigen (PKKKRKV) to the C terminus of NUB1L. B, immunofluorescence imaging of NUB1L and its M5 mutant. FLAG-tagged NUB1L or its M5 mutant was transfected into HEK 293T cells. NUB1L was stained with FLAG-antibody, whereas the nuclei were stained with Hoechst. Scale bar, 10 μm. C, co-localization of NUB1L-NLS with P97. FLAG-tagged NUB1L-NLS or its M5 mutant was co-transfected with HA-tagged P97 into HEK 293T cells. NUB1L or P97 was stained with FLAG- or HA-antibody, whereas the nuclei were stained with Hoechst. D, effect of NUB1L-NLS or its M5 mutant on the protein levels of NEDD8 and neddylation. HA-tagged NEDD8 and FLAG-tagged NUB1L, NUB1L-M5, NUB1L-NLS, or NUB1L-M5-NLS were co-transfected into HEK 293T cells. Around 24 h after transfection, the cell lysates were subjected to immunoblotting with respective antibodies.
NUB1L Promotes Proteasomal Degradation of NEDD8 by Interacting with P97
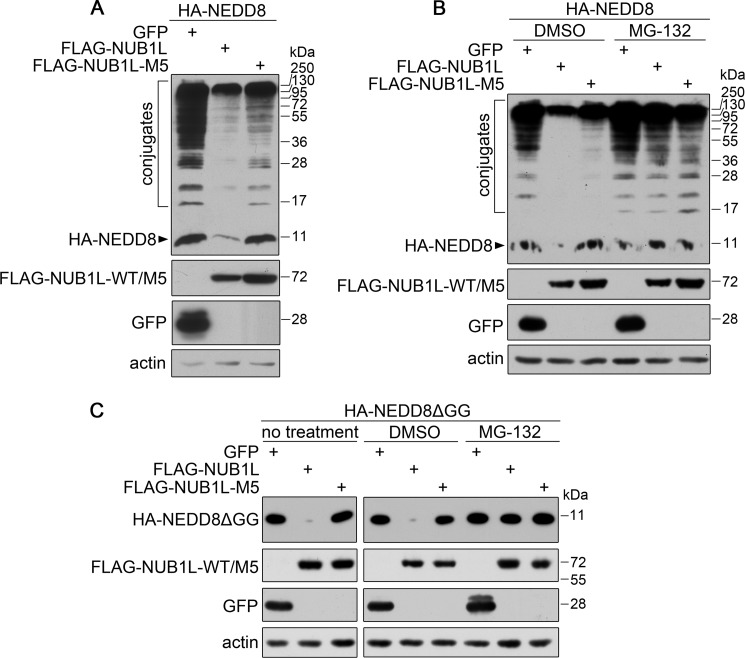
Because the multidomain protein NUB1L is able to bind NEDD8 and P97, we want to understand the biological significance of these interactions in cells. When co-expressed with NEDD8, NUB1L could efficiently reduce the amounts of NEDD8 and its conjugates, but its VBM mutant exhibited an alleviated effect (Fig. 5A). When cells were treated with MG132, a proteasome inhibitor, the action of NUB1L on the protein level of NEDD8 was significantly inhibited (Fig. 5B), as in the case of NUB1L mutant. This suggests that NUB1L promotes degradation of NEDD8 via the proteasome, and its interaction with P97 is required for this NEDD8 clearance pathway.
FIGURE 5.
NUB1L regulates the protein level of NEDD8 by interacting with P97. A, effect of NUB1L or its M5 mutant on the protein level of NEDD8. HA-NEDD8 was co-transfected with FLAG-tagged NUB1L or its M5 mutant into HEK 293T cells. The cell lysates were subjected to Western blotting using respective antibodies. B, as in A, in the presence of MG132. After transfection, the cells were treated with MG132 (DMSO as a control) for 12 h, and about 24 h after transfection, the cells were harvested and subjected to Western blotting. C, as in A and B, effect of NUB1L or its M5 mutant on the protein level of NEDD8ΔGG.
As shown in the previous experiments, both NEDD8 and neddylation levels are down-regulated in the presence of NUB1L. However, whether NUB1L can promote the degradation of free NEDD8 protein remains to be clarified. We prepared a C-terminal glycine-deleted mutant of NEDD8 (NEDD8ΔGG) that was unable to be conjugated to the substrate proteins. As in the case of wild-type NEDD8 (Fig. 5A), NUB1L could also reduce the amount of NEDD8ΔGG, whereas its M5 mutant exhibited no such effect (Fig. 5C, left panels). Similarly, MG132 treatment could abolish this effect (Fig. 5C, right panels). These data demonstrate that NUB1L can promote the proteasomal degradation of free NEDD8, and its interaction with P97 is essential to this action.
It was predicted that the VBM-motif region of NUB1L contains a nuclear localization signal (NLS) (20), mutation of which may cause mis-localization of the protein. To exclude the possibility that the loss of function of the NUB1L mutant is a result of its cellular mis-location, we examined the relationship between NUB1L localization and its effect on NEDD8 conjugates. Microscopy imaging showed that NUB1L localized predominantly in nucleus as well as distributed in cytoplasm, whereas its M5 mutant had a slight change in the localization distributed in the whole cell (Fig. 6B). This means that the mutation disrupting this potential NLS site alters the localization of NUB1L to a considerable extent. To overcome this difficulty, we fused an NLS sequence from large T-antigen of SV40 to the C terminus of NUB1L (Fig. 6A) to lead NUB1L and its mutant to localize in the nucleus (Fig. 6C). Still, both forms of wild-type NUB1L reduced the protein levels of NEDD8 and its conjugates, whereas the M5 mutant forms lost this ability (Fig. 6D) no matter whether the proteins were localized in nucleus or cytoplasm. It suggests that the region of NUB1L rich in basic residues is both a VBM motif for P97 binding and an NLS for nucleus localization. Thus, the loss of function of the M5 mutant is a result of disruption of the P97 binding but not originated from mis-localization of the protein.
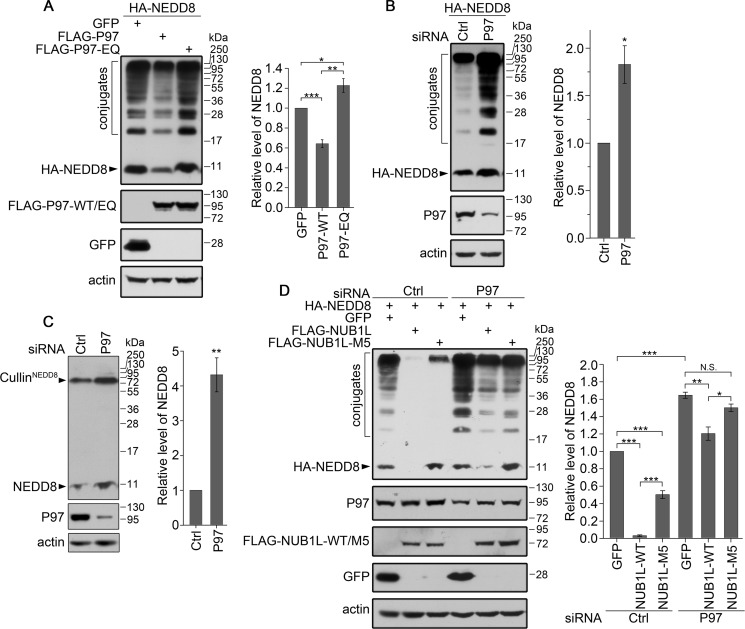
We then investigated whether P97 influenced the protein levels of NEDD8 and neddylation in cells. Co-expression of P97 caused a decrease of the overexpressed NEDD8 and its conjugates (Fig. 7A, second lane), whereas knockdown of P97 increased the amounts of both overexpressed NEDD8 and its conjugates (Fig. 7B). The amounts of endogenous NEDD8 and neddylated cullins were also raised through reducing P97 by siRNA (Fig. 7C). These data provide direct evidence that P97 is directly involved in the degradation of NEDD8 and its conjugates. Moreover, we examined whether the ATPase activity of P97 was required for this function. We replaced two active-site residues, Glu-305 and Glu-578, with Gln in the ATPase domains of P97 (P97-EQ) (61). Interestingly, P97-EQ significantly increased the amounts of NEDD8 and its conjugates (Fig. 7A, third lane). This is probably a result of the dominant-negative effect in which overexpression of the P97 mutant interferes with the normal function of endogenous P97 and consequently inhibits the degradation of NEDD8. Thus, the ATPase activity or ATP hydrolysis is required for P97 modulating the protein levels of NEDD8 and neddylation.
FIGURE 7.
P97 down-regulates the protein levels of NEDD8 and its conjugates. A, effect of P97 or its P97-EQ mutant on the protein level of overexpressed NEDD8. HA-NEDD8 was co-transfected with FLAG-tagged P97 or its P97-EQ mutant into HEK 293T cells. B and C, effect of P97 knockdown on the protein level of overexpressed (B) or endogenous NEDD8 (C). The P97 siRNA or its control was transfected into HEK 293T cells, and then its effect on the amount of overexpressed or endogenous NEDD8 was analyzed by Western blotting. D, effect of NUB1L and its M5 mutant on the protein level of NEDD8 under the circumstance of P97 silencing. The siRNA against P97 was transfected into HEK 293T cells, and 24 h later HA-NEDD8 was co-transfected with FLAG-tagged NUB1L or its M5 mutant. After another 24 h, the cells were collected and subjected to Western blotting. The protein levels of NEDD8 and its conjugates were quantitated and are presented as the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001; N.S., no significance.
To examine whether NUB1L coordinates with P97 in the turnover of NEDD8, we investigated the effect of NUB1L on the degradation of NEDD8 under the circumstance of P97 silencing. Similarly, knockdown of P97 caused the increases of NEDD8 and its conjugates (Fig. 7D, fourth lane). When P97 was down-regulated, NUB1L still had an ability to promote the degradation of NEDD8 but with a much less extent, whereas its M5 mutant lost this effect (Fig. 7D, fifth and sixth lanes). This indicates that NUB1L cooperates with P97 in regulating the degradation of NEDD8.
NUB1L Joins to the P97UFD1/NPL4 Complex
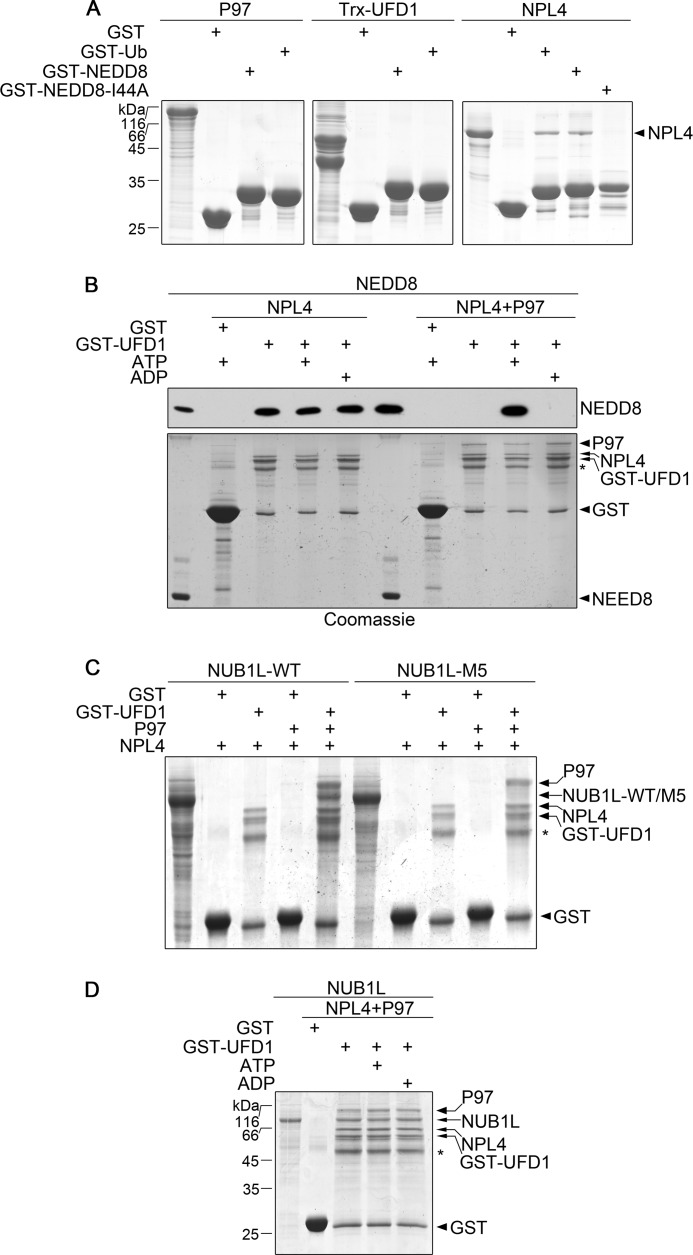
In the Ub-proteasome system, P97 is responsible for delivering ubiquitinated substrates by forming a tripartite complex with UFD1 and NPL4 (43, 62). Our data demonstrate that NUB1L promotes the proteasomal degradation of NEDD8 by interacting with P97. We wondered whether NUB1L coordinated with P97UFD1/NPL4 in the NEDD8-proteasome pathway. By using GST pulldown in vitro, we studied the interactions between P97UFD1/NPL4 and NEDD8. As in the case of Ub (50, 63), neither P97 nor UFD1 interacted with NEDD8 or Ub, but only NPL4 could bind NEDD8 as well as Ub (Fig. 8A). Moreover, the I44A mutant of NEDD8 did not interact with NPL4, indicating that the hydrophobic patch of NEDD8 centered with Ile-44 played a critical role in this NPL4 interaction. Without P97, UFD1/NPL4 could bind NEDD8 no matter whether ATP or ADP was present or not (Fig. 8B, third through fifth lanes). However, P97UFD1/NPL4 bound NEDD8 only in the presence of ATP, whereas it lost this ability when ADP was present (Fig. 8B, ninth and tenth lanes). This suggests that P97 regulates the binding of NPL4 with NEDD8 through conformational changes during ATP hydrolysis.
FIGURE 8.
NUB1L collaborates with P97UFD1/NPL4 for targeting NEDD8. A, examination of the interactions between the P97-complex components and NEDD8 or Ub. GST-tagged NEDD8 or Ub was used to pull down P97, Trx-UFD1, or NPL4. Trx-UFD1, a thioredoxin fusion of UFD1 for improving stability. B, interaction of P97UFD1/NPL4 with NEDD8 in the presence of ATP or ADP. GST-UFD1 was applied to pull down NEDD8 assisted by P97 and/or NPL4 in the presence of ATP or ADP, and NEDD8 was detected by Western blotting using an antibody against NEDD8. NEDD8 input (5% amount) was also loaded for control. The asterisk stands for a degradation band of GST-UFD1. C, NUB1L joins to the complex of P97UFD1/NPL4. GST-UFD1 was used to pull down NUB1L or its M5 mutant assisted by P97 and NPL4, and the proteins were detected by Coomassie Blue. The asterisk stands for a degradation band of GST-UFD1. D, as in C, in the presence ATP or ADP.
Because NUB1L interacts with P97 (Fig. 3D), we then investigated whether NUB1L was also involved in the P97UFD1/NPL4 complex. The result showed that P97UFD1/NPL4 could pull down NUB1L but not its M5 mutant (Fig. 8C, fifth and tenth lanes). This suggests that NUB1L joins to the P97UFD1/NPL4 complex by interacting with P97. Furthermore, formation of this complex was not affected by the nucleotide type (Fig. 8D). We propose that this quaternary complex might be the key element for NEDD8 transfer to proteasome.
NUB1L Coordinates P97UFD1/NPL4 to Down-regulate Neddylation of Cullin 1
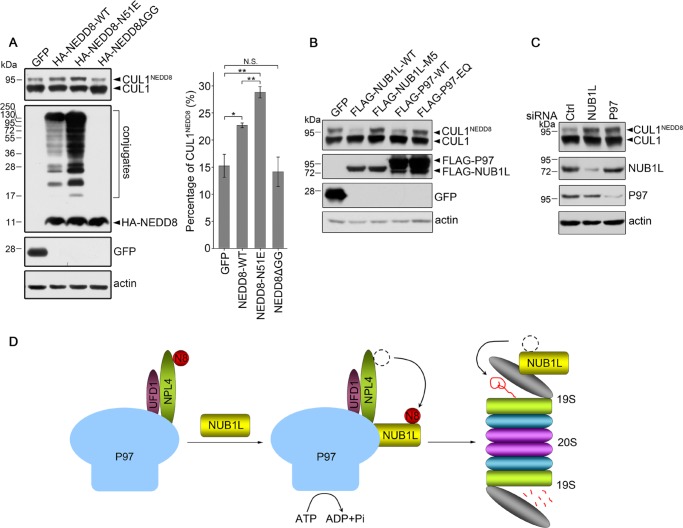
Our studies have revealed that NUB1L coordinates P97 functioning in the proteasomal degradation of NEDD8. However, whether the neddylated substrates are also delivered to the proteasome for degradation remains to be demonstrated. We employed cullin 1 (CUL1) as a model substrate to investigate the effect of NUB1L and P97 on its neddylation level (64, 65). By using an anti-CUL1 antibody, both free and neddylated CUL1 could be detected in a gel. As compared with wild-type NEDD8, overexpression of the N51E mutant significantly increased the neddylation of endogenous CUL1 (Fig. 9A). However, overexpression of the glycine-deleted mutant (NEDD8ΔGG) did not change the amount of CUL1NEDD8. This indicates that neddylation of CUL1 is probably influenced by interaction of NUB1L with NEDD8. Moreover, overexpression of wild-type NUB1L or P97 significantly decreased the neddylation level of CUL1 (Fig. 9B, second and fourth lanes), but it had no evident influence on the total protein level of CUL1. However, neither NUB1L-M5 nor P97-EQ mutants had any such effect (Fig. 9B, third and fifth lanes). On the other hand, knockdown of NUB1L or P97 significantly increased the amount of neddylated CUL1 (Fig. 9C). Together, these data demonstrate that both NUB1L and P97 are directly involved in regulating the neddylation level of CUL1 but not the total CUL1 protein.
FIGURE 9.
NUB1L or P97 suppresses the neddylation of CUL1. A, effect of NEDD8 and its mutants on the neddylation of CUL1. Transfection and Western blotting analysis were similar to that described in Fig. 1, A and B. Data were quantitated and presented as the means ± S.E. (n = 3). *, p < 0.05; **, p < 0.01; N.S., no significance. B, effect of NUB1L or P97 on the neddylation of CUL1. C, effect of knockdown of NUB1L or P97 on the neddylation of CUL1. The experimental condition is the same as in Fig. 1, C and D. E, schematic representation showing that NUB1L regulates proteasomal degradation of NEDD8 via the P97UFD1/NPL4 complex. The P97UFD1/NPL4 complex can recruit NEDD8 via NPL4 binding, whereas NUB1L forms a quaternary complex with P97UFD1/NPL4 by interacting with P97. In the complex the ATP hydrolysis by P97 triggers transfer of NEDD8 from NPL4 to NUB1L, by which NEDD8 is delivered to the proteasome for degradation.
DISCUSSION
NUB1L Specifically Recognizes NEDD8
We have identified a key residue of Asn-51 in NEDD8 critical for interacting with NUB1L, but the structural basis of NUB1L distinguishing NEDD8 from Ub remains to be elucidated. Analogously, the Asn-51 residue of NEDD8 also plays an important role in recognition by the NEDD8-specific proteases, such as DEN1 (66) and SENP8 (67). It was also reported that the N-terminal UBQ1 domain of FAT10, but not the C-terminal UBQ2 domain, interacts with NUB1L (24). By sequence alignment, the corresponding residues in UBQ1 and UBQ2 of FAT10 are lysine and glutamate, respectively. Our study indicates that NUB1L can interact with the N51Q mutant of NEDD8 but not with N51D or N51E (Fig. 2C). Therefore, we propose that the binding interfaces of NUB1L for NEDD8 or FAT10 might be negatively charged. If the corresponding residue in the Ub-like proteins (or domains) is also negatively charged, its interaction with NUB1L would be prohibited by charge repulsion. On the other hand, Tanaka et al. (39) reported that both UBA2 and PEST domains of NUB1L are responsible for NEDD8 binding. However, NMR titration did not detect the interaction between NEDD8 and UBA2 or PEST (data not shown). Our pulldown experiment indicated that the shortest fragment of NUB1L that is able to interact with NEDD8 is from Ala-148 to the C terminus. We propose that both of the UBA2 and PEST domains are important but not enough for binding with NEDD8. More detailed structural information is needed to reveal the interaction between NUB1L and NEDD8.
P97UFD1/NPL4 Is Involved in the Degradation of NEDD8
We have identified a VBM motif in NUB1L and demonstrated its specific interaction with P97. The interaction between NUB1L and P97UFD1/NPL4 is a key element of the NEDD8 degradation pathway. NEDD8 is recruited by NPL4 and then passed on to NUB1L. It is possible that P97 experiences significant conformational changes during hydrolysis of ATP by the ATPase domains of P97, by which the interaction between NPL4 and NEDD8 might be intervened. A number of studies have demonstrated the conformational changes of P97 during ATP hydrolysis (53, 54, 68–70). So far, little is known about how the conformational change of P97 affects the interactions of P97 cofactors with other proteins. Three-dimensional cryoEM reconstruction has revealed that the P97UFD1/NPL4 complex is highly dynamic, and UFD1/NPL4 shows distinct positions upon the addition of nucleotide (71). This research sheds light on the cooperation between P97 and its partners during ATP hydrolysis. Similarly, NUB1L is also reported responsible for the degradation of FAT10 (21, 24, 72) through interacting with the VWA domain of Rpn10 or Rpn1 (25). Thus, P97UFD1/NPL4 is probably involved in the degradation process of FAT10 as well.
NUB1L Functions in Delivering NEDD8 from the P97UFD1/NPL4 Complex to Proteasome for Degradation
A couple of studies have suggested that NUB1/NUB1L regulates the degradation of NEDD8 (19, 20, 39). Our studies have revealed that NUB1L joins to the P97UFD1/NPL4 complex and promotes transfer of NEDD8 for proteasomal degradation. Thus, a model that NUB1L functions in delivering NEDD8 from the P97UFD1/NPL4 complex to proteasome is proposed as follows (Fig. 9D). 1) P97UFD1/NPL4 recruits NEDD8 through the interaction between NPL4 and NEDD8. 2) NUB1L joins to the P97UFD1/NPL4 complex by interacting with P97. When hydrolysis of ATP by P97 triggers its conformational switch, the NPL4-bound NEDD8 is passed onto NUB1L. 3) By interacting with the subunits of proteasome, such as Rpn10 and Rpn1 (23, 25), NUB1L delivers NEDD8 to the proteasome for degradation. Thus, through modulating the turnover of NEDD8, the neddylation levels of NEDD8 substrate proteins are exquisitely regulated in cells.
Control of the NEDD8 and Neddylation Levels
The best known function of NEDD8 is activating cullin-RING ligases by conjugating to cullin proteins (9, 12, 26). Cullin-RING ligases control the turnover of a large number of substrate proteins, which are key elements of pivotal cellular processes, including cell-cycle progression, cell signaling, and so on (8). Therefore, it is very important to control the protein levels of NEDD8 and the neddylation. There is literature reporting that NUB1 mediates anti-proliferation and apoptosis in renal cell carcinoma cells (27) and suppression of mutant huntingtin toxicity in animal models (73). NUB1L (or NUB1) is an interferon-inducible protein whose expression is probably promoted under certain circumstances like infection and uncontrolled cell proliferation. By targeting NEDD8 for proteasomal degradation, NUB1L cooperates with P97UFD1/NPL4 to suppress the protein levels of NEDD8 and neddylation. Because down-regulation of protein neddylation (such as cullins) affects cell proliferation by inhibiting the Ub E3 ligase activity of SCF (SKP1-cullin1-F-box) complexes, inhibiting the NEDD8 conjugation pathway is considered to be a promising pharmaceutical intervention (74, 75).
Acknowledgments
We thank L. L. Jiang and C. J. Zhou for assisting in performing the experiments.
This work was supported by grants from the National Basic Research Program of China (2012CB911003), the National Natural Science Foundation of China (31270773), and the Sino-Swiss Joint Research Projects (GJHZ0909).
- Ub
- ubiquitin
- NLS
- nuclear localization signal
- CUL1
- cullin 1
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- VBM
- VCP binding motif.
REFERENCES
- 1. Kumar S., Yoshida Y., Noda M. (1993) Cloning of a cDNA which encodes a novel ubiquitin-like protein. Biochem. Biophys. Res. Commun. 195, 393–399 [DOI] [PubMed] [Google Scholar]
- 2. Kamitani T., Kito K., Nguyen H. P., Yeh E. T. (1997) Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272, 28557–28562 [DOI] [PubMed] [Google Scholar]
- 3. Liakopoulos D., Doenges G., Matuschewski K., Jentsch S. (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osaka F., Kawasaki H., Aida N., Saeki M., Chiba T., Kawashima S., Tanaka K., Kato S. (1998) A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walden H., Podgorski M. S., Huang D. T., Miller D. W., Howard R. J., Minor D. L., Jr., Holton J. M., Schulman B. A. (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol. Cell 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 6. Huang D. T., Ayrault O., Hunt H. W., Taherbhoy A. M., Duda D. M., Scott D. C., Borg L. A., Neale G., Murray P. J., Roussel M. F., Schulman B. A. (2009) E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell 33, 483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Petroski M. D., Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 8. Hua Z., Vierstra R. D. (2011) The cullin-RING ubiquitin-protein ligases. Annu. Rev. Plant Biol. 62, 299–334 [DOI] [PubMed] [Google Scholar]
- 9. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases. Conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldenberg S. J., Cascio T. C., Shumway S. D., Garbutt K. C., Liu J., Xiong Y., Zheng N. (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 [DOI] [PubMed] [Google Scholar]
- 11. Liu J., Furukawa M., Matsumoto T., Xiong Y. (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 12. Duda D. M., Scott D. C., Calabrese M. F., Zimmerman E. S., Zheng N., Schulman B. A. (2011) Structural regulation of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 21, 257–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarikas A., Hartmann T., Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D. A., Wei N., Shevchenko A., Deshaies R. J. (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385 [DOI] [PubMed] [Google Scholar]
- 15. Cope G. A., Suh G. S., Aravind L., Schwarz S. E., Zipursky S. L., Koonin E. V., Deshaies R. J. (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298, 608–611 [DOI] [PubMed] [Google Scholar]
- 16. Mendoza H. M., Shen L. N., Botting C., Lewis A., Chen J., Ink B., Hay R. T. (2003) NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 278, 25637–25643 [DOI] [PubMed] [Google Scholar]
- 17. Hemelaar J., Borodovsky A., Kessler B. M., Reverter D., Cook J., Kolli N., Gan-Erdene T., Wilkinson K. D., Gill G., Lima C. D., Ploegh H. L., Ovaa H. (2004) Specific and covalent targeting of conjugating and deconjugating enzymes of ubiquitin-like proteins. Mol. Cell. Biol. 24, 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gong L., Kamitani T., Millas S., Yeh E. T. (2000) Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J. Biol. Chem. 275, 14212–14216 [DOI] [PubMed] [Google Scholar]
- 19. Kamitani T., Kito K., Fukuda-Kamitani T., Yeh E. T. (2001) Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem. 276, 46655–46660 [DOI] [PubMed] [Google Scholar]
- 20. Kito K., Yeh E. T., Kamitani T. (2001) NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J. Biol. Chem. 276, 20603–20609 [DOI] [PubMed] [Google Scholar]
- 21. Hipp M. S., Raasi S., Groettrup M., Schmidtke G. (2004) NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J. Biol. Chem. 279, 16503–16510 [DOI] [PubMed] [Google Scholar]
- 22. Hipp M. S., Kalveram B., Raasi S., Groettrup M., Schmidtke G. (2005) FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol. Cell. Biol. 25, 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanji K., Tanaka T., Kamitani T. (2005) Interaction of NUB1 with the proteasome subunit S5a. Biochem. Biophys. Res. Commun. 337, 116–120 [DOI] [PubMed] [Google Scholar]
- 24. Schmidtke G., Kalveram B., Weber E., Bochtler P., Lukasiak S., Hipp M. S., Groettrup M. (2006) The UBA domains of NUB1L are required for binding but not for accelerated degradation of the ubiquitin-like modifier FAT10. J. Biol. Chem. 281, 20045–20054 [DOI] [PubMed] [Google Scholar]
- 25. Rani N., Aichem A., Schmidtke G., Kreft S. G., Groettrup M. (2012) FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis. Nat. Commun. 3, 749. [DOI] [PubMed] [Google Scholar]
- 26. Merlet J., Burger J., Gomes J. E., Pintard L. (2009) Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell. Mol. Life Sci. 66, 1924–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hosono T., Tanaka T., Tanji K., Nakatani T., Kamitani T. (2010) NUB1, an interferon-inducible protein, mediates anti-proliferative actions and apoptosis in renal cell carcinoma cells through cell-cycle regulation. Br. J. Cancer 102, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akey D. T., Zhu X., Dyer M., Li A., Sorensen A., Blackshaw S., Fukuda-Kamitani T., Daiger S. P., Craft C. M., Kamitani T., Sohocki M. M. (2002) The inherited blindness associated protein AIPL1 interacts with the cell cycle regulator protein NUB1. Hum. Mol. Genet. 11, 2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kanaya K., Sohocki M. M., Kamitani T. (2004) Abolished interaction of NUB1 with mutant AIPL1 involved in Leber congenital amaurosis. Biochem. Biophys. Res. Commun. 317, 768–773 [DOI] [PubMed] [Google Scholar]
- 30. van der Spuy J., Cheetham M. E. (2004) The Leber congenital amaurosis protein AIPL1 modulates the nuclear translocation of NUB1 and suppresses inclusion formation by NUB1 fragments. J. Biol. Chem. 279, 48038–48047 [DOI] [PubMed] [Google Scholar]
- 31. Hidalgo-de-Quintana J., Evans R. J., Cheetham M. E., van der Spuy J. (2008) The Leber congenital amaurosis protein AIPL1 functions as part of a chaperone heterocomplex. Invest. Ophthalmol. Vis. Sci. 49, 2878–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu G., Xirodimas D. P. (2010) NUB1 promotes cytoplasmic localization of p53 through cooperation of the NEDD8 and ubiquitin pathways. Oncogene 29, 2252–2261 [DOI] [PubMed] [Google Scholar]
- 33. Tanji K., Tanaka T., Mori F., Kito K., Takahashi H., Wakabayashi K., Kamitani T. (2006) NUB1 suppresses the formation of Lewy body-like inclusions by proteasomal degradation of synphilin-1. Am. J. Pathol. 169, 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Richet E., Pooler A. M., Rodriguez T., Novoselov S. S., Schmidtke G., Groettrup M., Hanger D. P., Cheetham M. E., van der Spuy J. (2012) NUB1 modulation of GSK3β reduces tau aggregation. Hum. Mol. Genet. 21, 5254–5267 [DOI] [PubMed] [Google Scholar]
- 35. Tanji K., Mori F., Kakita A., Zhang H., Kito K., Kamitani T., Takahashi H., Wakabayashi K. (2007) Immunohistochemical localization of NUB1, a synphilin-1-binding protein, in neurodegenerative disorders. Acta neuropathol. 114, 365–371 [DOI] [PubMed] [Google Scholar]
- 36. Mori F., Tanji K., Odagiri S., Hattori M., Hoshikawa Y., Kono C., Yasui K., Yokoi S., Hasegawa Y., Kamitani T., Yoshida M., Wakabayashi K. (2012) Ubiquitin-related proteins in neuronal and glial intranuclear inclusions in intranuclear inclusion body disease. Pathol. Int. 62, 407–411 [DOI] [PubMed] [Google Scholar]
- 37. Odagiri S., Tanji K., Mori F., Kakita A., Takahashi H., Kamitani T., Wakabayashi K. (2012) Immunohistochemical analysis of Marinesco bodies using antibodies against proteins implicated in the ubiquitin-proteasome system, autophagy, and aggresome formation. Neuropathology 32, 261–266 [DOI] [PubMed] [Google Scholar]
- 38. Tanji K., Mori F., Kito K., Kakita A., Mimura J., Itoh K., Takahashi H., Kamitani T., Wakabayashi K. (2011) Synphilin-1-binding protein NUB1 is colocalized with nonfibrillar, proteinase K-resistant α-synuclein in presynapses in Lewy body disease. J. Neuropathol. Exp. Neurol. 70, 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanaka T., Kawashima H., Yeh E. T., Kamitani T. (2003) Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J. Biol. Chem. 278, 32905–32913 [DOI] [PubMed] [Google Scholar]
- 40. Ye Y. (2006) Diverse functions with a common regulator. Ubiquitin takes command of an AAA ATPase. J. Struct. Biol. 156, 29–40 [DOI] [PubMed] [Google Scholar]
- 41. Raasi S., Wolf D. H. (2007) Ubiquitin receptors and ERAD. A network of pathways to the proteasome. Semin. Cell Dev. Biol. 18, 780–791 [DOI] [PubMed] [Google Scholar]
- 42. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48. A power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 43. Meyer H., Bug M., Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 44. Tucker P. A., Sallai L. (2007) The AAA+ superfamily. A myriad of motions. Curr. Opin. Struct. Biol. 17, 641–652 [DOI] [PubMed] [Google Scholar]
- 45. Wang Q., Song C., Li C. C. (2004) Molecular perspectives on p97-VCP. Progress in understanding its structure and diverse biological functions. J. Struct. Biol. 146, 44–57 [DOI] [PubMed] [Google Scholar]
- 46. Meyer H. H. (2005) Golgi reassembly after mitosis. The AAA family meets the ubiquitin family. Biochim. Biophys. Acta 1744, 481–492 [PubMed] [Google Scholar]
- 47. Ye Y., Meyer H. H., Rapoport T. A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 48. Ye Y., Meyer H. H., Rapoport T. A. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol. Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dai R. M., Li C. C. (2001) Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat. Cell Biol. 3, 740–744 [DOI] [PubMed] [Google Scholar]
- 50. Meyer H. H., Wang Y., Warren G. (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21, 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Richly H., Rape M., Braun S., Rumpf S., Hoege C., Jentsch S. (2005) A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120, 73–84 [DOI] [PubMed] [Google Scholar]
- 52. Rumpf S., Jentsch S. (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 21, 261–269 [DOI] [PubMed] [Google Scholar]
- 53. Davies J. M., Brunger A. T., Weis W. I. (2008) Improved structures of full-length p97, an AAA ATPase. Implications for mechanisms of nucleotide-dependent conformational change. Structure 16, 715–726 [DOI] [PubMed] [Google Scholar]
- 54. Davies J. M., Tsuruta H., May A. P., Weis W. I. (2005) Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-ray scattering. Structure 13, 183–195 [DOI] [PubMed] [Google Scholar]
- 55. DeLaBarre B., Brunger A. T. (2005) Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347, 437–452 [DOI] [PubMed] [Google Scholar]
- 56. Bao W. J., Gao Y. G., Chang Y. G., Zhang T. Y., Lin X. J., Yan X. Z., Hu H. Y. (2006) Highly efficient expression and purification system of small-size protein domains in Escherichia coli for biochemical characterization. Protein Expr. Purif. 47, 599–606 [DOI] [PubMed] [Google Scholar]
- 57. Chang Y. G., Song A. X., Gao Y. G., Shi Y. H., Lin X. J., Cao X. T., Lin D. H., Hu H. Y. (2006) Solution structure of the ubiquitin-associated domain of human BMSC-UbP and its complex with ubiquitin. Protein Sci. 15, 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morreale G., Conforti L., Coadwell J., Wilbrey A. L., Coleman M. P. (2009) Evolutionary divergence of valosin-containing protein/cell division cycle protein 48 binding interactions among endoplasmic reticulum-associated degradation proteins. FEBS J. 276, 1208–1220 [DOI] [PubMed] [Google Scholar]
- 59. Laser H., Conforti L., Morreale G., Mack T. G., Heyer M., Haley J. E., Wishart T. M., Beirowski B., Walker S. A., Haase G., Celik A., Adalbert R., Wagner D., Grumme D., Ribchester R. R., Plomann M., Coleman M. P. (2006) The slow Wallerian degeneration protein, WldS, binds directly to VCP/p97 and partially redistributes it within the nucleus. Mol. Biol. Cell 17, 1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Boeddrich A., Gaumer S., Haacke A., Tzvetkov N., Albrecht M., Evert B. O., Müller E. C., Lurz R., Breuer P., Schugardt N., Plassmann S., Xu K., Warrick J. M., Suopanki J., Wüllner U., Frank R., Hartl U. F., Bonini N. M., Wanker E. E. (2006) An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 25, 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dalal S., Rosser M. F., Cyr D. M., Hanson P. I. (2004) Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell 15, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolf D. H., Stolz A. (2012) The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta 1823, 117–124 [DOI] [PubMed] [Google Scholar]
- 63. Alam S. L., Sun J., Payne M., Welch B. D., Blake B. K., Davis D. R., Meyer H. H., Emr S. D., Sundquist W. I. (2004) Ubiquitin interactions of NZF zinc fingers. EMBO J. 23, 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hori T., Osaka F., Chiba T., Miyamoto C., Okabayashi K., Shimbara N., Kato S., Tanaka K. (1999) Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18, 6829–6834 [DOI] [PubMed] [Google Scholar]
- 65. Kamura T., Conrad M. N., Yan Q., Conaway R. C., Conaway J. W. (1999) The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13, 2928–2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Reverter D., Wu K., Erdene T. G., Pan Z. Q., Wilkinson K. D., Lima C. D. (2005) Structure of a complex between Nedd8 and the Ulp/Senp protease family member Den1. J. Mol. Biol. 345, 141–151 [DOI] [PubMed] [Google Scholar]
- 67. Shin Y. C., Tang S. J., Chen J. H., Liao P. H., Chang S. C. (2011) The molecular determinants of NEDD8 specific recognition by human SENP8. PLoS One 6, e27742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beuron F., Flynn T. C., Ma J., Kondo H., Zhang X., Freemont P. S. (2003) Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantized elastic deformational model. J. Mol. Biol. 327, 619–629 [DOI] [PubMed] [Google Scholar]
- 69. Rouiller I., Butel V. M., Latterich M., Milligan R. A., Wilson-Kubalek E. M. (2000) A major conformational change in p97 AAA ATPase upon ATP binding. Mol. Cell 6, 1485–1490 [DOI] [PubMed] [Google Scholar]
- 70. Rouiller I., DeLaBarre B., May A. P., Weis W. I., Brunger A. T., Milligan R. A., Wilson-Kubalek E. M. (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol. 9, 950–957 [DOI] [PubMed] [Google Scholar]
- 71. Bebeacua C., Förster A., McKeown C., Meyer H. H., Zhang X., Freemont P. S. (2012) Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schmidtke G., Kalveram B., Groettrup M. (2009) Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L. FEBS Lett. 583, 591–594 [DOI] [PubMed] [Google Scholar]
- 73. Lu B., Al-Ramahi I., Valencia A., Wang Q., Berenshteyn F., Yang H., Gallego-Flores T., Ichcho S., Lacoste A., Hild M., Difiglia M., Botas J., Palacino J. (2013) Identification of NUB1 as a suppressor of mutant Huntingtin toxicity via enhanced protein clearance. Nat. Neurosci. 16, 562–570 [DOI] [PubMed] [Google Scholar]
- 74. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 75. Tanaka T., Nakatani T., Kamitani T. (2012) Inhibition of NEDD8-conjugation pathway by novel molecules. Potential approaches to anticancer therapy. Mol. Oncol. 6, 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]