Background: Sirtuins regulate metabolism, genome maintenance, and stress responses.
Results: Long-chain free fatty acids stimulate SIRT6 deacetylase, and sirtuins display distinct but overlapping specificity for diverse acylated peptides.
Conclusion: SIRT6 is activated by biologically relevant fatty acids, and long-chain deacylation is a general feature of sirtuins.
Significance: Discovery of endogenous, small-molecule activators of SIRT6 demonstrates the therapeutic potential of compounds that promote SIRT6 function.
Keywords: Gene Regulation, Histone Deacetylase, Metabolism, Polyunsaturated Fatty Acids, Post-translational Modification, Sirtuins, coA, Enzymology, Fatty Acids
Abstract
Mammalian sirtuins (SIRT1 through SIRT7) are members of a highly conserved family of NAD+-dependent protein deacetylases that function in metabolism, genome maintenance, and stress responses. Emerging evidence suggests that some sirtuins display substrate specificity toward other acyl groups attached to the lysine ϵ-amine. SIRT6 was recently reported to preferentially hydrolyze long-chain fatty acyl groups over acetyl groups. Here we investigated the catalytic ability of all sirtuins to hydrolyze 13 different acyl groups from histone H3 peptides, ranging in carbon length, saturation, and chemical diversity. We find that long-chain deacylation is a general feature of mammalian sirtuins, that SIRT1 and SIRT2 act as efficient decrotonylases, and that SIRT1, SIRT2, SIRT3, and SIRT4 can remove lipoic acid. These results provide new insight into sirtuin function and a means for cellular removal of an expanding list of endogenous lysine modifications. Given that SIRT6 is a poor deacetylase in vitro, but binds and prefers to hydrolyze long-chain acylated peptides, we hypothesize that binding of certain free fatty acids (FFAs) could stimulate deacetylation activity. Indeed, we demonstrate that several biologically relevant FFAs (including myristic, oleic, and linoleic acids) at physiological concentrations induce up to a 35-fold increase in catalytic efficiency of SIRT6 but not SIRT1. The activation mechanism is consistent with fatty acid inducing a conformation that binds acetylated H3 with greater affinity. Binding of long-chain FFA and myristoylated H3 peptide is mutually exclusive. We discuss the implications of discovering endogenous, small-molecule activators of SIRT6.
Introduction
SIRT6 is one of seven members of the mammalian NAD+-dependent protein deacetylases that are evolutionarily ancient (1). Recent studies indicate that SIRT6 plays critical roles in intermediary metabolism and genomic stability (2). SIRT6-deficient mice have a striking degenerative phenotype leading to shortened lifespan and are associated with hypoglycemia and defects in DNA repair (2). Deletion of SIRT6 is sufficient to promote tumorigenesis independent of oncogene activation (3). SIRT6 overexpression lowers LDL and triglyceride levels, improves glucose tolerance (4), and increases lifespan of male mice (5). SIRT6 also plays a role in inflammatory pathways, exerting anti-inflammatory effects at the transcriptional level. However, other studies suggest a pro-inflammatory effect on intracellular signaling (6). Although it is clear that SIRT6 functions in metabolism, inflammation, and genome maintenance, its molecular functions remain enigmatic.
SIRT6 is grouped among the mammalian sirtuins (including SIRT4, SIRT5, and SIRT7) that display extremely low deacetylase activity in vitro (7). SIRT1–3 display robust protein deacetylase activity, and numerous studies have identified in vivo substrates (35, 36). The recent observation that SIRT5 prefers succinylated/malonylated substrates helps explain the observed low activity on acetylated substrates (8). Additional acyl-lysine modifications have been identified in histone and non-histone proteins, including propionyl, butyryl, myristoyl, and crotonyl (8–11), suggesting the existence of a very diverse acylation landscape in cells.
These observations have led the field to postulate that some sirtuins harbor unique acyl group specificities, distinct from deacetylase activity (7, 12). Consistent with this idea, Jiang et al. (13) demonstrated recently that SIRT6 preferentially hydrolyzes long-chain fatty acyl groups, including myristoyl and palmitoyl groups, from lysine residues. A crystal structure of SIRT6 bound to a myristoylated peptide supported this observation. However, several prior studies indicated that SIRT6 acted through deacetylation of histone marks H3K9Ac (14) and H3K56Ac (15), which modulated HIF1-α-dependent (16), NF-κB-dependent (17), and c-Myc-dependent (3) pathways. In cells, the ability of SIRT6 to promote deacetylation of histones and non-histone proteins (18) seemed at odds with the low in vitro deacetylation rates (13, 19) and with the recently described demyristoylation activity (13). X-ray structural analysis of SIRT6 suggested that the low deacetylase activity stems from a splayed configuration between the Rossmann fold domain and the zinc binding subdomain (19). Given this unique conformation, we proposed that SIRT6 could be activated if the two domains could be induced to the canonical active-site conformation (19).
To provide molecular understanding of the unique catalytic functions of SIRT6 and to resolve the apparent contradictory results discussed above, we addressed two major questions: 1) Is long-chain protein deacylation a general feature of mammalian sirtuins? and 2) Are there endogenous SIRT6 activators that stimulate deacetylation activity?
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant SIRT1-SIRT7
His-tagged SIRT1–7 were overexpressed in BL21(DE3) or BL21(DE3) ΔCobB Escherichia coli strains and purified as described (19, 20).
[32P]NAD+ TLC Assay
Assays were performed in 20 mm HEPES, pH 7.4, at 25 °C using [32P]NAD+ (American Radiolabeled Chemicals). Products were resolved by TLC and quantified using a Typhoon FLA 9500 (GE Healthcare) (12). TLC plates were analyzed by ImageQuant TL (GE Healthcare). Intensities were determined for [32P]NAD+ and O-[32P]acylADPr2 using spot edge average as a local background subtraction method.
HPLC Deacylation Assay
Deacylation reactions were analyzed by reversed phase HPLC on a Kinetex C18 column (100A, 100 × 4.6 mm, 2.6-μm, Phenomenex) by monitoring the formation of deacylated product at 214 nm, as described (21). Sirtuin (0.2 μm) or 0.8 μm SIRT6 was incubated with 40 μm H3K9Ac, H3K9Dodec, or H3K9Myr in the presence of 0.5 mm NAD+ and 3% DMSO at 37 °C. Product and substrate peaks were quantified, and deacylation rates were determined.
Deacetylase Activity in the Presence of Fatty Acids (FAs)
SIRT6 (4 μm) or 2 μm SIRT1 were incubated with 70 μm H3K9Ac peptide, 0.5 mm NAD+, 100 μm FA (dodecanoic, myristic, palmitic, oleic, linoleic, γ-linolenic, α-linolenic). Reactions were incubated for either 1 h (SIRT6) or 15 s (SIRT1) and quenched with TFA. To examine dose dependence, FA concentrations were varied from 0 to 1 mm for myristic acid and 0–300 μm for oleic and linoleic acid. FAs were incubated with 70 μm H3K9Ac peptide, 0.5 mm NAD+, and 4 μm SIRT6 at 37 °C.
Steady-state Kinetic Analyses
Steady-state rates were measured by varying H3K9Ac peptide (0–200 μm) in the presence of 0.9 μm SIRT6 and 2 mm NAD+ ± 400 μm myristic acid. Initial velocities were determined; data in the presence of myristic acid were fitted to the Michaelis-Menten equation, and data in the absence of myristic acid were fitted to a modified version of the equation (22).
Critical Micelle Concentration
-Fold increase in fluorescence of 1,6-dipheynl-1,3,5-hexatriene (DPH) was monitored in the presence of increasing concentration of FFAs (23). Reactions contained 20 mm potassium phosphate (pH 7.5), 6.7% DMSO, 5 μm DPH, and 0–2 mm myristic acid or 0–1.5 mm oleic and linoleic acid at 37 °C.
Myristic Acid Inhibition Kinetics
SIRT6 (2 μm) was incubated with 70 μm H3K9Myr peptide in the presence of 0.5 mm NAD+ and myristic acid (0–1 mm) for 1.5 min at 37 °C. To determine the inhibition constant, Kis, 2 μm SIRT6 and 0.5 mm NAD+ were incubated with 15–220 μm H3K9Myr peptide in the presence of 0–200 μm myristic acid. Data were fitted to an equation for competitive inhibition using KinetAsyst (IntelliKinetics, State College, PA).
RESULTS AND DISCUSSION
Long-chain Deacylase Activity Is an Intrinsic Activity of Most Sirtuins
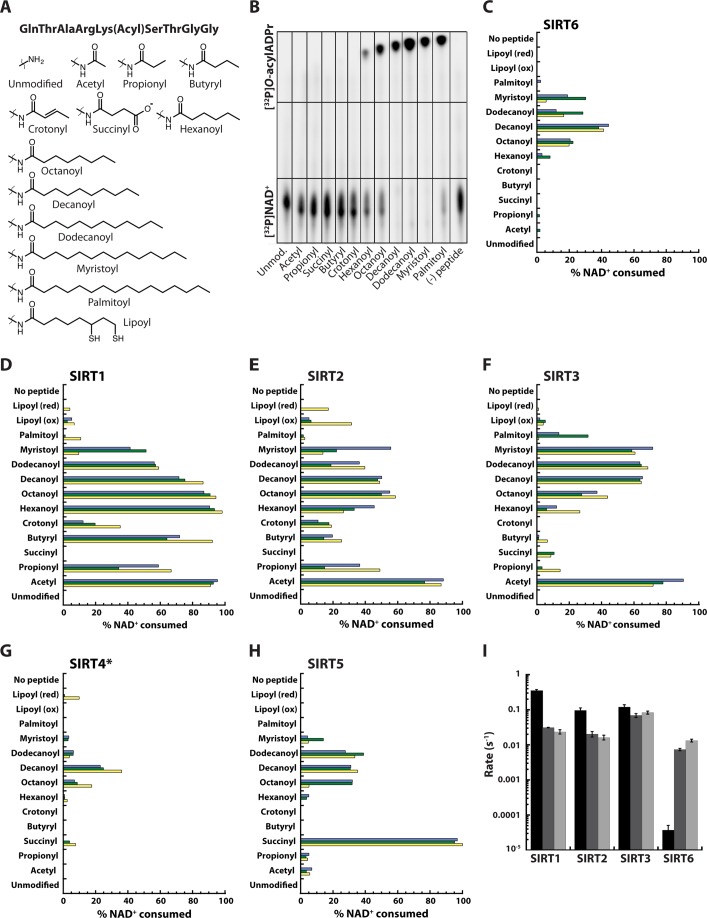
It was recently reported that SIRT6 preferentially hydrolyzes long-chain fatty acyl (myristoyl and palmitoyl groups), relative to deacetylation (13). Here we investigated whether such long-chain deacylation activity was specific to SIRT6 or whether this is a general feature of mammalian sirtuins. We synthesized a panel of acylated lysine peptides corresponding to amino acids 5–13 of histone H3 containing diverse acyl-ϵ-lysine modifications, including acetyl, propionyl, succinyl, butyryl, crotonyl, hexanoyl, octanoyl, decanoyl, dodecanoyl, myristoyl, or palmitoyl groups on Lys-9 (see supplemental Methods) (Fig. 1A). Human sirtuins 1 through 7 (SIRT1–7) were recombinantly expressed and purified, and their abilities to hydrolyze each acyl group were determined. First, the activity of SIRT6 was assessed using an excess of peptide (5 mm) and limiting [32P]NAD+ (0.18 μm). The [32P]NAD+ substrate and the various O-[32P]acylADPr products were separated by TLC and visualized by phosphorimaging (Fig. 1B).
FIGURE 1.
Deacylase activity of mammalian sirtuins. A, panel of acylated lysine peptides corresponding to amino acids 5–13 of histone H3 used in TLC assays. B, TLC assay monitoring the formation of O-[32P]acylADPr products. SIRT6 (2 μm) was incubated with 1 μCi of [32P]NAD+ (0.18 μm) and 5 mm acylated peptide for 1 h. Unmod, unmodified. C–H, quantification of TLC assay monitoring O-[32P]acylADPr formation for SIRT1, SIRT2, SIRT3, SIRT5, and SIRT6. Sirtuin (2 μm) was incubated with 5 mm acylated peptide and 12.25 μm NAD+ (1 μCi [32P]NAD+) for 1 h. *[SIRT4] = 6 μm. The percentage of NAD+ consumed was determined by measuring the intensities of the [32P]NAD+ and O-[32P]acylADPr spots. Three sets of experiments (yellow, green, blue) were performed on three separate days, using multiple enzyme preparations and peptide stocks. red, reduced; ox, oxidized. I, quantitative steady-state rates of deacetylation (black), dedodecanoylation (dark gray), and demyristoylation (light gray) for SIRT1–3 and SIRT6 determined by HPLC. Error bars represent S.D. of at least three replicates.
SIRT6 displayed a clear preference for longer acyl chains, including myristoyl and palmitoyl chains, consuming 98 and 81% of the [32P]NAD+ (Fig. 1B), respectively, consistent with a prior study (13). In addition, SIRT6 was able to deacylate the decanoyl (96%) and dodecanoyl (98%) groups from lysine residues. To a lesser degree, SIRT6 was able to hydrolyze the hexanoyl and octanoyl groups, consuming ∼23 and ∼48% of the [32P]NAD+, respectively. Under these assay conditions, SIRT6 was unable to remove detectable amounts of the acetyl group.
Next, we analyzed the deacylase activity of SIRT1 through SIRT7 using catalytic amounts of enzyme and a more expansive panel of acylated H3K9 peptides, including acetyl, propionyl, succinyl, butyryl, crotonyl, hexanoyl, octanoyl, decanoyl, dodecanoyl, myristoyl, palmitoyl, and reduced and oxidized lipoyl groups. No appreciable amounts of products were observed with SIRT7. As noted in the prior experiment (Fig. 1B), SIRT6 displayed a strong preference for long-chain fatty acyl groups (Fig. 1C). Under these conditions, octanoyl, decanoyl, dodecanoyl, and myristoyl groups were preferentially hydrolyzed. The corresponding O-[32P]acylADPr products from hexanoyl and palmitoyl were detectable, but the other acyl groups did not accumulate to quantifiable amounts.
Surprisingly, SIRT1, SIRT2, SIRT3, and SIRT5 also displayed strong deacylation activity against long-chain acyl groups. Particularly striking was the overall similar preference among these sirtuins for decanoyl and dodecanoyl, which was shared with SIRT6 (Fig. 1, C–H). As expected, SIRT1–3 efficiently hydrolyzed the acetyl group from lysine (>75% conversion), and to a lesser extent, the propionyl and butyryl groups (Fig. 1, D–F). SIRT5 was most efficient at catalyzing desuccinylation (>95% conversion) (Fig. 1H), as was reported previously (8). However, SIRT5 more efficiently hydrolyzed (2–3×) decanoyl and dodecanoyl peptides as compared with the corresponding acetylated form, which was hydrolyzed faster than the propionyl, octanoyl, and myristoyl groups. At 3× the protein concentration, SIRT4 displayed low, but measureable amounts of deoctanoylase and dedecanoylase activity (Fig. 1G).
Crotonyllysine was identified as a histone modification in several eukaryotic cells types and is associated with active promoters or enhancers (11). A product corresponding to [32P]O-crotonylADPr was observed when either SIRT1 or SIRT2 was reacted with NAD+ and the crotonylated H3K9 peptide, consuming ∼20% of the [32P]NAD+ (Fig. 1, D and E).
Among sirtuins displaying quantifiable activity, SIRT1 displayed the greatest promiscuity for diverse acyl groups, except for succinylated substrate, which neither SIRT1, SIRT2, nor SIRT6 hydrolyzed. SIRT3 and SIRT4 also exhibited activity against the succinylated substrate, but to a lesser extent than SIRT5 (Fig. 1, C–H). Interestingly, a specific mammalian delipoylase has not been identified (24). Here, SIRT2, and to a lesser extent, SIRT1, SIRT3, and SIRT4 were able to hydrolyze the removal of the lipoyl group from lysine (Fig. 1, D–G). Importantly, SIRT1–3 displayed more efficient conversion of many long-chain acylated peptides as compared with SIRT6. Even SIRT5, an enzyme reported to harbor exquisite selectivity for succinylated peptide, hydrolyzed long-chain acyl groups with similar efficiency to that of SIRT6.
To obtain true steady-state rates of deacylation and to ensure that the deacylase activity observed in the above experiments was not due to contamination by the E. coli sirtuin CobB, SIRT1, SIRT2, SIRT3, and SIRT6 were overexpressed and purified from a CobB knock-out BL21(DE3) strain. The purified enzymes were subjected to steady-state kinetic analysis, and the rates of deacetylation, Dedodecanoylation, and demyristoylation were determined. The resulting kinetic analysis yielded trends that were in excellent agreement with the qualitative TLC-based assay, indicating that the activities do not arise from contaminating CobB (Fig. 1I). SIRT1–3 catalyzed deacetylation more efficiently than long-chain deacylation, whereas SIRT6 exhibited the opposite trend. All the sirtuins displayed robust long-chain deacylase activity, with SIRT3 harboring the greatest activity toward H3K9Dodec and H3K9Myr peptides. SIRT3 was ∼2-, 3-, and 9× more active as a dedodecanoylase than SIRT1, SIRT2, and SIRT6, respectively. Likewise, SIRT3 was ∼3.5-, 5-, and 6× more active as a demyristoylase. Collectively, these results indicate that in addition to SIRT6, other mammalian sirtuins possess intrinsic deacylation activity toward long-chain acyl groups, although each sirtuin displays a unique substrate signature among the 13 acylated versions tested here.
The expanding identification of new acyl modifications and the ability of sirtuins to remove these suggest that other abundant acyl-CoAs might serve as donor molecules for lysine post-translational modification. The nutrient status of the cell may favor the addition of one acyl modification over another, which might rise and fall as a function of [acyl-CoA]. Acyl-CoA donors are intermediates in different central metabolic pathways, including glycolysis, the TCA cycle, β-oxidation, and FA synthesis (7). Many of these modifications occur on metabolic enzymes, suggesting a regulatory link between metabolism and lysine acylation. Our results suggest that sirtuins are well suited to function as the demodifying enzymes of such acylations.
SIRT6 Deacetylase Activity Is Stimulated by Long-chain FFAs
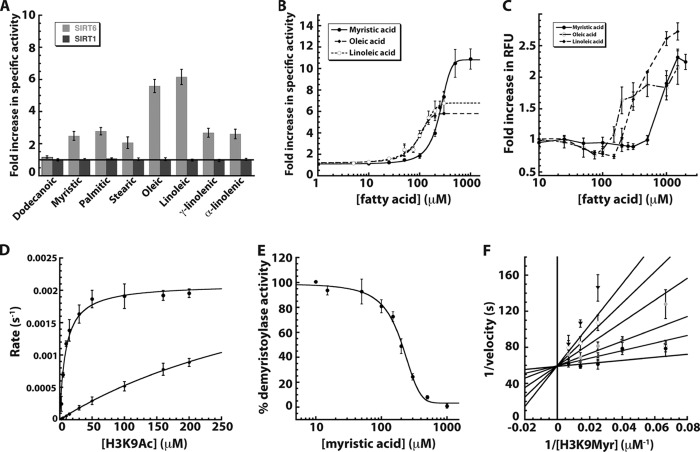
The fact that sirtuins other than SIRT6 displayed higher specific activity against long-chain acyl groups was surprising, especially given the prior observation of a co-crystal structure with bound myristoylated peptide. This structure revealed a long acyl chain binding cleft. Comparing SIRT6 crystal structures with bound and unbound myristoylated peptide revealed conformational differences, adopting a more canonical sirtuin fold when myristoylated peptide is bound (13, 19). We hypothesized that this acyl group binding pocket might bind FFAs, and if such binding could induce closure of the splayed SIRT6 subdomains, then SIRT6 might show stimulated deacetylation activity. To determine whether FFAs can stimulate SIRT6 deacetylase activity in vitro, SIRT6 was incubated with 70 μm H3K9Ac peptide, 0.5 mm NAD+, and 100 μm dodecanoic, myristic, palmitic, stearic, oleic (18:1, n-9), linoleic (18:2, n-6), γ-linolenic (18:3, n-6), or α-linolenic (18:3, n-3) acids. Separate experiments were performed with SIRT1 for comparison. Reaction substrates and products were separated by HPLC and quantified. Specific activities of SIRT6 and SIRT1 in the presence of different FAs were compared.
SIRT6-dependent deacetylation of H3K9Ac peptide exhibited 2–6× stimulation with 7 out of 8 FAs examined (Fig. 2A): oleic (5.6 ± 0.4 times) and linoleic acid (6.2 ± 0.5), myristic (2.5 ± 0.3), palmitic (2.8 ± 0.2), stearic (2.1 ± 0.4), γ-linolenic (2.7 ± 0.3), and α-linolenic (2.6 ± 0.3) acids. Dodecanoic acid was unable to stimulate SIRT6 activity, suggesting a minimum chain length for stimulating deacetylation. In stark contrast, SIRT1 displayed no effect with any FA on H3K9Ac deacetylation, suggesting that FA-stimulated deacetylation might be a unique feature of SIRT6.
FIGURE 2.
Sirtuin activity in the presence of free fatty acids. A, -fold change in SIRT1 and SIRT6 deacetylase activity was monitored in the presence of various FFAs and compared with a reaction without fatty acid. SIRT1 (dark gray) and SIRT6 (light gray) were incubated with 70 μm H3K9Ac peptide and 0.5 mm NAD+ in the presence of 100 μm fatty acid and analyzed by HPLC. B, -fold increase in SIRT6 deacetylase activity when 70 μm H3K9ac peptide was incubated with 0.5 mm NAD+ and 0–1 mm myristic (filled circles, solid line), 0–300 μm oleic (filled diamonds, long dashed lines), and linoleic acids (open circles, short dashed lines). C, critical micelle concentration determined by DPH (5 μm) assay (23) in 20 mm potassium phosphate (pH 7.5)/6.7% DMSO in the presence of varied myristic (filled circle, solid line), oleic (open circle, dash dot line), and linoleic acids (filled diamonds, long dashed line). D, steady-state kinetic analyses of SIRT6 deacetylation of 0–200 μm H3K9Ac peptide in the presence of 2 mm NAD+ with (filled circles) and without (filled diamonds) 400 μm myristic acid. E, percentage of demyristoylase activity of SIRT6 incubated with 70 μm H3K9myr peptide, 0.5 mm NAD+, and 0–1 mm myristic acid. F, myristic acid inhibition of SIRT6 demyristoylase activity displayed in double-reciprocal format. Reactions contained 2 μm SIRT6, 0.5 mm NAD+, and varied H3K9Myr peptide in the presence of 0 (filled circles), 25 (open circles), 50 (dark gray triangles), 100 (light shaded circles), 150 (open diamonds), and 200 (inverted triangles) μm myristic acid. Data were fitted (using nonlinear least squares) to the equation for competitive inhibition and yielded a Kis of 15 ± 9 μm. Error bars represent S.D. of at least three replicates.
Next, we determined the dose-dependent effects and the magnitude of the -fold activation by FFAs. Deacetylase activity of SIRT6 was analyzed at various concentrations of myristic, oleic, and linoleic acids, at fixed H3K9Ac peptide (70 μm) and NAD+ (500 μm). Myristic, oleic, and linoleic acid were all able to stimulate SIRT6 deacetylase activity in a concentration-dependent manner (Fig. 2B). Myristic acid stimulated deacetylase activity by a factor of 10.8 ± 0.2, with an EC50 of 246 ± 7 μm. Oleic and linoleic acid stimulated SIRT6 activity to a maximum -fold increase of 5.8 ± 0.2 and 6.8 ± 0.3 and yielded an EC50 of 90 ± 7 and 100 ± 9 μm, respectively. The EC50 values are within the range of postprandial levels of nonesterified FAs in mice (25) and humans (26).
To rule out the possibility that activation was due to micelle formation, the critical micelle concentration (CMC) for the FFAs was determined. The CMC for myristic acid is between 600 μm and 1 mm (Fig. 2C). The CMC for oleic acid is between 150 and 200 μm, and the CMC for linoleic acid is between 200 and 300 μm. Thus, the CMC is beyond the concentration needed for maximal activation of each FFA, suggesting that the ability to saturate the stimulated deacetylation results from specific binding between enzyme and FFA.
To investigate the activation mechanism, we performed steady-state kinetic analyses with increasing H3K9Ac peptide at saturating levels of NAD+ in the absence or presence of 400 μm myristic acid (Fig. 2D). The data were subjected to Michaelis-Menten analysis, and kcat/Km values were compared. The kcat/Km is considered the most physiologically relevant parameter because it reflects enzyme activity at low substrate levels, mimicking conditions typically encountered in vivo (27). Strikingly, the kcat/Km value for deacetylation with 400 μm myristic acid (230 ± 30 s−1 m−1) is ∼35 times faster than that for deacetylation in the absence of myristic acid (6.4 ± 2 s−1 m−1). Increased catalytic efficiency is due to a decrease in Km rather than a substantial increase in kcat. In the presence of myristic acid, the Km is 9 ± 1 μm, and in the absence of myristic acid, the Km is estimated to be ∼450 μm. Together, these results suggest that FFAs stimulate the deacetylase activity of SIRT6 by increasing the affinity of SIRT6 for an acetylated substrate by 35 times in the case of myristic acid.
To test the hypothesis that myristic acid binds in the same site as the fatty acid chain of a myristoylated lysine peptide, SIRT6 demyristoylase activity was measured in the presence of increasing myristic acid at fixed peptide substrate concentration. Myristic acid was able to inhibit the demyristoylase activity of SIRT6 with an IC50 of 190 ± 10 μm (Fig. 2E), a value in good agreement to the EC50 determined for myristic acid-stimulated deacetylation (246 ± 7 μm) and consistent with myristic acid binding to a single site shared with myristoylated peptide.
To provide additional evidence that myristic acid shares the same binding site with the fatty acid chain of a myristoylated peptide, a detailed steady-state inhibition analysis was performed. The concentration of H3K9Myr peptide was varied at several fixed concentrations of myristic acid. The data displayed clear competitive behavior and were fitted to a model of competitive inhibition and plotted in double-reciprocal format (Fig. 2F). The analysis yielded a Kis value of 15 ± 9 μm for myristic acid. The ability of myristic acid to competitively inhibit demyristoylation is consistent with free myristic acid sharing the same binding pocket with myristoylated peptide. FFA binding likely induces a conformation in SIRT6, similar to a myristoylated peptide, allowing for efficient deacetylation.
The observation that FFAs stimulate deacetylation provides a mechanism by which the low intrinsic activity of SIRT6 might be activated in vivo in response to elevated levels of particular FAs, e.g. from the diet or from fasting. Fatty acids or their metabolites, through various signaling pathways, modulate numerous metabolic and inflammatory processes (28–30). For instance, omega-3 fatty acids, including eicosapentaenoic acid (20:5, n-3) and docosahexaenoic acid (22:6, n-3), are reported to exert beneficial effects against cardiovascular disease and inflammation (29, 30). Similarly, SIRT6 protects the heart from cardiac hypertrophy by attenuating insulin-like growth factor-Akt signaling through H3K9Ac deacetylation at the promoters of insulin-like growth factor signaling-related genes (31).
SIRT6 was shown to negatively control the mRNA expression of glycolytic and lipogenic genes (16, 32). This observation is strikingly similar to the decreased mRNA levels of pyruvate kinase (PK) and fatty acid synthase (FAS) that are reported with PUFA treatment in primary hepatocytes (33). Importantly, a decrease in l-PK gene expression correlated with a reduction in H3 and H4 acetylation (34), indicating that FFAs have the ability to alter the histone acetylation status at specific genomic loci. We propose that certain FAs stimulate SIRT6 deacetylase activity in vivo, leading to targeted histone deacetylation and repression of glycolytic and lipogenic genes. Together with transcription factors peroxisome proliferator-activated receptor (PPARα, -β, and -γ), hepatic nuclear factor-4 (HNF-4), MAX-like factor X (MLX), and sterol regulatory element-binding proteins (SREBPs) 1 and 2 (28), these mechanisms permit tight control of carbohydrate and lipid metabolism pathways.
With the observation that SIRT6 demyristoylation is mutually exclusive with FFA binding, it is attractive to speculate that increased cellular levels of specific FAs trigger a switch from SIRT6-dependent demyristoylation to activated histone deacetylation. SIRT6 functions as an anti-inflammatory protein through H3K9Ac deacetylation at the promoter of genes regulated by NF-κB (17). On the other hand, SIRT6 enhances a pro-inflammatory response by promoting the secretion of TNF-α through lysine demyristoylation (13). Omega-3 fatty acids, including eicosapentaenoic acid, elicit an anti-inflammatory response in part by preventing translocation of NF-κB to the nucleus, thereby decreasing TNF-α expression (30). As an additional layer of control, free PUFAs might inhibit SIRT6-dependent demyristoylation of the intracellular pool of TNF-α while simultaneously enhancing SIRT6-dependent histone deacetylation of genes regulated by NF-κB.
The identification of newly discovered acyl modifications broadens the cellular landscape that can be targeted by sirtuins. The findings herein uncovered previously unknown enzymatic activities of the mammalian sirtuins, including long-chain FA deacylation by SIRT1–5, decrotonylation by SIRT1 and SIRT2, and delipoylation by SIRT1–4. These newly discovered enzymatic activities provide a roadmap to uncover the biological functions of diverse protein acyl modifications and the role played by sirtuins. Also, we demonstrate that SIRT6 is activated directly by biologically relevant FAs, several of which are linked to the health benefits of dietary PUFAs. Discovery of endogenous, small-molecule activators of SIRT6 reveals the therapeutic potential of compounds that promote SIRT6 function, particularly anti-inflammation and decreased tumorigenesis. Lastly, the precedent for FA-regulated SIRT6 activity might apply to other sirtuins. The fact that SIRT1 through SIRT5 bind and efficiently act as (e.g.) dedecanoylases suggests the possibility that other sirtuins are regulated by FAs.
Acknowledgment
We thank Jorge Escalante for providing the BL21(DE3) ΔCobB E. coli strain.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065386 (to J. M. D.), a predoctoral fellowship (to J. L. F.) from the American Heart Association (11PRE7300059) and a predoctoral fellowship (to J. B.) from the National Science Foundation (1256259).
This article was selected as a Paper of the Week.

This article contains supplemental Methods.
- O-acylADPr
- O-acyl-ADP-ribose
- FA
- fatty acid
- PUFA
- polyunsaturated fatty acid
- DPH
- 1,6-dipheynl-1,3,5-hexatriene
- CMC
- critical micelle concentration
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Michan S., Sinclair D. (2007) Sirtuins in mammals: insights into their biological function. Biochem. J. 404, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mostoslavsky R., Chua K. F., Lombard D. B., Pang W. W., Fischer M. R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M. M., Mills K. D., Patel P., Hsu J. T., Hong A. L., Ford E., Cheng H. L., Kennedy C., Nunez N., Bronson R., Frendewey D., Auerbach W., Valenzuela D., Karow M., Hottiger M. O., Hursting S., Barrett J. C., Guarente L., Mulligan R., Demple B., Yancopoulos G. D., Alt F. W. (2006) Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124, 315–329 [DOI] [PubMed] [Google Scholar]
- 3. Sebastián C., Zwaans B. M., Silberman D. M., Gymrek M., Goren A., Zhong L., Ram O., Truelove J., Guimaraes A. R., Toiber D., Cosentino C., Greenson J. K., MacDonald A. I., McGlynn L., Maxwell F., Edwards J., Giacosa S., Guccione E., Weissleder R., Bernstein B. E., Regev A., Shiels P. G., Lombard D. B., Mostoslavsky R. (2012) The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151, 1185–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kanfi Y., Peshti V., Gil R., Naiman S., Nahum L., Levin E., Kronfeld-Schor N., Cohen H. Y. (2010) SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell 9, 162–173 [DOI] [PubMed] [Google Scholar]
- 5. Kanfi Y., Naiman S., Amir G., Peshti V., Zinman G., Nahum L., Bar-Joseph Z., Cohen H. Y. (2012) The sirtuin SIRT6 regulates lifespan in male mice. Nature 483, 218–221 [DOI] [PubMed] [Google Scholar]
- 6. Preyat N., Leo O. (2013) Sirtuin deacylases: a molecular link between metabolism and immunity. J. Leukocyte Biol. 93, 669–680 [DOI] [PubMed] [Google Scholar]
- 7. Lin H., Su X., He B. (2012) Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol. 7, 947–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stevenson F. T., Bursten S. L., Locksley R. M., Lovett D. H. (1992) Myristyl acylation of the tumor necrosis factor α precursor on specific lysine residues. J. Exp. Med. 176, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu A. Y., Zhou Y., Khan S., Deitsch K. W., Hao Q., Lin H. (2012) Plasmodium falciparum Sir2A preferentially hydrolyzes medium and long chain fatty acyl lysine. ACS Chem. Biol. 7, 155–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C., Du J., Kim R., Ge E., Mostoslavsky R., Hang H. C., Hao Q., Lin H. (2013) SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michishita E., McCord R. A., Berber E., Kioi M., Padilla-Nash H., Damian M., Cheung P., Kusumoto R., Kawahara T. L., Barrett J. C., Chang H. Y., Bohr V. A., Ried T., Gozani O., Chua K. F. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang B., Zwaans B. M., Eckersdorff M., Lombard D. B. (2009) The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8, 2662–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhong L., D'Urso A., Toiber D., Sebastian C., Henry R. E., Vadysirisack D. D., Guimaraes A., Marinelli B., Wikstrom J. D., Nir T., Clish C. B., Vaitheesvaran B., Iliopoulos O., Kurland I., Dor Y., Weissleder R., Shirihai O. S., Ellisen L. W., Espinosa J. M., Mostoslavsky R. (2010) The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawahara T. L., Michishita E., Adler A. S., Damian M., Berber E., Lin M., McCord R. A., Ongaigui K. C., Boxer L. D., Chang H. Y., Chua K. F. (2009) SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaidi A., Weinert B. T., Choudhary C., Jackson S. P. (2010) Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 329, 1348–1353 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19. Pan P. W., Feldman J. L., Devries M. K., Dong A., Edwards A. M., Denu J. M. (2011) Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hallows W. C., Lee S., Denu J. M. (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borra M. T., Denu J. M. (2004) Quantitative assays for characterization of the Sir2 family of NAD+-dependent deacetylases. Methods Enzymol. 376, 171–187 [DOI] [PubMed] [Google Scholar]
- 22. Northrop D. B. (1983) Fitting enzyme-kinetic data to V/K. Anal. Biochem. 132, 457–461 [DOI] [PubMed] [Google Scholar]
- 23. Yuan C., Sidhu R. S., Kuklev D. V., Kado Y., Wada M., Song I., Smith W. L. (2009) Cyclooxygenase allosterism, fatty acid-mediated cross-talk between monomers of cyclooxygenase homodimers. J. Biol. Chem. 284, 10046–10055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spalding M. D., Prigge S. T. (2010) Lipoic acid metabolism in microbial pathogens. Microbiol. Mol. Biol. Rev. 74, 200–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gonçalves de Albuquerque C. F., Burth P., Younes Ibrahim M., Garcia D. G., Bozza P. T., Castro Faria Neto H. C., Castro Faria M. V. (2012) Reduced plasma nonesterified fatty acid levels and the advent of an acute lung injury in mice after intravenous or enteral oleic acid administration. Mediators Inflamm. 2012, 601032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tholstrup T., Sandström B., Bysted A., Hølmer G. (2001) Effect of 6 dietary fatty acids on the postprandial lipid profile, plasma fatty acids, lipoprotein lipase, and cholesterol ester transfer activities in healthy young men. Am. J. Clin. Nutr. 73, 198–208 [DOI] [PubMed] [Google Scholar]
- 27. Berndsen C. E., Denu J. M. (2005) Assays for mechanistic investigations of protein/histone acetyltransferases. Methods 36, 321–331 [DOI] [PubMed] [Google Scholar]
- 28. Jump D. B. (2002) Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr. Opin. Lipidol. 13, 155–164 [DOI] [PubMed] [Google Scholar]
- 29. Calder P. C. (2010) Omega-3 fatty acids and inflammatory processes. Nutrients 2, 355–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adkins Y., Kelley D. S. (2010) Mechanisms underlying the cardioprotective effects of omega-3 polyunsaturated fatty acids. J. Nutr. Biochem. 21, 781–792 [DOI] [PubMed] [Google Scholar]
- 31. Sundaresan N. R., Vasudevan P., Zhong L., Kim G., Samant S., Parekh V., Pillai V. B., Ravindra P. V., Gupta M., Jeevanandam V., Cunningham J. M., Deng C. X., Lombard D. B., Mostoslavsky R., Gupta M. P. (2012) The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat. Med. 18, 1643–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim H. S., Xiao C., Wang R. H., Lahusen T., Xu X., Vassilopoulos A., Vazquez-Ortiz G., Jeong W. I., Park O., Ki S. H., Gao B., Deng C. X. (2010) Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 12, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jump D. B., Clarke S. D., Thelen A., Liimatta M. (1994) Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J. Lipid Res. 35, 1076–1084 [PubMed] [Google Scholar]
- 34. Xu J., Christian B., Jump D. B. (2006) Regulation of rat hepatic l-pyruvate kinase promoter composition and activity by glucose, n-3 polyunsaturated fatty acids, and peroxisome proliferator-activated receptor-α agonist. J. Biol. Chem. 281, 18351–18362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakagawa T., Guarente L. (2011) Sirtuins at a glance. J. Cell Sci. 124, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feldman J. L., Dittenhafer-Reed K. E., Denu J. M. (2012) Sirtuin catalysis and regulation. J. Biol. Chem. 287, 42419–42427 [DOI] [PMC free article] [PubMed] [Google Scholar]