Abstract
Background
Recent molecular characterization studies have identified clinically relevant molecular subtypes to coexist within the same histological entities of glioma. Comparative studies between serum-supplemented and serum-free (SF) culture conditions have demonstrated that SF conditions select for glioma stem-like cells, which superiorly conserve genomic alterations. However, neither the representation of molecular subtypes within SF culture assays nor the molecular distinctions between successful and nonsuccessful attempts have been elucidated.
Methods
A cohort of 261 glioma samples from varying histological grades was documented for SF culture success and clinical outcome. Gene expression and single nucleotide polymorphism arrays were interrogated on a panel of tumors for comparative analysis of SF+ (successful cultures) and SF− (unsuccessful cultures).
Results
SF culture outcome was correlated with tumor grade, while no relation was found between SF+ and patient overall survival. Copy number–based hierarchical clustering revealed an absolute separation between SF+ and SF− parental tumors. All SF+ cultures are derived from tumors that are isocitrate dehydrogenase 1 (IDH1) wild type, chromosome 7 amplified, and chromosome 10q deleted. SF− cultures derived from IDH1 mutant tumors demonstrated a fade-out of mutated cells during the first passages. SF+ tumors were enriched for The Cancer Genome Atlas Classical subtype and intrinsic glioma subtype-18. Comparative gene ontology analysis between SF+ and SF− tumors demonstrated enrichment for modules associated with extracellular matrix composition, Hox-gene signaling, and inflammation.
Conclusion
SF cultures are derived from a subset of parental tumors with a shared molecular background including enrichment for extracellular matrix–associated gene modules. These results provide leads to develop enhanced culture protocols for glioma samples not propagatable under current SF conditions.
Keywords: extracellular matrix, glioma, IDH1, molecular subtype, serum-free cultures
Glial tumors consist of a heterogeneous group of primary CNS neoplasms. While the clinical outcome of these tumors varies substantially between different grades (World Health Organization [WHO] grades I–IV), only grade I tumors can be treated curatively.1 In the last decade, substantial effort has been put into characterizing the pathogenic mechanisms underlying this complex group of glial cell–derived tumors. These efforts have led to the establishment of several molecular subclasses of glioma, which differentiate types of gliomas based on their intrinsic molecular distinctions and similarities, rather than the conventional characteristics used for histology-based grading.2–5 Indeed, gene expression–based clustering of glioma from several grades within the WHO grading system proved more predictive of survival than the histological classification.4, 6 Thus far, however, this gain of insight has not led to improved clinical outcome for patients, since there are no specific treatment strategies designed to target specific subtypes of glioma as of yet. Consequently, the development of preclinical models that accurately reflect the molecular heterogeneity of glioma is imperative for both translationally relevant drug screening programs and the advancement of patient-tailored treatment options.
Commercially available cell cultures of glioma have been demonstrated to poorly mimic the molecular aberrations found in patient samples.7,8 Furthermore, the xenograft models derived from these cultures do not sufficiently recapitulate the histological hallmarks of glioma.9 With the advent of the cancer stem cell hypothesis, several groups have implemented serum-free (SF) cell culture regimens, originally developed for neural stem cell propagation, to establish glioma stem-like cell (GSC) cultures from fresh tumor tissue.10,11 By analyzing gene expression profiles of GSCs and serum supplemented (SS) cultures, a distinct separation between the aforementioned and the parental tumors was revealed by unsupervised clustering.7 Several groups have reported GSC cultures to be superior with regard to retaining the original patient gene expression signature and histological phenotype in xenografts.7,11,12 This has led to a wide variety of applications for these cell culture assays, ranging from inquiries into fundamental hypotheses13,14 to preclinical testing of novel agents.15–17
Several contradicting publications have since been published on the optimal cell culture methodology,18,19 mandatory molecular aberrations for successful propagation,20–22 and positive selection for gene expression signatures related to specific molecular subtypes in vitro.14, 23 Most of these previous publications have focused on the characterization of successfully propagated specimens of glioblastoma multiforme (GBM; WHO grade IV) from adults. Reports on the culture success of grade II and grade III gliomas are sparse.20,24 Therefore, we undertook a characterization study of a large cohort (N = 261), which addresses the distribution of glioma from all histological entities for the outcome of GSC culture attempt. Within equal WHO grades, correlations between cell culture outcome and patient overall survival were assessed. Tumor samples of both successful and unsuccessful cultures (n = 46 in total) were also subjected to molecular analysis, and a number of molecular traits that influence cell culture success rate were identified, as well as genes that may play a role in this process. These results emphasize the need for, and provide leads to, the development of improved culture protocols supporting growth of all subtypes of glioma. This is essential for implementation of this model in drug screening programs for personalized treatment strategies.
Materials and Methods
Glial Stem-like Cell Cultures and Serum-supplemented Cultures From Glioma Resection Specimens
A detailed protocol for SS and SF culture establishment from primary glioma samples is included in the supplementary information (Supplementary Methods and Materials). In short, tumor specimens were handled within 2 h postresection. Dissociated tumor cells were plated in Dulbecco's modified Eagle's medium (DMEM)–F12 with 1% penicillin/streptomycin, B27 (Invitrogen), human epidermal growth factor (EGF; 5 μg/mL), human basic fibroblast growth factor (FGF; 5 μg/mL) (both from Tebu-Bio), and heparin (5 mg/mL; Sigma-Aldrich). Passaging of proliferating GSC cultures was performed on growth factor reduced extracellular matrix (ECM)–coated plates (BD Biosciences). Tumor sphere formation was tested regularly by plating passaged cell cultures from coated to noncoated flasks. SS cultures were established in parallel with GSC cultures from 25%–50% of the total yield of cell pellet derived from the dissociation process, depending on total volume after visual inspection. For all samples, the use of patient tumor material was acquired with informed consent from patients as approved by the institutional review board of the Erasmus Medical Center, Rotterdam. Cell culture images were acquired on the Incucyte-FLR system (Essen Bioscience).
Nucleic Acid Isolation, cDNA Synthesis, and Array Hybridization From Tumor and Cell Culture Specimens
Samples were selected based on volume (for isolation of both DNA and RNA) and tissue viability (as verified by histological examination using conventional hematoxylin and eosin staining). Total RNA and genomic DNA were isolated from cell culture pellets or from fresh frozen tissue samples (DNeasy or RNeasy isolation kits [Qiagen]). DNA and RNA concentration thresholds were 25 ng/μL and 50 ng/μL, respectively. RNA quality was assessed on a Bioanalyzer (Agilent). RNA integrity numbers >6.5 were used for our experiments. Sample labeling, DNA amplification, and array hybridization for SNP6.0 arrays were performed at AROS Applied Biotechnology, according to standard array manufacturer’s protocol (Affymetrix) with 100–500 ng total DNA per sample.
The whole-genome expression cDNA-mediated annealing, selection, extension, and ligation (DASL) assay, HumanHT-12 v4 beadchip (Illumina), was performed at AROS Applied Biotechnology, according to Illumina instructions with a minimum of 400 ng total RNA per sample.
Copy Number Analysis on Tumor and Cell Culture Samples
After quality control inspection, raw data files in the .CEL format were loaded into Partek Genomics Suite vv6.5 and 6.6 and annotated for sample identification. Before allele intensities and copy number ratios were calculated, batch effects of separately run cohorts were removed by algorithms distributed by the software manufacturer. Samples were normalized and log2 transformed, and subsequently copy number intensities were calculated against the SNP6.0 hapmap reference file. Histograms were plotted for detected segments for both the SF+ and the SF−/SFnp groups. Unsupervised hierarchical clustering was performed on copy number intensities on Euclidian based algorithms developed by the manufacturer.
Loss of heterozygosity for 1p19q information was attained from microsatellite analysis or copy number intensity measurements on SNP6.0 arrays. Microsatellite analysis was performed as described previously.25 Direct sequencing of isocitrate dehydrogenase 1 (IDH1) exon 4 mutations was performed as described previously.4
Gene Expression Profiling
For comparative analysis, raw probe calls were log2 transformed and quantile normalized before processing. Differentially expressed genes were detected through 1-way ANOVAs with the SF result as the discriminating factor. Genes with a P-value <.05 and that were 2-fold higher or lower in the SF+ group were used for gene set enrichment studies. Gene ontology libraries used were supplied through DAVID.26 Biological function and cellular component libraries were checked for at least 3 genes per module and a P-value <.05.
Statistics
Apart from molecular characterization data, all clinical data statistical analysis was performed in SPSS Statistics v19 (IBM). P-values are provided in the legends or the text. Group proportion statistics are based on univariate analysis based on Fishers exact test. Survival statistics are based on Wilcoxon log-rank pairwise comparisons.
Results
Malignant Gliomas of All Histological Grades Can Be Propagated in Serum-free Medium
To evaluate the success rate of primary glioma in SF cultures, we set up a tissue handling routine for a systematic and reproducible throughput of primary tumor samples. After ruling out technical or contaminating sources for failure, we analyzed a panel of 261 individual specimens, covering the complete spectrum of histological grades of malignant glioma (Table 1). The ability to form tumor spheres after dissociation (n = 165, 63%) was merely indicative for initial selection of glioma-initiating cells. SF culture outcome was not unambiguous, since a subset of cases (35%, termed SFnp) did initially yield spheres, but these would senesce or expire within 5 passages. The SF+ group (28%) represents cultures that proliferated 6 passages or more, exhibiting long-term expansion suitable for reproducible experiments. The majority of these cultures were kept in culture past p10 (n = 55, 75%), demonstrating the capacity of a (sub)population of these cultures to drive self-renewal. The SF− group (37%) consisted of tumors that did not form any spheres after dissociation. These cultures failed in p0, despite transferring floating aggregates onto ECM coating.
Table 1.
SF culture results related to histological WHO grade
| Histology | SF+ | SF− | SFnp | Total, % |
|---|---|---|---|---|
| A | 1 | 9 | 10*** | 8 |
| OA | 1 | 0 | 4 | 2 |
| OD | 1 | 10 | 8 | 7 |
| Grade II, n (%) | 3 | 19 | 22 | 44 (17%) |
| AA | 3 | 10 | 7 | 8 |
| AOA | 3 | 4 | 4 | 4 |
| AOD | 3 | 7 | 7 | 7 |
| Grade III, n (%) | 9 | 21 | 18 | 48 (18%) |
| GBM | 54** | 51 | 41 | 56 |
| GBM-r | 7 | 4 | 5 | 6 |
| GBM-s | 0 | 2 | 5 | 3 |
| Grade IV, n (%) | 61* | 57 | 51 | 169 (65%) |
| Total (%) | 73 (28%) | 97 (37%) | 91 (35%) | 261 |
SF culture results in relation to histopathological diagnosis. Abbreviations: SF−, failed to form tumor spheres under SF conditions; SFnp, failed to proliferate for more than 5 passages under SF conditions; SF+, thrived past 5 passages under SF conditions; A, astrocytoma (WHO II); AA, anaplastic astrocytoma (WHO III); OD, oligodendroglioma (WHO II); AOD, anaplastic oligodendroglioma (WHO III); OA, oligoastrocytoma (WHO II); AOA, anaplastic oligoastrocytoma (WHO III); GBM-r, recurrence of primary GBM (WHO IV); GBM-s, GBM secondary to known grades II–III glioma. *P < .05 compared with SF+ grade 2 and grade 3. **P < .05 compared with GBM SF− and SFnp, ***P < .05 compared with A SF+.
As expected, the yield of SF+ within grades II and III glioma is substantially lower (n = 12 of 92 attempts [13%]) compared with grade IV (n = 61 of 169 [36%], P < .05). Grade II astrocytoma was significantly associated with SFnp (P < .05). As anticipated, for primary GBM, 37% were successfully cultured under SF conditions, which is in great contrast to secondary GBM (0/7). The proportion was even larger in recurrent GBM, where 7/16 (44%) succeeded. Taken together, SF culture media are suitable for propagation of all histological subtypes of glioma, except secondary GBM. However, in general, the probability of SF culture success increases with WHO grade.
Serum-free Culture Success Is Not Prognostic of Clinical Outcome
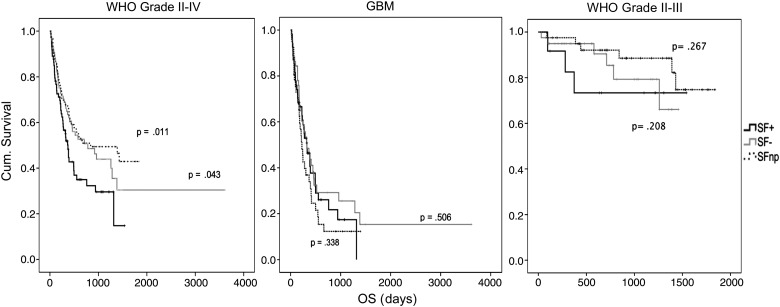
Other studies have suggested that sphere formation is prognostic for clinical outcome in both adult and pediatric glial tumors.24,27,28 Therefore, we compared overall survival of the 261 patients in this study with regard to SF culture outcome of patient-derived tumor tissue. Overall survival of SF+ (median = 373 days, 95% confidence interval [CI] = 301–445), SF− (median = 783 days, 95% CI = 301–1265), and SFnp (median = 841 days, 95% CI = 48–1634) was significantly different for SF+ versus SFnp samples (Wilcoxon log-rank P = .011) and for SF+ versus SF− (P = .043) (Fig. 1). However, comparison of these groups within the same histology demonstrated that the differences in overall survival were related mainly to tumor grade. Indeed, differences within the GBM samples were found to be nonsignificant for SF+ versus SF− (P = .506) and SF+ versus SFnp (P = .338). Within the grades II–III tumors, a trend was observed for dismal survival in the SF+ group. At the time of writing, a significant difference was not reached, since the majority of grades II–III patients are alive to date (SF+ vs SF− [P = .208] and SF+ vs SFnp [P = .267]).
Fig. 1.
SF culture outcome is not associated with worse clinical outcome in GBM. Kaplan–Meier-based overall survival (OS) curves for SF culture results for indicated histological entities of malignant glioma. SF+ (black), SF− (gray), and SFnp (dotted). Log-rank (Mantel–Cox) P-values are based on SF+ vs SF− or SF+ vs SFnp. Note a trend in decreased OS for SF+ grades II–III samples, while GBM samples show no difference.
SF+ Cultures Recapitulate TCGA-Defined Genotypic Hallmarks of Parental Tumor Tissue
The ability to retain driver mutations in vitro is crucial for the implementation of SF cultures for drug screening assays. To address the genotypic similarity of parental tumors and the derived in vitro progeny, a panel of SF+, SF−, and SFnp tumor samples (n = 27) was interrogated on single nucleotide polymorphism (SNP) whole-genome arrays. Samples were selected for being representative of SF culture outcome with regard to proliferation, morphology, and pattern of extinction in vitro. Frequently occurring copy number alterations (CNAs), as previously reported in The Cancer Genome Atlas (TCGA),29 were observed in both SF+ and SF−/SFnp parental tumors. Specifically, we found focal amplifications on segments containing EGF receptor (EGFR), cyclin-dependent kinase CDK)4, and murine double minute (MDM) 2, but not in platelet derived growth factor receptor–α (Supple-mentary data, Table S1). For 10 individual cases, we added early (before p4) and late passages (past p4, before p12) to evaluate genotypic stability in vitro under SF conditions (Supplementary data, Fig. S1). Copy number intensity analysis revealed that both early and late passages of SF+ cultures retain hallmark somatic CNAs.
Serum Cultures of SFnp Tumors, but Not SF− Tumors, Retain Parental Copy Number Alterations for a Number of Passages
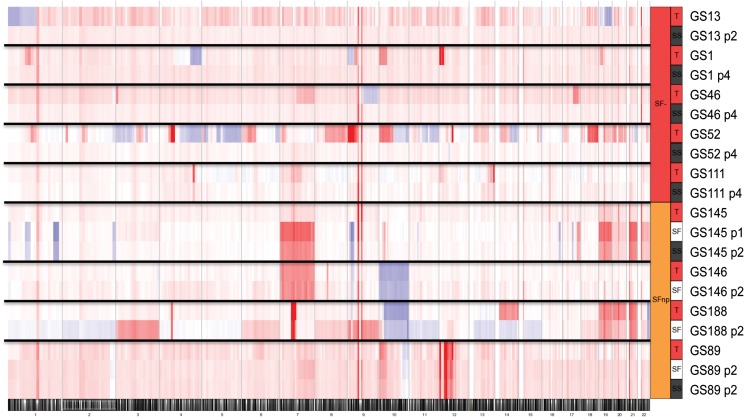
Since a considerable number of SFnp/SF− samples do proliferate under SS conditions, we investigated whether this platform could serve as an alternative culture model for this specific subgroup of tumors. It has been reported that serum cultures tend to drift from the original genotypic profile by either the emergence of de novo mutations or the total loss of cancer cells due to overgrowth by nonneoplastic cells.12 Indeed, for all SF− (n = 5) samples cultured under serum conditions, we noticed a loss of parental CNAs (Fig. 2), coinciding with morphological changes of the tumor cells from a heterogeneous and polarized phenotype to a more fibroblastic phenotype. On the contrary, parallel-established early passages of both SF and serum cultures derived from SFnp tumors (n = 4) were found to harbor mutations analogous to the parental tumor (Fig. 2). We therefore conclude that SS culture does not offer an alternative for culture of SF− tumors; however, it may support low passage drug screening experiments on SFnp samples, since these samples retain (at least for a limited number of passages) their genomic profile. In addition, there is an apparent molecular distinction between SF− and SFnp tumors with regard to the ability to proliferate as SS cultures.
Fig. 2.
Low passage cultures from SFnp tumors retain CNAs as found in parental tumors, while SS cultures from SF− tumors are overgrown by nonneoplastic cells. Heat map plots of copy number analysis results demonstrated loss of aberrations in SS cultures derived from SF− tumors. Each band is composed of the genomic data of 1 sample, as indicated in the legend on the right. Color-coding indicates blue for losses, red for gains, and white for nonaffected. Numbers below indicate chromosome number.
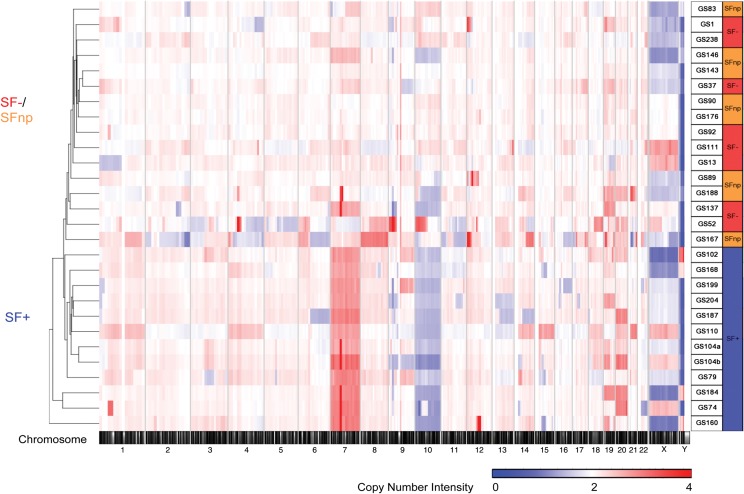
Molecular Characterization Based on Copy Number Alterations Identifies an Absolute Separation Between SF+ and SF−/SFnp Tumors
The distinction between SF+ versus SFnp/SF− tumors with regard to CNA conservation in vitro led us to hypothesize that a specific molecular mechanism dictates SF culture outcome. We therefore investigated differences in CNAs occurring between the 3 SF outcome groups, by copy number intensity–based unsupervised hierarchical genome clustering on patient tissue (n = 27). Strikingly, SF+ separated completely from SF− and SFnp tumors on the cluster dendrogram (Fig. 3). Furthermore, SF− and SFnp intermixed within the cluster dendrogram, illustrating less genomic variation between these groups individually than between the combined SF−/SFnp group and SF+ samples. Moreover, a principal component analysis of SNP allele intensities between SF+ and SF−/SFnp samples revealed a similar clear separation between the 2 groups (Supplementary data, Fig. S2).
Fig. 3.
Genomic analysis of parental tumors reveals complete separation of SF+ and SF−/SFnp samples. Copy number intensity–based unsupervised hierarchical clustering of SF+ and SF−/SFnp tumors. Heat map intensities are coded between copy number intensity varying 0–4n. Note the similarity between SF−/SFnp samples as indicated by the integration of the samples within the same cluster dendrogram tree.
Detailed analysis showed that all SF+ tumors (n = 11) demonstrated gains of chromosome 7 (7p+) and loss of heterozygosity of chromosome 10q or the entire chromosome 10 (SF− P = .0009 and SFnp P = .0027) (Fig. 3, Table 2). These 2 arms harbor the EGFR (chr.7p11.2) and phosphatase and tensin homolog (PTEN) (chr.10q23.31) loci, respectively, which were both found to be significantly discriminating between SF+ and SF−/SFnp cohorts (P < .001 in an ANOVA based on SNP allele intensity). However, Fishers exact tests based on EGFR amplification status demonstrated no significant difference between SF+ and SF−/SFnp cohorts. Although CNAs on CDKN2A/B were more frequently found in SF+ tumors, there was no significant difference between SF+ and SF− or SFnp. Other differentially affected segments were found on chr1p, 12q, and 15q, which do not harbor focal somatic mutations, as reported by TCGA, but may carry drivers of SF culture results because these segments are frequently affected in both low-grade and high-grade glioma.30
Table 2.
SF+ samples are positively corrected with CNA 7p10q, CLA and IGS-18 subtype and IDH1-WT tumors
| Group |
||||
|---|---|---|---|---|
| SF− | SFnp | SF+ | P | |
| n = 12 | n = 15 | n = 19 | ||
| TCGA | ||||
| ND | 1 | 2 | 2 | |
| CLA | 2 | 3 | 11 | .0086 |
| MES | 2 | 2 | 2 | |
| NEU | 1 | 2 | 2 | |
| PRO | 6 | 6 | 2 | .0183 |
| IGS | ||||
| ND | 1 | 2 | 2 | |
| 16 | 0 | 0 | 1 | |
| 17 | 3 | 6 | 3 | .296 |
| 18 | 3 | 1 | 10 | .0078 |
| 22 | 1 | 0 | 0 | |
| 23 | 1 | 3 | 3 | |
| 9 | 3 | 3 | 0 | .032 |
| IDH | ||||
| MUT | 7 | 6 | 0 | .00021 |
| WT | 5 | 9 | 19 | |
| 1p19q loss | ||||
| ND | 4 | 2 | 8 | |
| Loss | 4 | 3 | 0 | .066 |
| NC | 4 | 10 | 11 | |
| Chr7p10q | ||||
| ND | 6 | 10 | 8 | |
| Normal | 5 | 4 | 0 | |
| Present | 1 | 1 | 11 | .00022 |
| EGFR | ||||
| ND | 6 | 10 | 8 | |
| n < 4 | 5 | 3 | 7 | |
| n > 4 | 1 | 2 | 4 | |
| CDKN2A/B | ||||
| ND | 6 | 10 | 8 | |
| NC | 3 | 3 | 2 | |
| n < 2 | 3 | 2 | 9 | .18 |
Table illustrates an overview of the distribution of previously identified driver alterations and subtypes associated with glioma. Significance levels are derived from Fishers exact test–based comparison between SF+ vs SF−/SFnp combined cohort. Abbreviations (for group names similar to Table 1): ND, not determined; NC, no change; MES, mesenchymal; NEU, neuronal; PRO, proneural; CLA, classical. 1p19q signifies the combined loss of chromosome 1p and 19q. Chr7p10q, the combined gain of 7p and loss of 10q.
Gene Expression Profiling of SF+ Tumors Reveals Enrichment for the TCGA Classical and Intrinsic Glioma–18 Subtypes
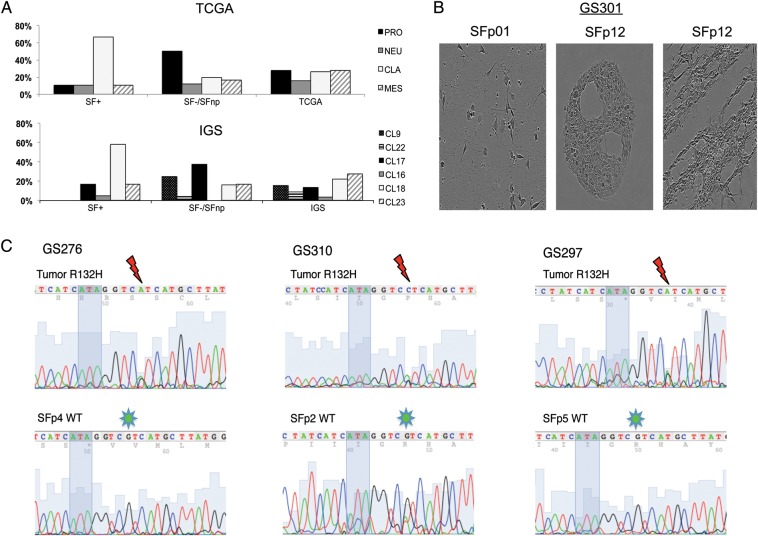
Gene expression–based molecular subtyping has been reported to correlate with specific CNA profiles in GBM and to have prognostic and predictive value.4,5,31 We therefore assessed whether the spectrum of reported subtypes was represented within the SF+ tumors. Gene signatures from 2 previously published molecular classifiers based on WHO grade IV glial tumors (referred to as the TCGA classifier)5 and WHO grades I–IV glial tumors (here referred to as the intrinsic glioma subtype [IGS] classifier4) were determined in parental tissue. We expanded the previously described cohort of samples used for SNP profiling with a selection of SF+, SF−, and SFnp samples (n = 41) (Supplementary data, Table S2). All TCGA subtypes were represented in the SF+ group, although a significant enrichment for the Classical (CLA) subtype was revealed (64% vs 27% incidence in the TCGA dataset). The SF−/SFnp cohort was enriched for PRO tumors, while the only SF+ PRO samples were histologically grade II-III samples (Fig. 4A and Supplementary data, Table S2).
Fig. 4.
Gene expression profiling and IDH1 sequencing data demonstrates SF culture selects for TCGA CLA/IGS-18 and IDH1 wild-type tumors. (A) Distribution of SF+ and SF− samples across the previously published TCGA and IGS classifiers. Samples were assigned according to the centroids as published in the original reports, on the right. Proneural samples are spread between IGS-9, -17, and -22. Noteworthy is the lack of IGS-9 and -22 in the SF+ group, as well as the overrepresentation of IGS-18/CLA samples in the SF+ population compared with the incidences reported in the original publications. (B) Sequencing illustrations of IDH1 codons 130 (ATA in shadow) through 134. Thundermarks indicate G to A or C mutation in codon R132, indicative of R132H. Note the loss of this mutation in GS276 and GS297 SF passages. In GS310 SFp2 the sequence reveals a minor A peak, demonstrating an ambiguous mutation pattern, indicative of mixed populations of mutated tumor cells and (nonneoplastic) IDH-WT cells. (C) Microscopic images of GS301 SS/SF cultures demonstrate loss of neoplastic cells. SF culture retains original morphology as found in p0 for a longer duration than SS; however, eventually (p12) proliferation of nonneoplastic stromal cells is abundant with typical accumulation of bands and halos suggestive for fibroblasts.
As our cohort contains tumors from all WHO grades, we also evaluated the distribution of the IGS classifier set within our SF+ cohort. This classifier has been reported to be prognostic of clinical outcome in gliomas of all WHO grades. Considerable overlap was demonstrated between IGS and TCGA subtypes,4 which is recapitulated within our cohort (Table 2). For instance, IGS-18 was found to be overrepresented in the SF+ group (58%) compared with the original IGS dataset (22.5%), which is congruent with the fact that this subtype resembles the TCGA CLA subtype. Additionally, the TCGA PRO subtype is associated with 2 separate IGSs, which were coined IGS-9 and IGS-17. Despite the fact that both subtypes are enriched for IDH1 mutated and cytosine–phosphate–guanine island methylator phenotype (CIMP) tumors, median survival between the 2 subtypes differed almost 2-fold (6.06 y in IGS-9 vs 3.3 y in IGS-17). Interestingly, IGS-9 tumors were not represented in our SF+ cohort, whereas IGS-17 samples were (n = 3), and both of these IGSs were overrepresented in our SF−/SFnp cohort (Fig. 4A, Supplementary data, Table S2).
Culture Success Is Not Solely Dependent on EGFR Copy Number or Expression Level
With the demonstrated enrichment for molecular subtypes enriched for EGFR gains and amplifications (TCGA CLA-mesenchymal-neuronal and IGS-18-23-16) in culture medium supplemented with a surfeit of ligand (EGF), we hypothesized that one of the driving mechanisms might originate from the dependency that EGFR-expressing cells have under EGF supplemented medium. However, when EGF was omitted from the medium of established SF+ cultures, no effect on proliferation or viability was found in 9 out of 12 SF+ cultures (Supplementary data, Fig. S3A-B). From these 12 cultures, 2 samples (GS184 and GS184rec), with known EGFR mutation in tissue, even demonstrated enhanced proliferation on fibroblast growth factor–only medium, which could be in line with an inhibitory effect of EGF on EGFR amplified GSCs as described by others.32 Together with the lack of a significant difference between the occurrence of EGFR amplifications (Table 2 and Supplementary data, Table S1) between the SF+ and SF−/SFnp cohorts, we conclude that the EGFR copy number is unlikely to dictate SF culture outcome.
Current Culture Protocols Select Against Propagation of IDH1-mutant Cells
Since grades II and III samples, and specifically IGS-9/PRO tumors, were found to be less prone to SF culture propagation, we looked into other characteristics of these subtypes in relation to SF success. As both IGS-9/17 and PRO subtypes are enriched for tumors that harbor IDH1/2 mutations (IDH-MUT),25,33 we investigated the IDH1 status of tumors and cultures.
None of the SF+ tumors we investigated (n = 19) were IDH-MUT, while 12 of 24 SF−/SFnp tumors were mutated (Supplementary data, Table S5). IDH1 mutation status was also assessed in early passages of SFnp as well as SS cultures from SF− tumors and revealed a gradual decrease or instant loss of the IDH-MUT sequencing peak, invariably resulting in cultures that contained solely IDH1 wild-type (IDH-WT) cells (Fig. 4B and C, Supplementary data, Table S5). SNP array–based karyograms and 1p19q loss of heterozygosity analysis of these cultures also revealed a loss of CNAs present in parental tumor (exemplified in Supplementary data, Fig. S4). We therefore conclude that current SF and SS culture protocols select against and do not sustain long-term proliferation of IDH-MUT cells.
Comparative Gene Expression Analysis of SF+ Versus SF− Tumors Identifies Enrichment for Extracellular Matrix–Associated Gene Modules in SF+ Tumors
Identification of genes or pathways that dictate SF culture outcome may perhaps provide leads to defining culture conditions for the SF−/SFnp tumors. We therefore analyzed DASL probe intensities of parental tumors from a cohort of 17 SF+ tumors and 24 SF−/SFnp tumors and performed an ANOVA-based statistical analysis for SF culture outcome. As a result, 999 genes were identified that differentiated between SF+ and SF−/SFnp on a 2-fold expression level (P < .05). Gene Ontology (GO) analysis revealed that Hox genes, cell cycling and division, inflammatory, chemotaxis, and migration clusters are upregulated, whereas neuronal development and adhesion clusters are downregulated in SF+ tumors (Supplementary data, Table S3–4). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis demonstrated SF+ tumors to be upregulated for cytokine-cytokine interaction, ECM and focal adhesion pathways, nucleotide-binding oligomerization domain–like receptor, and Janus kinase/signal transducers and activators of transcription signaling, among others. Downregulated modules in SF+ tumors compared with SF− tumors are neuroactive ligand receptor interaction genes, calcium signaling and a “cancer specific” module containing genes such as PI3KR1, RET, BCL2, BMP2, CTNNA3, and CDKN2B (Table 3). Altogether these results suggest that SF+ tumors share molecular features oriented around ECM interaction, which may drive SF outcome.
Table 3.
SF+ tumors with increased expression of genes implicating ECM signaling
| Term | Count | Genes |
|---|---|---|
| SF+ downregulated pathways | ||
| hsa04080: Neuroactive ligand-receptor interaction | 22 | GABRD, GABRG2, PARD3, GABRA1, GABRB3, GRIK2, GRIN1, PTH1R, GRIK4, HTR4, GRIA4, VIPR2, GRM4, GRM3, SSTR2, HRH3, GRIN2C, SSTR1, CHRM1, PRSS3, ADRA1A, GRID1 |
| hsa04020: Calcium signaling pathway | 13 | SLC8A3, SLC8A2, ERBB3, GRIN1, HTR4, PRKCB, GNAL, ATP2B3, GRIN2C, CHRM1, RYR1, ADRA1A, CACNA1A |
| hsa05200: Pathways in cancer | 15 | FGF8, BMP2, RET, HSP90AA1, BCR, FGF12, ZBTB16, CTNNA3, PRKCB, WNT7B, CDKN2B, NTRK1, BCL2, PTCH1, PIK3R1 |
| hsa05412: Arrhythmogenic right ventricular cardiomyopathy | 6 | CACNG6, SGCD, CACNG3, CACNG2, SGCA, CTNNA3 |
| hsa04144: Endocytosis | 10 | SH3GL3, RET, RAB11FIP2, PARD3, CLTA, CLTB, ERBB3, NTRK1, AGAP2, EPN2 |
| hsa05410: Hypertrophic cardiomyopathy | 6 | MYL3, CACNG6, SGCD, CACNG3, CACNG2, SGCA |
| hsa05414: Dilated cardiomyopathy | 6 | MYL3, CACNG6, SGCD, CACNG3, CACNG2, SGCA |
| SF+ upregulated pathways | ||
| hsa04060: Cytokine-cytokine receptor interaction | 18 | CSF3, CCL2, TNFRSF12A, IL7, IL21R, MET, CXCL2, CCL8, CD70, IL15, CCL27, TNFRSF11A, IL12RB1, CXCL14, CCL20, IL1RAP, VEGFA, IL1B |
| hsa04512: ECM-receptor interaction | 7 | IBSP, COL6A3, ITGA11, LAMC1, COL5A1, HMMR, FN1 |
| hsa04621: NOD-like receptor signaling pathway | 6 | CCL2, CXCL2, CCL8, IL1B, TNFAIP3, BIRC3 |
| hsa00360: Phenylalanine metabolism | 4 | ALDH1A3, ALDH3B1, ALDH3A1, HPD |
| hsa00350: Tyrosine metabolism | 5 | ALDH1A3, METTL2B, ALDH3B1, ALDH3A1, HPD |
| hsa00340: Histidine metabolism | 4 | ALDH1A3, METTL2B, ALDH3B1, ALDH3A1 |
| hsa04510: Focal adhesion | 10 | IBSP, RAC2, MET, VEGFA, COL6A3, ITGA11, LAMC1, BIRC3, COL5A1, FN1 |
| hsa04630: JAK-STAT signaling pathway | 8 | CSF3, SPRY1, IL12RB1, IL7, SOCS1, IL21R, IL15, IL13RA2 |
| hsa04114: Oocyte meiosis | 6 | INS, PLK1, SGOL1, CAMK2D, CDC20, AURKA |
| hsa04950: Maturity onset diabetes of the young | 3 | BHLHA15, INS, HNF4G |
| hsa04612: Antigen processing and presentation | 5 | CIITA, PDIA3, HSPA7, KIR2DL1, HLA-C |
Abbreviations: NOD, nucleotide-binding oligomerization domain; JAK-STAT, Janus kinase/signal transducers and activators of transcription. Overview of pathways differentially expressed between SF+ and SF−/SFnp tumors. KEGG pathway analysis on ANOVA-derived gene lists comparing SF+ with SF−/SFnp tumors were uploaded and mapped according to the KEGG annotation tool in DAVID. Significant pathways and associated genes are depicted by official gene symbol.
Discussion
Serum-free cultures of patient-derived GSCs have become a preferred platform for in vitro drug screening studies. The validity of this approach depends, however, on the representation of the whole spectrum of glioma subtypes within the SF culture assay. In order to address this issue, we examined the distribution of glioma subtypes within SF cultures based on histology and expression profiles, as well as on the presence of hallmark molecular characteristics described for grades II–IV glioma. To our knowledge, the presented series of 261 low- and high-grade gliomas is unique in its variation in histological diagnoses and cohort size.
Retrospective analysis of SF culture outcome within this cohort leads us to conclude that tumor sphere formation by itself is not a valid parameter to designate a culture as successful (SF+). The relatively high incidence of tumor spheres that cease to proliferate within a few passages (as reported by others before18) advocates for the introduction of a threshold differentiating between SF+ and samples that do not render practically useful cultures. One argument for the separation of SF+ and SF−/SFnp samples was provided by our genomic analysis, which demonstrated a separation of SFnp/SF− collectively from the SF+ samples. In addition, the enrichment of molecular subtypes between SF−/SFnp and SF+ samples suggests that these groups have distinct molecular backgrounds that drive SF culture outcome.
Within our investigated cohort, SF culture outcome was directly correlated with WHO tumor grade. Others have reported a correlation between the ability of patient-derived glioma cultures to form tumor spheres on SF culture medium and dismal clinical outcome.24,27,28 In accordance with these authors, we find the incidence of SF+ to increase with WHO grade and we see a significant decrease in overall survival for the SF+ group. However, within tumors of equal grade, we found no significant reduction in overall survival for SF+ compared with SF− or SFnp. Therefore, we have not been able to confirm the results of earlier publications in our large cohort. As for the grades II–III SF+ tumors, the observed tendency for dismal outcome may suggest a reflection of shared molecular features as found in SF+ GBM, resulting in a propensity to proliferate under these selective in vitro conditions.
The driving CNAs found in parental tumors are conserved in vitro under multiple passages on SF conditions, which has been demonstrated before.12,15 For SF− samples, we demonstrated that SS parallel cultures are rapidly overgrown with nonneoplastic cells, indicating that short-term serum cultures do not offer an alternative for in vitro propagation of these tumors. On the contrary, the apparent conservation of CNAs in low passages of SFnp cultures suggests that SS medium may serve as a suitable alternative for low-passage, high-throughput screening in selected cases; however, this warrants further study.
To date, 3 publications have reported propagation of IDH1-MUT patient-derived glioma cells under SF conditions.34–36 Of these, 2 groups of researchers published data suggestive of the loss of proliferative capacity in IDH-MUT cells during long-term in vitro propagation, which could be circumvented by serial passaging in xenografts.34,35 Recently, Rohle et al36 reported an anaplastic oligodendroglioma SF culture that sufficiently propagated to perform viability assays for drug screening, although data with regard to the longevity and passage numbers of this culture in vitro were not provided. Taking this into account, we conclude that IDH-MUT cell cultures are selected against, under the current SF and SS culture protocols applied within this study, for the derivation of reproducible and long-term expanding in vitro models.
Identifying biological drivers of SF culture may provide leads to adjustments in culture protocols that increase SF culture yield. By interrogating genomic and expression-based differences between samples of SF+ and SF−/SFnp groups, we identified a distinct molecular profile associated with SF propagation. Genomic profiling demonstrated the characteristic combination of chr7p gain and chr10q loss in all SF+ tumors. Loss of functional phosphatase and tensin homolog, which resides on chr10q, has been reported mandatory for successful SF cultures by Chen and colleagues14; however, these authors did not investigate the coincidence of chr7p gains. Gene expression profiling of parental tumors demonstrated that the SF+ population was significantly enriched for TCGA CLA subtype tumors and decreased for the PRO subtype. In accordance with these data, the 7p10q CNA combination has been demonstrated to be associated with the TCGA CLA subtype by Verhaak et al.5 Interestingly, Schulte et al23 reported superior conservation of PRO-related gene expression in GSC's compared with adherent SF or SS cultures. This phenomenon may be attributed to the neurosphere culture medium composition, which was initially developed to facilitate propagation of nonneoplastic neural stem cells and suggests that transcriptomal changes occur in vitro. In addition, we have found a molecular IGS (IGS-9) that is exclusively incompatible with in vitro propagation. This subtype is correlated with 1p19q codeletion, CIMP, and IDH1-MUT tumors.
Our gene expression–based ontology analysis revealed several discriminatory pathways for SF+ tumors (Table 3). Given the enrichment for genes that modulate the ECM (such as inflammation, neovascularization, etc), we hypothesize that the driving substrates of SF culture outcome are related to these modules. Further support for this assumption is provided by the differential expression of several tumor-associated ECM molecules and cell adhesion genes between SF+ and SF−/SFnp. The interplay between ECM and cancer stem cells has been demonstrated to play a crucial role in the switch between cell survival and programmed cell death,37,38 suggesting a possible link in the ability to proliferate as (floating) spheres. We speculate that this signature renders the GSCs in these tumors less susceptible to the activation of programmed cell death in the absence of components of the ECM that are mandatory for survival. In addition, the leading GO-module upregulated in SF+ tumors was highly enriched for Hox genes, which have recently been implicated in SF+ culture proliferation and maintenance.39 Future research into these differentially expressed gene modules is expected to provide insight into the underlying mechanisms involved in GSC survival under SF conditions.
In sum, we conclude that SF culture medium selects for a subgroup of grades II–IV gliomas that share a CNA trait consisting of chr7p10q alterations as well as gene expression modules that dictate the interaction between GSCs and the ECM. Follow-up studies are under way to validate genes identified in this study. These may ultimately provide leads to improve the yield and subtype distribution of glial tumors in SF culture assays. A next aim is the development of improved culture protocols based on a better understanding of the requirements of these tumors and may include alternative (combinations of) growth factors and ECM substrates, which better recapitulate the intratumoral environment. With regard to the testing of novel drugs and inquiries into canonical pathways driving glial tumors, we suggest that caution is warranted with conclusions drawn from SF culture models, as current protocols support only the cultures of glioma of the described subtypes.
Supplementary Material
Funding
Part of this work was funded by Foundation STOPBraintumors.org, Netherlands.
Conflict of interest statement. None declared.
Supplementary Material
Acknowledgment
We thank Dr W.N.M. Dinjens from the Erasmus MC Department of Pathology for 1p19q LOH analysis.
References
- 1.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 2.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102(16):5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 4.Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 5.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- 7.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 8.Li A, Walling J, Kotliarov Y, et al. Genomic changes and gene expression profiles reveal that established glioma cell lines are poorly representative of primary human gliomas. Mol Cancer Res. 2008;6(1):21–30. doi: 10.1158/1541-7786.MCR-07-0280. [DOI] [PubMed] [Google Scholar]
- 9.Huszthy PC, Daphu I, Niclou SP, et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol. 2012 doi: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 11.Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 12.Ernst A, Hofmann S, Ahmadi R, et al. Genomic and expression profiling of glioblastoma stem cell-like spheroid cultures identifies novel tumor-relevant genes associated with survival. Clin Cancer Res. 2009;15(21):6541–6550. doi: 10.1158/1078-0432.CCR-09-0695. [DOI] [PubMed] [Google Scholar]
- 13.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5(5):504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17(4):362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 15.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Wakimoto H, Kesari S, Farrell CJ, et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69(8):3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visnyei K, Onodera H, Damoiseaux R, et al. A molecular screening approach to identify and characterize inhibitors of glioblastoma stem cells. Mol Cancer Ther. 2011;10(10):1818–1828. doi: 10.1158/1535-7163.MCT-11-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard S, Clarke ID, Smith A, Dirks P. Brain cancer stem cells: a level playing field. Cell Stem Cell. 2009;5(5):468–469. [Google Scholar]
- 19.Reynolds BA, Vescovi AL. Brain cancer stem cells: think twice before going flat. Cell Stem Cell. 2009;5(5):466–467. doi: 10.1016/j.stem.2009.10.017. author reply 468–9. [DOI] [PubMed] [Google Scholar]
- 20.Patru C, Romao L, Varlet P, et al. CD133, CD15/SSEA-1, CD34 or side populations do not resume tumor-initiating properties of long-term cultured cancer stem cells from human malignant glio-neuronal tumors. BMC Cancer. 2010;10:66. doi: 10.1186/1471-2407-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan F, Zhang S, Xie R, et al. The utility and limitations of neurosphere assay, CD133 immunophenotyping and side population assay in glioma stem cell research. Brain Pathol. 2010;20(5):877–889. doi: 10.1111/j.1750-3639.2010.00379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(–) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 23.Schulte A, Gunther HS, Phillips HS, et al. A distinct subset of glioma cell lines with stem cell-like properties reflects the transcriptional phenotype of glioblastomas and overexpresses CXCR4 as therapeutic target. Glia. 2011;59(4):590–602. doi: 10.1002/glia.21127. [DOI] [PubMed] [Google Scholar]
- 24.Panosyan EH, Laks DR, Masterman-Smith M, et al. Clinical outcome in pediatric glial and embryonal brain tumors correlates with in vitro multi-passageable neurosphere formation. Pediatr Blood Cancer. 2010;55(4):644–651. doi: 10.1002/pbc.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Bent MJ, Gravendeel LA, Gorlia T, et al. A hypermethylated phenotype is a better predictor of survival than MGMT methylation in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. Clin Cancer Res. 2011;17(22):7148–7155. doi: 10.1158/1078-0432.CCR-11-1274. [DOI] [PubMed] [Google Scholar]
- 26.Dennis G, Jr., Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 27.Laks DR, Masterman-Smith M, Visnyei K, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27(4):980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pallini R, Ricci-Vitiani L, Banna GL, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14(24):8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302(3):261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erdem-Eraslan L, Gravendeel LA, de Rooi J, et al. Intrinsic molecular subtypes of glioma are prognostic and predict benefit from adjuvant procarbazine, lomustine, and vincristine chemotherapy in combination with other prognostic factors in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. J Clin Oncol. 2013;31(3):328–336. doi: 10.1200/JCO.2012.44.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulte A, Gunther HS, Martens T, et al. Glioblastoma stem-like cell lines with either maintenance or loss of high-level EGFR amplification, generated via modulation of ligand concentration. Clin Cancer Res. 2012;18(7):1901–1913. doi: 10.1158/1078-0432.CCR-11-3084. [DOI] [PubMed] [Google Scholar]
- 33.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klink B, Miletic H, Stieber D, et al. A novel, diffusely infiltrative xenograft model of human anaplastic oligodendroglioma with mutations in FUBP1, CIC, and IDH1. PLoS One. 2013;8(3):e59773. doi: 10.1371/journal.pone.0059773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luchman HA, Stechishin OD, Dang NH, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol. 2012;14(2):184–191. doi: 10.1093/neuonc/nor207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chrenek MA, Wong P, Weaver VM. Tumour-stromal interactions. Integrins and cell adhesions as modulators of mammary cell survival and transformation. Breast Cancer Res. 2001;3(4):224–229. doi: 10.1186/bcr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lathia JD, Li M, Hall PE, et al. Laminin alpha 2 enables glioblastoma stem cell growth. Ann Neurol. 2012;72(5):766–778. doi: 10.1002/ana.23674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallo M, Ho J, Coutinho FJ, et al. A tumorigenic MLL-homeobox network in human glioblastoma stem cells. Cancer Res. 2013;73(1):417–427. doi: 10.1158/0008-5472.CAN-12-1881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.