Abstract
Adaptations by hosts in response to parasitism are generally believed to reduce the susceptibility of the adapted individual. However, recent work on Escherichia coli showed that bacteria can fight deadly phage attacks by committing altruistic suicide upon infection, in order to prevent parasite transmission to nearby relatives. Here, we compare the efficiency of suicidal host defense with individual-based resistance. We show that in unstructured environments suicidal host defense is futile since suicide cannot preferentially protect relatives, whereas individual-based resistance is highly efficient in defying phages. In contrast, we found that in structured environments suicidal host defense and individual-based resistance were both efficient in withstanding phages, with the latter type performing slightly better. We propose that the putative lower efficiency of suicidal host defense might be compensated by the fact that suicidal systems usually do not bear pleiotropic costs of resistance, as it is usually the case for individual-based resistance mechanisms.
Keywords: altruistic suicide, resistance, structured population, parasite transmission, host-parasite interactions, E. coli, T4 phage
Organisms are consistently exposed to parasites causing morbidity and mortality. A key interest in parasitology is therefore to understand adaptive responses by hosts to evade or reduce detrimental parasitic effects.1 While research has traditionally focused on adaptations reducing the susceptibility of individuals,2 a valuable alternative seems that natural selection favors the expression of traits in infected individuals that protect others from becoming infected.3,4 For instance, it has been observed that infected individuals in social insect and aphid colonies leave the group to die in isolation.5-8 Such behavioral patterns suggest that infected individuals do not care for themselves, but remove themselves altruistically from the group (i.e., commit suicide) in order to prevent the spreading of disease among relatives. Although compelling, the idea of altruistic suicide as adaptive host-defense strategy has repeatedly been challenged,9,10 and has lead to a more general controversy about whether suicide can be adaptive.11-13
This fundamental controversy has now conclusively been solved through a number of studies, which examined abortive infection (Abi) systems in the bacterium Escherichia coli. Many bacterial species possess Abi systems, which respond to lethal phage attacks by causing infected cells to die together with the infecting phage.14-17 Abi mechanisms usually involve joint actions by a prophage (a phage integrated into the bacterial genome) or a plasmid that encodes the genes for the abortive infection, and the bacterium, which transcribes and synthesizes the abortive machinery. Although Abi systems have long been described, the key questions of whether they represent altruistic acts and have been selected for that purpose have long remained unaddressed. Recently, Fukuyo et al.,18 resolved the first question by showing that an engineered Abi system was exclusively selected for in structured environments where abortion preferentially protected clone mates (i.e., relatives) from becoming infected. This finding is compatible with social evolution theory, predicting that altruism can only evolve when its benefit accrues to relatives.19,20 The second question was subsequently addressed by Berngruber et al.,21 and Refardt et al.,22 who recovered similar evolutionary dynamics for naturally occurring Abi systems, which supports the idea that these systems have been selected for the purpose of committing suicide. Moreover, Refardt et al.,22 showed that altruistic host suicide could spread with ease across a wide range of ecological conditions. This was because suicide in their system incurred negligible costs since infected individuals were moribund, such that the commitment of suicide simply forestalled the inevitable phage-induced death.
While these novel findings have demonstrated that altruistic host suicide is important in combating parasites, they also raise the question of how effective altruistic host-defense is compared with classical individual-based resistance mechanisms. Here, we address this question by comparing the fitness benefits of the two strategies both in structured and unstructured environments.
Results and Discussion
In all experiments, we exposed bacterial strains to the phage T4rII, which targets lipopolysaccharides embedded in the bacterial outer membrane.23 We used three different strains of E. coli str. K-12 substr. MG1655. As our altruistic suicidal strain, we used E. coli carrying the prophage λ (henceforth E. coli λ), which encodes the Rex abortive infection system14 that is triggered by several phages including T4rII.24 As the phage-sensitive strain, we used E. coli carrying the Abi-free control prophage HK97 (henceforth E. coli HK97). Finally, we used an E. coli HK97 strain that has evolved resistance against T4rII (henceforth E. coli T4r). While we compared the performance of phage-sensitive E. coli HK97 vs. suicidal E. coli λ in our previous study,22 we here compare the competitive abilities of phage-sensitive E. coli HK97 vs. phage-resistant E. coli T4r, and relate it to our previous findings.
We first examined the performance of E. coli HK97 and E. coli T4r in an unstructured environment. We did this by growing the two strains together in shaken liquid medium at various frequencies (E. coli T4r varied from 0.1% to 99%) in the presence of T4rII (MOI = 0.001) and followed population growth and the change in relative strain frequency over time. We found that T4rII eradicated monocultures of E. coli HK97, whereas E. coli T4r could thrive even when initially rare (Fig. 1). Moreover, E. coli T4r significantly outcompeted E. coli HK97 under all conditions (Fig. 2, one-sample t-tests for fitness > 0: t5 > 6.9, p < 0.001). These results demonstrate that individual-based resistance mechanisms can easily spread in a completely unstructured environment, a finding that strongly contrasts with the suicidal host-defense strategy, which either could not prevent population collapse (with E. coli λ frequencies ≤ 90%) or did not result in fitness benefits (Fig. 2). This comparison reveals that individual-based resistance is the only way of how bacteria can defy bacteriophages in unstructured environments.
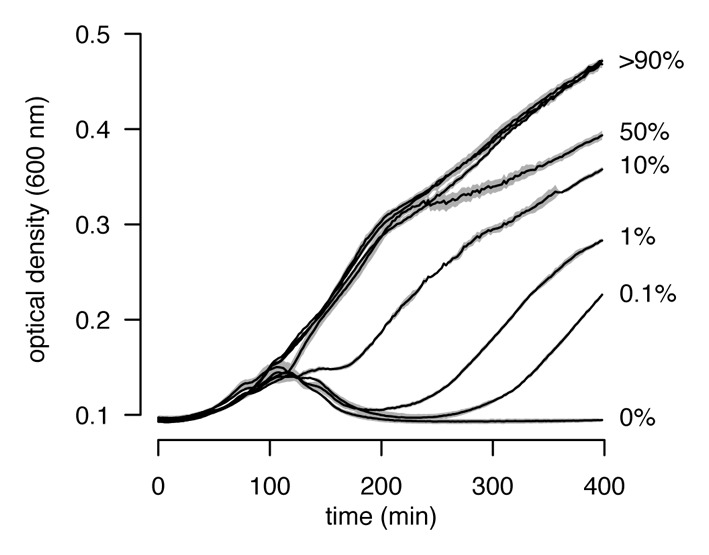
Figure 1. Population growth of mixed cultures containing E. coli T4r (resistant against the obligately lytic phage T4rII) and E. coli HK97 (T4rII sensitive) in shaken liquid medium. While phage T4rII eradicated monocultures of E. coli HK97, cultures containing E. coli T4r could always grow even when E. coli T4r was initially rare (percentage of E. coli T4r is indicated on the right).
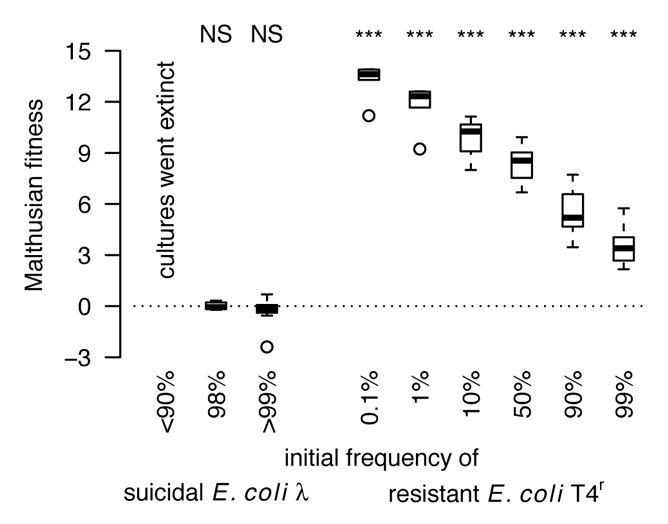
Figure 2. Comparing the performance of suicidal E. coli λ and resistant E. coli T4r in competition against E. coli HK97 in the presence of phage T4rII in an unstructured environment (shaken liquid medium). While E. coli T4r significantly outcompeted E. coli HK97 under all conditions (Malthusian fitness > 0, p < 0.001), E. coli λ either experienced no relative fitness benefit or the entire culture went extinct (these latter results were initially published in Refardt et al.,2210.1098/rspb.2012.3035). These findings demonstrate that individual-based resistance is the only way of how bacteria can defy bacteriophages in unstructured environments.
We found a different pattern when competitions were performed in a structured environment (Fig. 3). Specifically, when we added 1.5% agar to the medium to limit bacterial dispersal,25 E. coli HK97 was significantly outcompeted by both suicidal E. coli λ (one-sample t-test for fitness > 0: t29 = 11.17, p < 0.0001) and E. coli T4r (t11 = 9.52, p < 0.0001). This comparison shows that both altruistic host-defense and individual-based resistance mechanisms are efficient means to defy bacteriophages in a structured environment. However, E. coli T4r seemed to be slightly but significantly more efficient than E. coli λ (Fig. 3; two sample t-test: t40 = 4.20, p = 0.0001). A plausible explanation is that suicide always takes its toll, especially at high MOI, which reduces the efficiency of suicidal host defense compared with an individual-based resistance mechanism. Additionally, we also found that the relative fitness of E. coli T4r was greatly reduced in the structured compared with the unstructured environment, an observation that is compatible with the view that medium viscosity also reduces phage dispersal, such that some fraction of E. coli HK97 colonies remain unharmed.
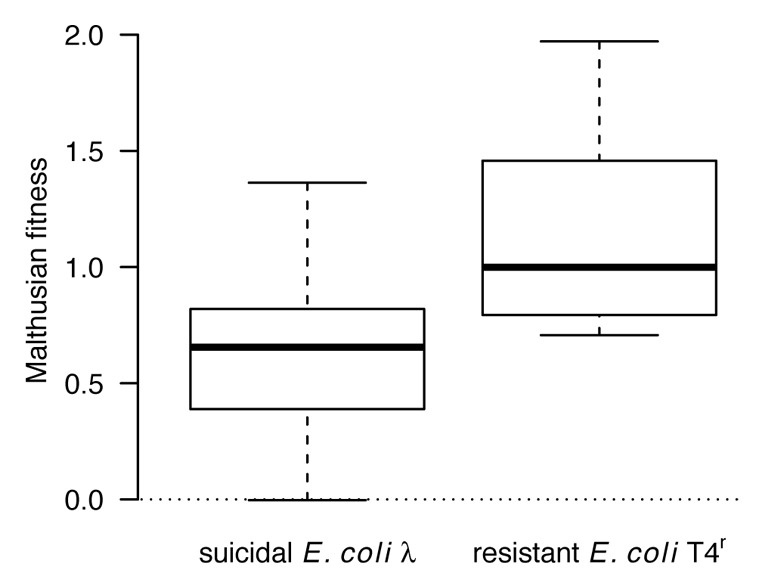
Figure 3. Comparing the performance of suicidal E. coli λ and resistant E. coli T4r in competition against E. coli HK97 in the presence of phage T4rII in a structured environment (1.5% agar plate). Both strains significantly outcompeted E. coli HK97, indicating that suicidal host defense as well as individual-based resistance were efficient in withstanding phages. A direct comparison between the two strategies reveals that individual-based resistance is slightly but significantly better under these conditions. But note that although the experiments were performed following the exact same protocol, they were performed at different dates, which might explain some of the differences.
To be able to put our findings in an ecological context, we need to become clear about the level of structuring that occurs in natural bacterial populations. While an unifying answer is not possible owing to the fact that bacteria populate an enormous range of different habitats, it seems justified to conclude that some degree of population structure always exists.26 Consequently, we can deduce that altruistic host defense likely represents an efficient strategy to withstand bacteriophages in natural habitats.
While our comparison revealed that both host-defense strategies can be efficient in laboratory settings, it remains unclear, which of the two strategies is more prevalent in nature. Individual-based resistance might be more common because it is easier to evolve, since a single point mutation can lead to resistance.27 In contrast, suicidal defense systems are often based on complex machineries17 that eventually have first evolved for other purposes, and have only later been coopted into suicidal systems.22 Although more complex, we propose that suicidal host-defense systems might be cheaper to maintain in the long run because they are often decoupled from other functions, and might therefore entail little pleiotropic cost. Individual-based resistance, on the other hand, is typically achieved by altering a molecule on the bacterial cell surface that is recognized by a phage and to which the phage adsorbs.17 Such an adaptation may impair the original function of the molecule and consequently reduce bacterial fitness (as observed in T4 phages28), even more so if further modifications are necessary to resist different phages, or to respond to phage counter-resistance.29-31 These potential cost differences might also affect the co-evolutionary dynamics between phages and bacteria. Moreover, it would be interesting to see whether both resistance mechanisms can occur in the same individual, or whether they are mutually exclusive, with one resistance mechanisms eventually replacing the other one over evolutionary time scales. While speculative at this stage, these considerations will hopefully stimulate future work.
Finally, another unresolved issue about Abi systems is that they often involve the action of prophages. A plausible explanation for this is that prophages already possess mechanisms that can kill bacteria during induction (i.e., when a prophage enters the lytic phase), which can eventually be coopted into suicide machineries. More sinister is the possibility that bacteria are not always free to choose their doom.11 For instance, is has been observed that competition between two prophages in the same bacterial cell leads some prophages to induce premature cell death, possibly to prevent the other prophage from replicating.32,33
Methods
To obtain E. coli T4r, we diluted overnight cultures of E. coli HK97 100-fold, and incubated 100 μl together with 104 pfu of phage T4rII overnight in a plate reader. Cultures whose growth pattern indicated the evolution of resistance were selected and serially passaged (three times) in 3 ml medium. Before every passage, we diluted cultures 1,000-fold and added 108 pfu of phage T4. This was done to select for full resistance, and to reduce possible resistance costs through compensatory mutations. We then streaked out cultures, and picked a single colony to establish a stock culture, stored at −80°C. We verified resistance by cross-streaking against lysates of both phages T4 and T4rII. We chose this procedure because we were interested in obtaining a strain that exhibits a different defense mechanism against phage T4rII, but otherwise grows comparably to E. coli λ and E. coli HK97 (which do not differ in their growth rate in the absence of phages22). Indeed, when grown in mixed cultures, we found that neither E. coli T4r nor E. coli HK97 experienced a selective advantage in the absence of phages (t4 = 0.48, p = 0.66, difference in Malthusian fitness = 0.02). The downside of this procedure is, however, that we cannot infer the initial cost of phage resistance.
We performed all experiments following the same protocol as described previously.22 In short, experiments shown in Figures 1 and 2 were performed in 96-well microtiter plates in 100 μl of medium. Initial concentration of both competing strains together was 5 × 107 cfu/ml, initial concentration of phage T4rII was 105 pfu/ml. Experiments for Figure 3 were performed in 24-well plates with wells filled with 1.5 ml LB 1.5% agar each. Between 10 and 106 cfu of E. coli λ and 104 cfu of E. coli T4r were added together with 104 pfu of phage T4rII.
To be able to distinguish bacterial strains in mixed competition, we marked all strains with plasmids pGFP and pDsRed carrying genes for fluorescent proteins under an inducible promoter as described previously.22 Competition assays in unstructured and structured environments followed protocols previously described,22 except that experiments in structured environments were performed on agar instead of agarose.
Acknowledgments
We thank John W. Little, Ing-Nang Wang, Benjamin Kerr, Bärbel Stecher, Martin Ackermann and Winfried Boos for providing bacteria and phage strains, the flow cytometry laboratory at the ETH Zürich, Anette Schütz and Olin Silander for help with flow cytometry, and the referee for comments. This work was supported by the Competence Center Environment and Sustainability of the ETH (to D.R.), the Swiss National Science Foundation and a Marie Curie Reintegration grant by the European Commission (to R.K.).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/25159
References
- 1.Schmid-Hempel P. Evolutionary parasitology - the integrated study of infections, immunology, ecology, and genetics. Oxford, UK: Oxford University Press, 2011. [Google Scholar]
- 2.Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–4. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 3.Débarre F, Lion S, van Baalen M, Gandon S. Evolution of host life-history traits in a spatially structured host-parasite system. Am Nat. 2012;179:52–63. doi: 10.1086/663199. [DOI] [PubMed] [Google Scholar]
- 4.Smith Trail DR. Behavioral interactions between parasites and hosts: host suicide and the evolution of complex life cycles. Am Nat. 1980;116:77–92. doi: 10.1086/283612. [DOI] [Google Scholar]
- 5.McAllister MK, Roitberg BD. Adaptive suicidal behaviour in pea aphids. Nature. 1987;328:797–9. doi: 10.1038/328797b0. [DOI] [Google Scholar]
- 6.Müller CB, Schmid-Hempel R. To die for host or parasite? Anim Behav. 1992;44:177–9. doi: 10.1016/S0003-3472(05)80770-5. [DOI] [Google Scholar]
- 7.Heinze J, Walter B. Moribund ants leave their nests to die in social isolation. Curr Biol. 2010;20:249–52. doi: 10.1016/j.cub.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Rueppell O, Hayworth MK, Ross NP. Altruistic self-removal of health-compromised honey bee workers from their hive. J Evol Biol. 2010;23:1538–46. doi: 10.1111/j.1420-9101.2010.02022.x. [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson I. Adaptive and non-adaptive suicide in aphids. Nature. 1987;330:701. doi: 10.1038/330701a0. [DOI] [Google Scholar]
- 10.Poulin R. “Adaptive” changes in the behaviour of parasitized animals: a critical review. Int J Parasitol. 1995;25:1371–83. doi: 10.1016/0020-7519(95)00100-X. [DOI] [PubMed] [Google Scholar]
- 11.Gardner A, Kümmerli R. Social evolution: this microbe will self-destruct. Curr Biol. 2008;18:R1021–3. doi: 10.1016/j.cub.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Nedelcu AM, Driscoll WW, Durand PM, Herron MD, Rashidi A. On the paradigm of altruistic suicide in the unicellular world. Evolution. 2011;65:3–20. doi: 10.1111/j.1558-5646.2010.01103.x. [DOI] [PubMed] [Google Scholar]
- 13.Reece SE, Pollitt LC, Colegrave N, Gardner A. The meaning of death: evolution and ecology of apoptosis in protozoan parasites. PLoS Pathog. 2011;7:e1002320. doi: 10.1371/journal.ppat.1002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parma DH, Snyder M, Sobolevski S, Nawroz M, Brody E, Gold L. The Rex system of bacteriophage λ: tolerance and altruistic cell death. Genes Dev. 1992;6:497–510. doi: 10.1101/gad.6.3.497. [DOI] [PubMed] [Google Scholar]
- 15.Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol Microbiol. 1995;15:415–20. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 16.Chopin M-C, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8:473–9. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 18.Fukuyo M, Sasaki A, Kobayashi I. Success of a suicidal defense strategy against infection in a structured habitat. Sci Rep. 2012;2:238. doi: 10.1038/srep00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 20.West SA, Griffin AS, Gardner A, Diggle SP. Social evolution theory for microorganisms. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 21.Berngruber TW, Lion S, Gandon S. Evolution of suicide as a defence strategy against pathogens in a spatially structured environment. Ecol Lett. 2013;16:446–53. doi: 10.1111/ele.12064. [DOI] [PubMed] [Google Scholar]
- 22.Refardt D, Bergmiller T, Kümmerli R. Altruism can evolve when relatedness is low: evidence from bacteria committing suicide upon phage infection. Proc Biol Sci. 2013;280:20123035. doi: 10.1098/rspb.2012.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu F, Mizushima S. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J Bacteriol. 1982;151:718–22. doi: 10.1128/jb.151.2.718-722.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Court D, Oppenheim AB. Phage λ’s accessory genes. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, eds. Lambda II. Cold Spring Harbor, NY: CSH Laboratory Press, 1983:251-77. [Google Scholar]
- 25.Kümmerli R, Griffin AS, West SA, Buckling A, Harrison F. Viscous medium promotes cooperation in the pathogenic bacterium Pseudomonas aeruginosa. Proc Biol Sci. 2009;276:3531–8. doi: 10.1098/rspb.2009.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Or D, Smets BF, Wraith JM, Dechesne A, Friedman SP. Physical constraints affecting bacterial habitats and activity in unsaturated porous media – a review. Adv Water Resour. 2007;30:1505–27. doi: 10.1016/j.advwatres.2006.05.025. [DOI] [Google Scholar]
- 27.Lenski RE. Two-step resistance by Escherichia coli B to bacteriophage T2. Genetics. 1984;107:1–7. doi: 10.1093/genetics/107.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quance MA, Travisano M. Effects of temperature on the fitness cost of resistance to bacteriophage T4 in Escherichia coli. Evolution. 2009;63:1406–16. doi: 10.1111/j.1558-5646.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- 29.Bohannan BJM, Lenski RE. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett. 2000;3:362–77. doi: 10.1046/j.1461-0248.2000.00161.x. [DOI] [Google Scholar]
- 30.Hall AR, Scanlan PD, Morgan AD, Buckling A. Host-parasite coevolutionary arms races give way to fluctuating selection. Ecol Lett. 2011;14:635–42. doi: 10.1111/j.1461-0248.2011.01624.x. [DOI] [PubMed] [Google Scholar]
- 31.Koskella B, Lin DM, Buckling A, Thompson JN. The costs of evolving resistance in heterogeneous parasite environments. Proc Biol Sci. 2012;279:1896–903. doi: 10.1098/rspb.2011.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Refardt D. Within-host competition determines reproductive success of temperate bacteriophages. ISME J. 2011;5:1451–60. doi: 10.1038/ismej.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Refardt D. Real-time quantitative PCR to discriminate and quantify lambdoid bacteriophages of Escherichia coli K-12. Bacteriophage. 2012;2:98–104. doi: 10.4161/bact.20092. [DOI] [PMC free article] [PubMed] [Google Scholar]


