Abstract
Recycling endosomes are key platforms for endocytic recycling that return internalized molecules back to the plasma membrane. To determine how recycling endosomes perform their functions, searching for proteins and lipids that specifically localized at recycling endosomes has often been performed by colocalization analyses between candidate molecules and conventional recycling endosome markers. However, it remains unclear whether all the conventional markers have identical localizations. Here we report finding that three well-known recycling endosome markers, i.e., Arf6, Rab11 and transferrin receptor (TfR), have different intracellular localizations in PC12 cells. The results of immunofluorescence analyses showed that the signals of endogenous Arf6, Rab11 and TfR in nerve growth factor-stimulated PC12 cells generally differed, although there was some overlapping. Our findings provide new information about recycling endosome markers, and they highlight the heterogeneity of recycling endosomes.
Keywords: Arf6, Rab11, transferrin receptor, Rab35, recycling endosome
Endocytic recycling is an intracellular process that returns internalized molecules back to the plasma membrane,1,2 and it plays crucial roles not only in the reuse of receptor molecules but in remodeling of the protein and lipid composition of the plasma membrane. Thus, endocytic recycling is required for a variety of cellular events, including cell migration, cytokinesis and neurite outgrowth.3-5 One of the platforms that plays a central role in endocytic recycling is the recycling endosome.6-8 Recycling endosomes are usually localized in the perinuclear area, and they function as endosomal hubs that connect endocytic and exocytic membrane trafficking by receiving internalized molecules from early endosomes and returning them to the plasma membrane. However, the molecular mechanisms by which they perform their functions are not fully understood.
To identify the molecular mechanisms by which recycling endosomes perform their functions, searching for proteins and lipids that specifically localized at recycling endosomes has often been performed by colocalization analyses between candidate molecules and conventional recycling endosome markers, e.g., the Arf family small GTPase Arf6, the Rab family small GTPase Rab11 and the transferrin receptor (TfR).2,9,10 Thus, candidate molecules that colocalize with Arf6, Rab11 or TfR have been simply referred to as “recycling endosomal” molecules in the literature. However, it is unclear whether the intracellular localizations of Arf6, Rab11 or TfR are the same or not.
We previously reported finding that Rab35, which regulates endocytic recycling and promotes neurite outgrowth,11-14 was highly colocalized with Arf6 in the pericentrosomal area of nerve growth factor (NGF)-stimulated PC12 cells,15 indicating that Rab35 also localizes at recycling endosomes. However, the clear difference between the distribution of Rab35 signals and Rab11 signals in NGF-stimulated PC12 cells15 led us to hypothesize that the intracellular localizations of conventional recycling endosomal proteins are not necessarily the same. In the present study, we investigated whether the three well-known recycling endosome marker proteins Arf6, Rab11 and TfR have different intracellular localizations by visualizing endogenous proteins with specific antibodies.
First, we performed colocalization analyses of Rab35 and the three recycling endosome marker proteins in NGF-stimulated PC12 cells. Consistent with the findings in our previous report, the results showed that Rab35 was highly colocalized with Arf6 in the pericentrosomal area (Fig. 1A and B), but that it was only partially colocalized with Rab11 and hardly colocalized with TfR (Fig. 1C and D). We then performed colocalization analyses of the three recycling endosome marker proteins in NGF-stimulated PC12 cells. The results showed generally distinct intracellular localizations of Arf6, Rab11 and TfR, although there was some overlapping, and Arf6, Rab11 and TfR were generally concentrically localized in the perinuclear area, with Arf6 localized closest to the centrosome and TfR farthest from the centrosome (Fig. 2; Fig. S1).
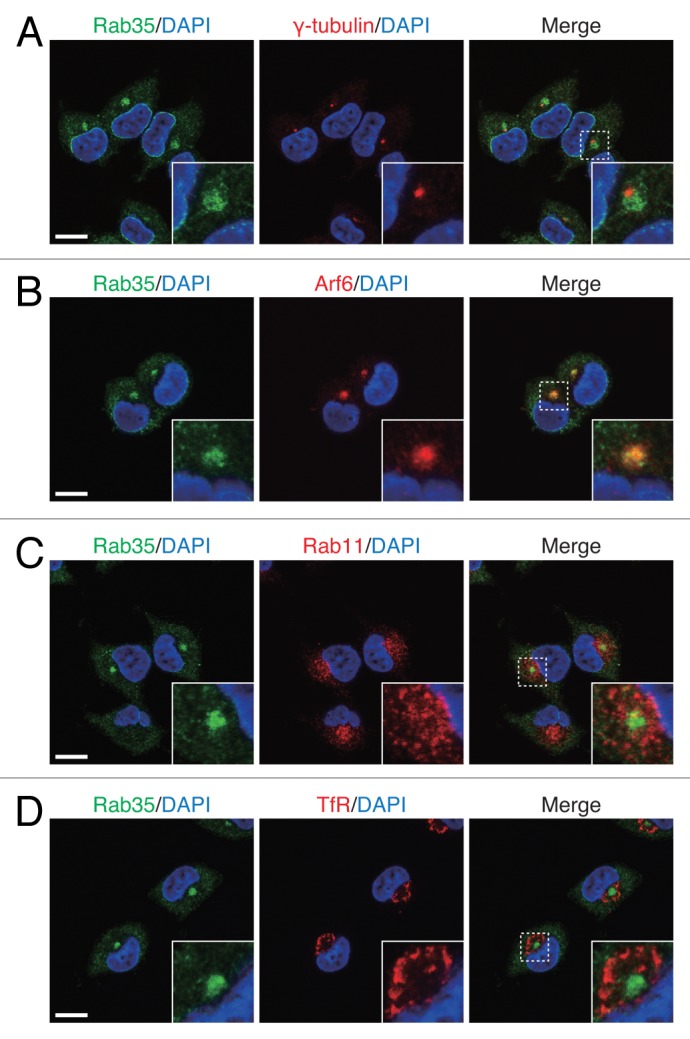
Figure 1. Rab35 highly colocalized with Arf6, but only partially colocalized with Rab11 and hardly colocalized with TfR. After stimulating PC12 cells with NGF for 6 h, the cells were fixed and then stained with anti-Rab35 antibody and DAPI together with anti-γ-tubulin antibody (A), anti-Arf6 antibody (B), anti-Rab11 antibody (C) or anti-TfR antibody (D). The insets show magnified views of the boxed areas. Scale bars, 10 μm.
Figure 2. Arf6, Rab11 and TfR have distinct intracellular localizations. After stimulating PC12 cells with NGF for 6 h, the cells were fixed and stained with antibodies against the recycling endosome marker proteins indicated and with DAPI. The panels on the right are magnified views of the boxed areas in the panels on the left. Scale bars, 10 μm.
Since TfR continuously recycles between the plasma membrane and recycling endosomes through early endosomes, and newly synthesized TfR passes through the Golgi en route to the plasma membrane, the distinct intracellular localization of TfR and Rab11 (or Arf6) can be explained by the majority of TfR proteins being localized on early endosomes and/or the Golgi. However, because only small amounts of TfR were found to be colocalized with EEA-1 (an early endosome marker) and GM130 (a Golgi marker), the majority of TfR-positive compartments should be regarded as recycling endosomes (Fig. S2). We therefore concluded that the distinct intracellular localizations of Arf6, Rab11 and TfR reflect the heterogeneity of recycling endosomes.
Interestingly, the immunofluorescence signals of Arf6, but not of Rab11 or TfR, were changed in response to NGF stimulation. In the absence of NGF stimulation, only faint Arf6 signals were observed in the perinuclear area, whereas the Arf6 signals increased in response to NGF stimulation (Fig. 3A). By contrast, the Rab11 signals and TfR signals did not undergo any marked changes during NGF stimulation. Because the level of Arf6 expression was unchanged by NGF stimulation (Fig. 3B), the formation of Arf6-positive recycling endosomes is likely to be promoted by NGF signals. We speculate that Arf6-positive recycling endosomes have a specific role in endocytic recycling that facilitates neurite outgrowth of PC12 cells.
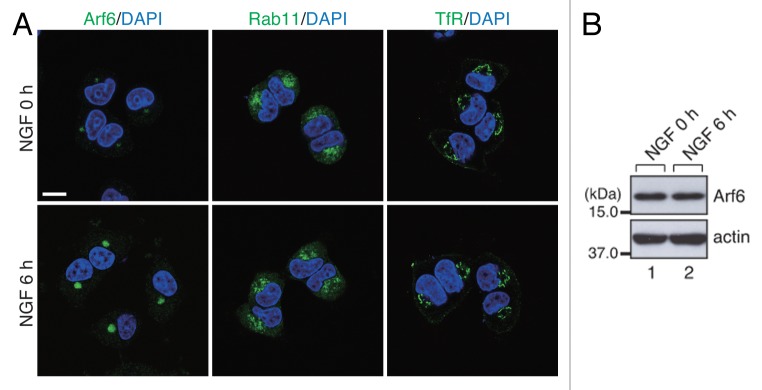
Figure 3. Accumulation of Arf6 signals in the pericentrosomal area in response to NGF stimulation. (A) After NGF stimulation for 0 h (no NGF-treatment) and 6 h, PC12 cells were fixed and stained with anti-Arf6 antibody, anti-Rab11 antibody and anti-TfR antibody together with DAPI. Scale bar, 10 μm. (B) Unaltered expression of Arf6 in PC12 cells during NGF stimulation. After NGF stimulation for 0 h and 6 h, cell lysates of PC12 cells were immunoblotted with anti-Arf6 antibody and anti-actin antibody.
Our findings provide new information about recycling endosome markers, i.e., the localizations of Arf6, Rab11 and TfR only partially overlap in PC12 cells. Thus, a combination of recycling endosome markers should be used to determine whether a candidate molecule localizes to recycling endosomes. Our findings also highlight the heterogeneity of recycling endosomes in PC12 cells (Fig. 4). Recycling endosomes have been reported to regulate not only endocytic recycling but also exocytic and retrograde membrane trafficking,16,17 indicating that recycling endosomes have multiple sorting functions. Therefore, the different intracellular localizations of the recycling endosome marker proteins may reflect the presence of functionally distinct populations of recycling endosomes. In the future, endogenous recycling endosomal proteins should be visualized in cell types other then PC12 cells to determine whether recycling endosome heterogeneity exists in many cell types. In any event, extensive research will be necessary to understand the physiological significance of the heterogeneity of recycling endosomes.
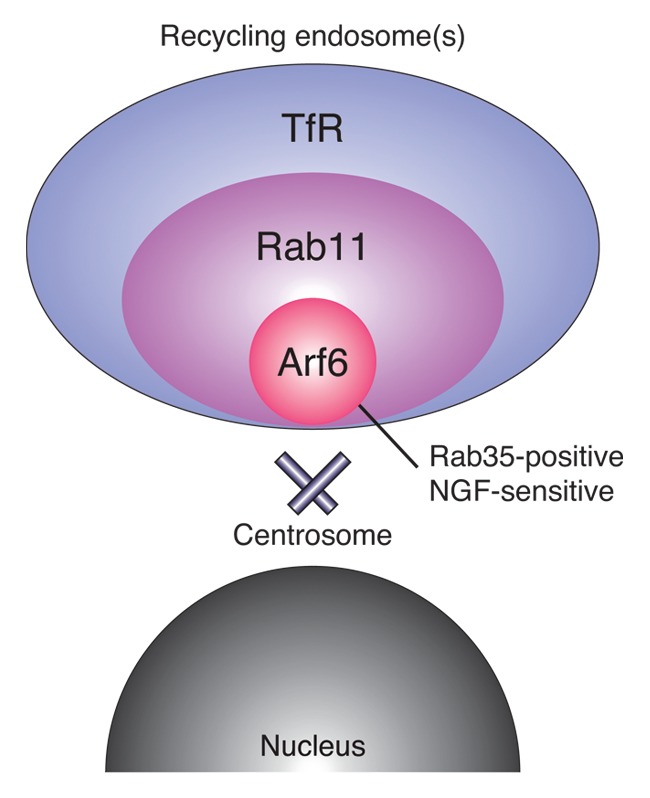
Figure 4. A model of the intracellular localizations of recycling endosomal proteins in NGF-stimulated PC12 cells. Arf6, Rab11 and TfR are localized in the perinuclear area, with Arf6 localized closest to the centrosome and TfR farthest from the centrosome, suggesting heterogeneity of the recycling endosomes.
Materials and Methods
Antibodies
Anti-Rab35 antibody was prepared as described previously.15,18 Anti-γ-tubulin mouse monoclonal antibody (Sigma-Aldrich Corp), anti-Arf6 mouse monoclonal antibody (Santa Cruz Biotechnology, Inc), anti-Rab11 rabbit polyclonal antibody, anti-TfR mouse monoclonal antibody (Invitrogen Corp), anti-EEA-1 mouse monoclonal antibody and anti-GM130 mouse monoclonal antibody (BD Biosciences) were obtained commercially. The immunostaining in Figs. S1 and S2 was performed with a fluorophore-conjugated anti-TfR antibody that was generated by using the Zenon Antibody Labeling Kit (Invitrogen Corp). Using this antibody slightly enhanced the TfR signals at the plasma membrane (Figs. S1 and S2).
Cell culture, immunoblotting and immunofluorescence analyses
All of the procedures used to perform cell culture, immunoblotting and the immunofluorescence analyses have been described elsewhere.15,18
Supplementary Material
Acknowledgments
We thank Megumi Aizawa for technical assistance and members of the Fukuda Laboratory for valuable discussions. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports and Technology (MEXT) of Japan (to M.F.) and by a grant from the Daiichi-Sankyo Foundation of Life Science (to M.F.). H.K. was supported by the Japan Society for the Promotion of Science (JSPS) and by the International Advanced Research and Education Organization of Tohoku University (IAREO).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/25036
References
- 1.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–32. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 2.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones MC, Caswell PT, Norman JC. Endocytic recycling pathways: emerging regulators of cell migration. Curr Opin Cell Biol. 2006;18:549–57. doi: 10.1016/j.ceb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Echard A. Membrane traffic and polarization of lipid domains during cytokinesis. Biochem Soc Trans. 2008;36:395–9. doi: 10.1042/BST0360395. [DOI] [PubMed] [Google Scholar]
- 5.Sann S, Wang Z, Brown H, Jin Y. Roles of endosomal trafficking in neurite outgrowth and guidance. Trends Cell Biol. 2009;19:317–24. doi: 10.1016/j.tcb.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 6.van IJzendoorn SCD. Recycling endosomes. J Cell Sci. 2006;119:1679–81. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
- 7.Golachowska MR, Hoekstra D, van IJzendoorn SCD. Recycling endosomes in apical plasma membrane domain formation and epithelial cell polarity. Trends Cell Biol. 2010;20:618–26. doi: 10.1016/j.tcb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 8.Hsu VW, Prekeris R. Transport at the recycling endosome. Curr Opin Cell Biol. 2010;22:528–34. doi: 10.1016/j.ceb.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–58. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 10.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–25. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 11.Kouranti I, Sachse M, Arouche N, Goud B, Echard A. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr Biol. 2006;16:1719–25. doi: 10.1016/j.cub.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 12.Sato M, Sato K, Liou W, Pant S, Harada A, Grant BD. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2008;27:1183–96. doi: 10.1038/emboj.2008.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanno E, Ishibashi K, Kobayashi H, Matsui T, Ohbayashi N, Fukuda M. Comprehensive screening for novel Rab-binding proteins by GST pull-down assay using 60 different mammalian Rabs. Traffic. 2010;11:491–507. doi: 10.1111/j.1600-0854.2010.01038.x. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M, Kobayashi H, Ishibashi K, Ohbayashi N. Genome-wide investigation of the Rab binding activity of RUN domains: development of a novel tool that specifically traps GTP-Rab35. Cell Struct Funct. 2011;36:155–70. doi: 10.1247/csf.11001. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi H, Fukuda M. Rab35 regulates Arf6 activity through centaurin-β2 (ACAP2) during neurite outgrowth. J Cell Sci. 2012;125:2235–43. doi: 10.1242/jcs.098657. [DOI] [PubMed] [Google Scholar]
- 16.Ang AL, Taguchi T, Francis S, Fölsch H, Murrells LJ, Pypaert M, et al. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J Cell Biol. 2004;167:531–43. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchida Y, Hasegawa J, Chinnapen D, Inoue T, Okazaki S, Kato R, et al. Intracellular phosphatidylserine is essential for retrograde membrane traffic through endosomes. Proc Natl Acad Sci USA. 2011;108:15846–51. doi: 10.1073/pnas.1109101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi H, Fukuda M. Rab35 establishes the EHD1-association site by coordinating two distinct effectors during neurite outgrowth. J Cell Sci. 2013 doi: 10.1242/jcs.117846. In press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



