Abstract
Manual segmentation from magnetic resonance imaging (MR) is the gold standard for evaluating hippocampal atrophy in Alzheimer’s disease (AD). Nonetheless, different segmentation protocols provide up to 2.5-fold volume differences. Here we surveyed the most frequently used segmentation protocols in the AD literature as a preliminary step for international harmonization. The anatomical landmarks (anteriormost and posteriormost slices, superior, inferior, medial, and lateral borders) were identified from 12 published protocols for hippocampal manual segmentation ([Abbreviation] first author, publication year: [B] Bartzokis, 1998; [C] Convit, 1997; [dTM] deToledo-Morrell, 2004; [H] Haller, 1997; [J] Jack, 1994; [K] Killiany, 1993; [L] Lehericy, 1994; [M] Malykhin, 2007; [Pa] Pantel, 2000; [Pr] Pruessner, 2000; [S] Soininen, 1994; [W] Watson, 1992). The hippocampi of one healthy control and one AD patient taken from the 1.5T MR ADNI database were segmented by a single rater according to each protocol. The accuracy of the protocols’ interpretation and translation into practice was checked with lead authors of protocols through individual interactive web conferences. Semantically harmonized landmarks and differences were then extracted, regarding: (a) the posteriormost slice, protocol [B] being the most restrictive, and [H, M, Pa, Pr, S] the most inclusive; (b) inclusion [C, dTM, J, L, M, Pr, W] or exclusion [B, H, K, Pa, S] of alveus/fimbria; (c) separation from the parahippocampal gyrus, [C] being the most restrictive, [B, dTM, H, J, Pa, S] the most inclusive. There were no substantial differences in the definition of the anteriormost slice. This survey will allow us to operationalize differences among protocols into tracing units, measure their impact on the repeatability and diagnostic accuracy of manual hippocampal segmentation, and finally develop a harmonized protocol.
Keywords: Hippocampus, manual segmentation protocol, harmonization, anatomical landmark, Alzheimer’s disease, manual tracing, medial temporal lobes, atrophy, degeneration, MRI
INTRODUCTION
Hippocampal volumetry is a marker sensitive to disease state and progression in Alzheimer’s disease (AD). The proposal for revised diagnostic criteria [1] posits that, even in the preclinical stages of the disease, the presence of hippocampal atrophy on magnetic resonance imaging (MR) is a marker suggestive of AD, the others being temporo-parietal hypometabolism on FDG PET, abnormal CSF tau and Abeta42 proteins, cerebral amyloidosis on molecular PET imaging. Currently, hippocampal volumetry is included as a secondary outcome measure in several clinical trials of disease modifying drugs to support the claim of disease modification [2].
Manual outlining on MR images by trained raters is presently the most accurate, validated and used procedure to measure hippocampal volumes [3–6], and the gold standard for the validation of automated segmentation algorithms [7–12]. However, a large number of protocols for the manual segmentation of the hippocampus is available and adopted in different fields of neuroscience research, including those investigating a variety of psychiatric and neurodegenerative conditions [13, 14]. These segmentation protocols differ in their definition of anatomical boundaries and tracing procedures, thus originating hippocampal volume estimates that cannot be straightforwardly compared. Indeed, the mean volume for a normal hippocampus can range from 2 to 5.3 cm3 [14] across laboratories worldwide. Even if individual differences in head size are taken into account, this range is far too broad to accept hippocampal volumetry as a valid marker for any neurological condition. Although these differences are in part caused by heterogeneities in image acquisition and preprocessing, heterogeneities in the landmark definitions are major contributors. Heterogeneity was found in the definition of the most rostral and most caudal slices, in the criteria for inclusion or exclusion of hippocampal white matter (alveus and fimbria), in the definition of boundary lines with adjacent anatomical structures [13, 14].
This heterogeneity prevents a direct comparison of the outcome of different studies, and slows down the transfer of the marker from the research laboratory to the clinical setting. Standard operational procedures (SOPs) are clearly required for manual hippocampal volumetry to be transferred to routine diagnostic settings and gain status as a surrogate outcome in clinical trials for disease modifying drugs. SOPs will promote the large use of hippocampal atrophy measurements for the early diagnosis of AD and allow the comparison of the effect of different drugs in clinical trials. Moreover, the automated approaches need to be validated using a gold standard, for a given clinical population. SOPs may represent the gold standard for the many automated algorithms aiming to extract hippocampal volume with minimal or no human input that are presently under development [15, 16].
The aim of this study is to survey a selection of the most popular protocols for hippocampal segmentation used in AD research, in order to extract commonalities and differences. Importantly, we sought explicit input from protocol authors to check for proper understanding of their work. This survey is the first step of a larger project aiming to develop an internationally harmonized protocol for hippocampal segmentation.
MATERIALS AND METHODS
In this work we surveyed the landmark definitions provided for the manual segmentation of the hippocampus from MR images. A pilot survey was first carried out on 5 protocols belonging to the repertoire routinely used at LENITEM as tracing criteria, or for study purposes. The study design was then extended to a wider set of protocols including all of the most commonly used ones within the AD literature (see ‘Selection of segmentation protocols’ below). Hippocampal manual segmentation was performed by a single tracer on the left hippocampus of two subjects based on each protocol (see ‘Selection of scans for the prototypical tracings’, ‘Image processing’, and ‘Prototypical tracings’ sections below), and tracings were then checked for correctness with the lead authors of the protocols (‘Authors’ check’) to obtain certified segmentations. We then extracted differences among protocols through the semantic harmonization of terms (see ‘Extraction of similarities and differences’ below).
Selection of segmentation protocols
The pilot survey was carried out on five protocols routinely used in our laboratory for learning and as tracing criteria [17–21]. In the experimental phase, the selection was expanded to include all the most widely cited protocols in the AD literature (Table 1).
Table 1.
Surveyed protocols for hippocampal segmentation.
| Protocol | Citations in the AD literature |
Cited by Konrad and colleagues |
Cited by Geuze and colleagues |
Compliance with selection criteria |
Inclusion stage |
Selected for harmonization |
Ref |
|---|---|---|---|---|---|---|---|
| Killiany, 1993 | 173 | No | No | Satisfying criteria | Experimental | Yes | [24] |
| Convit, 1997 | 143 | Yes | Yes | Satisfying criteria | Experimental | Yes | [26] |
| Watson, 1992 | 122 | Yes | Yes | Satisfying criteria | Experimental | Yes | [30] |
| Soininen, 1994 | 118 | Yes | Yes | Satisfying criteria | Experimental | Yes | [28] |
| Sheline, 1996 | 79 | Yes | No | Author not available | Experimental | No | [31] |
| Lehericy, 1994 | 78 | No | No | Satisfying criteria | Experimental | Yes | [25] |
| Pruessner, 2000 | 78 | Yes | No | Satisfying criteria | Pilot | Yes | [18] |
| Bremner, 1995 | 53 | Yes | Yes | Only hippocampal body included |
Experimental | No | [32] |
| deToledo-Morrell, 2004 | 50 | No | No | Satisfying criteria | Experimental | Yes | [23] |
| Haller, 1997 | 44 | Yes | No | Satisfying criteria | Experimental | Yes | [27] |
| Cook, 1992 | 44 | Yes | Yes | Plexus choroideus included |
Experimental | No | [33] |
| Bigler, 1997 | 38 | Yes | Yes | Less than 40 citations | Experimental | No | [34] |
| Shenton, 1992 | 33 | Yes | Yes | Less than 40 citations | Experimental | No | [35] |
| MacQueen, 2003 | 30 | Yes | No | Less than 40 citations | Experimental | No | [36] |
| Bogerts, 1993 | 30 | No | Yes | Less than 40 citations | Experimental | No | [22] |
| Narr, 2004 | 25 | Yes | No | Less than 40 citations | Experimental | No | [37] |
| Mervaala, 2000 | 21 | Yes | No | Less than 40 citations | Experimental | No | [38] |
| Steffens, 2002 | 20 | Yes | No | Less than 40 citations | Experimental | No | [39] |
| Jack, 1994 | 18 | Yes | Yes | Satisfying criteria except citations number |
Pilot | Yes | [17] |
| O’Brien, 2004 | 18 | Yes | No | Less than 40 citations | Experimental | No | [40] |
| Ashtari, 1999 | 17 | Yes | No | Less than 40 citations | Experimental | No | [41] |
| Bartzokis, 1993 | 17 | Yes | Yes | Satisfying criteria except citations number (<40 in AD literature) |
Pilot | Yes | [21] |
| Pantel, 2000 | 17 | No | No | Satisfying criteria except citations number (<40 in AD literature) |
Pilot | Yes | [19] |
| Zipursky, 1994 | 16 | Yes | Yes | Less than 40 citations | Experimental | No | [42] |
| Giedd, 1996 | 16 | Yes | Yes | Less than 40 citations | Experimental | No | [43] |
| Kates, 1997 | 16 | Yes | No | Less than 40 citations | Experimental | No | [44] |
| Honeycutt, 1998 | 15 | Yes | Yes | Less than 40 citations | Experimental | No | [45] |
| von Gunten, 2000 | 13 | Yes | No | Less than 40 citations | Experimental | No | [46] |
| Vythilingam, 2004 | 10 | Yes | No | Less than 40 citations | Experimental | No | [47] |
| Hastings, 2004 | 9 | Yes | No | Less than 40 citations | Experimental | No | [48] |
| Niemann, 2000 | 9 | Yes | No | Less than 40 citations | Experimental | No | [49] |
| Lobnig, 2006 | 7 | Yes | No | Less than 40 citations | Experimental | No | [50] |
| Lloyd, 2004 | 6 | Yes | No | Less than 40 citations | Experimental | No | [51] |
| Driessen, 2000 | 6 | Yes | No | Less than 40 citations | Experimental | No | [52] |
| Barr, 1997 | 5 | Yes | No | Less than 40 citations | Experimental | No | [53] |
| Neumeister, 2005 | 5 | Yes | No | Less than 40 citations | Experimental | No | [54] |
| Caetano, 2004 | 5 | Yes | No | Less than 40 citations | Experimental | No | [55] |
| Frodl, 2004 | 5 | Yes | No | Less than 40 citations | Experimental | No | [56] |
| Rusch, 2001 | 4 | Yes | No | Less than 40 citations | Experimental | No | [57] |
| Nakano, 2002 | 4 | Yes | No | Less than 40 citations | Experimental | No | [58] |
| MacMillan, 2003 | 3 | Yes | No | Less than 40 citations | Experimental | No | [59] |
| Yucel, 2007 | 3 | Yes | No | Less than 40 citations | Experimental | No | [60] |
| Brambilla, 2003 | 3 | Yes | No | Less than 40 citations | Experimental | No | [61] |
| Saylam, 2006 | 3 | Yes | No | Less than 40 citations | Experimental | No | [62] |
| MacMaster & Kusumakar, 2004 | 2 | Yes | No | Less than 40 citations | Experimental | No | [63] |
| Malykhin, 2007 | 2 | No | No | Satisfying criteria except citations number (<40 in AD literature) |
Pilot | Yes | [20] |
| Scott, 2004 | 2 | Yes | No | Less than 40 citations | Experimental | No | [64] |
| Chen, 2004 | 2 | Yes | No | Less than 40 citations | Experimental | No | [65] |
| Vermetten, 2006 | 1 | Yes | No | Less than 40 citations | Experimental | No | [66] |
| Arango, 2003 | 1 | Yes | No | Less than 40 citations | Experimental | No | [67] |
| Chang, 2005 | 1 | Yes | No | Less than 40 citations | Experimental | No | [68] |
| Rosso, 2005 | 1 | Yes | No | Less than 40 citations | Experimental | No | [69] |
| Xia, 2004 | 1 | Yes | No | Less than 40 citations | Experimental | No | [70] |
| Frazier, 2005 | 1 | Yes | No | Less than 40 citations | Experimental | No | [71] |
| Bossini, 2008 | 1 | Yes | No | Less than 40 citations | Experimental | No | [72] |
| Starkman, 2007 | 0 | Yes | No | Less than 40 citations | Experimental | No | [73] |
Protocols are sorted in order of citation in the AD literature (first the most cited)
The protocols mentioned in the two available reviews on manual segmentation of the hippocampus were examined first [13,14]. Original protocols were drawn from the recent paper by Konrad and colleagues [13] and the review by Geuze and colleagues [14]. The review by Konrad and colleagues’ included 71 protocols for hippocampal segmentation. Of these, 50 protocols provided an original description of landmarks for segmentation and were retained in the present survey, while the remaining 21 redirected the reader to previously published protocols and, thus, were excluded from this survey. All but one [22] of the protocols reviewed by Geuze and colleagues were included among these 50; the one that was not [22] was retained in the present survey.
Two of the 5 protocols of the pilot study [19,20] were not among those reviewed by Konrad and colleagues or Geuze and colleagues, and were retained in the present survey.
In order to ascertain whether the 53 protocols retained in the present survey included all of the most cited protocols for hippocampal segmentation in the AD literature, we repeated the same search carried out by Konrad, using the key-words “Alzheimer*”, “AD” or “dementia” instead of Konrad’s “depression”, “major depression” or “unipolar depression”. This search lead to the inclusion of 3 more protocols [23–25] with a high rate of citations in the AD literature (Table 1), leading to a total of 56 protocols retained in the present survey.
To be selected for harmonization, protocols had to satisfy five criteria, considered in the following hierarchical order: i) Number of citations in the AD literature greater than 40. Literature citations of the 56 protocols were identified using the ISI Web of Science portal, and the keywords “(Alzheimer* OR dementia) AND hippo*” were used to compute citations only within the AD literature (Table 1, “Citation in the AD literature”). Papers with less than 40 citations on December 31, 2009 were excluded; ii) at least the head and body of the hippocampus were included in the segmentation; iii) adjacent structures such as the amygdala, the choroid plexus, and major portions of the parahippocampal cortex were excluded from the segmentation; iv) three-dimensional (3D) T1-weighted MR sequences with slice thickness smaller or equal to 3 mm were used; v) MR scans were acquired using a scanner with field strength greater than 1 Tesla; vi) the availability of a lead author to perform a one hour web-conference was required, to check the correctness of the tracings according to the specific protocol (Table 1, “Compliance with selection criteria”).
Finally, we included 12 protocols (Table 1, First author, publication year, in alphabetical order): [B] Bartzokis, 1998 [21]; [C] Convit, 1997 [26]; [dTM] deToledo-Morrell, 2004 [23]; [H] Haller, 1997 [27]; [J] Jack, 1994 [17]; [K] Killiany, 1993 [24]; [L] Lehericy, 1994 [25]; [M] Malykhin, 2007 [20]; [Pa] Pantel, 2000 [19]; [Pr] Pruessner, 2000 [18]; [S] Soininen, 1994 [28]; [W] Watson, 1992 [29].
Selection of scans for the prototypical tracings
Tracings of the left hippocampus were carried out on images of one healthy control and one AD patient taken from the3DT1-weighted structural ADNI dataset [74], following the landmarks of each of the 12 protocols. The accuracy of the application of the protocols as described in the manuscripts was checked with the pertinent lead author.
The AD patient (ID: 021 S 0642) was selected from those with moderate to severe medial temporal atrophy (MTA) (score of 3 on Scheltens’s visual rating scale, ranging from 0 to 4) [75], was 85 years old, had MMSE score of 25/30, and Clinical Dementia Rating (CDR) of 1. The control (ID: 023 S 0058) was chosen for having minimum atrophy (score of 1 on Scheltens’s visual rating scale). The control was 70 years of age, had a MMSE score of 30/30, a CDR of 0 [75].
Image processing
A combination of several freely available tools was used to prepare the raw MR ADNI images for manual segmentation. DICOM images were converted to Analyze/NIFTI format using the MRIcron software (V.8.0, http://www.cabiatl.com/mricro/). Prior to prototypical tracing, the 3D image was manually reoriented based on the requirement of the corresponding protocol. Seven of the protocols considered in this study required the reorientation of the image to the long axis of the hippocampus, 5 required aligning the image to the line which passes through the anterior and posterior commissures of the brain (AC-PC line). These reorientation steps were performed using the 3D-Slicer software (V.3.2, http://www.slicer.org/). All MR images were analyzed in native space.
Prototypical tracings
A single tracer (RG) segmented the left hippocampus of the two subjects according to each of the 12 protocols, after reorienting the image as required by each protocol. The intra-rater (0.94) and inter-rater (0.89) correlation coefficients of the tracer were computed previously, on a sample of 20 healthy controls, not including the ones examined for this study, and in comparison to another expert tracer within the laboratory [76]. The protocol used for computing the correlation coefficients was that by Pruessner and colleagues [18]. Tracings were performed using Multitracer (http://air.bmap.ucla.edu/MultiTracer/) developed at the Laboratory Of NeuroImaging (LONI) at UCLA (Los Angeles, USA), allowing simultaneous 3D navigation in the axial and sagittal planes (Fig. 1). Hippocampal boundaries based on each protocol were traced on approximately 20 1.2 mm thick coronal slices.
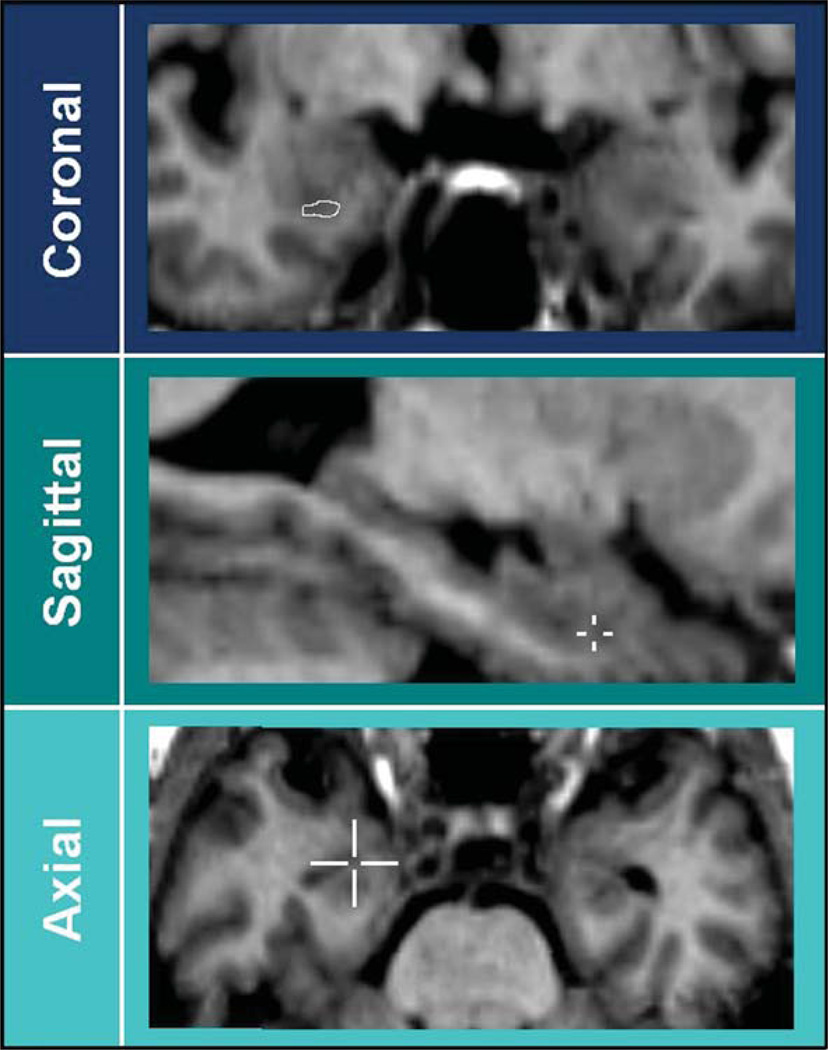
Fig. 1.
Most anterior slice. Differences in the definition of the most anterior slice are overcome due to currently used software for 3D brain navigation. The visualization of the cursor position in 3D provides unequivocal information about its location in the amygdale or in the hippocampus, even if a single coronal section may not provide sufficiently clear information.
Authors’ check
Hippocampal tracings were certified as compliant with the original protocols using a three-stage check procedure: check with the lead author of the protocol, trace editing, and final check.
A Power Point presentation was provided to the lead author showing native and segmented slices of the two sample subjects. These slices were also paired with pictures of corresponding histological cuts, and text notes were added to describe tracings, landmarks and questions on unclear issues. Power Point presentations begun with a survey table, summarizing the explicit landmarks for that protocol, the main included and excluded structures, and other possibly specific key features of each protocol (Power Point presentations can be accessed at http://www.hippocampal-protocol.net/SOPs/investigatedprotocols.html). The lead author was asked to correct the survey table, examine the tracings, and take note of the unclear points that he/she would be asked to discuss subsequently. The appropriateness of our understanding of criteria was verified with the lead author at both the semantic and the practical levels, by means of individual teleconferences (TCs). TCs were carried out with a web-seminar system that allowed all participants to share the same desktop view, scroll the slides, control the cursor and carry out tracings on the same image viewed by all concurrently. TCs were recorded (available upon request), to ensure information availability over time. The tracings were verified, and unclear points were elucidated, by working on the power point presentations and with the use of Multitracer when necessary. It should be noted that the correction stage included the update to advances in tracing method, such as the use of 3D visualization tools, or other changes occurring in the tracing methods over time. After the TC, the tracings were edited according to the author’s input and resent to the author for further check. This procedure was iteratively repeated until the author was satisfied that tracings had been performed according to his/her original description. These presentations, including the final summary table and the final “certified” tracings, are available at http://www.hippocampal-protocol.net/SOPs/investigatedprotocols.html.
Extraction of similarities and differences
A compendium of the survey tables of all protocols was created, by reporting the features for each segmentation criterion as it was certified by the lead authors (http://www.hippocampal-protocol.net/SOPs/LINKPAGE/anatomical-landmarks-certified-12.xls). A semantic harmonization of the features was necessary in order to ensure the correct comparison among these heterogeneous criteria, since the very same landmark was indeed named and described using different words in different protocols (http://www.hippocampal-protocol.net/SOPs/LINK PAGE/harmonized-anatomical-la ndmarks-12.xls). This phase of the survey can also be considered as the first step in the operationalization of the differences among the examined protocols. By operationalization, we mean the process of reducing the variability among protocols into a finite number of differences, sufficiently well defined to lend themselves to quantitative investigation. In practice, the wide heterogeneity in landmarks would be reduced into a finite number of elementary tracing units, which could be measured and tested.
The harmonized definitions for each landmark were chosen by two of us (MB and RG) after having checked our comprehension in the teleconferences, based on the ability of definitions to unequivocally identify the landmark, and assuming that tracing would be carried out on coronal slices from rostral to caudal.
After the semantic harmonization, the extraction of differences among the author-certified protocols was carried out for each of the examined portion of the hippocampus where different criteria could be applied, i.e., most anterior and most posterior slices, superior border (i.e., inclusion or exclusion of alveus and fimbria), and medial border at the level of the body. Each different criterion adopted for each of these portions denotes a different definition of landmarks, which outline specific regions of hippocampal tissue. This means that, based on these criteria, well defined regions of hippocampal tissue can be isolated, and included or excluded in the segmentation of the hippocampus on MR images, depending on the adopted protocol. Therefore, this phase was also the basis for the operationalization of differences among protocols into a limited number of concrete and elementary segmentation units, to allow quantitative investigation in the next stage of this project.
The semantic harmonization and the extraction of similarities and differences also implied a hierarchy-based selection. Landmarks internal (e.g., alveus) or adjacent (e.g., CSF, parahippocampal white matter) to the hippocampus were considered as having higher value than those external to the hippocampus (i.e., pulvinar), and, whenever possible, were chosen for the definition and first operationalization stage, since these are invariant to the plane of orientation of the 3D MR images.
RESULTS
Protocols were uniform as to magnetic field strength of MR (1.5T) and reproducibility values. Moreover, they were uniform on use of 3D visualization, and in the exclusion of non-hippocampal tissue (amygdala, choroid plexus). Instead, differences could be detected in the direction of segmentation (i.e., rostral to caudal), as well as in the population used for validating the procedure (Table 2).
Table 2.
General features of the 12 selected protocols
| Protocol | Slice | Direction of tracing | MR Sequence | Validation Sample | Included Hippocampal tissue | Reproducibility measures |
|---|---|---|---|---|---|---|
| Bartzokis et al., 1998 [21] | coronal | from rostral to caudal | T1 | healthy controls | Head, body, and tail until visualization of colliculi |
5% error |
| Convit et al., 1997 [26] | coronal | from rostral to caudal | T1 | AD | Head, body, and tail until visualization of crus/crura of fornix/ces in full profile |
0.85 inter-rater reliability (ICC) |
| deToledo-Morrell et al., 2004 [23] |
coronal | from rostral to caudal | T1 | MCI | Head, body, and tail until visualization of crus/crura of fornix/ces in full profile |
0.97 intra-rater and inter-rater reliability (ICC) |
| Haller et al., 1997 [27] | coronal, sagittal and axial |
from caudal to rostral | T1 | schizophrenia | Head, body, and whole tail | 77.9% overlap of voxels |
| Jack et al., 1994 [17] | coronal | from caudal to rostral | T1 | epilepsy | Head, body, and tail until visualization of crus/crura of fornix/ces in full profile |
1.2% and 3.4% intra- and inter-observer variability (5% error with slices below a thickness of 3mm) |
| Killiany et al., 1993 [24] | coronal | from rostral to caudal | T1 | AD | Head, body, and tail until visualization of crus/crura of fornix/ces in full profile |
0.91 and 0.92 intra- and inter-rater reliability (ICC) |
| Lehericy et al., 1994 [25] | coronal | from caudal to rostral | T1 | AD | Head, body, and tail ntil visualization of crus/crura of fornix/ces in full profile |
7% inter-rater relative error on volume [81] |
| Malykhin et al., 2007 [20] | coronal; sagittal at the level of the head |
from the hippocampal body to the tail, then from the posterior part of head to its rostral part |
T1 | Parkinson’s disease; depression |
Head, body, and whole tail | 0.86 and 0.96 intra- and inter-rater reliability (ICC) |
| Pantel et al., 2000 [19] | coronal | from rostral to caudal | T1+ T2 | healthy controls | Head, body, and whole tail | intraclass R = 0.78 |
| Pruessner et al., 2000 [18] | coronal | from caudal to rostral | T1 | healthy controls | Head, body, and whole tail | 0.92 and 0.99 intra- and inter-rater reliability (ICC) |
| Soininen et al., 1994 [28] | coronal | from rostral to caudal | T1 | MCI | Head, body, and tail until visualization of crus/crura of fornix/ces in full profile |
0.95 intra-rater reliability (ICC) |
| Watson et al., 1992 [29] | coronal | from rostral to caudal | T1 | healthy controls | Head, body, and tail until visualization of crus/crura of fornix/ces in full profile |
intra-rater score range between 0.88 and 0.99 |
The survey tables for each protocol are available at: www.hippocampal-protocol.net. As illustrated in the summary survey table, following semantic harmonization of landmarks (http://www.hippocampal-protocolnet/SOPs/LINK PAGE/harmonized-anatomical-land marks-12.xls.), differences between the protocols that most likely had an impact on the volumetric estimates concerned heterogeneities in the definition of (a) the orientation of the images; (b) the most posterior slice; (c) the superior border; (d) the separation from the parahippocampal gyrus at the level of the subiculum, in the hippocampal body (Table 3). Heterogeneities in the definition of (e) the most anterior slice are not shown in Table 3 for reasons that will be explained below.
Table 3.
Differences of anatomical landmarks among the 12 selected protocols, after semantic harmonization
| a) Plane of tracing | |||
| Axis of hippocampus [B, C, dTM, J, L, S, W] |
AC-PC line [H, K, M, Pa, Pr] | ||
| b) Most posterior slice | |||
| Where inferior and superior colliculi are jointly visualized [B] |
Where crus/crura of fornix/ces is/are visible in full profile [C, dTM, J, K, L, S, W] |
Where gray matter is visible inferomedially to the trigone of the lateral ventricle [H, M, Pa, Pr] |
|
| c) Superior border | |||
| Lower border of alveus/fimbria [B, H, K, Pa, S] |
Upper border of alveus/fimbria [C, dTM, J, L, M, Pr, W] |
||
|
d) Medial border at subiculum level |
|||
| vertical line from the CA to the WM of the parahippocampal gyrus [C] |
Oblique line with same inclination of parahippocampal WM, connecting the inferior part of the subiculum to the quadrigeminal cistern [K, L, M, Pr, W] |
Horizontal line from the highest medial point of the parahippocampal WM to the cistern [B, dTM, H] |
Line outlining the contour of white matter of parahippocampal gyrus [J, Pa, S] |
AC = anterior commissure; PC=posterior commissure; CA =cornu Ammonis; WM = white matter; [B] Bartzokis et al., 1998, [C] Convit et al.,1997, [dTM] deToledo-Morrell et al., 2004; [H] Haller et al., 1997, [J] Jack et al., 1994, [K] Killiany et al., 1993, [L] Lehericy et al., 1994, [M] Malykhin et al., 2007, [Pa] Pantel et al., 2000, [Pr] Pruessner et al., 2000, [S] Soininen et al., 1994, [W] Watson et al., 1992.
a) Plane of tracing. Seven of the protocols reoriented the images along the long axis of the hippocampus [17, 21, 23, 25, 26, 28, 29], and 5 used images oriented along the AC-PC line [18–20, 24, 27]. Among these, 3 are the most recently published protocols [18–20], consistently with a trend to taking advantage of the greater availability of AC-PC automatic registration algorithms [77–80], that minimize and facilitate human work in the preprocessing stages.
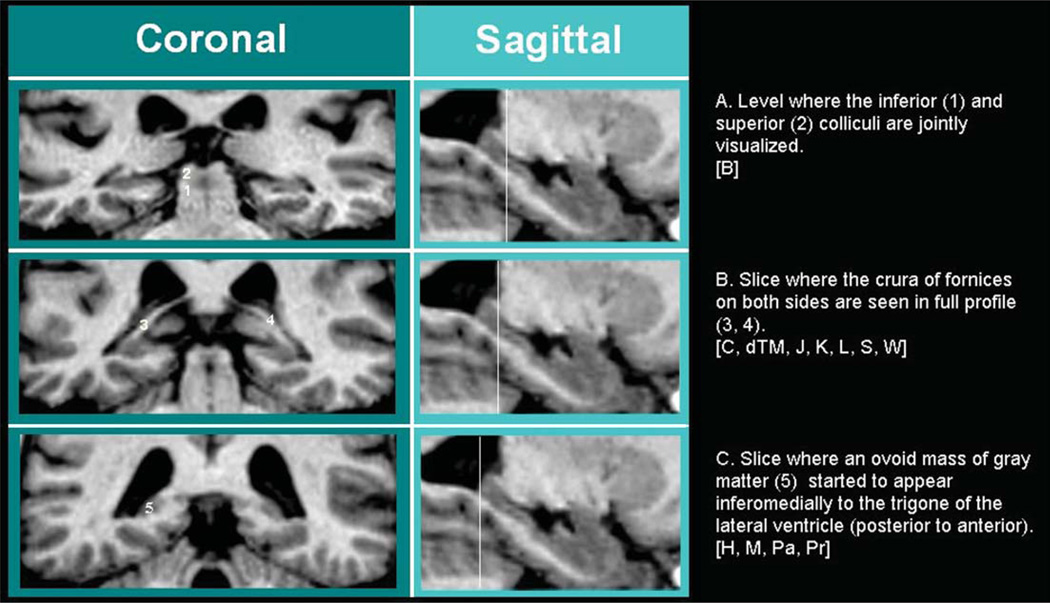
b) Definition of the most posterior slice. Important differences characterize the protocols as to the definition of the most posterior slice where hippocampal tissue is segmented. The most restrictive protocol stops tracing at the level where both the inferior and superior colliculi are visible (Table 3; Fig. 2A). Less restrictive protocols stop when the crus or both crura of the fornices are seen in full profile, these two criteria differing often by one single slice (Fig. 2B). The less restrictive ones trace as long as they can detect hippocampal gray matter on the coronal slices (Fig. 2C), the only difference among these consisting in the attempt to exclude gray matter belonging to the vestigial hippocampal tissue of the Andrea Retzius and fasciolar gyri (see point c, Definition of the superior border).
Fig. 2.
Most posterior slice. The criterion in A) is followed by protocol [B], B) by protocols [C, dTM, J, K, L, S, W], C) by [H, M, Pa, Pr].
c) Definition of the superior border. The definition of the superior border concerns the inclusion or exclusion of hippocampal white matter (i.e., alveus and fimbria) along the structure and, for most caudal slices, the inclusion or exclusion of the vestigial gray matter (i.e., fasciolar gyrus and Andrea Retzius gyrus) that extends dorso-medially (Fig. 3, last column).
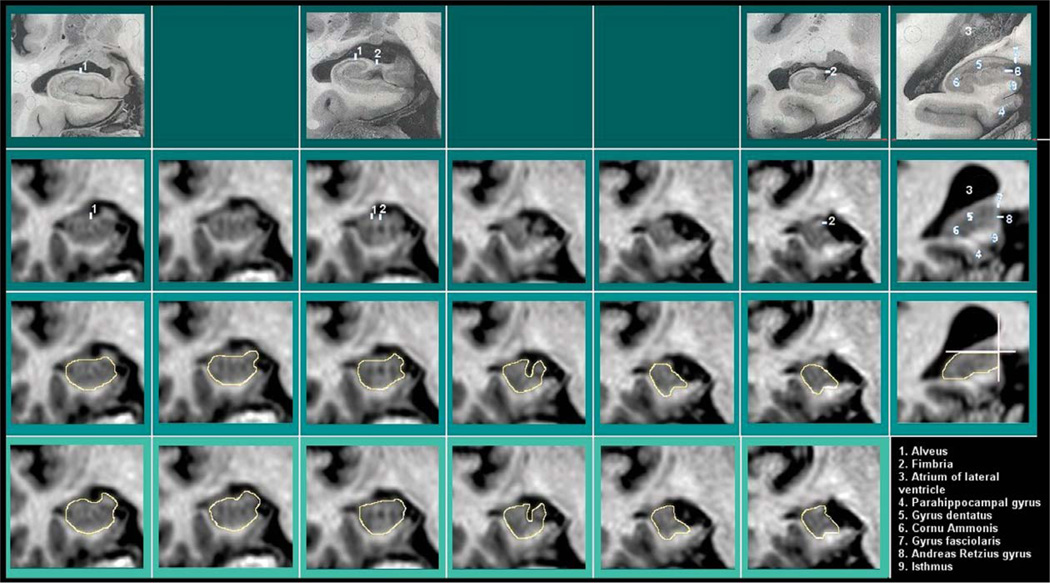
Fig. 3.
Superior border. The superior border concerns the hippocampal white matter, i.e. the alveus and fimbria, that can be excluded [B, H, K, Pa, S] or included in different tracing protocols [C, dTM, J, L, M, Pr, W]. Additional criteria for separation of vestigial gray matter tissue in posterior-most slices are reported in [Pr] (last column). In the first line, histological pictures corresponding to the MRI slice of the same column are presented. Line II: MRI images without tracings. Lines III-IV: same MRI images with tracing example of exclusion (III) and inclusion (IV) of the hippocampal white matter.
Seven of the protocols included alveus and fimbria in the segmentation (Fig. 3, bottom line), and five excluded these white matter layers (Table 3; Fig. 3, 3rd line), at least whenever visible.
As to the vestigial gray matter located on the most caudal portion of the hippocampus, arbitrary linear demarcations are adopted to separate the uppermost tissue belonging to the fasciolar and Andrea Retzius gyri [Pr] (Fig. 3, last column), or a superior portion is limited, by excluding a little layer of gray matter below the cingulate gyrus and the isthmus of the cingulum [M]. As in the case of the most anterior slice, the separation from vestigial gray matter may benefit from the 3D visualization allowed by recent software.
d) Definition of the medial border. The definition of the medial border was not problematic for the head and tail. Instead, its definition differed through the protocols for the level of the body, where the subicular region of the hippocampus joins the entorhinal cortex within the parahippocampal gray matter.
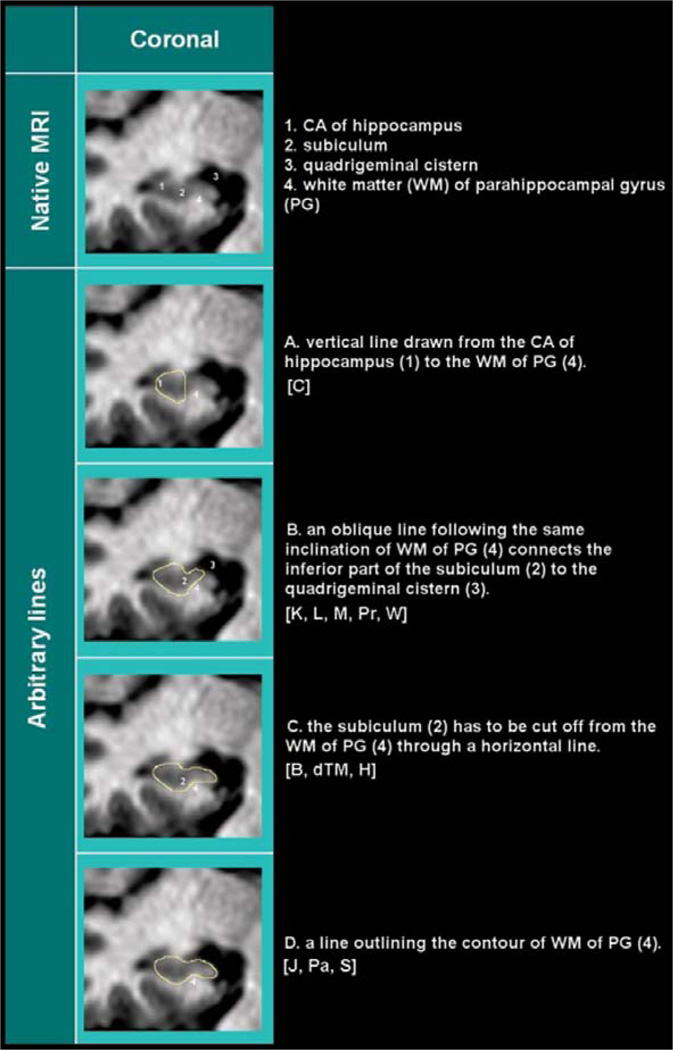
Five different methods can be found across the 12 protocols to separate the hippocampal body from the adjacent parahippocampal gray matter (Table 3; Fig. 4). Arbitrary linear demarcations are adopted by most of them. The most restrictive protocol draws a vertical line from the most medial point of the cornu Ammonis (CA) gray matter, on the dorsal aspect of the hippocampus, down to the parahippocampal white matter (Fig. 4A). Less restrictive protocols use oblique lines with different angles. These oblique lines are drawn from the lowest point of the parahippocampal white matter, and proceed medially to the liquor of the cistern, with an angle of 45°, or with a similar angle as the parahippocampal white matter below (Fig. 4B). Three protocols used a horizontal line connecting the highest point in the medial parahippocampal white matter to the CSF (Fig. 4C). Finally, three protocols segment the hippocampal gray matter relying on the visible morphology determined by the white matter shape and possible gray matter signal (Fig. 4D).
Fig. 4.
Medial border. Five different methods are described across protocols to separate the subiculum, medially, from the rest of the parahippocampal gyrus. CA=cornu Ammonis; WM=white matter; PG=parahippocampal gyrus. The method in A is adopted by [C], that in B by [K, L, M, Pr, W], C by [B, dTM, H], D by [J, Pa, S].
e) Definition of the most anterior slice. Heterogeneities in the definition of the boundary with the amygdala were found (http://www.hippocampal-protocol.net/SOPs/LINK PAGE/anatomical-landmarks-cer tified-12.xls), but these can be overcome due to the currently available software allowing 3D navigation (Fig. 1). This common approach allows direct visualization of the exact cursor location and of the tracing in different planes simultaneously; i.e., within the hippocampal head, within the amygdala, or in any neighboring or boundary region, the cursor can be seen from orthogonal visualization planes, where the spatial relationships among neighboring structures can be seen in different perspectives, thus providing additional complementary information. This possibility to visualize in 3D the position of the cursor allows to disambiguate whether the gray matter belongs to the hippocampus or to the amygdala in the most anterior slice. Thus, for this slice, the definition of anatomical landmarks in the coronal plane, which may differ among protocols, and which is less reliable than the direct visualization in 3D, can be considered less relevant. The consequence of this approach was that the only difference that applied to the segmentation of the hippocampal head consisted in the inclusion or exclusion of the hippocampal white matter (alveus/fimbria) when visible, i.e. in the definition of its superior border (see point c).
DISCUSSION
In this work, we have extracted similarities and differences among 12 protocols for hippocampal segmentation widely used in the field of Alzheimer’s disease, in order to capture the source of volume variability that can be ascribed to heterogeneity in landmark definition across protocols. This extraction is the basis for an operationalization procedure aiming to achieve a finite number of well-defined units representing differences among protocols, that enable the gathering of quantitative information on these differences. Quantitative investigation will assess their impact on re-trace reliability, and on the volume differences due to AD. This and other information will help a panel of experts to make evidence-based decisions about the specific features that should be included in a harmonized protocol, using a Delphi procedure, within an international project that is currently ongoing (“A harmonized protocol for hippocampal volumetry: an EADC-ADNI effort”, Alzheimer’s Association funding number 174022, www.hippocampal-protocol.net).
Heterogeneities across protocols
The main differences between protocols concerned the definition of the most caudal slice, of the medial border at the level of the hippocampal body, and the inclusion or exclusion of the alveus and fimbria. The definition of the most rostral slice was considered to be no longer relevant as current software packages for 3D visualization can clarify the anatomical localization of the cursor on the MR image. It is possible that this tool may also solve the separation of the vestigial fasciolar and Andrea-Retzius gyri. There is consensus that their exclusion from proper hippocampal tissue should be recommended, and care in excluding them was indeed used by two protocols [M, Pr]. Most of the protocols [B, C, dTM, J, K, L, S, W] did not trace any hippocampal tissue located caudal to the slice where the crus or both crura of the fornices could be seen in full profile. This led to an a priori exclusion of the vestigial hippocampal tissue, but also to sacrificing a large portion of the hippocampal tail, which may convey relevant information about AD [76].
Different arbitrary linear demarcations were used to trace the medial border, to separate hippocampal tissue from the rest of the parahippocampal gyrus, at the level of the subiculum. No anatomical details have ever been certified as valid landmarks for the tracing of this boundary [13]. The next stage of the project will evaluate whether arbitrary linear demarcations are more reliable than tracings based on the visual morphology apparent on the MR images.
Results in the context of the literature
Two reviews on protocols for hippocampal segmentation were previously carried out. The first [14], examining most cited protocols up to December 2003, provided an extensive description of heterogeneities in all components of tracing protocols, including image acquisition parameters, pre-processing procedures, landmark definitions, and provided recommendations to carry out optimal hippocampal segmentation. The second [13] focused mostly on landmark definitions. Although we extracted protocols from these reviews, we evaluated a much lower number of reports than those examined in the review by Konrad. Nonetheless, our results are similar to this larger review. Across the 71 protocols examined, Konrad found heterogeneities in the definition of the most anterior and most posterior slices, of the in feromedial border of hippocampal body, in the inclusion or exclusion of the alveus and fimbria, and on the use of arbitrary linear demarcations when anatomical detail provided by the imaging sequence was unclear [13]. All these sources of variance were also detected among the 12 protocols examined in our study. This increases the confidence that, although our study was carried out on a limited number of protocols, they were representative of the variability of landmarks present in the literature on hippocampal segmentation, without missing anatomical landmark specifications that may affect a harmonization project. Furthermore, we obtained the authors’ certification of appropriate comprehension and execution of the tracing. This is an important point because the complexity of the anatomical structure of the hippocampal formation requires clarification of protocol details from the expert protocol designers with first-hand tracing experience.
Study limitations
We adopted the criterion of selecting protocols widely used in the literature of AD although different selection criteria could have been used. Nevertheless, our results were entirely in line with the previous larger review.
Another possible limitation is the inclusion of both recent and older protocols. These may differ in landmark definition since the older protocols are based on the view of coronal slices. Moreover, our categorization of landmarks for these protocols includes subsequent modifications that the authors carried out in more recent years to benefit of more recent tools like the 3D visualization, and that we ascertained through the individual teleconferences. Other subsequent changes to protocols were dictated by focusing on different study targets than those expressed in the paper reporting the original protocols. One example is the desire (or lack thereof) to accurately segment adjacent regions, like the parahippocampal gyrus, that overlaps in part with hippocampal tissue from the point of view of its anatomical definition. Moreover, alternative criteria may be used by the same author, depending on image quality. For example, the medial border may be segmented following visible morphology or through arbitrary linear demarcations depending on visibility of any boundary on the MR slices [J], and the inclusion of alveus and fimbria could be an acceptable criterion if their exclusion was made difficult due to poor visibility or contrast [B, K, Pa]. In cases like these, we gave priority to the criterion that the author would adopt in cases where anatomical structures are fully visible, even though more than one criterion may be appropriately reported for the same author.
The next steps towards the development of a harmonized protocol
The global project, described and updated on www.hippocampal-protocol.net, joins experts from the ADNI and EADC consortia, and other centres with advisory role. It is aimed to achieve and validate a homogeneous protocol, and implement its standard learning and use across laboratories. Standard hip-pocampal segmentation is indeed required in clinical trials and as a gold standard in the development and validation of more automated approaches to define hippocampal volumes, as they demonstrate utility. The development of a standard approach that relies on now commonly used T1-weighted high resolution images will allow a broader comparison across studies. The immediate next step of the project will be the completion of the operationalization of differences among protocols in order to model and quantify them. Information about differences in reliability, across different tracers, and in test re-test assessments will be provided for each of the relevant differences among protocols detected by this survey. Moreover, their value to inform on AD pathology will be computed on an adequate sample of representative patients with a diagnosis of AD. A panel of experts will then be provided with quantitative information that would support decisions on which features should be included in a harmonized protocol.
This effort is particularly relevant in light of the new diagnostic clinical and pre-clinical criteria for AD, which can be found at http://www.alz.org/research/diagnostic criteria. In line with the 2007 research criteria by Dubois et al. [1], these criteria place emphasis on hippocampal volumetry, stating that, although not specific for this kind of disorder, this biomarker better correlates with disease progression than the more specific molecular biomarkers, such as the Abeta and Tau CSF concentration levels [1, 82]. A uniform method for the computation of hippocampal volume would improve AD diagnosis across laboratories, and hopefully provide a comparable hippocampal outcome measure in clinical trials for disease modifying drugs.
ACKNOWLEDGMENTS
The Alzheimer’s Association has provided logistic support for the following project meetings: “Harmonization of Protocols for the Manual Segmentation of the Hippocampus”, held in Toronto, April 12, 2010; “First update on the EADC-ADNI Harmonization of Protocols for Hippocampal Segmentation”, held in Honolulu, July 14, 2010.
Wyeth, a part of the Pfizer group, and Lilly have provided unrestricted grants in support of the work reported in this paper.
A follow-up project has been funded by the Alzheimer’s Association.
We also thank Cristina Scarpazza for logistic help in the initial phase of this work.
REFERENCES
- 1.Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, Delacourte A, Galasko D, Gauthier S, Jicha G, Meguro K, rsquo O’brien J, Pasquier F, Robert P, Rossor M, Salloway S, Stern Y, Visser PJ, Scheltens P. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6:734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 2.Gauthier S, Aisen PS, Ferris SH, Saumier D, Duong A, Haine D, Garceau D, Suhy J, Oh J, Lau W, Sampalis J. Effect of tramiprosate in patients with mild-to-moderate Alzheimer’s disease: exploratory analyses of the MRI subgroup of the Alphase study. J Nutr Health Aging. 2009;13:550–557. doi: 10.1007/s12603-009-0106-x. [DOI] [PubMed] [Google Scholar]
- 3.Rodionov R, Chupin M, Williams E, Hammers A, Kesavadas C, Lemieux L. Evaluation of atlas-based segmentation of hippocampi in healthy humans. Magn Reson Imaging. 2009;27:1104–1109. doi: 10.1016/j.mri.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy KM, Erickson KI, Rodrigue KM, Voss MW, Colcombe SJ, Kramer AF, Acker JD, Raz N. Age-related differences in regional brain volumes: a comparison of optimized voxel-based morphometry to manual volumetry. Neurobiol Aging. 2009;30:1657–1676. doi: 10.1016/j.neurobiolaging.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasan KM, Pedraza O. Improving the reliability of manual and automated methods for hippocampal and amygdala volume measurements. Neuroimage. 2009;48:497–498. doi: 10.1016/j.neuroimage.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosscher L, Scheltens P. Functional imaging in dementia. In: Qizilbash N, Schneider LS, Chui H, Tariot P, Brodaty H, Kaye J, Erkinjuntti T, editors. In Evidence-Based Dementia Practice. Blackwell: Oxford, UK; 2002. pp. 162–169. [Google Scholar]
- 7.Duchesne S, Pruessner J, Collins DL. Appearancebased segmentation of medial temporal lobe structures. Neuroimage. 2002;17:515–531. [PubMed] [Google Scholar]
- 8.Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC. A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage. 2008;40:1655–1671. doi: 10.1016/j.neuroimage.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR, Jr., Weiner MW, Thompson PM. Alzheimer’s Disease Neuroimaging, Initiative Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer’s disease mild cognitive impairment, and elderly controls. Neuroimage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brewer JB, Magda S, Airriess C, Smith ME. Fullyautomated quantification of regional brain volumes for improved detection of focal atrophy in Alzheimer disease. AJNR Am J Neuroradiol. 2009;30:578–580. doi: 10.3174/ajnr.A1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colliot O, Chetelat G, Chupin M, Desgranges B, Magnin B, Benali H, Dubois B, Garnero L, Eustache F, Lehericy S. Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology. 2008;248:194–201. doi: 10.1148/radiol.2481070876. [DOI] [PubMed] [Google Scholar]
- 12.Collins DL, Pruessner JC. Towards accurate, automatic segmentation of the hippocampus and amygdala from MRI by augmenting ANIMAL with a template library and label fusion. Neuroimage. 2010;52:1355–1366. doi: 10.1016/j.neuroimage.2010.04.193. [DOI] [PubMed] [Google Scholar]
- 13.Konrad C, Ukas T, Nebel C, Arolt V, Toga AW, Narr KL. Defining the human hippocampus in cerebral magnetic resonance images-an overview of current segmentation protocols. Neuroimage. 2009;47:1185–1195. doi: 10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geuze E, Vermetten E, Bremner JD. MR-based in vivo hippocampal volumetrics: 1. Review of methodologies currently employed. Mol Psychiatry. 2005;10:147–159. doi: 10.1038/sj.mp.4001580. [DOI] [PubMed] [Google Scholar]
- 15.Frisoni GB, Fox NC, Jack CR, Jr., Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: A mandatory step for wide clinical use. Alzheimer’s & Dementia. 2010 doi: 10.1016/j.jalz.2010.06.007. in press. [DOI] [PubMed] [Google Scholar]
- 17.Jack CR. MRI-based hippocampal volume measurements in epilepsy. Epilepsia. 1994;35:S21–S29. doi: 10.1111/j.1528-1157.1994.tb05986.x. [DOI] [PubMed] [Google Scholar]
- 18.Pruessner JC, Li LM, Serles W, Pruessner M, Collins DL, Kabani N, Lupien S, Evans AC. Volumetry of hippocampus and amygdala with high-resolution MRI and three-dimensional analysis software: minimizing the discrepancies between laboratories. Cereb Cortex. 2000;10:433–442. doi: 10.1093/cercor/10.4.433. [DOI] [PubMed] [Google Scholar]
- 19.Pantel J, O’Leary DS, Cretsinger K, Bockholt HJ, Keefe H, Magnotta VA, Andreasen NC. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus. 2000;10:752–758. doi: 10.1002/1098-1063(2000)10:6<752::AID-HIPO1012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 20.Malykhin NV, Bouchard TP, Ogilvie CJ, Coupland NJ, Seres P, Camicioli R. Three-dimensional volumetric analysis and reconstruction of amygdala and hippocampal head, body and tail. Psychiatry Res. 2007;155:155–165. doi: 10.1016/j.pscychresns.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Bartzokis G, Altshuler LL, Greider T, Curran J, Keen B, Dixon WJ. Reliability of medial temporal lobe volume measurements using reformatted 3D images. Psychiatry Res. 1998;82:11–24. doi: 10.1016/s0925-4927(98)00007-9. [DOI] [PubMed] [Google Scholar]
- 22.Bogerts B, Lieberman JA, Ashtari M, Bilder RM, Degreef G, Lerner G, Johns C, Masiar S. Hippocampus-amygdala volumes and psychopathology in chronic schizophrenia. Biol Psychiatry. 1993;33:236–246. doi: 10.1016/0006-3223(93)90289-p. [DOI] [PubMed] [Google Scholar]
- 23.deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25:1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Killiany RJ, Moss MB, Albert MS, Sandor T, Tieman J, Jolesz F. Temporal lobe regions on magnetic resonance imaging identify patients with early Alzheimer’s disease. Arch Neurol. 1993;50:949–954. doi: 10.1001/archneur.1993.00540090052010. [DOI] [PubMed] [Google Scholar]
- 25.Lehericy S, Baulac M, Chiras J, Pierot L, Martin N, Pillon B, Deweer B, Dubois B, Marsault C. Amygdalohip-pocampal MR volume measurements in the early stages of Alzheimer disease. AJNR Am J Neuroradiol. 1994;15:929–937. [PMC free article] [PubMed] [Google Scholar]
- 26.Convit A, De Leon MJ, Tarshish C, De Santi S, Tsui W, Rusinek H, George A. Specific hippocampal volume reductions in individuals at risk for Alzheimer’s disease. Neurobiol Aging. 1997;18:131–138. doi: 10.1016/s0197-4580(97)00001-8. [DOI] [PubMed] [Google Scholar]
- 27.Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, Sheline Y, Vannier MW, Csernansky JG. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- 28.Soininen HS, Partanen K, Pitkanen A, Vainio P, Hanninen T, Hallikainen M, Koivisto K, Riekkinen PJS. Volumetric MRI analysis of the amygdala and the hippocampus in subjects with age-associated memory impairment: correlation to visual and verbal memory. Neurology. 1994;44:1660–1668. doi: 10.1212/wnl.44.9.1660. [DOI] [PubMed] [Google Scholar]
- 29.Watson C, Andermann F, Gloor P, Jones-Gotman M, Peters T, Evans A, Olivier A, Melanson D, Leroux G. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42:1743–1750. doi: 10.1212/wnl.42.9.1743. [DOI] [PubMed] [Google Scholar]
- 30.Watson C, Jack CR, Jr., Cendes F. Volumetric magnetic resonance imaging. Clinical applications and contributions to the understanding of temporal lobe epilepsy. Arch Neurol. 1997;54:1521–1531. doi: 10.1001/archneur.1997.00550240071015. [DOI] [PubMed] [Google Scholar]
- 31.Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci U S A. 1996;93:3908–3913. doi: 10.1073/pnas.93.9.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bremner JD, Randall P, Scott TM, Bronen RA, Seibyl JP, Southwick SM, Delaney RC, McCarthy G, Charney DS, Innis RB. MRI-based measurement of hippocampal volume in patients with combat-related posttraumatic stress disorder. Am J Psychiatry. 1995;152:973–981. doi: 10.1176/ajp.152.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook MJ, Fish DR, Shorvon SD, Straughan K, Stevens JM. Hippocampal volumetric and morphometric studies in frontal and temporal lobe epilepsy. Brain. 1992;115:1001–1015. doi: 10.1093/brain/115.4.1001. [DOI] [PubMed] [Google Scholar]
- 34.Bigler ED, Blatter DD, Anderson CV, Johnson SC, Gale SD, Hopkins RO, Burnett B. Hippocampal volume in normal aging and traumatic brain injury. AJNR Am J Neuroradiol. 1997;18:11–23. [PMC free article] [PubMed] [Google Scholar]
- 35.Shenton ME, Kikinis R, Jolesz FA, Pollak SD, LeMay M, Wible CG, Hokama H, Martin J, Metcalf D, Coleman M. Abnormalities of the left temporal lobe and thought disorder in schizophrenia. A quantitative magnetic resonance imaging study. N Engl J Med. 1992;327:604–612. doi: 10.1056/NEJM199208273270905. [DOI] [PubMed] [Google Scholar]
- 36.MacQueen GM, Campbell S, McEwen BS, Macdonald K, Amano S, Joffe RT, Nahmias C, Young LT. Course of illness, hippocampal function, and hippocampal volume in major depression. Proc Natl Acad Sci U S A. 2003;100:1387–1392. doi: 10.1073/pnas.0337481100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narr KL, Thompson PM, Szeszko P, Robinson D, Jang S, Woods RP, Kim S, Hayashi KM, Asunction D, Toga AW, Bilder RM. Regional specificity of hippocampal volume reductions in first-episode schizophrenia. Neuroimage. 2004;21:1563–1575. doi: 10.1016/j.neuroimage.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 38.Mervaala E, Fohr J, Kononen M, Valkonen-Korhonen M, Vainio P, Partanen K, Partanen J, Tiihonen J, Viinamaki H, Karjalainen AK, Lehtonen J. Quantitative MRI of the hippocampus and amygdala in severe depression. Psychol Med. 2000;30:117–125. doi: 10.1017/s0033291799001567. [DOI] [PubMed] [Google Scholar]
- 39.Steffens DC, Byrum CE, McQuoid DR, Greenberg DL, Payne ME, Blitchington TF, MacFall JR, Krishnan KR. Hip-pocampal volume in geriatric depression. Biol Psychiatry. 2000;48:301–309. doi: 10.1016/s0006-3223(00)00829-5. [DOI] [PubMed] [Google Scholar]
- 40.O’Brien JT, Lloyd A, McKeith I, Gholkar A, Ferrier N. A longitudinal study of hippocampal volume, cortisol levels, and cognition in older depressed subjects. Am J Psychiatry. 2004;161:2081–2090. doi: 10.1176/appi.ajp.161.11.2081. [DOI] [PubMed] [Google Scholar]
- 41.Ashtari M, Greenwald BS, Kramer-Ginsberg E, Hu J, Wu H, Patel M, Aupperle P, Pollack S. Hippocampal/ amygdala volumes in geriatric depression. Psychol Med. 1999;29:629–638. doi: 10.1017/s0033291799008405. [DOI] [PubMed] [Google Scholar]
- 42.Zipursky RB, Marsh L, Lim KO, DeMent S, Shear PK, Sullivan EV, Murphy GM, Csernansky JG, Pfefferbaum A. Volumetric MRI assessment of temporal lobe structures in schizophrenia. Biol Psychiatry. 1994;35:501–516. doi: 10.1016/0006-3223(94)90097-3. [DOI] [PubMed] [Google Scholar]
- 43.Giedd JN, Vaituzis AC, Hamburger SD, Lange N, Rajapakse JC, Kaysen D, Vauss YC, Rapoport JL. Quantitative MRI of the temporal lobe, amygdala, and hippocampus in normal human development: ages 4–18 years. J Comp Neurol. 1996;366:223–230. doi: 10.1002/(SICI)1096-9861(19960304)366:2<223::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 44.Kates WR, Abrams MT, Kaufmann WE, Breiter SN, Reiss AL. Reliability and validity of MRI measurement of the amygdala and hippocampus in children with fragile X syndrome. Psychiatry Res. 1997;75:31–48. doi: 10.1016/s0925-4927(97)00019-x. [DOI] [PubMed] [Google Scholar]
- 45.Honeycutt NA, Smith PD, Aylward E, Li Q, Chan M, Barta PE, Pearlson GD. Mesial temporal lobe measurements on magnetic resonance imaging scans. Psychiatry Res. 1998;83:85–94. doi: 10.1016/s0925-4927(98)00035-3. [DOI] [PubMed] [Google Scholar]
- 46.von Gunten A, Fox NC, Cipolotti L, Ron MA. A volumetric study of hippocampus and amygdala in depressed patients with subjective memory problems. J Neuropsychiatry Clin Neurosci. 2000;12:493–498. doi: 10.1176/jnp.12.4.493. [DOI] [PubMed] [Google Scholar]
- 47.Vythilingam M, Vermetten E, Anderson GM, Luckenbaugh D, Anderson ER, Snow J, Staib LH, Charney DS, Bremner JD. Hippocampal volume, memory, and cortisol status in major depressive disorder: effects of treatment. Biol Psychiatry. 2004;56:101–112. doi: 10.1016/j.biopsych.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Hastings RS, Parsey RV, Oquendo MA, Arango V, Mann JJ. Volumetric analysis of the prefrontal cortex, amygdala, and hippocampus in major depression. Neuropsychopharmacology. 2004;29:952–959. doi: 10.1038/sj.npp.1300371. [DOI] [PubMed] [Google Scholar]
- 49.Niemann K, Hammers A, Coenen VA, Thron A, Klosterkotter J. Evidence of a smaller left hippocampus and left temporal horn in both patients with first episode schizophrenia and normal control subjects. Psychiatry Res. 2000;99:93–110. doi: 10.1016/s0925-4927(00)00059-7. [DOI] [PubMed] [Google Scholar]
- 50.Lobnig BM, Kromeke O, Optenhostert-Porst C, Wolf OT. Hippocampal volume and cognitive performance in long-standing Type 1 diabetic patients without macrovascular complications. Diabet Med. 2006;23:32–39. doi: 10.1111/j.1464-5491.2005.01716.x. [DOI] [PubMed] [Google Scholar]
- 51.Lloyd AJ, Ferrier IN, Barber R, Gholkar A, Young AH, O’Brien JT. Hippocampal volume change in depression: late- and early-onset illness compared. Br J Psychiatry. 2004;184:488–495. doi: 10.1192/bjp.184.6.488. [DOI] [PubMed] [Google Scholar]
- 52.Driessen M, Herrmann J, Stahl K, Zwaan M, Meier S, Hill A, Osterheider M, Petersen D. Magnetic resonance imaging volumes of the hippocampus and the amygdala in women with borderline personality disorder and early traumatization. Arch Gen Psychiatry. 2000;57:1115–1122. doi: 10.1001/archpsyc.57.12.1115. [DOI] [PubMed] [Google Scholar]
- 53.Barr WB, Ashtari M, Schaul N. Bilateral reductions in hippocampal volume in adults with epilepsy and a history of febrile seizures. J Neurol Neurosurg Psychiatry. 1997;63:461–467. doi: 10.1136/jnnp.63.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neumeister A, Wood S, Bonne O, Nugent AC, Lucken-baugh DA, Young T, Bain EE, Charney DS, Drevets WC. Reduced hippocampal volume in unmedicated, remitted patients with major depression versus control subjects. Biol Psychiatry. 2005;57:935–937. doi: 10.1016/j.biopsych.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Caetano SC, Hatch JP, Brambilla P, Sassi RB, Nicoletti M, Mallinger AG, Frank E, Kupfer DJ, Keshavan MS, Soares JC. Anatomical MRI study of hippocampus and amygdala in patients with current and remitted major depression. Psychiatry Res. 2004;132:141–147. doi: 10.1016/j.pscychresns.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 56.Frodl T, Meisenzahl EM, Zill P, Baghai T, Rujescu D, Leinsinger G, Bottlender R, Schule C, Zwanzger P, Engel RR, Rupprecht R, Bondy B, Reiser M, Moller HJ. Reduced hippocampal volumes associated with the long variant of the serotonin transporter polymorphism in major depression. Arch Gen Psychiatry. 2004;61:177–183. doi: 10.1001/archpsyc.61.2.177. [DOI] [PubMed] [Google Scholar]
- 57.Rusch BD, Abercrombie HC, Oakes TR, Schaefer SM, Davidson RJ. Hippocampal morphometry in depressed patients and control subjects: relations to anxiety symptoms. Biol Psychiatry. 2001;50:960–964. doi: 10.1016/s0006-3223(01)01248-3. [DOI] [PubMed] [Google Scholar]
- 58.Nakano T, Wenner M, Inagaki M, Kugaya A, Akechi T, Matsuoka Y, Sugahara Y, Imoto S, Murakami K, Uchitomi Y. Relationship between distressing cancer-related recollections and hippocampal volume in cancer survivors. Am J Psychiatry. 2002;159:2087–2093. doi: 10.1176/appi.ajp.159.12.2087. [DOI] [PubMed] [Google Scholar]
- 59.MacMillan S, Szeszko PR, Moore GJ, Madden R, Lorch E, Ivey J, Banerjee SP, Rosenberg DR. Increased amygdala: hippocampal volume ratios associated with severity of anxiety in pediatric major depression. J Child Adolesc Psy-chopharmacol. 2003;13:65–73. doi: 10.1089/104454603321666207. [DOI] [PubMed] [Google Scholar]
- 60.Yucel K, Taylor VH, McKinnon MC, Macdonald K, Alda M, Young LT, MacQueen GM. Bilateral hippocampal volume increase in patients with bipolar disorder and shortterm lithium treatment. Neuropsychopharmacology. 2008;33:361–367. doi: 10.1038/sj.npp.1301405. [DOI] [PubMed] [Google Scholar]
- 61.Brambilla P, Harenski K, Nicoletti M, Sassi RB, Mallinger AG, Frank E, Kupfer DJ, Keshavan MS, Soares JC. MRI investigation of temporal lobe structures in bipolar patients. J Psychiatr Res. 2003;37:287–295. doi: 10.1016/s0022-3956(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 62.Saylam C, Ucerler H, Kitis O, Ozand E, Gonul AS. Reduced hippocampal volume in drug-free depressed patients. Surg Radiol Anat. 2006;28:82–87. doi: 10.1007/s00276-005-0050-3. [DOI] [PubMed] [Google Scholar]
- 63.MacMaster FP, Kusumakar V. Hippocampal volume in early onset depression. BMC Me. 2004;2:2. doi: 10.1186/1741-7015-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott TM, Tucker KL, Bhadelia A, Benjamin B, Patz S, Bhadelia R, Liebson E, Price LL, Griffith J, Rosenberg I, Fol-stein MF. Homocysteine and B vitamins relate to brain volume and whitematter changes in geriatric patients with psychiatric disorders. Am J Geriatr Psychiatry. 2004;12:631–638. doi: 10.1176/appi.ajgp.12.6.631. [DOI] [PubMed] [Google Scholar]
- 65.Chen BK, Sassi R, Axelson D, Hatch JP, Sanches M, Nicoletti M, Brambilla P, Keshavan MS, Ryan ND, Birmaher B, Soares JC. Cross-sectional study ofabnormal amygdala development in adolescents and young adults with bipolar disorder. Biol Psychiatry. 2004;56:399–405. doi: 10.1016/j.biopsych.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 66.Vermetten E, Schmahl C, Lindner S, Loewenstein RJ, Bremner JD. Hippocampal and amygdalar volumes in dissociative identity disorder. Am J Psychiatry. 2006;163:630–636. doi: 10.1176/appi.ajp.163.4.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arango C, Breier A, McMahon R, Carpenter WT, Buchanan RW. The relationship of clozapine and haloperidol treatment response to prefrontal, hippocampal, and caudate brain volumes. Am J Psychiatry. 2003;160:1421–1427. doi: 10.1176/appi.ajp.160.8.1421. [DOI] [PubMed] [Google Scholar]
- 68.Chang K, Karchemskiy A, Barnea-Goraly N, Garrett A, Sime-onova DI, Reiss A. Reduced amygdalar gray matter volume in familial pediatric bipolar disorder. J Am Acad Child Adolesc Psychiatry. 2005;44:565–573. doi: 10.1097/01.chi.0000159948.75136.0d. [DOI] [PubMed] [Google Scholar]
- 69.Rosso IM, Cintron CM, Steingard RJ, Renshaw PF, Young AD, Yurgelun-Todd DA. Amygdala and hippocampus volumes in pediatric major depression. Biol Psychiatry. 2005;57:21–26. doi: 10.1016/j.biopsych.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 70.Xia J, Chen J, Zhou Y, Zhang J, Yang B, Xia L, Wang C. Volumetric MRI analysis of the amygdala and hippocampus in subjects with major depression. J Huazhong Univ Sci Technolog Med Sci. 2004;24:500–502. doi: 10.1007/BF02831120. 506. [DOI] [PubMed] [Google Scholar]
- 71.Frazier JA, Chiu S, Breeze JL, Makris N, Lange N, Kennedy DN, Herbert MR, Bent EK, Koneru VK, Dieterich ME, Hodge SM, Rauch SL, Grant PE, Cohen BM, Seidman LJ, Caviness VS, Biederman J. Structural brain magnetic resonance imaging of limbic and thalamic volumes in pediatric bipolar disorder. Am J Psychiatry. 2005;162:1256–1265. doi: 10.1176/appi.ajp.162.7.1256. [DOI] [PubMed] [Google Scholar]
- 72.Bossini L, Tavanti M, Calossi S, Lombardelli A, Polizzotto NR, Galli R, Vatti G, Pieraccini F, Castrogiovanni P. Magnetic resonance imaging volumes of the hippocampus in drug-naive patients with post-traumatic stress disorder without comorbidity conditions. J Psychiatr Res. 2008;42:752–762. doi: 10.1016/j.jpsychires.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 73.Starkman MN, Giordani B, Gebarski SS, Schteingart DE. Improvement in mood and ideation associated with increase in right caudate volume. J Affect Disord. 2007;101:139–147. doi: 10.1016/j.jad.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 74.Jack CR, Jr., Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DL, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scheltens P, Leys D, Barkhof F, Huglo D, Weinstein HC, Vermersch P, Kuiper M, Steinling M, Wolters EC, Valk J. Atrophy of medial temporal lobes on MRI in “probable” Alzheimer’s disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992;55:967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frisoni GB, Ganzola R, Canu E, Rub U, Pizzini FB, Alessandrini F, Zoccatelli G, Beltramello A, Caltagirone C, Thompson PM. Mapping local hippocampal changes in Alzheimer’s disease and normal ageing with MRI at 3 Tesla. Brain. 2008;131:3266–3276. doi: 10.1093/brain/awn280. [DOI] [PubMed] [Google Scholar]
- 77.Pallavaram S, Yu H, Spooner J, D’Haese PF, Koyama T, Bodenheimer B, Konrad PE, Dawant BM. Automated selection of anterior and posterior commissures based on a deformable atlas and its evaluation based on manual selections by neurosurgeons. SPIE. 2007 [Google Scholar]
- 78.Carone DA, Benedict RH, Dwyer MG, Cookfair DL, Srinivasaraghavan B, Tjoa CW, Zivadinov R. Semiautomatic brain region extraction (SABRE) reveals superior cortical and deep gray matter atrophy in MS. Neuroimage. 2006;29:505–514. doi: 10.1016/j.neuroimage.2005.07.053. [DOI] [PubMed] [Google Scholar]
- 79.Anbazhagan P, Carass A, Bazin PL, Prince JL. Automatic estimation of midsagittal plane and AC-PC alignment based on non-rigid registration. IEEE. 2006 [Google Scholar]
- 80.Han Y, Park HW. Automatic Brain MR Image Registration Based on Talairach Reference System. ICIP. 2003;1:I1097. doi: 10.1002/jmri.20168. [DOI] [PubMed] [Google Scholar]
- 81.Chupin M, Mukuna-Bantumbakulu AR, Hasboun D, Bardinet E, Baillet S, Kinkingnehun S, Lemieux L, Dubois B, Garnero L. Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: Method and validation on controls and patients with Alzheimer’s disease. Neuroimage. 2007;34:996–1019. doi: 10.1016/j.neuroimage.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 82.Dubois B, Feldman HH, Jacova C, Cummings JL, Dekosky ST, Barberger-Gateau P, Delacourte A, Frisoni G, Fox NC, Galasko D, Gauthier S, Hampel H, Jicha GA, Meguro K, O’Brien J, Pasquier F, Robert P, Rossor M, Salloway S, Sarazin M, de Souza LC, Stern Y, Visser PJ, Scheltens P. Revising the definition of Alzheimer’s disease: a new lexicon. Lancet Neurol. 2010 doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]