Abstract
Background
Glioblastoma multiforme (GBM) is a high-grade glioma with poor prognosis. Identification of new biomarkers specific to GBM could help in disease diagnosis. We have developed and validated a bioinformatics method to predict proteins likely to be suitable as glioma biomarkers via a global microarray meta-analysis to identify uncharacterized genes consistently coexpressed with known glioma-associated genes.
Methods
A novel bioinformatics method was implemented called global microarray meta-analysis, using ∼16 000 microarray experiments to identify uncharacterized genes consistently coexpressed with known glioma-associated genes. These novel biomarkers were validated as proteins highly expressed in human gliomas varying in tumor grades using immunohistochemistry. Glioma gene databases were used to assess delineation of expression of these markers in varying glioma grades and subtypes of GBM.
Results
We have identified 5 potential biomarkers—spondin1, Plexin-B2, SLIT3, fibulin-1, and LINGO1—that were validated as proteins highly expressed on the surface of human gliomas using immunohistochemistry. Expression of spondin1, Plexin-B2, and SLIT3 was significantly higher (P < .01) in high-grade gliomas than in low-grade gliomas. These biomarkers were significant discriminators in grade IV gliomas compared with either grade III or II tumors and also distinguished between GBM subclasses.
Conclusions
This study strongly suggests that this type of bioinformatics approach has high translational potential to rapidly discern which poorly characterized proteins may be of clinical relevance.
Keywords: biomarkers, gene databases, gliomas, glioblastoma multiforme, immunohistochemistry
An important issue to be resolved with regard to malignant gliomas is whether there are new relevant biomarkers that can help increase the specificity for the diagnosis of high- and low-grade gliomas. Forty percent of all primary central nervous system tumors are diagnosed as gliomas, of which glioblastoma multiforme (GBM) is the most malignant; the mean survival time is ∼15 months for patients diagnosed with GBM.1 The malignant nature of high-grade gliomas makes them one of the leading causes of cancer death. Information regarding tumor behavior, including cell proliferation (cellularity and mitotic activity), nuclear atypia, neovascularization, and the presence of necrosis and/or apoptotic regions, can be obtained from grading and identification criteria.2 To classify tumors such as adult malignant gliomas and to assess prognosis, differences in molecular composition among tissue types, or “biomarkers,” can be used diagnostically.2–7 The most useful are markers that can guide clinical decisions and predict response to certain therapies.
Recent biomarkers have been found from genome-wide surveys associating somatic mutations with risk of glioma development. The molecular biomarkers most commonly used to evaluate adult malignant gliomas from biopsies include 1p/19q codeletion, methylation of the O6-methylguanine-DNA methyltransferase gene promoter, alterations in the epidermal growth factor receptor pathway, and isocitrate dehydrogenase 1 and 2 gene mutations.2–8 Several proteomics-based approaches have been used to find proteins unique to gliomas,9 but these have been severely limited by issues of sample size, the ability to detect low-abundance proteins, and data reproducibility. It is also important to note that many of these studies have generated hundreds and even thousands of putative candidates, yet not many have been evaluated with subsequent validation and characterization approaches.
Our motivation was to identify novel glioma biomarkers, in the hopes that they would be of value for clinical diagnostics, prognostics, or therapeutics. An approach we previously developed that entails a global microarray meta-analysis (GAMMA) of genes differentially expressed across 3651 human 2-color microarray experiments was used to identify gene–gene coexpression patterns that were consistent and specific across heterogeneous microarray experiments.10 The significance and reproducibility of the GAMMA predictions from the 2-color array data have been corroborated by normalization11 and meta-analysis of 13 000 additional 1-color human microarrays,12 as well as 700 RNA sequence experiments (unpublished observations). This “guilt by association” approach identifies gene sets that are likely to be associated in biologically relevant ways such as phenotype, disease, and genetic network. GAMMA has been used successfully to identify the mitotic role of a formerly uncharacterized gene called C13ORF3 (now Ska3),13 to detect a role in coagulation for C6ORF105 (now androgen-dependent tissue factor pathway inhibitor–regulating protein)1,4 and to identify olfactomedin 4 as a novel neutrophil subset marker associated with granule secretion.15 Recently, we used GAMMA to identify the biomarker ELTD1 (epidermal growth factor, latrophilin, and 7 transmembrane domain-containing 1 on chromosome 1) as a putative glioma-associated marker with immunohistochemistry (IHC) validation in human and rodent gliomas and in vivo molecular-targeted MRI in rodent gliomas.1,6
With the GAMMA approach, genes are not analyzed directly, but the top 20 genes most consistently coexpressed with them are analyzed for what they have in common in the peer-reviewed literature using a large-scale computational analysis.17,18 This way, even if a protein has no known function, its function can be inferred. In this case, its role in gliomas can be inferred by its consistent patterns of coexpressions with other glioma-associated genes. Then, using the Human Protein Reference Database19 and other experimental sources on protein cellular localizations, we can screen this list of predicted glioma-associated proteins for those that are extracellular or membrane bound. It is thought that extracellular or membrane-bound proteins could be potential ideal therapeutic targets and could be reached for in vivo targeted imaging with fluorescence-labeled antibodies. After identifying putative glioma-associated markers, we used IHC to experimentally validate their presence in human gliomas. For IHC, the novel biomarkers can be compared with traditional IHC markers for human gliomas, including vascular endothelial growth factor (VEGF), glucose transporter–1 (GLUT-1), and hypoxia inducible factor–1α (HIF-1α). Expression of the novel biomarkers in human gliomas will be further evaluated from gene expression databases to establish whether these biomarkers are differentially expressed in varying glioma grades and GBM subtypes.
Materials and Methods
Bioinformatics
Using bioinformatics methods developed in our group to pair coexpression data with literature-based analysis of protein commonalities,10,17,18,20 we conducted a global meta-analysis of ∼16 000 microarray experiments (1- and 2-color) obtained from the database of the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO). For each human gene, the meta-analysis identified a set of 20 genes that were the most consistently and specifically coexpressed with it across the heterogeneous conditions analyzed. Each gene, whether it had any published literature describing its function or not, had its function inferred by analysis of what these 20 coexpressed genes had in common. This was done via an automated, large-scale analysis of the peer-reviewed literature17,20 to identify genes that were consistently transcribed with established glioma-related genes but that have themselves never been associated with gliomas in the literature. This circumvents a problem inherent in the lists of expressed genes derived by microarrays, which identify only those genes that are being actively transcribed at the time of the experiment without detecting proteins that are present but not actively transcribed. That is, GAMMA associates genes that are frequently cotranscribed regardless of the condition, and then if a statistically significant set of genes has been reported as glioma associated in the literature, these associations need not be transcriptional to be identified by GAMMA (eg, they could be from proteomics or genome-wide association studies). The enormous sample size of both microarray data and analyzed abstracts enables us to screen out genes that do not pass a threshold of statistical significance. This associative method works for glioma-derived literature-based associations as well as for searches on associated processes (such as angiogenesis, apoptosis, and cell migration), helping to corroborate any putative roles in tumorigenesis that we uncover. For each association, we calculated mutual information (a measure of variable dependency) between literature terms to prioritize the strength of association between each protein and a role in gliomas.18 Finally, we obtained increased confidence in the predictions because GAMMA also successfully predicted many established glioma-related genes (eg, epidermal growth factor receptor, matrix metalloproteinase–2, glial fibrillary acidic protein, fibroblast growth factor 2). These identifications serve as positive controls for predictive capacity.
Immunohistochemistry
The human tissue sample portion of the study was conducted in compliance with the University of Utah Health Sciences Center Institutional Review Board. For IHC analysis, tissue from 50 patients with high-grade gliomas (21 female, 29 male), including 40 GBM, 6 anaplastic astrocytomas, and 4 anaplastic oligodendrogliomas, was compared with tissue from 21 patients (10 female, 11 male) with tumors classified as low-grade gliomas (11 benign oligodendrogliomas, 10 low-grade astrocytomas). Antibodies (Abs) to spondin1, Plexin-B2, Slit homolog (SLIT3), fibulin-1, and leucine-rich repeat and immunoglobulin domain–containing Nogo receptor interacting protein 1 (LINGO1) were available commercially (F-spondin Ab [S-17], Plexin-B2 Ab [I-16], SLIT3 Ab [F-15], and fibulin-1 Ab [H-190] were all obtained from Santa Cruz Biotechnology. LINGO1 [LRRN6A]-S596 [C-term] Ab was obtained from Abgent. All human Abs were assessed and found to provide similar results; dilutions were 1:500). A toluidine blue (0.1%) counterstain was used (15 sec). IHC was performed using the Vectastain ABC Kit (Vector Laboratories). Negative controls were performed by replacing the primary Ab with nonimmune serum. Slides were examined using an Olympus BX41 microscope. Under 200× magnification (10 ocular × 20 objective), slides were scored by 2 investigators blinded to the specimen tumor grade or patient information. A score of 0–4 was assigned based on the percentage of cells stained in a given field: 0 = 0%–25%, 1 = 25%–50%, 2 = 50%–75%, 3 = 75%–100%, and 4 = 100%. In prior papers, we have demonstrated that this method is very reproducible, with good interrater reliability (P = .99, 95% confidence interval [CI] = 0.99–1.00) and intrarater reliability (P = .96, 95% CI = 0.92–0.99).21 Each investigator independently reviewed the slide at low power and at random high-power fields when determining the IHC score. Positive expression was considered for scores of 2–4, whereas negative expression was considered for scores of 0 and 1. Percent survival values for the GBM, anaplastic astrocytoma, and anaplastic oligodendroglioma patients were 0.0%, 0.0%, and 50.0%, respectively. Percent survivals for benign oligodendroglioma and low-grade astrocytoma patients were 27.3% and 70.0%, respectively.
Gene Expression Analysis
For the glioblastoma expression microarray analysis, raw Affymetric .cel files and level 3 Agilent expression data were downloaded from The Cancer Genome Atlas (TCGA)—529 and 594 GBM samples, respectively; data were also retrieved from the Repository for Molecular Brain Neoplasia Data (REMBRANDT; 229 total astrocytomas, of which 125 were GBM) and Erasmus (NCBI GEO Series GSE16011; a total of 187 astrocytomas, of which 159 were GBM), as well as the corresponding clinical annotations for each. The .cel files were then processed using R and Bioconductor, in a custom computable document format file, with background correction, log transformation, and quantile normalization performed using the robust multi-array algorithm implemented in R.
For mesenchymal and proneural gene signature definition, we used a composite of signatures from the Phillips et al.2,2 and Verhaak et al.23 studies. For a given tumor, the metagene mesenchymal and proneural signature scores were both calculated. Within a data set, the mesenchymal and proneural metascores were z-scores corrected to allow comparison between the two. Tumors were then assigned to one of the signatures based on the higher-expressing metagene.
Statistical Analyses
Statistical differences for IHC scoring and expression statistical differences were compared between groups using the Welch 2-sample t test (unpaired, 2-sided), with P < .05 considered to be significant.
Results
Bioinformatics
Using our GAMMA procedure, we identified membrane-bound proteins that had not yet been associated with gliomas but whose expression consistently correlated with genes reported to be associated with gliomas. We identified 195 putative candidate markers, all genes predicted or known to be membrane bound and not appearing in any MedLine article that mentioned gliomas (or synonymous terms). Of these 195, only 75 had commercial antibodies at the time of analysis that we could use to validate with IHC. Fibulin-1, LINGO1, spondin1, SLIT3, and Plexin-B2 were chosen from among this list of 75 because they all had high scores. With this analysis set to stringent thresholds, we empirically observed that the FBLN1, LINGO1, SPON1, SLIT3, and PLXNB2 genes were all found to be consistently transcribed with known glioma-associated genes. Supplementary Figs S1–S5 show how these genes are connected to other glioma-associated genes via protein–protein interactions (obtained from the Human Protein Reference Database).24 FBLN1is associated with several extracellular matrix remodeling genes (eg, collagen genes such as COL1A1/2, COL2A1, COL3A1, COL4A1/2/3, COL5A1, COL6A3, COL7A1, COL18A1; transforming growth factor [TGFβ1]; matrix metalloproteinases such as MMP2, MMP9, and MMP14) and other cell invasion- and migration-related genes, such as CD44, which are characteristic of the invasive nature of GBM.25–28 SPON1 is also associated with genes related to cell invasion or migration, such as various collagen genes, MMPs, CD44, and TGFβ1; endothelial cells (integrin ITGB1/3), and epidermal growth factor receptor, all found to be affected in GBM.25–30 PLXB2 is associated with genes that regulate extracellular matrix remodeling (COL2A1, COL1A1/2, MMP1, TGFβ1) and angiogenesis (VEGF-α and the integrin binding protein IGFBP3). LINGO1 is associated with tight junction protein 1 (also known as zonula occludens 1), which regulates membrane-type 1 MMP expression (involved in tumor cell invasion).31 Supplementary Fig. S6 includes gene association data for ELTD1.16 Functional relationships for biomarkers are outlined in Table 1.
Table 1.
GAMMA-associated relationships of potential novel glioma biomarkers (highest GAMMA scores) with cancer growth characteristics and other cancers
| Biomarker | Functional Relationships | Ref. |
|---|---|---|
| SLIT-13 | Growth factor, angiogenesis/VEGF, cell proliferation, cell migration, extracellular matrix proteins, epigenetic (hypermethylation) inactivation of SLIT1–3 genes in human cancers, melanoma, breast cancer, ovarian cancer, colorectal cancer, gastric cancer | 39,50,51 |
| SPON1 (spondin1) | Extracellular matrix protein, promotes cell attachment, cell adhesion, angiogenesis/VEGF, cell migration, cell growth; colorectal cancer, ovarian cancer | 40–43 |
| FBLN1 (fibulin-1) | Extracellular matrix organization (proteins, components), integrin, basement membrane, TGFβ, growth factor, cell adhesion, cell proliferation, cell growth, cell migration, angiogenesis, matrix metalloproteinases; overexpressed in other cancers, melanoma, breast cancer, hepatocellular carcinoma, prostate cancer | 45–49 |
| PLXNB2 (Plexin-B2) | Cell proliferation, angiogenesis, cell motility, TGFβ, pro-inflammatory cytokines | 52–56 |
| LINGO1 | Neurogenesis, brain development, epidermal growth factor receptor binding, astrocytes, glial cells | 57–60 |
GAMMA-predicted associations and their scores are based on a combination of (i) how many genes out of the top 20 coexpressed genes that were analyzed showed associations with gliomas based on published reports and (ii) their statistical significance based on random network simulations to estimate the probability that a set of equally frequent terms would associate with gliomas. Only proteins with P < .01 were selected as potential candidates. A flow diagram of the GAMMA approach is illustrated in Fig. 1. All predicted associations for these genes are given in Supplementary Tables S1–S5.
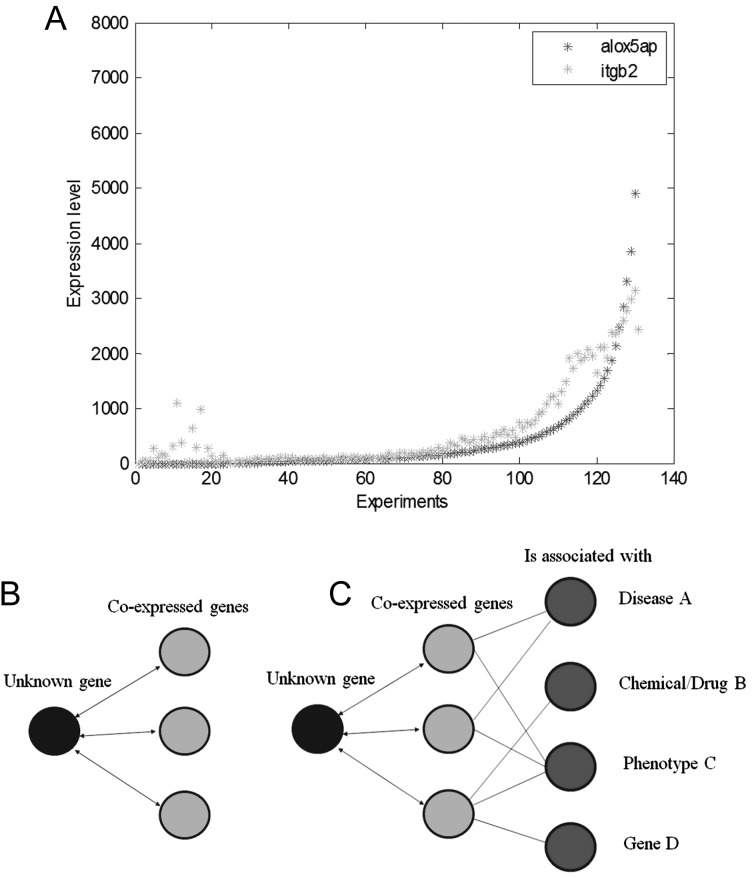
Fig. 1.
The global microarray meta-analysis (GAMMA) bioinformatics approach. (A) For all human genes, the first step is to identify their coexpression trends. Previous experiments show that genes positively correlated in their coexpression levels tend to be related by function, phenotype, and disease relevance. For example, for every experiment where 2 genes, alox5ap and itgb2, are expressed, this is quantified by Pearson correlation; however, not all genes have expression data. (B) For genes of unknown function, the second step is to identify the 20 genes with the strongest coexpression correlation. In this example, only 3 genes are shown for simplicity. (C) The third step is to identify what the coexpressed genes have in common. Text-mining software (IRIDESCENT) is used to analyze what diseases, phenotypes, chemicals, and other genes appear in the literature with each of the coexpressed genes. These are the inferred associations for the unknown gene. The set of coexpressed genes is searched for reported commonalities in the peer-reviewed literature. After all of the genes are analyzed, the list is shortened to include those with certain characteristics. In this example, we include only those that are inferred to be associated with gliomas, are plasma-membrane bound, and have commercial antibodies available for validation experiments to be conducted to establish whether these proteins are differentially present in gliomas.
Immunohistochemistry
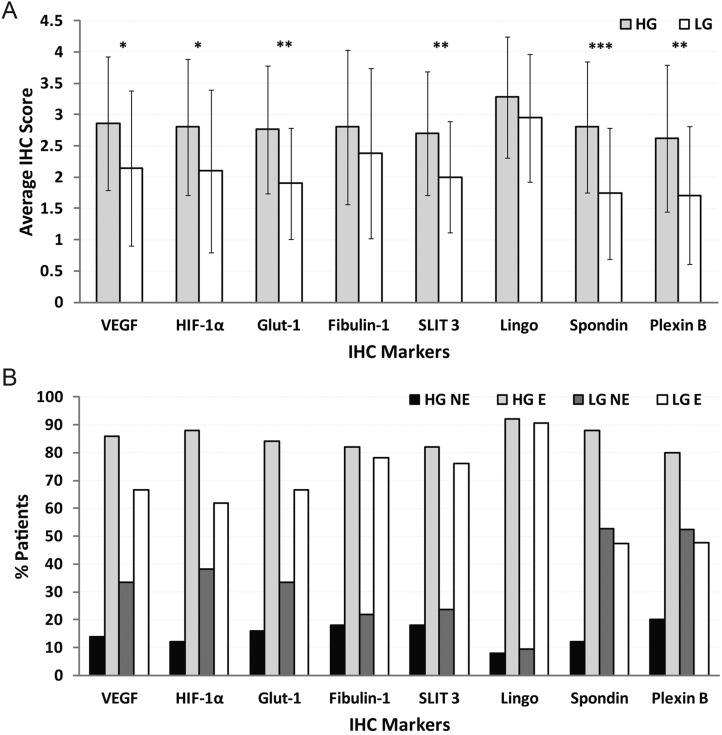
Fibulin-1, SLIT3, LINGO1, spondin1, and Plexin-B2 were all found to be expressed in gliomas in general (Fig. 2). Fibulin-1 and LINGO1 were not found to have significantly higher expression (P > .05 for both) when comparing high-grade and low-grade gliomas, whereas all other biomarkers (traditional biomarkers including VEGF, HIF-1α, and GLUT-1 had P < .05, P < .05, and P < .01, respectively), including SLIT3 (P < .01), spondin1 (P < .001), and Plexin-B2 (P < .01), had significantly higher expression in high-grade compared with low-grade gliomas (Fig. 2A). All 5 novel biomarkers compared well with VEGF, HIF-1α, or GLUT-1, although LINGO1 had a higher average IHC score in both high-grade and low-grade gliomas compared with traditional markers. In low-grade gliomas (low-grade astrocytomas and oligodendrogliomas), LINGO1 had the highest IHC score, followed by fibulin-1. All of the novel biomarkers were ≥80% positive in high-grade glioma patients, and fibulin-1, SLIT3, and LINGO1 were >70% positive in low-grade glioma patients (Fig. 2B).
Fig. 2.
Spondin1, SLIT3, and Plexin-B2 levels are higher in high-grade (HG) gliomas compared with the levels in low-grade (LG) gliomas. (A) Graph showing average IHC scores for biomarkers VEGF, HIF-1α, GLUT-1, fibulin-1, SLIT3, spondin1, LINGO1, or Plexin-B2 in high-grade gliomas (50 patients: 21 female and 29 male) and low-grade gliomas (21 patients: 10 female and 11 male). Scores were obtained using the following grading criteria—0: 0%; 1: 0–<25%; 2: 25–<50%; 3: 50–<75%; 4: 75–100% detection of IHC stain. Significant differences in marker levels between HG and LG were established when *P < .05, **P < .01, or ***P < .001. Actual P-values for each biomarker are: .0161 for VEGF, .0227 for HIF-1α, .0013 for GLUT-1, .2081 for fibulin-1, .0069 for SLIT3, .2054 for LINGO1, .0004 for spondin1, and .0035 for Plexin-B2. (B) Graph showing the percentage of patients that expressed biomarkers stained by IHC in high-grade gliomas (50 patients) and low-grade gliomas (21 patients). A negative expression (NE) result was attributed to IHC scores of 0 or 1. A positive expression (E) result was attributed to IHC scores of 2–4.
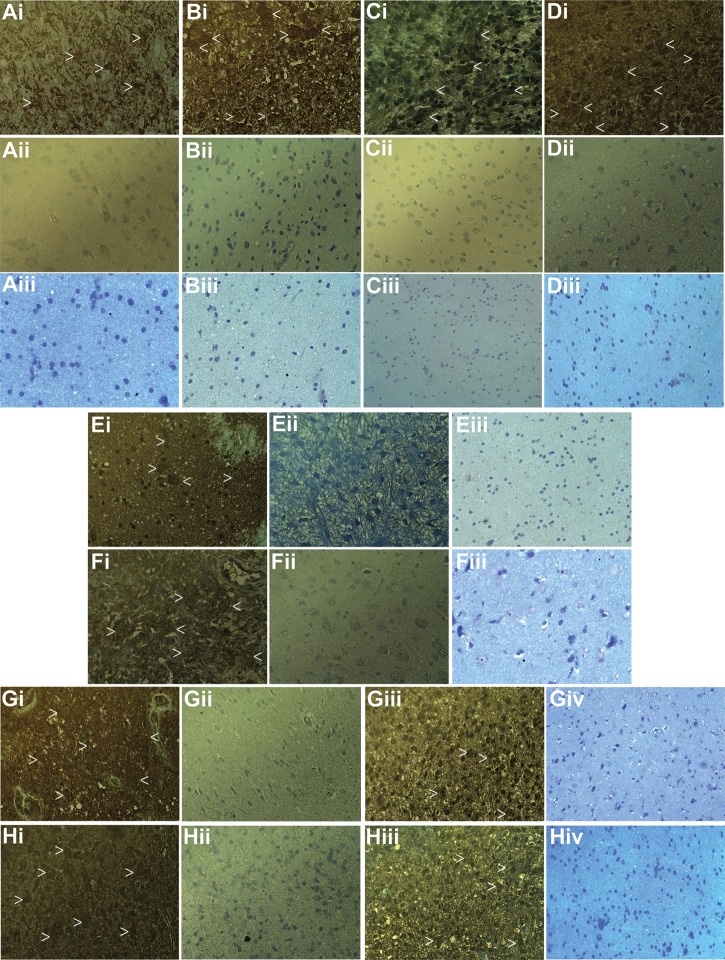
Figure 3 depicts representative IHC staining for VEGF, HIF-1α, GLUT-1, SLIT3, spondin1, Plexin-B2, fibulin-1, and LINGO1 in human high-grade and low-grade gliomas, indicating that high-grade gliomas had substantially higher levels of VEGF, HIF-1α, GLUT-1, SLIT3, spondin1, and Plexin-B2 compared with corresponding low-grade gliomas. In addition to examples depicting low levels in low-grade gliomas (Fig. 3, Gii and Hii), both fibulin-1 and LINGO1 had samples with high levels of staining in low-grade gliomas (Fig. 3, Giii and Hiii, respectively).
Fig. 3.
VEGF, HIF-1α, GLUT-1, SLIT3, spondin1, and Plexin-B2 levels are elevated in high-grade gliomas compared with low-grade gliomas. (A) Representative IHC staining for VEGF in a GBM (Ai), an oligodendroglioma (Aii), and normal brain (Aiii). (B) Representative IHC staining for HIF-1α in a GBM (Bi), a low-grade astrocytoma (LGA) (Bii), and normal brain (Biii). (C) Representative IHC staining for GLUT-1 in a GBM (Ci), an oligodendroglioma (Cii), and normal brain (Ciii). (D) Representative IHC staining for SLIT3 in a GBM (Di), an LGA (Dii), and normal brain (Diii). (E) Representative IHC staining for spondin1 in a GBM (Ei), an LGA (Eii), and normal brain (Eiii). (F) Representative IHC staining for Plexin-B2 in a GBM (Fi), an oligodendroglioma (Fii), and normal brain (Fiii). (G) Representative IHC staining for fibulin-1 in a GBM (Gi), an LGA (Gii, an example of low expression), an oligodendroglioma (Giii, an example of high expression), and normal brain (Giv). (H) Representative IHC staining for LINGO in a GBM (Hi), 2 oligodendrogliomas (Hii, an example of low expression; and Hiii, an example of high expression), and normal brain (Hiv). White arrow heads depict regions that highly stain for each biomarker. Magnification of all panels is 40×.
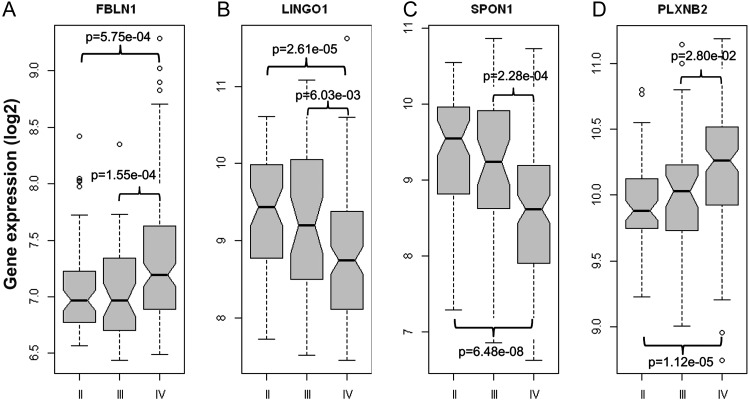
Figure 4 illustrates that gene expression levels of fibulin-1, LINGO1, spondin1, and Plexin-B2 are all significantly different in grade III or grade IV gliomas compared with grade II gliomas. Gene expression data for SLIT3 was found to be poorly represented on the Affymetrix platforms and did not provide conclusive differences in tumor grades in the Agilent TCGA data. Both fibulin-1 and Plexin-B2 were found to be expressed significantly more in grade IV gliomas than in grade II (P < .001 and P < .0001, respectively) or grade III (P < .001 and P < .05, respectively), whereas LINGO1 and spondin1 were found to have significantly lower expression in grade IV tumors than in grade II (P < .0001 for both) or grade III (P < .01 and P < .001, respectively).
Fig. 4.
Increased expression of fibulin-1, LINGO1, spondin1, or Plexin-B2 biomarkers is associated with higher grade in gliomas in the REMBRANDT gene expression database. Fibulin-1 and Plexin-B2 expressions were found to be significantly higher in grade IV gliomas, and conversely LINGO1 and spondin1 expressions were significantly lower compared with grade II and grade III glioma data sets. Significant differences between grades II and III or grades II and IV are shown for each marker. For statistical analysis, a Welch's 2-sample t-test was used.
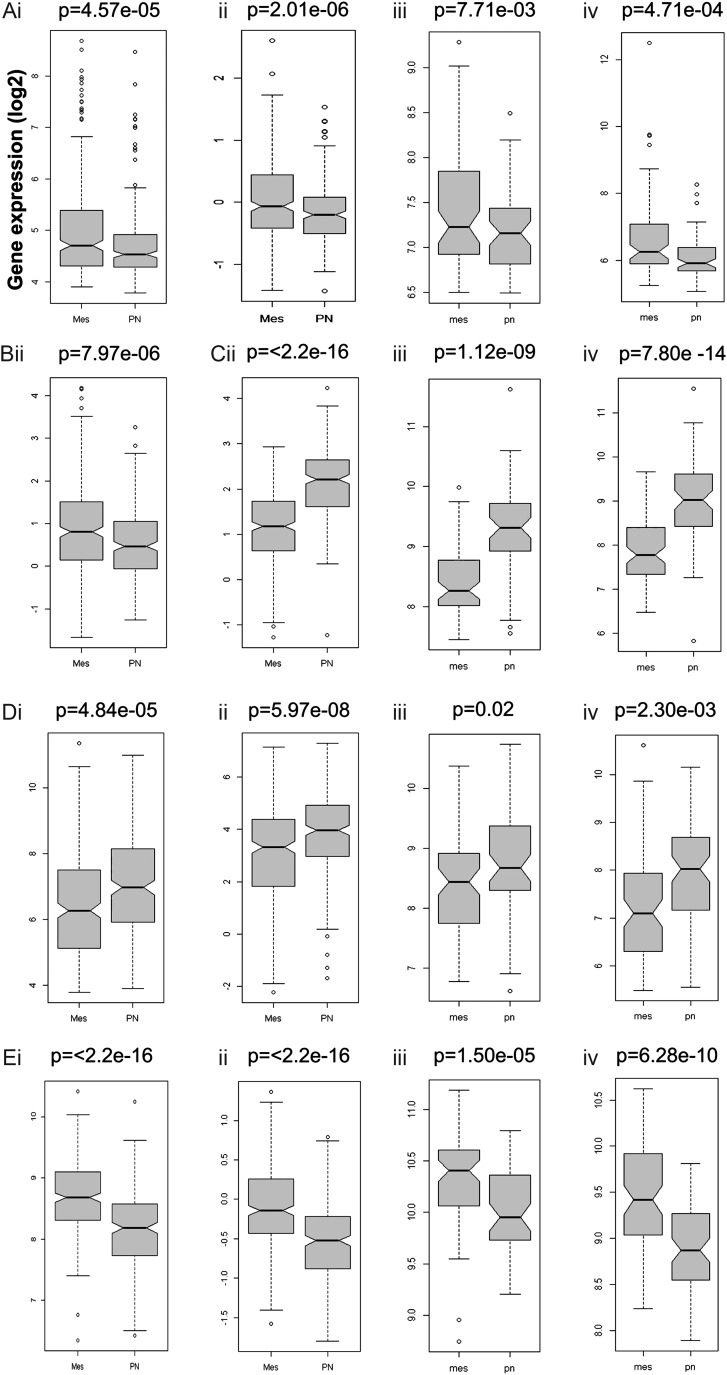
Figure 5 shows how each of the biomarkers can be used to differentiate between mesenchymal and proneural GBM subtypes, as measured in 4 gene expression databases (TCGA Affymetrix platform, TCGA Agilent platform, REMBRANDT, and Erasmus). Data for SLIT3 were available only in the TCGA Agilent database, and LINGO1 data were not available in the TCGA Affymetrix database. In 3 gene databases, fibulin-1 expression was found to be significantly higher (P < .01) in mesenchymal GBM, whereas LINGO1 expression was found to be significantly higher (P < .0001) in proneural GBM. In all 4 databases, spondin1 expression was found to be significantly higher (P < .05) in proneural GBM, whereas Plexin-B2 expression was found to be significantly higher (P < .0001) in mesenchymal GBM. SLIT3 expression was found to be significantly higher (P < .0001) in mesenchymal GBM compared with proneural GBM.
Fig. 5.
Biomarker gene expressions in GBM subtypes. (A) Fibulin-1 (FBLN1), (B) SLIT3, (C) LINGO1, (D) spondin1 (SPON1), and (E) Plexin-B2 (PLXNB2) expression for mesenchymal (Mes) (left bar) or proneural (PN) (right bar) GBM subtypes obtained from TCGA—either (i) Affymetrix or (ii) Agilent, (iii) Rembrandt, or (iv) Erasmus databases. Increased fibulin-1 (A), SLIT3 (B), and Plexin-B2 (E) expressions are associated with mesenchymal phenotype in GBM (compared with the proneural subtype), whereas increased LINGO1 (C) and spondin1 (D) expressions are associated with the proneural subtype. P-values for each marker are shown for each database. For statistical analysis, a Welch's 2-sample t-test was used.
Discussion
We found that each of the algorithmically predicted biomarkers was found in high levels within human gliomas in general, as assessed by IHC, and that some of the biomarkers, such as SLIT3, spondin1, and Plexin-B2, were expressed at significantly higher levels in high-grade gliomas than in low-grade gliomas (Fig. 2A) and may be useful diagnostic markers for high-grade gliomas (Fig. 3). Both fibulin-1 and LINGO1 were found to be positively expressed in both high-grade and low-grade gliomas with high IHC scores (Fig. 2B), although they did not significantly discriminate between the grades, and these 2 markers may serve as general glioma markers for all tumor grades. Examples of both high and low levels of these markers are depicted in Fig. 3. SLIT3, spondin1, and Plexin-B2 also all fare well in comparison with more traditional IHC markers currently used to diagnose GBM, in that the expression levels of these 3 biomarkers were similar to those of currently investigated glioma markers, including VEGF, HIF-1α, and GLUT-1. HIF-1α has been well documented to be an important diagnostic marker for gliomas, and this marker can be targeted for therapeutic intervention.32–38
From the gene expression results, we have also demonstrated that there was a strong association with expression of fibulin-1, LINGO1, spondin1, and Plexin-B2 in grade IV gliomas compared with either grade II or grade III gliomas (Fig. 4). Interestingly, both fibulin-1 and Plexin-B2 were found to be more highly expressed in grade IV gliomas, whereas LINGO1 and spondin1 were found to be less expressed in grade IV tumors compared with either grade II or grade III gliomas. In addition, when we looked at the GBM tumor subtypes, it looked like there was a significant increase with gene expression of fibulin-1, Plexin-B2, and SLIT3 for the mesenchymal subtype versus the proneural subtype, which was better associated with increased gene expression of LINGO1 and spondin1 (Fig. 5). These findings were consistent in 3 gene databases for fibulin-1, LINGO1, spondin1, and Plexin-B2. Unfortunately, SLIT3 gene data were usable in only the TCGA Agilent database. It is reasonable to conclude that spondin1 and Plexin-B2 expression data in particular are both strong biomarkers of grade (also supported by the IHC data), with Plexin-B2 increased in the mesenchymal subtype and spondin1 elevated in the proneural subtype.
None of the 5 proteins had been documented as discriminately present on the surface of glioma cells, although SLIT3 was found in a previous study to be hypermethylated in glioma and colorectal cancer cell lines.39 Spondin1, SLIT3, and fibulin-1 also had previously reported associations with different cancers. Spondin1 was previously found to be overexpressed in ovarian/peritoneal carcinomas,40–43 and SLIT3 was widely expressed in human hepatocellular carcinomas.44 Fibulin-1 was associated with gastric,45 breast,46 colon,47 and prostate48 cancers, and its promoter hypermethylation was associated with tumor progression in human hepatocellular carcinomas.49 SLIT3 is a predominant ligand transcribed in the early mouse heart and is expressed in the ventral wall of the linear heart tube and subsequently in the chamber.50 The SLIT3 gene at human chromosome 5q34-q35.1 is involved in encoding large secreted proteins functioning as ligands for Roundabout (ROBO) receptors, and the SLIT-ROBO signaling pathway is implicated in angiogenesis and endothelial cell migration.51
Neither Plexin-B2 nor LINGO1 has been previously associated with cancer, although some function is known for these genes. Plexins are a family of genes that are expressed in several organ systems and have been implicated in cell movement and cell–cell interaction.52 Plexin-B2 has been reported to be associated with the negative regulation of interleukin-12/interleukin-23p40 in dendritic cells.52 Plexins are cell surface receptors widely studied in the nervous system, where they mediate migration and morphogenesis through the Rho family of small GTPases.53 Plexin-B2 is highly expressed on cells of the innate immune system in the mouse, including macrophages and dendritic cells, and may serve as a negative regulator of basal cell motility.52 Although Plexin-B2 has not been associated with cancers, Plexin-B1 has been reported to be involved as a tumor suppressor in melanoma cells,54 and Plexin-B in general is involved in invasive growth55 and angiogenesis.56 LINGO1 has been found to be a potent regulator of neural stem cell maturation to neurons, and inhibition of LINGO1 during the first days of neural stem cell differentiation results in decreased neuronal maturation.57,58 LINGO1 is a central nervous system transmembrane protein that simultaneously interacts with the Nogo-66 receptor and p75 neurotrophin receptor or tumor necrosis factor receptor superfamily member 1 (TROY) on neurons to form a receptor complex responsible for myelin-mediated neurite outgrowth inhibition59 and thus is a negative regulator of myelination and repair of damaged axons.60
The results presented strongly suggest that the associative transcriptional network analysis method used in this study was able to accurately identify fibulin-1, LINGO1, spondin1, Plexin-B2, and SLIT3 as glioma-associated biomarkers. SLIT3, FBLN1, and SPON1 are predicted to influence glioma progression by their role in the extracellular matrix. SLIT3 has established associations with angiogenesis and cell migration, and several genes with known roles in extracellular matrix remodeling, such as SPARC and vascular-endothelial–cadherin, are predicted to be relevant to the SLIT3' network. Fibulin-1 has predicted associations with cell motility and invasion, and spondin1 is specifically predicted to exert its influence via collagen matrix attachments. PLXNB2, however, is predicted to be more relevant to cell proliferation and to be relevant to the Wnt/β-catenin pathway. LINGO1 is probably the most neural-specific protein of the group and, based on the IHC scores, appears to be increasingly important as tumor grade increases. Each marker has either known or predicted associations with different aspects of glioma tumor growth, and each marker or combination could provide valuable diagnostic information for gliomas.
Supplementary Material
Conflict of interest statement. None declared.
Funding
This work was supported by the Oklahoma Medical Research Foundation (OMRF) (to R.A.T.), the Chapman Foundation (to J.D.W.), and NIH grants 1P20GM103636 and 8P20GM103456-09 (to J.D.W.).
Supplementary Material
Acknowledgments
We thank Kristin Kraus, MSc, for editorial assistance with the paper.
References
- 1.Central Brain Tumor Registry of the United States. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004-2007. Hinsdale, IL: Author; 2011. http://www.cbtrus.org/2011-NPCR-SEER/WEB-0407-Report-3-3-2011.pdf. Accessed 17 July, 2013. [Google Scholar]
- 2.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas (TCGA) Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riemenschneider MJ, Jeuken JW, Wesseling P, Reifenberger G. Molecular diagnostics of gliomas: state of the art. Acta Neuropathol. 2010;120(5):567–584. doi: 10.1007/s00401-010-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9(7):717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colman H, Zhang L, Sulman EP, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farias-Eisner G, Bank AM, Hwang BY, et al. Glioblastoma biomarkers from bench to bedside: advances and challenges. Br J Neurosurg. 2012;26(2):189–194. doi: 10.3109/02688697.2011.629698. [DOI] [PubMed] [Google Scholar]
- 8.Silber JR, Bobola MS, Blank A, Chamberlain MC. O(6)-methylguanine-DNA methyltransferase in glioma therapy: promise and problems. Biochim Biophys Acta. 2012;1826(1):71–82. doi: 10.1016/j.bbcan.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niclou SP, Fack F, Rajcevic U. Glioma proteomics: status and perspectives. J Proteomics. 2010;73(10):1823–1838. doi: 10.1016/j.jprot.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Wren JD. A global meta-analysis of microarray expression data to predict unknown gene functions and estimate the literature-data divide. Bioinformatics. 2009;25(13):1694–1701. doi: 10.1093/bioinformatics/btp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dozmorov MG, Wren JD. High-throughput processing and normalization of one-color microarrays for transcriptional meta-analyses. BMC Bioinformatics. 2011;12(Suppl 10):S2. doi: 10.1186/1471-2105-12-S10-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dozmorov MG, Giles CB, Wren JD. Predicting gene ontology from a global meta-analysis of 1-color microarray experiments. BMC Bioinformatics. 2011;12(Suppl 10):S14. doi: 10.1186/1471-2105-12-S10-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daum JR, Wren JD, Daniel JJ, et al. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19(17):1467–1472. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupu C, Zhu H, Popescu NI, Wren JD, Lupu F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood. 2011;118(16):4463–4471. doi: 10.1182/blood-2011-05-355370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemmensen SN, Bohr CT, Rorvig S, et al. Olfactomedin 4 defines a subset of human neutrophils. J Leukoc Biol. 2012;91(3):495–500. doi: 10.1189/jlb.0811417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Towner RA, Jensen RL, Colman H, et al. ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013 doi: 10.1227/NEU.0b013e318276b29d. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics. 2004;20(2):191–198. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 18.Wren JD. Extending the mutual information measure to rank inferred literature relationships. BMC Bioinformatics. 2004;5:145. doi: 10.1186/1471-2105-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goel R, Muthusamy B, Pandey A, Prasad TS. Human Protein Reference Database and human proteinpedia as discovery resources for molecular biotechnology. Mol Biotechnol. 2011;48(1):87–95. doi: 10.1007/s12033-010-9336-8. [DOI] [PubMed] [Google Scholar]
- 20.Giles CB, Wren JD. Large-scale directional relationship extraction and resolution. BMC Bioinformatics. 2008;9(Suppl 9):S11. doi: 10.1186/1471-2105-9-S9-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen RL. Brain tumor hypoxia: tumorigenesis, angiogenesis, imaging, pseudoprogression, and as a therapeutic target. J Neurooncol. 2009;92(3):317–335. doi: 10.1007/s11060-009-9827-2. [DOI] [PubMed] [Google Scholar]
- 22.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2009;37(database issue):D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical regidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69(10):4167–74. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veeravalli KK, Rao JS. MMP-9 and uPAR regulated glioma cell migration. Cell Adh Migr. 2012;6(6):509–12. doi: 10.4161/cam.21673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chetty C, Vanamala SK, Gondi CS, et al. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell Signal. 2012;24(2):549–59. doi: 10.1016/j.cellsig.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Wick W, Platten M, Weller M. Glioma cell invasion: regulation of metalloproteinase activity by TGF-β. J Neuro-Oncol. 2001;53:177–185. doi: 10.1023/a:1012209518843. [DOI] [PubMed] [Google Scholar]
- 29.Fowler A, Thomson D, Giles K, et al. miR-124a is frequently down-regulated in glioblastoma and is involved in migration and invasion. Eur J Cancer. 2011;47(6):953–63. doi: 10.1016/j.ejca.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 30.Taylor TE, Furnari FB, Cavenee WK. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets. 2012;12(3):197–209. doi: 10.2174/156800912799277557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collen A, Hanemaaijer R, Lupu F, Quax PH, et al. Membrane-type matrix metalloproteinase-mediated angiogenesis in a fibrin-collagen matrix. Blood. 2003;101(5):1810–7. doi: 10.1182/blood-2002-05-1593. [DOI] [PubMed] [Google Scholar]
- 32.Flynn JR, Wang L, Gillespie DL, et al. Hypoxia-regulated protein expression, patient characteristics, and preoperative imaging as predictors of survival in adults with glioblastoma multiforme. Cancer. 2008;113(5):1032–1042. doi: 10.1002/cncr.23678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillespie DL, Flynn JR, Ragel BT, et al. Silencing of HIF-1alpha by RNA interference in human glioma cells in vitro and in vivo. Methods Mol Biol. 2009;487:283–301. doi: 10.1007/978-1-60327-547-7_14. [DOI] [PubMed] [Google Scholar]
- 34.Ragel BT, Couldwell WT, Gillespie DL, Jensen RL. Identification of hypoxia-induced genes in a malignant glioma cell line (U-251) by cDNA microarray analysis. Neurosurg Rev. 2007;30(3):181–187. doi: 10.1007/s10143-007-0070-z. discussion 187. [DOI] [PubMed] [Google Scholar]
- 35.Gillespie DL, Whang K, Ragel BT, Flynn JR, Kelly DA, Jensen RL. Silencing of hypoxia inducible factor-1alpha by RNA interference attenuates human glioma cell growth in vivo. Clin Cancer Res. 2007;13(8):2441–2448. doi: 10.1158/1078-0432.CCR-06-2692. [DOI] [PubMed] [Google Scholar]
- 36.Rong Y, Hu F, Huang R, et al. Early growth response gene-1 regulates hypoxia-induced expression of tissue factor in glioblastoma multiforme through hypoxia-inducible factor-1-independent mechanisms. Cancer Res. 2006;66(14):7067–7074. doi: 10.1158/0008-5472.CAN-06-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen RL. Hypoxia in the tumorigenesis of gliomas and as a potential target for therapeutic measures. Neurosurg Focus. 2006;20(4):E24. doi: 10.3171/foc.2006.20.4.16. [DOI] [PubMed] [Google Scholar]
- 38.Jensen RL, Ragel BT, Whang K, Gillespie D. Inhibition of hypoxia inducible factor-1alpha (HIF-1alpha) decreases vascular endothelial growth factor (VEGF) secretion and tumor growth in malignant gliomas. J Neurooncol. 2006;78(3):233–247. doi: 10.1007/s11060-005-9103-z. [DOI] [PubMed] [Google Scholar]
- 39.Dickinson RE, Dallol A, Bieche I, et al. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Cancer. 2004;91(12):2071–2078. doi: 10.1038/sj.bjc.6602222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson B, Stavnes HT, Holth A, et al. Gene expression signatures differentiate ovarian/peritoneal serous carcinoma from breast carcinoma in effusions. J Cell Mol Med. 2011;15(3):535–544. doi: 10.1111/j.1582-4934.2010.01019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gyorffy B, Dietel M, Fekete T, Lage H. A snapshot of microarray-generated gene expression signatures associated with ovarian carcinoma. Int J Gynecol Cancer. 2008;18(6):1215–1233. doi: 10.1111/j.1525-1438.2007.01169.x. [DOI] [PubMed] [Google Scholar]
- 42.Kobel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5(12):e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyle-Chenault RA, Stolk JA, Molesh DA, et al. VSGP/F-spondin: a new ovarian cancer marker. Tumour Biol. 2005;26(5):245–257. doi: 10.1159/000087379. [DOI] [PubMed] [Google Scholar]
- 44.Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66(6):454–458. doi: 10.1016/j.biopha.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 45.Cheng YY, Jin H, Liu X, et al. Fibulin 1 is downregulated through promoter hypermethylation in gastric cancer. Br J Cancer. 2008;99(12):2083–2087. doi: 10.1038/sj.bjc.6604760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pupa SM, Argraves WS, Forti S, et al. Immunological and pathobiological roles of fibulin-1 in breast cancer. Oncogene. 2004;23(12):2153–2160. doi: 10.1038/sj.onc.1207323. [DOI] [PubMed] [Google Scholar]
- 47.Wen Y, Giardina SF, Hamming D, et al. GROalpha is highly expressed in adenocarcinoma of the colon and down-regulates fibulin-1. Clin Cancer Res. 2006;12(20 Pt 1):5951–5959. doi: 10.1158/1078-0432.CCR-06-0736. [DOI] [PubMed] [Google Scholar]
- 48.Wlazlinski A, Engers R, Hoffmann MJ, et al. Downregulation of several fibulin genes in prostate cancer. Prostate. 2007;67(16):1770–1780. doi: 10.1002/pros.20667. [DOI] [PubMed] [Google Scholar]
- 49.Kanda M, Nomoto S, Okamura Y, et al. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol Carcinog. 2011;50(8):571–579. doi: 10.1002/mc.20735. [DOI] [PubMed] [Google Scholar]
- 50.Medioni C, Bertrand N, Mesbah K, et al. Expression of Slit and Robo genes in the developing mouse heart. Dev Dyn. 2010;239(12):3303–3311. doi: 10.1002/dvdy.22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh Y, Katoh M. Comparative genomics on SLIT1, SLIT2, and SLIT3 orthologs. Onco. Rep. 2005;14(5):1351–1355. [PubMed] [Google Scholar]
- 52.Holl EK, Roney KE, Allen IC, et al. Plexin-B2 and Plexin-D1 in dendritic cells: expression and IL-12/IL-23p40 production. PLoS One. 2012;7(8):e43333. doi: 10.1371/journal.pone.0043333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roney KE, O'Connor BP, Wen H, et al. Plexin-B2 negatively regulates macrophage motility, Rac, and Cdc42 activation. PLoS One. 2011;6(9):e24795. doi: 10.1371/journal.pone.0024795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Argast GM, Croy CH, Couts KL, et al. Plexin B1 is repressed by oncogenic B-Raf signaling and functions as a tumor suppressor in melanoma cells. Oncogene. 2009;28(30):2697–2709. doi: 10.1038/onc.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Conrotto P, Corso S, Gamberini S, Comoglio PM, Giordano S. Interplay between scatter factor receptors and B plexins controls invasive growth. Oncogene. 2004;23(30):5131–5137. doi: 10.1038/sj.onc.1207650. [DOI] [PubMed] [Google Scholar]
- 56.Basile JR, Barac A, Zhu T, Guan KL, Gutkind JS. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer Res. 2004;64(15):5212–5224. doi: 10.1158/0008-5472.CAN-04-0126. [DOI] [PubMed] [Google Scholar]
- 57.Zhang Z, Xu X, Zhang Y, Zhou J, Yu Z, He C. LINGO-1 interacts with WNK1 to regulate nogo-induced inhibition of neurite extension. J Biol Chem. 2009;284(23):15717–15728. doi: 10.1074/jbc.M808751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Loov C, Fernqvist M, Walmsley A, Marklund N, Erlandsson A. Neutralization of LINGO-1 during in vitro differentiation of neural stem cells results in proliferation of immature neurons. PLoS One. 2012;7(1):e29771. doi: 10.1371/journal.pone.0029771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stein T, Walmsley AR. The leucine-rich repeats of LINGO-1 are not required for self-interaction or interaction with the amyloid precursor protein. Neurosci Lett. 2012;509(1):9–12. doi: 10.1016/j.neulet.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 60.Pepinsky RB, Shao Z, Ji B, et al. Exposure levels of anti-LINGO-1 Li81 antibody in the central nervous system and dose-efficacy relationships in rat spinal cord remyelination models after systemic administration. J Pharmacol Exp Ther. 2011;339(2):519–529. doi: 10.1124/jpet.111.183483. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.