Abstract
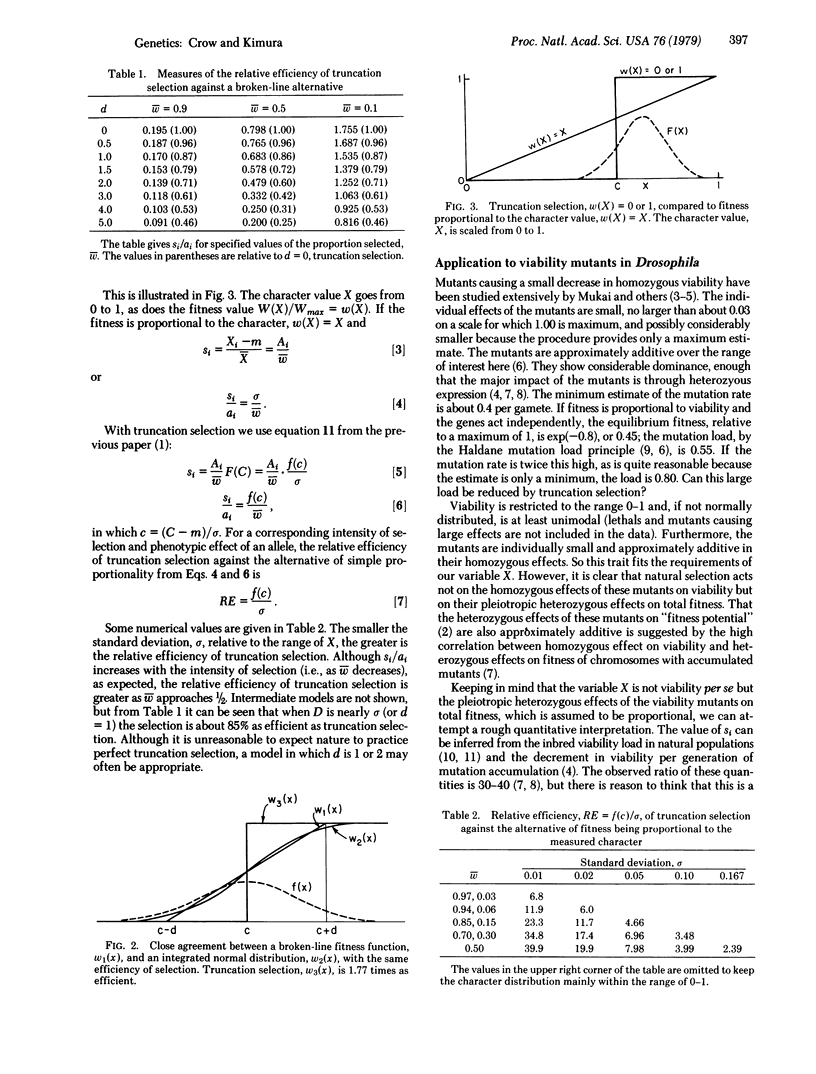
Truncation selection is known to be the most efficient form of directional selection. When this is modified so that the fitness increases linearly over a range of one or two standard deviations of the value of the selected character, the efficiency is reduced, but not greatly. When truncation selection is compared to a system in which fitness is strictly proportional to the character value, the relative efficiency of truncation selection is given by f(c)/σ, in which f(c) is the ordinate of the frequency distribution at the truncation point and σ is the standard deviation of the character. It is shown, for mutations affecting viability in Drosophila, that truncation selection or reasonable departures therefrom can reduce the mutation load greatly. This may be one way to reconcile the very high mutation rate of such genes with a small mutation load. The truncation model with directional selection is appropriate for this situation because of the approximate additivity of these mutations. On the other hand, it is doubtful that this simple model can be applied to all genes affecting fitness, for which there are intermediate optima and antagonistic selection among components with negative correlations. Whether nature ranks and truncates, or approximates this behavior, is an empirical question, yet to be answered.
Keywords: rank-order selection, fitness potential, mutation load, viability, fitness
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Greenberg R, Crow J F. A Comparison of the Effect of Lethal and Detrimental Chromosomes from Drosophila Populations. Genetics. 1960 Aug;45(8):1153–1168. doi: 10.1093/genetics/45.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRAIZUMI Y. Negative correction between rate of development and female fertility in Drosophila melanogaster. Genetics. 1961 Jun;46:615–624. doi: 10.1093/genetics/46.6.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Crow J. F. Effect of overall phenotypic selection on genetic change at individual loci. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6168–6171. doi: 10.1073/pnas.75.12.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Maruyama T. The mutational load with epistatic gene interactions in fitness. Genetics. 1966 Dec;54(6):1337–1351. doi: 10.1093/genetics/54.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. L. Continuously distributed factors affecting fitness. Genetics. 1967 Mar;55(3):483–492. doi: 10.1093/genetics/55.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MUKAI T. THE GENETIC STRUCTURE OF NATURAL POPULATIONS OF DROSOPHILA MELANOGASTER. I. SPONTANEOUS MUTATION RATE OF POLYGENES CONTROLLING VIABILITY. Genetics. 1964 Jul;50:1–19. doi: 10.1093/genetics/50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R. D. Heterosis as a major cause of heterozygosity in nature. Genetics. 1967 Mar;55(3):493–495. doi: 10.1093/genetics/55.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milkman R. Selection differentials and selection coefficients. Genetics. 1978 Feb;88(2):391–403. doi: 10.1093/genetics/88.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. A. Fitness effects of EMS-induced mutations on the X chromosome of Drosophila melanogaster. I. Viability effects and heterozygous fitness effects. Genetics. 1977 Dec;87(4):763–774. doi: 10.1093/genetics/87.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. A., Simmons M. J. Fitness effects of EMS-induced mutations on the X chromosome of Drosophila melanogaster. II. Hemizygous fitness effects. Genetics. 1977 Dec;87(4):775–783. doi: 10.1093/genetics/87.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai T., Chigusa S. I., Mettler L. E., Crow J. F. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics. 1972 Oct;72(2):335–355. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi O. Spontaneous and ethyl methanesulfonate-induced mutations controlling viability in Drosophila melanogaster. II. Homozygous effect of polygenic mutations. Genetics. 1977 Nov;87(3):529–545. doi: 10.1093/genetics/87.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M. J., Crow J. F. Mutations affecting fitness in Drosophila populations. Annu Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- Simmons M. J., Sheldon E. W., Crow J. F. Heterozygous effects on fitness of EMS-treated chromosomes in Drosophila melanogaster. Genetics. 1978 Mar;88(3):575–590. doi: 10.1093/genetics/88.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J. A. An estimate of heterosis in Drosophila melanogaster. Genet Res. 1971 Aug;18(1):97–105. doi: 10.1017/s0016672300012453. [DOI] [PubMed] [Google Scholar]
- Sved J. A., Ayala F. J. A population cage test for heterosis in Drosophila pseudoobscura. Genetics. 1970 Sep;66(1):97–113. doi: 10.1093/genetics/66.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J. A., Reed T. E., Bodmer W. F. The number of balanced polymorphisms that can be maintained in a natural population. Genetics. 1967 Mar;55(3):469–481. doi: 10.1093/genetics/55.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin R. G., Meyer H. U., Dawson P. S., Crow J. F. The influence of epistasis on homozygous viability depression in Drosophila melanogaster. Genetics. 1969 Feb;61(2):497–519. doi: 10.1093/genetics/61.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills C. Rank-order selection is capable of maintaining all genetic polymorphisms. Genetics. 1978 Jun;89(2):403–417. doi: 10.1093/genetics/89.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]



