Abstract
Arsenic species patterns in urine are associated with risk for cancer and cardiovascular diseases. The organic anion transporter coded by the gene SLCO1B1 may transport arsenic species, but its association with arsenic metabolites in human urine has not yet been studied. The objective of this study is to evaluate associations of urine arsenic metabolites with variants in the candidate gene SLCO1B1 in adults from the Strong Heart Family Study. We estimated associations between % arsenic species biomarker traits and 5 single-nucleotide polymorphisms (SNPs) in the SLCO1B1 gene in 157 participants, assuming additive genetics. Linear regression models for each SNP accounted for kinships and were adjusted for sex, body mass index, and study center. The minor allele of rs1564370 was associated with lower %MMA (p = .0003) and higher %DMA (p = .0002), accounting for 8% of the variance for %MMA and 9% for %DMA. The rs1564370 minor allele homozygote frequency was 17% and the heterozygote frequency was 43%. The minor allele of rs2291075 was associated with lower %MMA (p = .0006) and higher %DMA (p = .0014), accounting for 7% of the variance for %MMA and 5% for %DMA. The frequency of rs2291075 minor allele homozygotes was 1% and of heterozygotes was 15%. Common variants in SLCO1B1 were associated with differences in arsenic metabolites in a preliminary candidate gene study. Replication of this finding in other populations and analyses with respect to disease outcomes are needed to determine whether this novel candidate gene is important for arsenic-associated disease risks.
Key Words: American Indians, arsenic metabolism, arsenic species, SLCO1B1, OATPC, Strong Heart Study.
Arsenic exposure is a possible risk factor for diabetes (Maull et al., 2012) and cardiovascular disease (Moon et al., 2012), as well as a known human carcinogen (IARC, 2004). Arsenic metabolism involves a series of methylation and reduction reactions (Thomas, 2009), which may contribute to toxicokinetic variability across individuals (Vahter and Concha, 2001) and varying susceptibility to arsenic health effects (Chen et al., 2003a,b; Huang et al., 2008, 2009). Substantial effort has gone into characterizing the genetic basis of arsenic metabolism processes (Engstrom et al., 2011, 2013; Hernández et al., 2008a,b; Wood et al., 2006) and the diversity of probable metabolic variants across human populations (Fujihara et al., 2007, 2008, 2009, 2010, 2011). More recently, genome-wide screens have been conducted (Pierce et al., 2012; Tellez-Plaza et al., 2013). Less research, however, has focused on the potential role of transporters for arsenic toxicokinetics in humans (Hernández and Marcos, 2008). Transporters play an important role for arsenic kinetics in bacteria (Achour et al., 2007), plants (Briat, 2010; Catarecha et al., 2007; Zhao et al., 2010), fish (Hamdi et al., 2009), amphibians (Villa-Bellosta and Sorribas, 2010), and rodents (Carbrey et al., 2009; Kala et al., 2000; Kojima et al., 2006; Villa-Bellosta and Sorribas, 2008; Wang et al., 2009; Xie et al., 2004). Based on in vitro studies in human cells, transporters are also thought to be central to human arsenic toxicokinetics (Calatayud et al., 2010, 2012; Carew and Leslie, 2010; Chavan et al., 2011; Drobná et al., 2010; Lee et al., 2006). Some experiments have also shown the potential of human transporters such as aquaporins to transport arsenic when introduced into other organisms such as yeast (Liu et al., 2002) or frog (McDermott et al., 2010) models. However, given the complexity and context dependence of transporter biology (Kindla et al., 2011; König et al., 2013; Müller and Fromm, 2011) and the particular challenges for generalizing arsenic findings across toxicological models (States et al., 2011), laboratory results from other models should be regarded as hypothesis-generating for human toxicology. Human data, including genetic epidemiology studies and studies quantifying transporters such as by mass spectrometry (Cutler and Choo, 2011), remain important. Epidemiological studies are needed to clarify the role of transporters for arsenic susceptibility in human populations.
One of the best characterized transporters in pharmacology is the organic anion transporter coded by the Solute Carrier Organic Anion Transporter family member 1B1 (SLCO1B1) gene. Perhaps the most clinically important finding is the interaction of rs4149056 with simvastatin that causes myopathy (Wilke et al., 2012), but many other small anionic molecules besides simvastatin are also substrates for this transporter, including arsenicals. The protein coded by SLCO1B1 has been associated with transport of arsenic in a transformed HEK-293 kidney cell line (Lu et al. 2006), but the role of this transporter has not been evaluated in an epidemiological study of arsenic.
The objective of this study was to evaluate the association of variants in SLCO1B1 with arsenic metabolite patterns in urine of participants from the Strong Heart Study (SHS), a cohort recruited from rural communities of the Southwestern and Midwestern United States exposed to low-to-moderate arsenic levels in drinking water. Epidemiological studies often summarize the pattern of arsenic metabolites in urine as proportions of the 3 main inorganic arsenic metabolites (inorganic arsenic “iAs,” monomethylarsonate “MMA,” and dimethylarsinate “DMA”) contributing to their sum. In the SHS, these % arsenic species appear to be stable within individuals over a 10-year interval (Navas-Acien et al., 2009), highly heritable (Tellez-Plaza et al., 2013), and largely independent of arsenic exposure levels (Tellez-Plaza et al., 2013).
MATERIALS AND METHODS
Study population and measures.
The SHS is a prospective cohort study for cardiovascular disease risk factors relevant for American Indian communities from 3 regions: Arizona, Oklahoma, and North and South Dakota (Lee et al., 1990). The study visit included a questionnaire, a physical exam, and biological specimen collection by centrally trained and certified staff (Lee et al., 1990). In the SHS, urine arsenic species were measured in the 1989–1991 baseline visit as a possible cardiovascular disease and diabetes risk factor (Gribble et al., 2012; Scheer et al., 2012). Arsenic species were determined by anion-exchange high-performance liquid chromatography (Agilent 1100, Agilent Technologies, Waldbronn, Germany)-inductively coupled plasma mass spectrometry (Agilent 7700x) and had a detection limit of 0.1 µg/l (Scheer et al., 2012). We excluded 2 participants missing data on total arsenic or arsenic species levels in urine, 222 participants with at least 1 arsenic species below the limit of detection, and 16 participants missing data on body mass index.
The SHS has a genetic component called the Strong Heart Family Study (SHFS), a large pedigree study that includes some of the original SHS participants and their family members (North et al., 2003). Genetic data happened to be available in our candidate gene SLCO1B1 because a previous study in the SHFS identified a linkage region of interest in Chromosome 12 for left ventricular mass, and subsequent fine mapping of that region genotyped several variants in the SLCO1B1 gene in SHFS participants. Single-nucleotide polymorphisms (SNPs) were genotyped at the SHFS Genetic Center using the multiplex VeraCode technology from Illumina according to the manufacturer’s protocol (Illumina, San Diego, California). Details of the technique are reported elsewhere (Voruganti et al., 2010). Cluster calls were checked for accuracy, and genotypes were exported as text files for further use in association analysis. Replica samples were included as controls for genotyping and allele calling consistency. There was an overlap of 157 SHFS participants with urine arsenic species measures above detection limit, data on correlates of % arsenic species, and SLCO1B1 genotypes available for this analysis, coming from Arizona, North Dakota, and South Dakota communities. In general, participants in this subset were slightly older, less likely to be current drinkers or smokers, more often female, and had higher blood pressure than in the full SHFS sample recruited from Arizona, North Dakota, and South Dakota communities (see Supplementary data).
Statistical analysis.
An allelic association test at each locus, with residual polygenic variance component estimated from family relationships (Boerwinkle et al., 1986), was employed to estimate the associations between the dose of each SLCO1B1 polymorphism with the mean level and variance of % arsenic species quantitative traits. Genotype frequencies were calculated for each SNP and tested for departures from Hardy-Weinberg equilibrium. Estimates of linkage disequilibrium (LD) between SNPs were determined by calculating pairwise r 2 statistics. We adjusted for the major correlates of arsenic metabolite patterns in this population: Study center, sex, and body mass index. The % arsenic species traits conformed to normal distributions and were not transformed for association analysis. All analyses were performed in the Sequential Oligogenic Linkage Analysis Routines (SOLAR) software (Almasy and Blangero, 1998). We tested for population stratification in each analysis and as a sensitivity analysis estimated a test statistic robust to population stratification, the quantitative transmission disequilibrium test (QTDT) (Abecasis et al., 2000a,b). The QTDT works by partitioning between-family and within-family associations of the genotype with the trait mean; models constraining the within-family association to 0 are compared with models allowing flexible estimation of the within-family association for a test of association that is robust to population stratification because population stratification has minimum influence on within-family comparisons.
RESULTS
Genetic Variants and Phenotypes
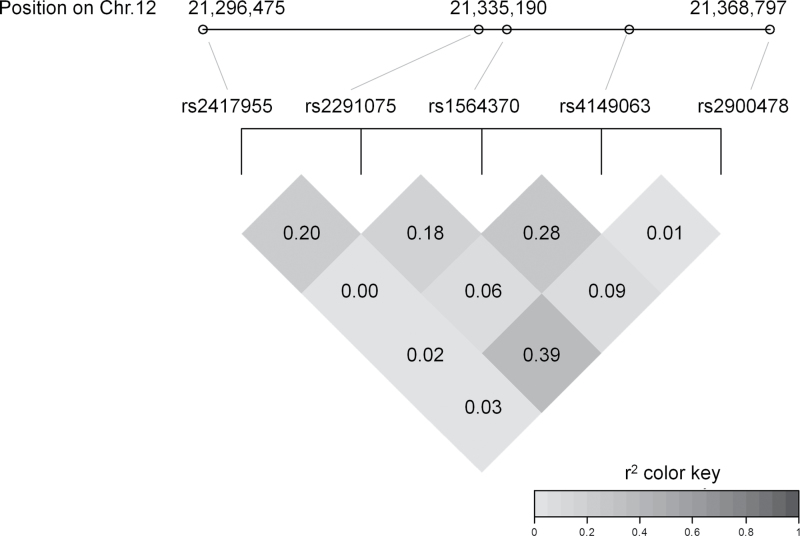
Genotype frequencies are shown in Table 1. One participant failed genotyping at reference SNP 2900478 (rs2900478). We calculated the effective number of SNPs accounting for LD to be 4.3 using the method of Moskvina and Schmidt (2008) and therefore set the alpha (adjusting for multiple testing) at .012. The LD patterns for these SNPs are shown in Figure 1; correlations range between .63 and −.24. The % arsenic species biomarker distribution summaries are presented according to genotype in Table 2.
TABLE 1.
Genotype Frequencies for SLCO1B1 Polymorphisms
| SNP | Genotype | N | Frequency | HWE p Value |
|---|---|---|---|---|
| rs1564370 | C/C | 26 | 0.17 | .90 |
| C/G | 68 | 0.43 | ||
| G/G | 63 | 0.40 | ||
| rs2291075 | A/A | 2 | 0.01 | .42 |
| A/G | 24 | 0.15 | ||
| G/G | 131 | 0.83 | ||
| rs2417955 | A/A | 96 | 0.61 | .93 |
| A/T | 56 | 0.36 | ||
| T/T | 5 | 0.03 | ||
| rs2900478 | Missing | 1 | 0.01 | .71 |
| A/A | 144 | 0.92 | ||
| A/T | 12 | 0.08 | ||
| T/T | 0 | < 0.01 | ||
| rs4149063 | A/A | 4 | 0.03 | .90 |
| A/C | 56 | 0.36 | ||
| C/C | 97 | 0.62 |
Note. No markers showed significant departure from HWE. Abbreviation: HWE, Hardy-Weinberg equilibrium.
FIG. 1.
Linkage disequilibrium (r2) among SNPs in SLCO1B1.
TABLE 2.
Urine Arsenic Metabolite Patterns (% Species) by Genotype
| SNP | Genotype | Trait | Mean | SD | Minimum | Maximum | N |
|---|---|---|---|---|---|---|---|
| Overall | — | %iAs | 8.8 | 4.9 | 1.1 | 31.9 | 157 |
| — | %MMA | 14.7 | 5.2 | 5.1 | 30.7 | 157 | |
| — | %DMA | 76.4 | 8.6 | 31.3 | 91.5 | 157 | |
| rs1564370 | C/C | %iAs | 8.2 | 3.7 | 3.0 | 16.9 | 26 |
| C/G | %iAs | 8.2 | 4.6 | 1.1 | 23.9 | 68 | |
| G/G | %iAs | 9.8 | 5.6 | 1.1 | 38.0 | 63 | |
| C/C | %MMA | 13.9 | 5.2 | 7.4 | 25.4 | 26 | |
| C/G | %MMA | 13.5 | 4.5 | 5.1 | 29.5 | 68 | |
| G/G | %MMA | 16.4 | 5.4 | 8.1 | 30.7 | 63 | |
| C/C | %DMA | 77.9 | 7.7 | 61.8 | 89.5 | 26 | |
| C/G | %DMA | 78.3 | 7.2 | 60.7 | 91.5 | 68 | |
| G/G | %DMA | 73.8 | 9.7 | 31.3 | 87.6 | 63 | |
| rs2291075 | A/A | %iAs | 6.2 | 4.4 | 3.1 | 9.4 | 2 |
| A/G | %iAs | 7.4 | 3.6 | 2.2 | 16.9 | 24 | |
| G/G | %iAs | 9.2 | 5.1 | 1.1 | 38.0 | 131 | |
| A/A | %MMA | 8.6 | 1.6 | 7.4 | 9.8 | 2 | |
| A/G | %MMA | 12.3 | 3.8 | 5.6 | 21.2 | 24 | |
| G/G | %MMA | 15.2 | 5.2 | 5.1 | 30.7 | 131 | |
| A/A | %DMA | 85.2 | 6.1 | 80.9 | 89.5 | 2 | |
| A/G | %DMA | 80.3 | 5.8 | 66.2 | 88.6 | 24 | |
| G/G | %DMA | 75.6 | 8.8 | 31.3 | 91.5 | 131 | |
| rs2417955 | A/A | %iAs | 8.8 | 4.6 | 1.1 | 23.9 | 96 |
| A/T | %iAs | 9.3 | 5.6 | 1.8 | 38.0 | 56 | |
| T/T | %iAs | 5.9 | 2.3 | 3.1 | 9.4 | 5 | |
| A/A | %MMA | 15.1 | 4.9 | 5.8 | 30.3 | 96 | |
| A/T | %MMA | 14.6 | 5.5 | 5.1 | 30.7 | 56 | |
| T/T | %MMA | 9.1 | 2.0 | 7.4 | 12.2 | 5 | |
| A/A | %DMA | 76.2 | 7.8 | 54.8 | 91.5 | 96 | |
| A/T | %DMA | 76.1 | 9.8 | 31.3 | 88.6 | 56 | |
| T/T | %DMA | 85.1 | 3.9 | 80.9 | 89.5 | 5 | |
| rs2900478 | A/A | %iAs | 8.9 | 5.0 | 1.1 | 38.0 | 144 |
| A/T | %iAs | 7.5 | 4.2 | 2.2 | 16.9 | 12 | |
| A/A | %MMA | 15.0 | 5.2 | 5.1 | 30.7 | 144 | |
| A/T | %MMA | 12.2 | 3.5 | 7.9 | 17.2 | 12 | |
| A/A | %DMA | 76.1 | 8.7 | 31.3 | 91.5 | 144 | |
| A/T | %DMA | 80.3 | 6.8 | 66.2 | 87.6 | 12 | |
| rs4149063 | A/A | %iAs | 11.1 | 1.7 | 9.3 | 12.9 | 4 |
| A/C | %iAs | 9.7 | 4.7 | 1.9 | 23.9 | 56 | |
| C/C | %iAs | 8.2 | 5.1 | 1.1 | 38.0 | 97 | |
| A/A | %MMA | 17.8 | 6.3 | 10.0 | 25.4 | 4 | |
| A/C | %MMA | 15.0 | 4.7 | 6.7 | 29.5 | 56 | |
| C/C | %MMA | 14.4 | 5.4 | 5.1 | 30.7 | 97 | |
| A/A | %DMA | 71.1 | 7.8 | 61.8 | 80.7 | 4 | |
| A/C | %DMA | 75.3 | 7.4 | 60.7 | 88.3 | 56 | |
| C/C | %DMA | 77.3 | 9.1 | 31.3 | 91.5 | 97 |
Association Analysis
There were 2 SNPs in SLCO1B1 that were significantly associated with variability in %MMA and %DMA in additive genetic models after adjusting for study center, sex, and body mass index: rs1564370 and rs2291075 (Table 3). Minor alleles of rs1564370 were associated with lower %MMA and higher %DMA, accounting for 9% of the variance for %DMA (p = .0002) and 8% for %MMA (p = .0003) (Table 3). The rs1564370 minor allele homozygote frequency was 17% and the heterozygote frequency was 43%. Minor alleles of rs2291075 were associated with lower %MMA and higher %DMA, accounting for 5% of the residual variance for %DMA (p = .0014) and 7% for %MMA (p = .0006) (Table 3). The rs2291075 association with %MMA had evidence for confounding by population stratification. However, the QTDT yielded the same inferences for association as the main analysis (QTDT p value < .001). The nonsignificant association between rs2417955 and %MMA was possibly also confounded by population stratification, but the QTDT again yielded the same inference (p = .017) for an alpha of .012.
TABLE 3.
Associations of SLCO1B1 Genotypes With % Arsenic Species From Measured Genotype Analysis
| SNP | Trait | p Value | h 2 m | μAA | SE(μAA) | μAB | SE(μAB) | μBB | SE(μBB) | p for Population Stratification |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1564370 | %iAs | .010 | 0.04 | 12.8 | 0.9 | 11.5 | 0.8 | 10.2 | 1.0 | 1.00 |
| %MMA | < .001 | 0.08 | 17.3 | 0.9 | 15.4 | 0.7 | 13.5 | 0.9 | .44 | |
| %DMA | < .001 | 0.09 | 70.0 | 1.4 | 73.3 | 1.2 | 76.6 | 1.6 | .89 | |
| rs2291075 | %iAs | .124 | 0.01 | 11.8 | 0.8 | 10.3 | 1.1 | 8.9 | 1.9 | .61 |
| %MMA | < .001 | 0.07 | 16.0 | 0.7 | 12.8 | 1.1 | 9.6 | 1.9 | .03 | |
| %DMA | .001 | 0.05 | 72.2 | 1.3 | 77.3 | 1.9 | 82.4 | 3.2 | .24 | |
| rs2417955 | %iAs | .607 | < 0.01 | 11.8 | 0.8 | 11.5 | 0.9 | 11.1 | 1.3 | 1.00 |
| %MMA | .185 | 0.01 | 16.0 | 0.8 | 15.0 | 0.8 | 14.1 | 1.3 | .05 | |
| %DMA | .225 | < 0.01 | 72.3 | 1.4 | 73.7 | 1.5 | 75.1 | 2.3 | .09 | |
| rs2900478 | %iAs | .092 | 0.02 | 11.9 | 0.8 | 9.5 | 1.5 | 7.2 | 2.7 | .48 |
| %MMA | .120 | 0.02 | 15.8 | 0.7 | 13.6 | 1.5 | 11.4 | 2.8 | .77 | |
| %DMA | .075 | 0.02 | 72.4 | 1.3 | 76.7 | 2.5 | 81.0 | 4.7 | .84 | |
| rs4149063 | %iAs | .492 | < 0.01 | 11.4 | 0.9 | 11.9 | 0.8 | 12.3 | 1.2 | 1.00 |
| %MMA | .317 | < 0.01 | 15.1 | 0.9 | 15.9 | 0.8 | 16.6 | 1.3 | .76 | |
| %DMA | .372 | < 0.01 | 73.5 | 1.5 | 72.4 | 1.4 | 71.3 | 2.1 | .58 |
Note. The h 2 m is the proportion of the residual phenotypic variance explained by the minor allele. Means and standard errors for each % arsenic species biomarker are presented according to genotype, adjusting for study center, sex, and body mass index, along with a p value for significance of the association and p value for evidence for population stratification. “A” corresponds to the major allele and “B” corresponds to the minor allele for each SNP. Abbreviations: µ, Mean.
DISCUSSION
In this preliminary study, common variants in the SLCO1B1 gene accounted for a substantial amount of phenotypic variance in % arsenic species in urine after controlling for study region, sex, and body mass index. These findings are preliminary evidence in support of the hypothesis that SLCO1B1 may be an important gene for arsenic toxicokinetics in human populations.
One implication from our study is that the % arsenic species biomarkers commonly used to draw conclusions about arsenic metabolism (Engstrom et al., 2013; Gardner et al., 2011; Li et al., 2011) may reflect genetic influences beyond the assumed metabolic drivers. For example, the most commonly studied arsenic metabolism gene, AS3MT, encoding arsenic (III) methyltransferase, is approximately 3018kb (Smith, 2008; Thorisson et al., 2005; HapMap Data Rel 28 phase II + III, August 10, on NCBI 36 assembly, dbSNP b126) from the ABCC2 transporter gene whose protein (MRP2) has strong experimental evidence for arsenic transport (Drobná et al., 2010). Also, AS3MT is in a conserved haplotype block with a metal transporter gene CNNM2 (Engstrom et al., 2013; Gomez-Rubio et al., 2010). Recent analyses suggest that haplotypes of this region are associated with DNA methylation in CNNM2, which in turn corresponds with expression of AS3MT and CNNM2 (Chen Engstrom et al., 2013), but the relationship of CNNM2 has not been evaluated yet with urine % arsenic species. Genetic signals ascribed to methylation in some cases might be confounded by genetic signals coming from nearby transporters. Underestimating the role of transporters may neglect important exposure-exposure interactions between transporter-affecting chemicals and toxicants (Epel et al., 2008). No previous studies have specifically evaluated the association between SLCO1B1 variants and arsenic patterns in urine. Understanding the genetic architecture of arsenic susceptibility more comprehensively might provide additional targets for possible interventions to reduce arsenic toxicity. The potential relevance of SLCO1B1 variation to human population arsenic toxicokinetics does not preclude other transporter genes, perhaps including CNNM2 (Engstrom et al., 2013) or AQP3 (Tellez-Plaza et al., 2013), from also being important. The field of arsenic toxicology is ready for further study of transporters (Hernández and Marcos, 2008).
Although the results of this preliminary study are intriguing, our study has several limitations. The sample size was only 157 participants, which was adequate to detect such a major effect with a limited number of hypothesis tests, but may also increase our chance of a false positive. It is important to replicate our finding in other populations, including other participants of the SHFS. Recent research in Northern Mexico has been exploring the genetic epidemiology of arsenic toxicokinetics (Gomez-Rubio et al., 2010). The comparison of our findings with their study populations, which are more similar in body mass index (Gomez-Rubio et al., 2011) and possibly other attributes relevant for arsenic metabolism than, for instance, populations in Bangladesh (Ahsan et al., 2006; Pierce et al., 2012), would be important. However, we note that the recent genome-wide association scan for urine arsenic species patterns in Bangladesh found an intriguing, but nonsignificant, signal on Chromosome 12 that might reflect the same genetic basis as in our population (Pierce et al., 2012). A second limitation of our study is that it only examined 5 variants in 1 candidate gene. Genotyping additional markers in this region of Chromosome 12 might allow us to better understand the source of the genetic association in future analyses. Neither of the 2 significant SNPs in our study is a known functional mutation. Although we do not know the causal variant, we can speculate about the role SLCO1B1 might play in the toxicokinetics of arsenic. If this liver uptake transporter affects bilary elimination of arsenic species differentially according to species, then differences in the transporter’s function could change the blood distribution of arsenic species, which could affect the arsenic species available to be eliminated at the kidneys into the urine. A third limitation of our study is that we did not measure arsenic species intake or blood arsenic species, so knowledge of the biology of the arsenic transport within the body is limited. However, these limitations do not reduce the potential value of SLCO1B1 for characterizing susceptibility differences to arsenic across individuals. These are common variants with large estimated effect sizes. It would be useful to examine possible modification of the association of arsenic with clinical outcomes by rs1564370 and rs2291075; even if its biological importance is challenging to clarify, the utility for refining risk assessment for susceptible subpopulations could be major.
In conclusion, this preliminary candidate gene study identified novel, substantial, and significant associations between common SLCO1B1 variants and the pattern of arsenic metabolites in urine of a subset of participants in the SHFS. Future research is needed to replicate these associations and examine the possible importance for arsenic-associated disease risks.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Heart Lung and Blood Institute (R01HL090863, SHS grants HL41642, HL41652, HL41654, HL065520, HL65521); National Institute of Environmental Health Sciences (R01ES021367, P30ES03819); National Institute of Diabetes and Digestive and Kidney Diseases (5T32DK062707-10); National Center for Research Resources (C06 RR13556, C06 RR017515).
Supplementary Material
ACKNOWLEDGMENTS
The authors have no conflict of interest to declare.
REFERENCES
- Abecasis G. R., Cardon L. R., Cookson W. O. (2000a). A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 66, 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis G. R., Cookson W. O., Cardon L. R. (2000b). Pedigree tests of transmission disequilibrium. Eur. J. Hum. Genet. 8, 545–551 [DOI] [PubMed] [Google Scholar]
- Achour A. R., Bauda P., Billard P. (2007). Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 158, 128–137 [DOI] [PubMed] [Google Scholar]
- Ahsan H., Chen Y., Parvez F., Argos M., Hussain A. I., Momotaj H., Levy D., van Geen A., Howe G., Graziano J. (2006). Health Effects of Arsenic Longitudinal Study (HEALS): Description of a multidisciplinary epidemiologic investigation. J. Expo. Sci. Environ. Epidemiol. 16, 191–205 [DOI] [PubMed] [Google Scholar]
- Almasy L., Blangero J. (1998). Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62, 1198–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E., Chakraborty R., Sing C. F. (1986). The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann. Hum. Genet. 50(Pt 2), 181–194 [DOI] [PubMed] [Google Scholar]
- Briat J. F. (2010). Arsenic tolerance in plants: “Pas de deux” between phytochelatin synthesis and ABCC vacuolar transporters. Proc. Natl. Acad. Sci. U.S.A. 107, 20853–20854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calatayud M., Barrios J. A., Velez D., Devesa V. (2012). In vitro study of transporters involved in intestinal absorption of inorganic arsenic. Chem. Res. Toxicol. 25, 446–453 [DOI] [PubMed] [Google Scholar]
- Calatayud M., Gimeno J., Vélez D., Devesa V., Montoro R. (2010). Characterization of the intestinal absorption of arsenate, monomethylarsonic acid, and dimethylarsinic acid using the Caco-2 cell line. Chem. Res. Toxicol. 23, 547–556 [DOI] [PubMed] [Google Scholar]
- Carbrey J. M., Song L., Zhou Y., Yoshinaga M., Rojek A., Wang Y., Liu Y., Lujan H. L., DiCarlo S. E., Nielsen S., et al. (2009). Reduced arsenic clearance and increased toxicity in aquaglyceroporin-9-null mice. Proc. Natl. Acad. Sci. U.S.A. 106, 15956–15960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew M. W., Leslie E. M. (2010). Selenium-dependent and -independent transport of arsenic by the human multidrug resistance protein 2 (MRP2/ABCC2): Implications for the mutual detoxification of arsenic and selenium. Carcinogenesis 31, 1450–1455 [DOI] [PubMed] [Google Scholar]
- Catarecha P., Segura M. D., Franco-Zorrilla J. M., García-Ponce B., Lanza M., Solano R., Paz-Ares J., Leyva A. (2007). A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell 19, 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan H., Oruganti M., Krishnamurthy P. (2011). The ATP-binding cassette transporter ABCB6 is induced by arsenic and protects against arsenic cytotoxicity. Toxicol. Sci. 120, 519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. C., Guo Y. L., Su H. J., Hsueh Y. M., Smith T. J., Ryan L. M., Lee M. S., Chao S. C., Lee J. Y., Christiani D. C. (2003a). Arsenic methylation and skin cancer risk in southwestern Taiwan. J. Occup. Environ. Med. 45, 241–248 [DOI] [PubMed] [Google Scholar]
- Chen Y. C., Su H. J., Guo Y. L., Hsueh Y. M., Smith T. J., Ryan L. M., Lee M. S., Christiani D. C. (2003b). Arsenic methylation and bladder cancer risk in Taiwan. Cancer Cause Control 14, 303–310 [DOI] [PubMed] [Google Scholar]
- Cutler M. J., Choo E. F. (2011). Overview of SLC22A and SLCO families of drug uptake transporters in the context of cancer treatments. Curr. Drug Metab. 12, 793–807 [DOI] [PubMed] [Google Scholar]
- Drobná Z., Walton F. S., Paul D. S., Xing W., Thomas D. J., Stýblo M. (2010). Metabolism of arsenic in human liver: The role of membrane transporters. Arch. Toxicol. 84, 3–16 [DOI] [PubMed] [Google Scholar]
- Engström K., Vahter M., Mlakar S. J., Concha G., Nermell B., Raqib R., Cardozo A., Broberg K. (2011). Polymorphisms in arsenic(+III oxidation state) methyltransferase (AS3MT) predict gene expression of AS3MT as well as arsenic metabolism. Environ. Health Perspect. 119, 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström K. S., Hossain M. B., Lauss M., Ahmed S., Raqib R., Vahter M., Broberg K. (2013). Efficient arsenic metabolism–the AS3MT haplotype is associated with DNA methylation and expression of multiple genes around AS3MT. PLoS One 8, e53732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel D., Luckenbach T., Stevenson C. N., Macmanus-Spencer L. A., Hamdoun A., Smital T. (2008). Efflux transporters: Newly appreciated roles in protection against pollutants. Environ. Sci. Technol. 42, 3914–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara J., Fujii Y., Agusa T., Kunito T., Yasuda T., Moritani T., Takeshita H. (2009). Ethnic differences in five intronic polymorphisms associated with arsenic metabolism within human arsenic (+3 oxidation state) methyltransferase (AS3MT) gene. Toxicol. Appl. Pharmacol. 234, 41–46 [DOI] [PubMed] [Google Scholar]
- Fujihara J., Kunito T., Agusa T., Yasuda T., Iida R., Fujii Y., Takeshita H. (2007). Population differences in the human arsenic (+3 oxidation state) methyltransferase (AS3MT) gene polymorphism detected by using genotyping method. Toxicol. Appl. Pharmacol. 225, 251–254 [DOI] [PubMed] [Google Scholar]
- Fujihara J., Soejima M., Koda Y., Kunito T., Takeshita H. (2008). Asian specific low mutation frequencies of the M287T polymorphism in the human arsenic (+3 oxidation state) methyltransferase (AS3MT) gene. Mutat. Res. 654, 158–161 [DOI] [PubMed] [Google Scholar]
- Fujihara J., Soejima M., Yasuda T., Koda Y., Agusa T., Kunito T., Tongu M., Yamada T., Takeshita H. (2010). Global analysis of genetic variation in human arsenic (+3 oxidation state) methyltransferase (AS3MT). Toxicol. Appl. Pharmacol. 243, 292–299 [DOI] [PubMed] [Google Scholar]
- Fujihara J., Yasuda T., Kato H., Yuasa I., Panduro A., Kunito T., Takeshita H. (2011). Genetic variants associated with arsenic metabolism within human arsenic (+3 oxidation state) methyltransferase show wide variation across multiple populations. Arch. Toxicol. 85, 119–125 [DOI] [PubMed] [Google Scholar]
- Gardner R. M., Nermell B., Kippler M., Grandér M., Li L., Ekström E. C., Rahman A., Lönnerdal B., Hoque A. M., Vahter M. (2011). Arsenic methylation efficiency increases during the first trimester of pregnancy independent of folate status. Reprod. Toxicol. 31, 210–218 [DOI] [PubMed] [Google Scholar]
- Gomez-Rubio P., Meza-Montenegro M. M., Cantu-Soto E., Klimecki W. T. (2010). Genetic association between intronic variants in AS3MT and arsenic methylation efficiency is focused on a large linkage disequilibrium cluster in chromosome 10. J. Appl. Toxicol. 30, 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rubio P., Roberge J., Arendell L., Harris R. B., O’Rourke M. K., Chen Z., Cantu-Soto E., Meza-Montenegro M. M., Billheimer D., Lu Z., et al. (2011). Association between body mass index and arsenic methylation efficiency in adult women from southwest U.S. and northwest Mexico. Toxicol. Appl. Pharmacol. 252, 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribble M. O., Howard B. V., Umans J. G., Shara N. M., Francesconi K. A., Goessler W., Crainiceanu C. M., Silbergeld E. K., Guallar E., Navas-Acien A. (2012). Arsenic exposure, diabetes prevalence, and diabetes control in the Strong Heart Study. Am. J. Epidemiol. 176, 865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdi M., Sanchez M. A., Beene L. C., Liu Q., Landfear S. M., Rosen B. P., Liu Z. (2009). Arsenic transport by zebrafish aquaglyceroporins. BMC Mol. Biol. 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández A., Marcos R. (2008). Genetic variations associated with interindividual sensitivity in the response to arsenic exposure. Pharmacogenomics 9, 1113–1132 [DOI] [PubMed] [Google Scholar]
- Hernández A., Xamena N., Sekaran C., Tokunaga H., Sampayo-Reyes A., Quinteros D., Creus A., Marcos R. (2008a). High arsenic metabolic efficiency in AS3MT287Thr allele carriers. Pharmacogenet. Genomics 18, 349–355 [DOI] [PubMed] [Google Scholar]
- Hernández A., Xamena N., Surrallés J., Sekaran C., Tokunaga H., Quinteros D., Creus A., Marcos R. (2008b). Role of the Met(287)Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutat. Res. 637, 80–92 [DOI] [PubMed] [Google Scholar]
- Huang Y. K., Huang Y. L., Hsueh Y. M., Yang M. H., Wu M. M., Chen S. Y., Hsu L. I., Chen C. J. (2008). Arsenic exposure, urinary arsenic speciation, and the incidence of urothelial carcinoma: A twelve-year follow-up study. Cancer Cause Control 19, 829–839 [DOI] [PubMed] [Google Scholar]
- Huang Y. L., Hsueh Y. M., Huang Y. K., Yip P. K., Yang M. H., Chen C. J. (2009). Urinary arsenic methylation capability and carotid atherosclerosis risk in subjects living in arsenicosis-hyperendemic areas in southwestern Taiwan. Sci. Total Environ. 407, 2608–2614 [DOI] [PubMed] [Google Scholar]
- IARC (2004). Some Drinking-Water Disinfectants and Contaminants, Including Arsenic. (IARC Monographs on the Evaluation of Carcinogenic Risks to Humans). IARC, Lyon, France: [PMC free article] [PubMed] [Google Scholar]
- Kala S. V., Neely M. W., Kala G., Prater C. I., Atwood D. W., Rice J. S., Lieberman M. W. (2000). The MRP2/cMOAT transporter and arsenic-glutathione complex formation are required for biliary excretion of arsenic. J. Biol. Chem. 275, 33404–33408 [DOI] [PubMed] [Google Scholar]
- Kindla J., Müller F., Mieth M., Fromm M. F., König J. (2011). Influence of non-steroidal anti-inflammatory drugs on organic anion transporting polypeptide (OATP) 1B1- and OATP1B3-mediated drug transport. Drug Metab. Dispos. 39, 1047–1053 [DOI] [PubMed] [Google Scholar]
- Kojima C., Qu W., Waalkes M. P., Himeno S., Sakurai T. (2006). Chronic exposure to methylated arsenicals stimulates arsenic excretion pathways and induces arsenic tolerance in rat liver cells. Toxicol. Sci. 91, 70–81 [DOI] [PubMed] [Google Scholar]
- König J., Müller F., Fromm M. F. (2013). Transporters and drug-drug interactions: Important determinants of drug disposition and effects. Pharmacol. Rev. 65, 944–966 [DOI] [PubMed] [Google Scholar]
- Lee E. T., Welty T. K., Fabsitz R., Cowan L. D., Le N. A., Oopik A. J., Cucchiara A. J., Savage P. J., Howard B. V. (1990). The Strong Heart Study. A study of cardiovascular disease in American Indians: Design and methods. Am. J. Epidemiol. 132, 1141–1155 [DOI] [PubMed] [Google Scholar]
- Lee T. C., Ho I. C., Lu W. J., Huang J. D. (2006). Enhanced expression of multidrug resistance-associated protein 2 and reduced expression of aquaglyceroporin 3 in an arsenic-resistant human cell line. J. Biol. Chem. 281, 18401–18407 [DOI] [PubMed] [Google Scholar]
- Li X., Li B., Xu Y., Wang Y., Jin Y., Itoh T., Yoshida T., Sun G. (2011). Arsenic methylation capacity and its correlation with skin lesions induced by contaminated drinking water consumption in residents of chronic arsenicosis area. Environ. Toxicol. 26, 118–123 [DOI] [PubMed] [Google Scholar]
- Liu Z., Shen J., Carbrey J. M., Mukhopadhyay R., Agre P., Rosen B. P. (2002). Arsenite transport by mammalian aquaglyceroporins AQP7 and AQP9. Proc. Natl. Acad. Sci. U.S.A. 99, 6053–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W. J., Tamai I., Nezu J., Lai M. L., Huang J. D. (2006). Organic anion transporting polypeptide-C mediates arsenic uptake in HEK-293 cells. J. Biomed. Sci. 13, 525–533 [DOI] [PubMed] [Google Scholar]
- Maull E. A., Ahsan H., Edwards J., Longnecker M. P., Navas-Acien A., Pi J., Silbergeld E. K., Styblo M., Tseng C. H., Thayer K. A., et al. (2012). Evaluation of the association between arsenic and diabetes: A National Toxicology Program workshop review. Environ. Health Perspect. 120, 1658–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott J. R., Jiang X., Beene L. C., Rosen B. P., Liu Z. (2010). Pentavalent methylated arsenicals are substrates of human AQP9. Biometals 23, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K., Guallar E., Navas-Acien A. (2012). Arsenic exposure and cardiovascular disease: An updated systematic review. Curr. Atheroscler. Rep. 14, 542–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina V., Schmidt K. M. (2008). On multiple-testing correction in genome-wide association studies. Genet. Epidemiol. 32, 567–573 [DOI] [PubMed] [Google Scholar]
- Müller F., Fromm M. F. (2011). Transporter-mediated drug-drug interactions. Pharmacogenomics 12, 1017–1037 [DOI] [PubMed] [Google Scholar]
- Navas-Acien A., Umans J. G., Howard B. V., Goessler W., Francesconi K. A., Crainiceanu C. M., Silbergeld E. K., Guallar E. (2009). Urine arsenic concentrations and species excretion patterns in American Indian communities over a 10-year period: The Strong Heart Study. Environ. Health Perspect. 117, 1428–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North K. E., Howard B. V., Welty T. K., Best L. G., Lee E. T., Yeh J. L., Fabsitz R. R., Roman M. J., MacCluer J. W. (2003). Genetic and environmental contributions to cardiovascular disease risk in American Indians: The strong heart family study. Am. J. Epidemiol. 157, 303–314 [DOI] [PubMed] [Google Scholar]
- Pierce B. L., Kibriya M. G., Tong L., Jasmine F., Argos M., Roy S., Paul-Brutus R., Rahaman R., Rakibuz-Zaman M., Parvez F., et al. (2012). Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet. 8, e1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J., Findenig S., Goessler W., Francesconi K. A., Howard B., Umans J. G., Pollak J., Tellez-Plaza M., Silbergeld E. K., Guallar E., et al. (2012). Arsenic species and selected metals in human urine: Validation of HPLC/ICPMS and ICPMS procedures for a long-term population-based epidemiological study. Anal. Methods 4, 406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. V. (2008). Browsing HapMap data using the Genome Browser. CSH Protoc. 2008, pdb.prot5023. [DOI] [PubMed] [Google Scholar]
- States J. C., Barchowsky A., Cartwright I. L., Reichard J. F., Futscher B. W., Lantz R. C. (2011). Arsenic toxicology: Translating between experimental models and human pathology. Environ. Health Perspect. 119, 1356–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez-Plaza M., Gribble M. O., Voruganti V. S., Francesconi K. A., Goessler W., Umans J. G., Silbergeld E. K., Guallar E., Franceschini N., North K. E., et al. (2013). Heritability and preliminary genome-wide linkage analysis of arsenic metabolites in urine. Environ. Health Perspect. 121, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. J. (2009). Unraveling arsenic–glutathione connections. Toxicol. Sci. 107, 309–311 [DOI] [PubMed] [Google Scholar]
- Thorisson G. A., Smith A. V., Krishnan L., Stein L. D. (2005). The International HapMap Project Web site. Genome Res. 15, 1592–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahter M., Concha G. (2001). Role of metabolism in arsenic toxicity. Pharmacol. Toxicol. 89, 1–5 [DOI] [PubMed] [Google Scholar]
- Villa-Bellosta R., Sorribas V. (2008). Role of rat sodium/phosphate cotransporters in the cell membrane transport of arsenate. Toxicol. Appl. Pharmacol. 232, 125–134 [DOI] [PubMed] [Google Scholar]
- Villa-Bellosta R., Sorribas V. (2010). Arsenate transport by sodium/phosphate cotransporter type IIb. Toxicol. Appl. Pharmacol. 247, 36–40 [DOI] [PubMed] [Google Scholar]
- Voruganti V. S., Cole S. A., Ebbesson S. O., Göring H. H., Haack K., Laston S., Wenger C. R., Tejero M. E., Devereux R. B., Fabsitz R. R., et al. (2010). Genetic variation in APOJ, LPL, and TNFRSF10B affects plasma fatty acid distribution in Alaskan Eskimos. Am. J. Clin. Nutr. 91, 1574–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chen G., Jiang J., Qiu L., Hosoi K., Yao C. (2009). Aquaglyceroporins are involved in uptake of arsenite into murine gastrointestinal tissues. J. Med. Invest. 56(Suppl), 343–346 [DOI] [PubMed] [Google Scholar]
- Wilke R. A., Ramsey L. B., Johnson S. G., Maxwell W. D., McLeod H. L., Voora D., Krauss R. M., Roden D. M., Feng Q., Cooper-Dehoff R. M., et al. Clinical Pharmacogenomics Implementation Consortium (CPIC) (2012). The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin. Pharmacol. Ther. 92, 112–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T. C., Salavagionne O. E., Mukherjee B., Wang L., Klumpp A. F., Thomae B. A., Eckloff B. W., Schaid D. J., Wieben E. D., Weinshilboum R. M. (2006). Human arsenic methyltransferase (AS3MT) pharmacogenetics: Gene resequencing and functional genomics studies. J. Biol. Chem. 281, 7364–7373 [DOI] [PubMed] [Google Scholar]
- Xie Y., Liu J., Liu Y., Klaassen C. D., Waalkes M. P. (2004). Toxicokinetic and genomic analysis of chronic arsenic exposure in multidrug-resistance mdr1a/1b(-/-) double knockout mice. Mol. Cell. Biochem. 255, 11–18 [DOI] [PubMed] [Google Scholar]
- Zhao F. J., Ago Y., Mitani N., Li R. Y., Su Y. H., Yamaji N., McGrath S. P., Ma J. F. (2010). The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol. 186, 392–399 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.