Abstract
Characterization of prophages in sequenced bacterial genomes is important for virulence assessment, evolutionary analysis, and phage application development. The objective of this study was to identify complete, inducible prophages in the cystic fibrosis (CF) clinical isolate Burkholderia cenocepacia H111. Using the prophage-finding program PHAge Search Tool (PHAST), we identified three putative intact prophages in the H111 sequence. Virions were readily isolated from H111 culture supernatants following extended incubation. Using shotgun cloning and sequencing, one of these virions (designated ϕH111-1 [vB_BceM_ϕH111-1]) was identified as the infective particle of a PHAST-detected intact prophage. ϕH111-1 has an extremely broad host range with respect to B. cenocepacia strains and is predicted to use lipopolysaccharide (LPS) as a receptor. Bioinformatics analysis indicates that the prophage is 42,972 base pairs in length, encodes 54 proteins, and shows relatedness to the virion morphogenesis modules of AcaML1 and “Vhmllikevirus” myoviruses. As ϕH111-1 is active against a broad panel of clinical strains and encodes no putative virulence factors, it may be therapeutically effective for Burkholderia infections.
Keywords: prophage identification, PHAST, bioinformatics, phage therapy, Burkholderia cepacia complex
Introduction
The development and continual improvement of next-generation sequencing technologies now allows for the rapid genomic analysis of diverse populations of previously uncharacterized bacteria. Although not necessarily a focus of such studies, the characterization of prophage sequences within these genomes can provide important insights into mechanisms of virulence, horizontal transfer, and the evolution of both host and phage.1 Furthermore, these newly sequenced genomes (along with their embedded prophage sequences) have the potential to be a repository of novel inducible phages that could be exploited for biotechnological and/or medical applications.
Within the genus Burkholderia, prophages have been intensively studied to assess their contribution to host virulence and evolution and to identify appropriate inducible phage candidates for diagnostic or therapeutic use.2-5 For the Burkholderia cepacia complex (BCC)—a group of opportunistic pathogens infecting cystic fibrosis (CF) patients—characterization studies generally focus on the potential for medical application of a specific phage. Of particular importance are phages infecting B. cenocepacia due to the clinical predominance and virulence of this species.4 Although the therapeutic use of temperate phages is generally discouraged, the limited availability of obligately lytic BCC phages has necessitated the use of confirmed, putative, or modified temperate phages for several in vivo efficacy studies.6-8 The use of such phages against Burkholderia is arguably safer than it is against many other pathogens because virulence factors in this genus have not been discovered to be encoded by temperate phages.2
One of the challenges of prophage identification is the differentiation of inducible prophages from defective prophage remnants, a distinction with both evolutionary and practical implications. From an evolutionary standpoint, inducible prophages can transfer bacterial or phage genes through transduction, while prophage remnants do not actively facilitate horizontal exchange (with some exceptions, such as gene transfer agents).1,9 From a practical standpoint, the ability to independently propagate, characterize, modify, and utilize temperate phages is extremely limited if the prophage cannot be induced. PHAge Search Tool (PHAST) is a newly developed prophage identification program developed in part to address this challenge.10 It can predict if a prophage is intact, incomplete, or questionable. Here, we use PHAST to facilitate the identification and further classical and molecular characterization of an inducible prophage in the newly sequenced genome of the CF isolate Burkholderia cenocepacia H111. This work demonstrates how the integration of improved bioinformatics tools with next-generation DNA sequencing can greatly accelerate the identification and isolation of biomedically important inducible phages.
Results and Discussion
B. cenocepacia H111 prophage screening and isolation
Preliminary sequence analysis of B. cenocepacia H111 contigs identified several regions containing prophage-like genes. To determine which regions might contain complete prophages, PHAST was used to analyze the 71 available H111 contigs (NZ_CAFQ01000001.1–NZ_CAFQ01000071.1). This program identified three potential intact prophages in contigs NZ_CAFQ01000015.1 (C15), NZ_CAFQ01000032.1 (C32), and NZ_CAFQ01000043.1 (C43). In C15, the region identified is 47.0 kilobase pairs (kbp) in length and shows similarity to proteins of the Shigella myovirus SfV and other phages (including Burkholderia phages AH2, Bcep176, BcepNazgul, Bcep22, ϕ644-2, ϕ1026b, ϕE125, BcepF1, and KS5). In C32, the prophage region is shorter (33.2 kbp) and shows extensive similarity at the protein level to the P2-like myovirus ϕE202, a prophage of Burkholderia thailandensis.3 In C43, the region identified is 26.0 kbp in length and shows similarity to proteins of the Vibrio myovirus vB_VpaM_MAR and other phages infecting species such as Burkholderia, Ralstonia, Erwinia, Salmonella, Hemophilus, Streptomyces, and Escherichia.
Based on the PHAST prediction that intact prophages were present in the H111 genome, we assayed H111 culture supernatants for spontaneous phage release following extended incubation. When filter-sterilized supernatant was plated with B. cenocepacia C6433 (a common BCC phage host) in soft-agar overlays, many small (1–2 mm) identical plaques with turbid centers and very turbid halos were observed. A single phage plaque designated as ϕH111-1 (or vB_BceM_ϕH111-1) was subsequently picked and propagated to high titer on C6433. The following analyses were performed on the single phage type isolated from a single plaque. In order to potentially isolate any other putative prophages from this strain, alternative screening procedures that vary the mode of induction and/or the propagation host may be required.
ϕH111-1 morphology, receptor, and host range
ϕH111-1 is a myovirus with a capsid diameter of approximately 65 nm (Fig. 1). To identify the putative phage receptor, we tested the ability of ϕH111-1 to infect B. cenocepacia K56-2 strains with lipopolysaccharide (LPS) mutations.11,12 The majority of strains remained susceptible to phage infection excluding the truncated inner core wabO and waaC mutants (Table S1), indicating that ϕH111-1 likely interacts with moieties in the LPS inner core.
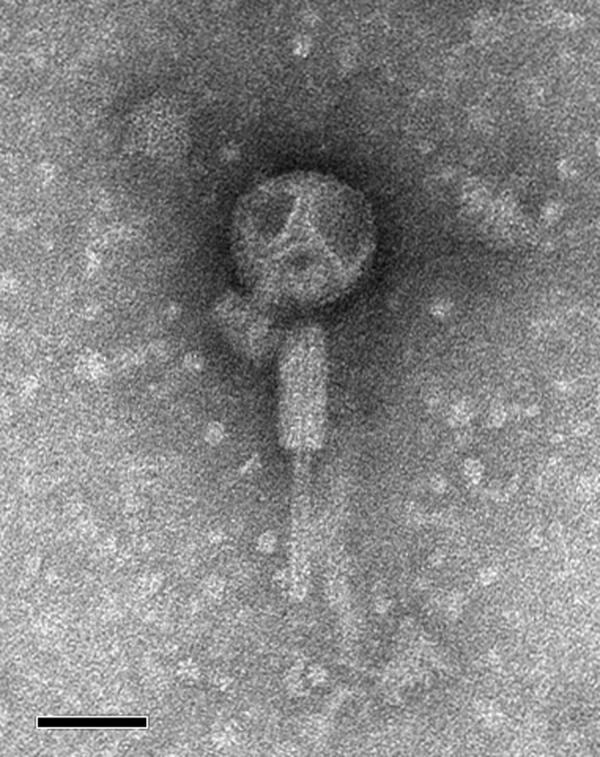
Figure 1. Transmission electron micrograph of a ϕH111-1 virion stained with phosphotungstic acid. The micrograph was taken at 140,000-fold magnification; scale bar represents 50 nm.
When tested against a panel of nine B. cenocepacia strains (all of which were originally isolated from CF patients), ϕH111-1 was able to infect seven of these strains: C6433, 715J, J2315, K56-2, C1257, C5424, and PC184 (Table S1). The broad host range with respect to characterized B. cenocepacia CF strains suggested that ϕH111-1 could be active against a wide variety of clinical isolates. To confirm this prediction, we tested ϕH111-1 against a B. cenocepacia panel acquired from the University of Alberta Hospital Cystic Fibrosis Clinic.13 ϕH111-1 was able to infect all 13 strains tested (Table S1), providing further evidence that this phage may be an appropriate candidate for therapeutic use (particularly if the lysogeny module were deleted).8 Excluding B. cenocepacia, the ϕH111-1 host range was found to be relatively narrow as only B. multivorans ATCC 17616 and C5274 were susceptible to phage infection from a panel of 18 other Burkholderia strains tested (representing 8 additional BCC species) (Table S1).
ϕH111-1 genome sequence
To determine if ϕH111-1 represented one of the intact prophages predicted by PHAST, we isolated phage DNA and performed shotgun cloning. Two randomly chosen EcoRI clones were partially sequenced and compared with the H111 reference sequence using BLASTN. One clone matched with C43 bp 172,986–176,784 while the other matched to C43 bp 179,518–181,496. Both of these sequences fall within the prophage region in C43 predicted by PHAST (159,103–185,124), indicating that this program correctly identified both the locus and the intact nature of the prophage.
To identify the exact prophage boundaries attL and attR, we screened for direct repeats 20 kbp upstream and downstream of the cloned sequences (from 152,986–201,496 in C43) using two-sequence BLASTN alignment. An imperfect 24 bp direct repeat (Fig. 2) was identified that flanked sequences consistent with prophage genes: one copy was found upstream of a series of hypothetical protein genes (starting with I35_4470) and one copy was found at the 5′ end of an arginine tRNA gene (I35_4520). To confirm that these repeats represented the attL and attR sequences, we designed primers (downstream of I35_4470 and upstream of I35_4520) and verified by PCR and sequencing that these regions became adjacent within packaged virion DNA, forming the attP overlap region (Fig. 2). Based on restriction analysis, DNA in the ϕH111-1 virion is predicted to be linear without cohesive ends.

Figure 2. Alignment of the ϕH111-1 attL (above), attP overlap region (center), and attR (below). The 24 base pair region common to all three sequences is underlined.
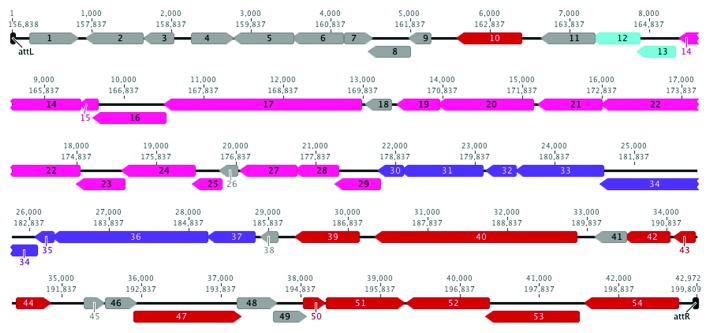
The ϕH111-1 prophage is 42,972 bp in length (including both attL and attR), has a 62% GC content (lower than the H111 GC content of 67%), and integrates at an arginine tRNA gene (as noted above). Based on GeneMark predictions, this prophage sequence contains 54 open reading frames (Fig. 3, Table 1). Putative functional annotations were assigned to these proteins based on BLASTP (Table 1) and HHpred (Table S2) analysis. No putative toxin genes were identified using BTXpred. As shown in Figure 3, ϕH111-1 genes are arranged in function-specific modules involved in DNA binding, lysis, tail morphogenesis, and capsid morphogenesis/DNA packaging (discussed below).
Figure 3. Map of the ϕH111-1 prophage. Arrows indicate gene transcription in the forward or reverse direction. Small numbers indicate base pairs within the prophage (above) or H111 contig NZ_CAFQ01000043.1 (below). Black, attL, or attR; gray, unknown function; red, DNA binding; blue, lysis; pink, tail morphogenesis; purple, capsid morphogenesis and DNA packaging.
Table 1. ϕH111-1 genome annotation.
| Gene | Prophage start | Prophage end | H111 contig start | H111 contig end | Strand | Length (amino acids) | Putative function | Closest relative (excluding H111 proteins) | BLASTP alignment region (amino acids) | Percent identity | Organism | GenBank accession number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
attL |
1 |
24 |
156838 |
156861 |
|
|
|
|
|
|
|
|
|
1 |
243 |
809 |
157080 |
157646 |
+ |
188 |
hypothetical |
hypothetical protein PMI16_01842 |
19–200/207 |
37 |
Herbaspirillum sp CF444 |
ZP_10720925.1 |
|
2 |
939 |
1628 |
157776 |
158465 |
- |
229 |
hypothetical |
hypothetical protein BuboB_19482 |
1–218/220 |
64 |
Burkholderia ubonensis Bu |
ZP_02379924.1 |
|
3 |
1668 |
2009 |
158505 |
158846 |
- |
113 |
hypothetical |
hypothetical protein bgla_2p0890 |
1–113/135 |
42 |
Burkholderia gladioli BSR3 |
YP_004351005.1 |
|
4 |
2266 |
2766 |
159103 |
159603 |
+ |
166 |
hypothetical |
gp31 |
2–144/153 |
48 |
Burkholderia pseudomallei 1710a |
ZP_04953065.1 |
|
5 |
2797 |
3513 |
159634 |
160350 |
- |
238 |
hypothetical |
hypothetical protein bgla_1g11110 |
1–238/238 |
61 |
Burkholderia gladioli BSR3 |
YP_004359751.1 |
|
6 |
3526 |
4137 |
160363 |
160974 |
- |
203 |
hypothetical |
hypothetical protein BP1026B_I2070 |
1–200/200 |
58 |
Burkholderia pseudomallei 1026b |
YP_006275083.1 |
|
7 |
4187 |
4510 |
161024 |
161347 |
+ |
107 |
hypothetical |
peptidase M23 |
74–132/432 |
37 |
Ferrimonas balearica DSM 9799 |
YP_003914362.1 |
|
8 |
4485 |
4982 |
161322 |
161819 |
- |
165 |
hypothetical |
hypothetical protein bgla_1g11080 |
1–165/165 |
67 |
Burkholderia gladioli BSR3 |
YP_004359748.1 |
|
9 |
4989 |
5237 |
161826 |
162074 |
- |
82 |
hypothetical |
hypothetical protein bgla_1g11070 |
62–142/143 |
62 |
Burkholderia gladioli BSR3 |
YP_004359747.1 |
|
10 |
5592 |
6377 |
162429 |
163214 |
- |
261 |
DNA adenine methylase |
D12 class N6 adenine-specific DNA methyltransferase |
1–262/262 |
84 |
Burkholderia ambifaria AMMD |
YP_773740.1 |
|
11 |
6655 |
7296 |
163492 |
164133 |
- |
213 |
hypothetical |
hypothetical protein bgla_1g27190 |
3–198/209 |
89 |
Burkholderia gladioli BSR3 |
YP_004361292.1 |
|
12 |
7299 |
7865 |
164136 |
164702 |
- |
188 |
endolysin |
hypothetical protein bgla_1g27200 |
1–188/188 |
89 |
Burkholderia gladioli BSR3 |
YP_004361293.1 |
|
13 |
7862 |
8311 |
164699 |
165148 |
- |
149 |
holin |
putative kinetochore protein spc25 protein |
80–200/259 |
26 |
Neofusicoccum parvum UCRNP2 |
EOD48207.1 |
|
14 |
8388 |
9437 |
165225 |
166274 |
- |
349 |
tail protein D |
phage late control D family protein |
1–349/349 |
91 |
Burkholderia glumae BGR1 |
YP_002912485.1 |
|
15 |
9447 |
9653 |
166284 |
166490 |
- |
68 |
tail protein X |
phage tail X family protein |
1–68/68 |
96 |
Burkholderia ambifaria AMMD |
YP_773745.1 |
|
16 |
9628 |
10506 |
166465 |
167343 |
- |
292 |
tail protein U |
phage P2 GpU family protein |
1–292/292 |
90 |
Burkholderia ambifaria AMMD |
YP_773746.1 |
|
17 |
10517 |
12958 |
167354 |
169795 |
- |
813 |
tail tape measure protein T |
pyocin R2_PP, tail length determination protein |
1–814/814 |
89 |
Burkholderia ambifaria AMMD |
YP_773747.1 |
|
18 |
13039 |
13341 |
169876 |
170178 |
- |
100 |
hypothetical |
hypothetical protein Bamb_1858 |
1–100/100 |
82 |
Burkholderia ambifaria AMMD |
YP_773748.1 |
|
19 |
13439 |
13942 |
170276 |
170779 |
- |
167 |
tail tube protein FII |
phage major tail tube protein |
1–167/167 |
92 |
Burkholderia ambifaria AMMD |
YP_773749.1 |
|
20 |
13953 |
15122 |
170790 |
171959 |
- |
389 |
tail sheath protein FI |
phage tail sheath protein |
1–389/389 |
93 |
Burkholderia ambifaria AMMD |
YP_773750.1 |
|
21 |
15207 |
15986 |
172044 |
172823 |
- |
259 |
tail fiber assembly protein G |
bacteriophage-acquired protein |
1–231/233 |
60 |
Burkholderia thailandensis TXDOH |
ZP_02376040.1 |
|
22 |
16002 |
18020 |
172839 |
174857 |
- |
672 |
tail fiber protein |
phage tail fiber protein |
1–670/670 |
59 |
Burkholderia glumae BGR1 |
YP_002912494.1 |
|
23 |
18008 |
18586 |
174845 |
175423 |
- |
192 |
baseplate assembly protein I |
phage tail protein I |
1–192/192 |
92 |
Burkholderia ambifaria AMMD |
YP_773753.1 |
|
24 |
18576 |
19472 |
175413 |
176309 |
- |
298 |
baseplate assembly protein J |
baseplate J family protein |
1–298/298 |
88 |
Burkholderia ambifaria AMMD |
YP_773754.1 |
|
25 |
19469 |
19804 |
176306 |
176641 |
- |
111 |
baseplate assembly protein W |
GPW/gp25 family protein |
1–111/111 |
93 |
Burkholderia ambifaria AMMD |
YP_773755.1 |
|
26 |
19804 |
20004 |
176641 |
176841 |
- |
66 |
hypothetical |
hypothetical protein Bamb_1866 |
1–66/66 |
85 |
Burkholderia ambifaria AMMD |
YP_773756.1 |
|
27 |
20064 |
20744 |
176901 |
177581 |
- |
226 |
baseplate assembly protein V |
phage baseplate assembly protein V |
1–226/226 |
90 |
Burkholderia ambifaria AMMD |
YP_773758.1 |
|
28 |
20748 |
21272 |
177585 |
178109 |
- |
174 |
tail protein |
hypothetical protein Bamb_1869 |
1–174/174 |
81 |
Burkholderia ambifaria AMMD |
YP_773759.1 |
|
29 |
21262 |
21792 |
178099 |
178629 |
- |
176 |
tail protein |
hypothetical protein Bamb_1870 |
1–176/176 |
88 |
Burkholderia ambifaria AMMD |
YP_773760.1 |
|
30 |
21795 |
22082 |
178632 |
178919 |
- |
95 |
head-tail joining protein |
hypothetical protein |
1–95/96 |
77 |
Burkholderia glumae BGR1 |
YP_002912503.1 |
|
31 |
22084 |
23079 |
178921 |
179916 |
- |
331 |
major capsid protein |
hypothetical protein Bamb_1872 |
1–331/331 |
92 |
Burkholderia ambifaria AMMD |
YP_773762.1 |
|
32 |
23153 |
23497 |
179990 |
180334 |
- |
114 |
head decoration protein |
hypothetical protein BURMUCF1_2022 |
1–114/114 |
91 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15920090.1 |
|
33 |
23528 |
24595 |
180365 |
181432 |
- |
355 |
Clp protease |
ATP-dependent Clp endopeptidase, proteolytic subunit ClpP |
1–355/357 |
84 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15921695.1 |
|
34 |
24592 |
26085 |
181429 |
182922 |
- |
497 |
portal protein |
phage portal protein, lambda family |
1–485/496 |
90 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15921707.1 |
|
35 |
26082 |
26288 |
182919 |
183125 |
- |
68 |
head-tail joining protein |
hypothetical protein Bcenmc03_1109 |
1–68/68 |
100 |
Burkholderia cenocepacia MC0–3 |
YP_001764407.1 |
|
36 |
26302 |
28221 |
183139 |
185058 |
- |
639 |
terminase large subunit |
terminase GpA |
1–639/639 |
97 |
Burkholderia cenocepacia MC0–3 |
YP_001764406.1 |
|
37 |
28250 |
28819 |
185087 |
185656 |
- |
189 |
terminase small subunit |
hypothetical protein Bcenmc03_1107 |
1–189/189 |
94 |
Burkholderia cenocepacia MC0–3 |
YP_001764405.1 |
|
38 |
28910 |
29104 |
185747 |
185941 |
- |
64 |
hypothetical |
hypothetical protein Bcenmc03_1105 |
1–64/64 |
86 |
Burkholderia cenocepacia MC0–3 |
YP_001764403.1 |
|
39 |
29352 |
30125 |
186189 |
186962 |
- |
257 |
DnaJ chaperone |
hypothetical protein BURMUCF1_2377 |
1–257/257 |
99 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15916016.1 |
|
40 |
30346 |
32853 |
187183 |
189690 |
- |
835 |
DNA primase |
virulence-associated E family protein |
1–835/835 |
95 |
Burkholderia ambifaria AMMD |
YP_773770.1 |
|
41 |
33114 |
33476 |
189951 |
190313 |
- |
120 |
hypothetical |
hypothetical protein Bamb_1881 |
8–127/127 |
90 |
Burkholderia ambifaria AMMD |
YP_773771.1 |
|
42 |
33484 |
34026 |
190321 |
190863 |
- |
180 |
transcriptional regulator |
hypothetical protein Bamb_1882 |
15–194/194 |
93 |
Burkholderia ambifaria AMMD |
YP_773772.1 |
|
43 |
34108 |
34344 |
190945 |
191181 |
- |
78 |
transcriptional regulator |
hypothetical protein PLA107_31961 |
3–62/74 |
57 |
Pseudomonas syringae pv lachrymans str. M301315 |
ZP_16673553.1 |
|
44 |
34448 |
34843 |
191285 |
191680 |
+ |
131 |
transcriptional regulator |
XRE family transcriptional regulator |
12–141/143 |
92 |
Burkholderia ambifaria AMMD |
YP_773773.1 |
|
45 |
35300 |
35518 |
192137 |
192355 |
+ |
72 |
hypothetical |
hypothetical protein BURMUCF1_2052 |
1–72/72 |
79 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15921714.1 |
|
46 |
35568 |
35930 |
192405 |
192767 |
+ |
120 |
hypothetical |
hypothetical protein BURMUCF1_2384 |
1–120/120 |
82 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15916012.1 |
|
47 |
35930 |
37228 |
192767 |
194065 |
+ |
432 |
ParB-like protein |
hypothetical protein BURMUCF1_2385 |
1–434/434 |
64 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15916017.1 |
|
48 |
37225 |
37689 |
194062 |
194526 |
+ |
154 |
hypothetical |
hypothetical protein BURMUCF1_2386 |
1–152/152 |
89 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15916020.1 |
|
49 |
37682 |
38059 |
194519 |
194896 |
+ |
125 |
hypothetical |
hypothetical protein |
408–466/532 |
39 |
Burkholderia glumae BGR1 |
YP_002911887.1 |
|
50 |
38056 |
38289 |
194893 |
195126 |
+ |
77 |
excisionase |
hypothetical protein Bpse14_41058 |
1–76/76 |
88 |
Burkholderia pseudomallei 14 |
ZP_02417306.1 |
|
51 |
38342 |
39295 |
195179 |
196132 |
+ |
317 |
DNA cytosine methylase |
DNA-cytosine methyltransferase |
1–317/317 |
91 |
Burkholderia phytofirmans PsJN |
YP_001894795.1 |
|
52 |
39341 |
40351 |
196178 |
197188 |
- |
336 |
restriction endonuclease |
hypothetical protein Bphyt_1154 |
1–336/336 |
75 |
Burkholderia phytofirmans PsJN |
YP_001894794.1 |
|
53 |
40341 |
41480 |
197178 |
198317 |
- |
379 |
ParB-like protein |
hypothetical protein Bphyt_1153 |
1–379/379 |
84 |
Burkholderia phytofirmans PsJN |
YP_001894793.1 |
|
54 |
41575 |
42732 |
198412 |
199569 |
- |
385 |
integrase |
site-specific recombinase, phage integrase family |
15–372/379 |
96 |
Burkholderia multivorans ATCC BAA-247 |
ZP_15916025.1 |
| attR | 42949 | 42972 | 199786 | 199809 |
H111 contig start and end values correspond with B. cenocepacia H111 accession number NZ_CAFQ01000043.1.
Sequence analysis
At the nucleotide level, ϕH111-1 is most similar to putative prophage elements in chromosome 1 of Burkholderia gladioli BSR3, Burkholderia glumae BGR1, Burkholderia ambifaria AMMD, and Burkholderia pseudomallei BPC006, 1026b, and MSHR346. Based on a BLASTN comparison, these sequences share 62–78% coverage with the ϕH111-1 prophage (Table S3). As PHAST analysis predicts that each of these regions represents an intact prophage (Table S3), ϕH111-1 may be the first isolated representative of a group of closely related but broadly distributed temperate phages in the genus Burkholderia.
To assess protein relatedness, we used CoreGenesUniqueGenes (CGUG) to compare ϕH111-1 to previously sequenced phages. This program assesses the percentage of proteins that are shared between a genome of interest and a reference genome (determined based on a defined BLASTP threshold).14 Based on BLASTP analysis, the ϕH111-1 tail proteins show similarity to those of enterobacteria phage P2 (NC_001895.1) and other phages in the genus P2likevirus. Comparing ϕH111-1 and P2 with CGUG (Table 2), the proteomes are 25.58% similar with respect to P2, placing ϕH111-1 in the same subfamily (Peduovirinae) but a different genus.15 We could not identify any previously characterized phages with ≥ 40% similarity that would belong to the same genus as ϕH111-1. Currently, phages with the most similar proteomes are AcaML1 of Acidithiobacillus caldus (JX507079.1; 28.17% similar) and the “Vhmllikevirus” phages VHML of Vibrio harveyi (NC_004456.1; 28.07% similar) and both VP58.5 (FN297812.1; 31.03% similar) and vB_VpaM_MAR (NC_019722.1; 29.03% similar) of Vibrio parahemolyticus.16-19 Although these phages share subfamily-level similarity, they are very distinct with respect to aspects such as host, gene content, and lifestyle. AcaML1 lysogenizes a thermophilic and acidophilic γ-proteobacterium and has a significantly larger 59 kbp genome with two insertion sequences.16 The “Vhmllikevirus” phages VHML, VP58.5, and vB_VpaM_MAR have similar genome sizes to ϕH111-1 (41–43 kbp) but are thought to lysogenize as linear plasmids with telomeres in Vibrio species.17-19
Table 2. CoreGenesUniqueGenes comparison of ϕH111-1, P2, AcaML1, and VHML.
| ϕH111-1 protein | Putative function | Similar protein in P2 | Similar protein in AcaML1 | Similar protein in VHML |
|---|---|---|---|---|
| gp1 |
Hypothetical |
None |
None |
None |
| gp2 |
Hypothetical |
None |
None |
None |
| gp3 |
Hypothetical |
None |
None |
None |
| gp4 |
Hypothetical |
None |
None |
None |
| gp5 |
Hypothetical |
None |
None |
None |
| gp6 |
Hypothetical |
None |
None |
None |
| gp7 |
hypothetical |
None |
None |
None |
| gp8 |
hypothetical |
None |
None |
None |
| gp9 |
hypothetical |
None |
None |
None |
| gp10 |
DNA adenine methylase |
None |
None |
None |
| gp11 |
hypothetical |
None |
None |
None |
| gp12 |
endolysin |
None |
None |
ORF19 |
| gp13 |
holin |
None |
None |
None |
| gp14 |
tail protein D |
tail protein |
phage tail protein X |
tail protein |
| gp15 |
tail protein X |
gpX |
phage late control protein D |
ORF45 |
| gp16 |
tail protein U |
gpU |
phage tail formation protein U |
ORF44 |
| gp17 |
tail tape measure protein T |
gpT |
phage tail length tape measure protein |
ORF43 |
| gp18 |
hypothetical |
None |
hypothetical protein |
None |
| gp19 |
tail tube protein FII |
major tail tube protein |
phage tail tube protein FII |
major tail tube protein |
| gp20 |
tail sheath protein FI |
major tail sheath protein |
phage tail sheath protein FI |
major tail sheath protein |
| gp21 |
tail fiber assembly protein G |
gpG |
None |
None |
| gp22 |
tail fiber protein |
None |
None |
None |
| gp23 |
baseplate assembly protein I |
gpI |
phage baseplate assembly protein gpI |
ORF33 |
| gp24 |
baseplate assembly protein J |
baseplate assembly protein |
phage baseplate assembly protein gpJ |
baseplate assembly protein |
| gp25 |
baseplate assembly protein W |
baseplate wedge subunit |
phage baseplate assembly protein gpW |
baseplate wedge subunit |
| gp26 |
hypothetical |
None |
None |
None |
| gp27 |
baseplate assembly protein V |
gpV |
phage baseplate assembly protein gpV |
ORF30 |
| gp28 |
tail protein |
None |
None |
ORF29 |
| gp29 |
tail protein |
None |
None |
ORF28 |
| gp30 |
head-tail joining protein |
None |
hypothetical protein |
None |
| gp31 |
major capsid protein |
None |
major capsid protein |
ORF26 |
| gp32 |
head decoration protein |
None |
hypothetical protein |
None |
| gp33 |
Clp protease |
None |
periplasmic serine proteases (ClpP class) |
None |
| gp34 |
portal protein |
None |
portal protein |
ORF23 |
| gp35 |
head-tail joining protein |
None |
hypothetical protein |
None |
| gp36 |
terminase large subunit |
None |
packaging terminase large subunit gpA |
ORF22 |
| gp37 |
terminase small subunit |
None |
None |
None |
| gp38 |
hypothetical |
None |
None |
None |
| gp39 |
DnaJ chaperone |
None |
None |
None |
| gp40 |
DNA primase |
None |
None |
None |
| gp41 |
hypothetical |
None |
None |
None |
| gp42 |
transcriptional regulator |
None |
None |
None |
| gp43 |
transcriptional regulator |
None |
None |
None |
| gp44 |
transcriptional regulator |
None |
None |
None |
| gp45 |
hypothetical |
None |
None |
None |
| gp46 |
hypothetical |
None |
None |
None |
| gp47 |
ParB-like protein |
None |
site specific recombinase large subunit |
None |
| gp48 |
hypothetical |
None |
None |
None |
| gp49 |
hypothetical |
None |
None |
None |
| gp50 |
excisionase |
None |
None |
None |
| gp51 |
DNA cytosine methylase |
None |
site specific DNA modification methylase Dcm |
None |
| gp52 |
restriction endonuclease |
None |
None |
None |
| gp53 |
ParB-like protein |
None |
None |
None |
| gp54 | integrase | None | None | None |
CoreGenesUniqueGenes analysis was performed using a cutoff score of 75.
The commonalities among ϕH111-1, AcaML1, and the “Vhmllikevirus” phages are largely restricted to the morphogenesis genes. These phages all have P2-like tail proteins, but encode capsid morphogenesis/DNA packaging and accessory proteins that are unrelated to those of P2 (Table 2). The ϕH111-1 tail morphogenesis module extends from gene 14-29, encoding only three proteins that lack homologs in either AcaML1 or VHML: tail fiber assembly protein gp21, tail fiber protein gp22, and hypothetical protein gp26 (Table 2). The dissimilarity of the tail fiber protein (predicted to be the phage anti-receptor) is expected based on the differences in host specificity.20 Based on CGUG analysis, ϕH111-1 encodes a protein similar to each P2 tail protein excluding E/E+E′, H, R, and S (Table 2). The ϕH111-1 capsid morphogenesis and DNA packaging proteins are more closely related to those of AcaML1 than the “Vhmllikevirus” phages. This module includes genes 30–37, encoding the head-tail joining proteins, major capsid protein, head decoration protein, Clp protease, portal protein, and terminase subunits (Table 1). AcaML1 encodes proteins similar to each of these (excluding the terminase small subunit), while VHML only encodes similar major capsid, portal, and terminase large subunit proteins (Table 2).
The predicted ϕH111-1 lysis and DNA binding proteins are largely unrelated to those of AcaML1 and the “Vhmllikevirus” phages (Table 2). A putative holin and N-acetylmuramidase endolysin are encoded proximal to the tail morphogenesis module, with the latter being similar to a VHML protein (Table 2). The predicted DNA binding proteins of ϕH111-1 have a range of functions based on HHpred analysis (Table S2): adenine and cytosine methylases, DnaJ chaperone, primase, transcriptional regulators, ParB-like proteins, excisionase, restriction endonuclease, and integrase (Table 1). Excluding the DNA adenine methylase gp10, each of these proteins is encoded near the right end of the prophage (Fig. 3). Only the gp47 ParB-like protein and gp51 DNA cytosine methylase are similar to AcaML1 proteins (Table 2). There is no evidence that ϕH111-1 carries AcaML1-type transposase or “Vhmllikevirus”-type protelomerase genes.
Conclusions
In order for phage therapy to be a viable alternative to antibiotic treatment for Burkholderia infections, phages must be identified that have activity against an array of clinical isolates without encoding potential virulence factors. By using the PHAST program as a rapid screening tool prior to classical and molecular characterization, we were able to identify such a phage in the chromosome of B. cenocepacia H111. ϕH111-1 has a broad B. cenocepacia host range and lacks genes associated with pathogenicity, making it one of the most clinically promising BCC temperate phages isolated to date. While the results of this study were highly informative with respect to ϕH111-1 morphology, host range, receptor binding, and genetic content, further analyses are required to characterize related prophages and to assess the safety and activity of this phage in vivo.
Materials and Methods
Bacterial strains
B. cenocepacia H111 was originally isolated from a CF patient in Germany.21 Strains from the original and updated BCC experimental strain panels,22,23B. cenocepacia K56-2 LPS mutants,11,12 and clinical isolates from the University of Alberta Hospital Cystic Fibrosis Clinic13 were used for phage isolation, propagation, and host range testing. Strains were grown aerobically overnight at 30 °C in half-strength Luria-Bertani (½ LB) broth or solid medium (containing agar or, for DNA isolation, agarose). LPS mutants were grown similarly but supplemented with 100 μg/ml trimethoprim.
Phage isolation and analysis
For ϕH111-1 isolation, a 10 ml broth culture of H111 was grown aerobically with shaking for 48 h at 30 °C. One milliliter of the culture was pelleted 2 min at 10,000 rcf and the supernatant was filter-sterilized using a Millex-HA 0.45 μm syringe-driven filter unit (Millipore). The supernatant was diluted in modified suspension medium (modified SM; 50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgSO4), plated in soft agar overlays with B. cenocepacia C6433, and incubated overnight at 30 °C. A single plaque was picked using a sterile Pasteur pipette and suspended in modified SM. To collect a high titer lysate, the single plaque stock was replated with C6433, overlaid with modified SM following overnight incubation, pelleted, and filter-sterilized as above. Phage stocks were stored at 4 °C.
For host range analysis (BCC panel strains, clinical isolates, and K56-2 LPS mutants), strains were screened with both overlays and spot testing (10 μl spots of diluted lysate on overlays of the host strain). Electron microscopy grids were prepared by incubating filter-sterilized (0.22 μm) lysate on a carbon-coated copper grid for 5 min followed by phosphotungstic acid staining for 30 s. A Philips/FEI (Morgagni) transmission electron microscope with charge-coupled device camera was used to capture images with the assistance of the University of Alberta Department of Biological Sciences Advanced Microscopy Facility. Phage DNA was isolated, digested, shotgun cloned, and partially sequenced as described previously.24 Screening for cohesive sites was also performed as described previously.25
Bioinformatics
Prophage regions were identified in H111 using PHAge Search Tool (PHAST) analysis of contigs NZ_CAFQ01000001.1–NZ_CAFQ01000071.1.10 Putative prophage boundaries (i.e., flanking direct repeats attL and attR) were identified using two-sequence BLASTN.26 ϕH111-1 lysate was PCR amplified (I35_4519F: TTGCTATACTC TGTCCCCGCCG; I35_4471R: CAACCATTTCGT CAGCCGGATAG) and sequenced to verify that the attL and attR sequences were found in a single copy in the phage DNA (as the attP overlap region). The prophage sequence was reannotated from the original record using GeneMark.hmm for prokaryotes.27 We were unable to definitively identify either a translationally frameshifted tail protein or an Rz/Rz1 pair following manual annotation.28,29 BLASTP, HHpred, CD-Search, and BTXpred were used to predict protein function.26,30-32 Genome and proteome relatedness were assessed using BLASTN and CoreGenesUniqueGenes (CGUG) with a cutoff score of 75, respectively.14,15,26 The genome map was constructed using Geneious.33 The ϕH111-1 prophage sequence can be found in the GenBank database under B. cenocepacia H111 accession number NZ_CAFQ01000043.1 (bp 156,838–199,809).
Supplementary Material
Acknowledgments
The authors would like to thank Miguel Valvano (Queen’s University Belfast) for supplying K56-2 LPS mutants, the University of Alberta Hospital Cystic Fibrosis Clinic for supplying clinical isolates, Arlene Oatway (University of Alberta Department of Biological Sciences Advanced Microscopy Facility), and the University of Alberta Department of Biological Sciences Molecular Biology Service Unit. JJD thanks the Canadian Institutes of Health Research and Cystic Fibrosis Canada for operating grant funding.
Glossary
Abbreviations:
- PHAST
PHAge Search Tool
- BCC
Burkholderia cepacia complex
- CF
cystic fibrosis
- bp
base pairs
- LPS
lipopolysaccharide
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/bacteriophage/article/26649
Footnotes
Previously published online: www.landesbioscience.com/journals/bacteriophage/article/26649
References
- 1.Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:1–12. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summer EJ, Gill JJ, Upton C, Gonzalez CF, Young R. Role of phages in the pathogenesis of Burkholderia, or ‘Where are the toxin genes in Burkholderia phages?’. Curr Opin Microbiol. 2007;10:410–7. doi: 10.1016/j.mib.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ronning CM, Losada L, Brinkac L, Inman J, Ulrich RL, Schell M, Nierman WC, Deshazer D. Genetic and phenotypic diversity in Burkholderia: contributions by prophage and phage-like elements. BMC Microbiol. 2010;10:202. doi: 10.1186/1471-2180-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch KH, Dennis JJ. Cangene gold medal award lecture - Genomic analysis and modification of Burkholderia cepacia complex bacteriophages. Can J Microbiol. 2012;58:221–35. doi: 10.1139/w11-135. [DOI] [PubMed] [Google Scholar]
- 5.Lynch KH, Dennis JJ. Genomics of Burkholderia phages. In: Coenye T, Mahenthiralingam E, eds. Burkholderia: From Genomes to Function. Hethersett, Norwich, UK: Horizon Scientific Press, in press. [Google Scholar]
- 6.Seed KD, Dennis JJ. Experimental bacteriophage therapy increases survival of Galleria mellonella larvae infected with clinically relevant strains of the Burkholderia cepacia complex. Antimicrob Agents Chemother. 2009;53:2205–8. doi: 10.1128/AAC.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmody LA, Gill JJ, Summer EJ, Sajjan US, Gonzalez CF, Young RF, LiPuma JJ. Efficacy of bacteriophage therapy in a model of Burkholderia cenocepacia pulmonary infection. J Infect Dis. 2010;201:264–71. doi: 10.1086/649227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch KH, Seed KD, Stothard P, Dennis JJ. Inactivation of Burkholderia cepacia complex phage KS9 gp41 identifies the phage repressor and generates lytic virions. J Virol. 2010;84:1276–88. doi: 10.1128/JVI.01843-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. Phage as agents of lateral gene transfer. Curr Opin Microbiol. 2003;6:417–24. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39(Web Server issue):W347-52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol. 2006;188:2073–80. doi: 10.1128/JB.188.6.2073-2080.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ortega X, Silipo A, Saldías MS, Bates CC, Molinaro A, Valvano MA. Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: truncation of the core oligosaccharide leads to increased binding and sensitivity to polymyxin B. J Biol Chem. 2009;284:21738–51. doi: 10.1074/jbc.M109.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch KH, Dennis JJ. Development of a species-specific fur gene-based method for identification of the Burkholderia cepacia complex. J Clin Microbiol. 2008;46:447–55. doi: 10.1128/JCM.01460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahadevan P, King JF, Seto D. CGUG: in silico proteome and genome parsing tool for the determination of “core” and unique genes in the analysis of genomes up to ca. 1.9 Mb. BMC Res Notes. 2009;2:168. doi: 10.1186/1756-0500-2-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009;9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tapia P, Flores FM, Covarrubias PC, Acuña LG, Holmes DS, Quatrini R. Complete genome sequence of temperate bacteriophage AcaML1 from the extreme acidophile Acidithiobacillus caldus ATCC 51756. J Virol. 2012;86:12452–3. doi: 10.1128/JVI.02261-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oakey HJ, Cullen BR, Owens L. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J Appl Microbiol. 2002;93:1089–98. doi: 10.1046/j.1365-2672.2002.01776.x. [DOI] [PubMed] [Google Scholar]
- 18.Zabala B, Hammerl JA, Espejo RT, Hertwig S. The linear plasmid prophage Vp58.5 of Vibrio parahaemolyticus is closely related to the integrating phage VHML and constitutes a new incompatibility group of telomere phages. J Virol. 2009;83:9313–20. doi: 10.1128/JVI.00672-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alanis Villa A, Kropinski AM, Abbasifar R, Griffiths MW. Complete genome sequence of Vibrio parahaemolyticus bacteriophage vB_VpaM_MAR. J Virol. 2012;86:13138–9. doi: 10.1128/JVI.02518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch KH, Stothard P, Dennis JJ. Genomic analysis and relatedness of P2-like phages of the Burkholderia cepacia complex. BMC Genomics. 2010;11:599. doi: 10.1186/1471-2164-11-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Römling U, Fiedler B, Bosshammer J, Grothues D, Greipel J, von der Hardt H, Tümmler B. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994;170:1616–21. doi: 10.1093/infdis/170.6.1616. [DOI] [PubMed] [Google Scholar]
- 22.Mahenthiralingam E, Coenye T, Chung JW, Speert DP, Govan JRW, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–3. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coenye T, Vandamme P, LiPuma JJ, Govan JR, Mahenthiralingam E. Updated version of the Burkholderia cepacia complex experimental strain panel. J Clin Microbiol. 2003;41:2797–8. doi: 10.1128/JCM.41.6.2797-2798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynch KH, Abdu AH, Schobert M, Dennis JJ. Genomic characterization of JG068, a novel virulent podovirus active against Burkholderia cenocepacia. BMC Genomics. 2013;14:574. doi: 10.1186/1471-2164-14-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch KH, Stothard P, Dennis JJ. Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genomics. 2012;13:223. doi: 10.1186/1471-2164-13-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–15. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Summer EJ, Berry J, Tran TAT, Niu L, Struck DK, Young R. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J Mol Biol. 2007;373:1098–112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 30.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(Web Server issue):W327-31. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saha S, Raghava GP. BTXpred: prediction of bacterial toxins. In Silico Biol. 2007;7:405–12. [PubMed] [Google Scholar]
- 33.Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, et al. Geneious v5.6. Available from http://www.geneious.com
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.