Abstract
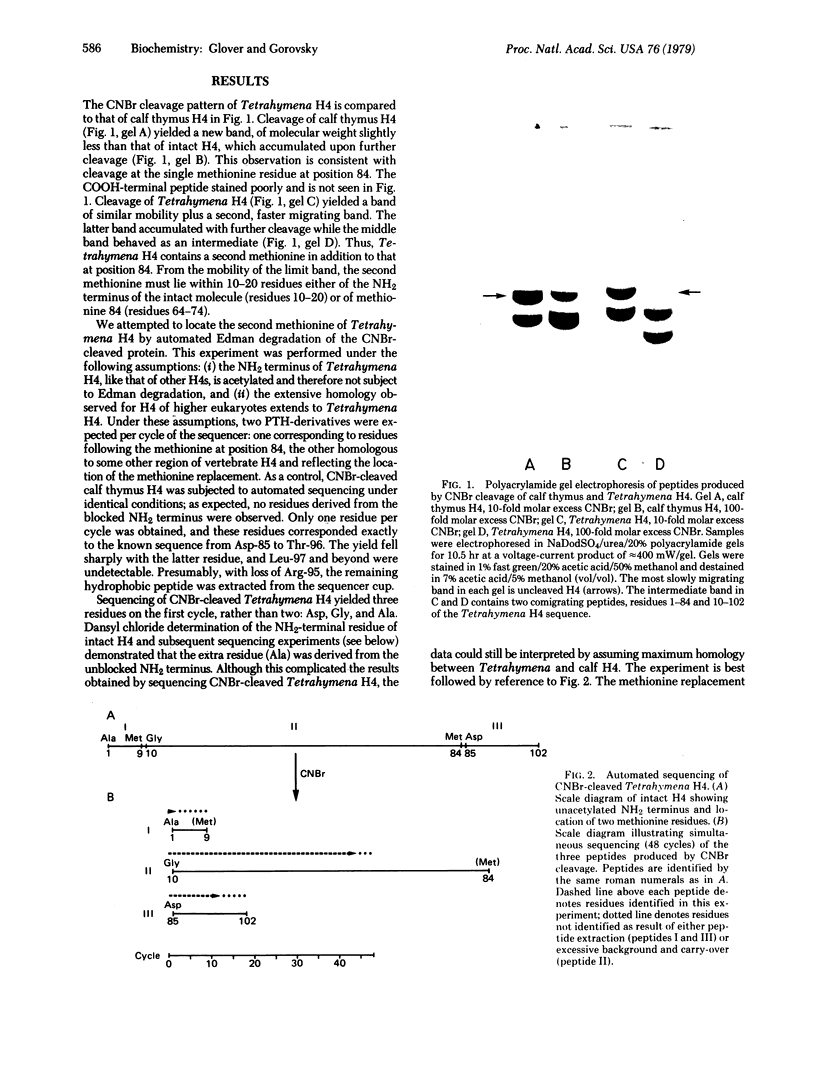
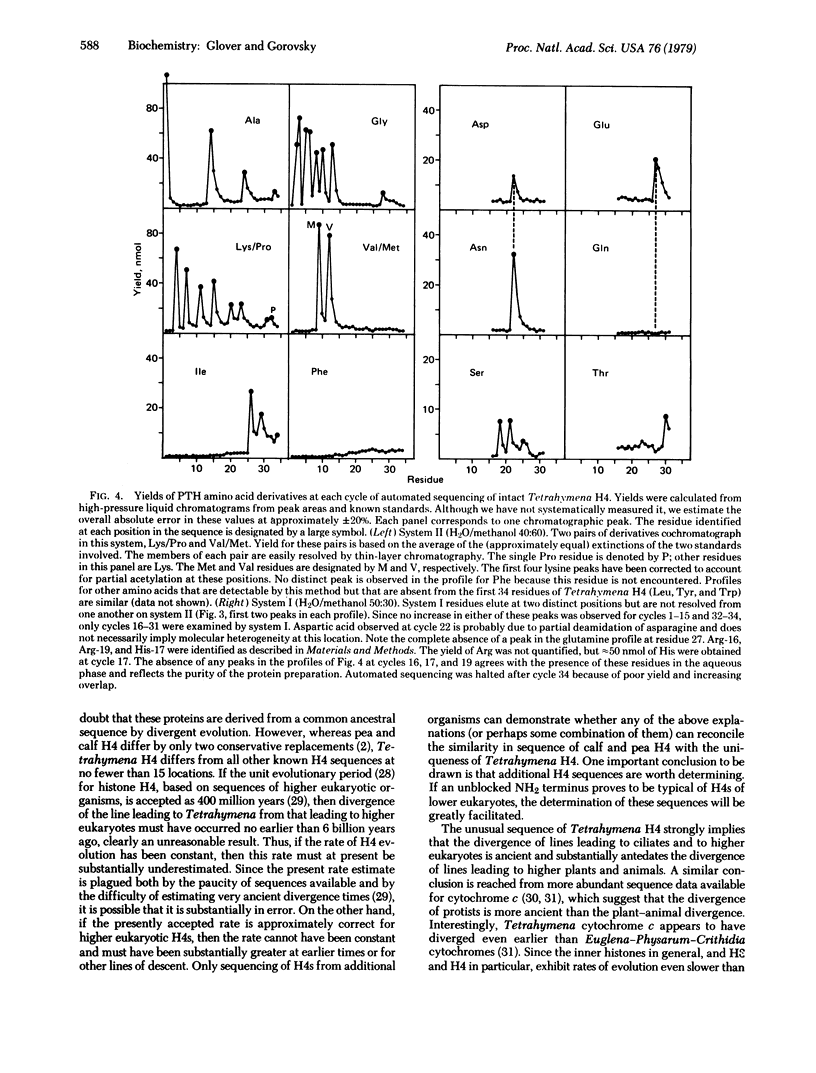
A partial amino-acid sequence of Tetrahymena histone H4 has been determined and differs significantly from the sequence of calf or pea H4. The amino terminus of Tetrahymena H4, unlike that of other H4s so far examined, is not acetylated. Of 66 residues determined, one is a single-residue insertion, one a single-residue deletion, and thirteen are amino-acid replacements with respect to the calf thymus H4 sequence. Most of the amino-acid replacements are nonconservative and are distributed nonrandomly, with a strong concentration in the amino-terminal arm. The first four lysines are partially acetylated. All but two of the replacements can be explained by single nucleotide substitutions at the level of the gene. The similarity in sequence of calf and pea H4 coupled with the substantial differences displayed by Tetrahymena H4 suggest that the divergence of protozoa and higher eukaryotes substantially antedates the divergence of plants and animals. Furthermore, quantitative analysis of the data requires either that the rate of H4 evolution be considerably more rapid than previously thought or that the rate be different for different periods of evolution or for different lines of descent.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfageme C. R., Zweidler A., Mahowald A., Cohen L. H. Histones of Drosophila embryos. Electrophoretic isolation and structural studies. J Biol Chem. 1974 Jun 25;249(12):3729–3736. [PubMed] [Google Scholar]
- Charlesworth M. C., Parish R. W. The isolation of nuclei and basic nucleoproteins from the cellular slime mold Dictyostelium discoideum. Eur J Biochem. 1975 May;54(1):307–316. doi: 10.1111/j.1432-1033.1975.tb04141.x. [DOI] [PubMed] [Google Scholar]
- DeLange R. J., Fambrough D. M., Smith E. L., Bonner J. Calf and pea histone IV. 3. Complete amino acid sequence of pea seedling histone IV; comparison with the homologous calf thymus histone. J Biol Chem. 1969 Oct 25;244(20):5669–5679. [PubMed] [Google Scholar]
- DeLange R. J., Fambrough D. M., Smith E. L., Bonner J. Calf and pea histone IV. II. The complete amino acid sequence of calf thymus histone IV; presence of epsilon-N-acetyllysine. J Biol Chem. 1969 Jan 25;244(2):319–334. [PubMed] [Google Scholar]
- Desai L., Ogawa Y., Mauritzen C. M., Taylor C. W., Starbuck W. C. Carboxyl-terminal sequence of the glycine-arginine-rich histone from bovine lymphosarcoma, Novikoff hepatoma and fetal calf thymus. Biochim Biophys Acta. 1969 May;181(1):146–153. doi: 10.1016/0005-2795(69)90234-7. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. The structures of cytochrome c and the rates of molecular evolution. J Mol Evol. 1971;1(1):26–45. doi: 10.1007/BF01659392. [DOI] [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Felden R. A., Sanders M. M., Morris N. R. Presence of histones in Aspergillus nidulans. J Cell Biol. 1976 Mar;68(3):430–439. doi: 10.1083/jcb.68.3.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff C. G. Histones of Neurospora crassa. J Biol Chem. 1976 Jul 10;251(13):4131–4138. [PubMed] [Google Scholar]
- Gorovsky M. A., Glover C., Johmann C. A., Keevert J. B., Mathis D. J., Samuelson M. Histones and chromatin structure in Tetrahymena macro- and micronuclei. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):493–503. doi: 10.1101/sqb.1978.042.01.052. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Johmann C. A., Gorovsky M. A. Purification and characterization of the histones associated with the macronucleus of Tetrahymena. Biochemistry. 1976 Mar 23;15(6):1249–1256. doi: 10.1021/bi00651a012. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D., Thomas J. O. Chromatin structure; oligomers of the histones. Science. 1974 May 24;184(4139):865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- Kulbe K. D. Micropolyamide thin-layer chromatography of phenylthiohydantoin amino acids (PTH) at subnanomolar level. A rapid microtechnique for simultaneous multisample identification after automated Edman degradations. Anal Biochem. 1974 Jun;59(2):564–573. doi: 10.1016/0003-2697(74)90310-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Panyim S., Bilek D., Chalkley R. An electrophoretic comparison of vertebrate histones. J Biol Chem. 1971 Jul 10;246(13):4206–4215. [PubMed] [Google Scholar]
- Podell D. N., Abraham G. N. A technique for the removal of pyroglutamic acid from the amino terminus of proteins using calf liver pyroglutamate amino peptidase. Biochem Biophys Res Commun. 1978 Mar 15;81(1):176–185. doi: 10.1016/0006-291x(78)91646-7. [DOI] [PubMed] [Google Scholar]
- Sautiere P., Tyrou D., Moschetto Y., Biserte G. Structure primaire de l'histone riche en glycine et en arginine isolée de la tumeur de chloroleucémic du rat. Biochimie. 1971;53(4):479–483. doi: 10.1016/s0300-9084(71)80165-7. [DOI] [PubMed] [Google Scholar]
- Sautière P., Lambelin-Breynaert M. D., Moschetto Y., Biserte G. Histone riche en glycine et en arginine du thymus de porc: étude des peptides trypsiques et séquence complète. Biochimie. 1971;53(5):711–715. [PubMed] [Google Scholar]
- Sawyer L., Shotton D. M., Campbell J. W., Wendell P. L., Muirhead H., Watson H. C. The atomic structure of crystalline porcine pancreatic elastase at 2.5 A resolution: comparisons with the structure of alpha-chymotrypsin. J Mol Biol. 1978 Jan 15;118(2):137–208. doi: 10.1016/0022-2836(78)90412-6. [DOI] [PubMed] [Google Scholar]
- Spiker S., Isenberg I. Cross-complexing pattern of plant histones. Biochemistry. 1977 May 3;16(9):1819–1826. doi: 10.1021/bi00628a009. [DOI] [PubMed] [Google Scholar]
- Strickland M., Strickland W. N., Brandt W. F., Von Holt C. Sequence of the cysteine-containing portion of histone F2al from the sea urchin Parechinus angulosus. FEBS Lett. 1974 Apr 1;40(2):346–348. doi: 10.1016/0014-5793(74)80260-7. [DOI] [PubMed] [Google Scholar]
- Summers M. R., Smythers G. W., Oroszlan S. Thin-layer chromatography of sub-nanomole amounts of phenylthiohydantoin (PTH) amino acids on polyamide sheets. Anal Biochem. 1973 Jun;53(2):624–628. doi: 10.1016/0003-2697(73)90114-0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Carlson S. S., White T. J. Biochemical evolution. Annu Rev Biochem. 1977;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- Yamada S., Itano H. Phenanthrenequinone as an analytical reagent for arginine and other monosubstituted guanidines. Biochim Biophys Acta. 1966 Dec 28;130(2):538–540. doi: 10.1016/0304-4165(66)90256-x. [DOI] [PubMed] [Google Scholar]
- van der Westhuyzen D. R., von Holt C. A new procedure for the isolation and fractionation of histones. FEBS Lett. 1971 May 20;14(5):333–337. doi: 10.1016/0014-5793(71)80294-6. [DOI] [PubMed] [Google Scholar]




