Abstract
Cold shock proteins (CSPs) of bacteria are produced in response to cold and function as RNA chaperones that are essential for cold adaptation. Arabidopsis thaliana COLD SHOCK DOMAIN PROTEIN 3 (AtCSP3) shares a domain with bacterial CSPs and is involved in acquisition of freezing tolerance. Our previous study revealed that many of the genes that are down regulated in an AtCSP3 knockout mutant (atcsp3–2) are functionally associated with responses to salt and drought as well as cold. Here, we examined the involvement of AtCSP3 in salt and drought stress tolerance. We found that AtCSP3 is induced during salt and drought stresses, and is regulated by ABA. A knockout mutant of AtCSP3 (atcsp3–2) showed lower survival rates after salt and drought stress treatments. Conversely, the AtCSP3-overexpressing plants displayed higher survival rates after treatment with these stresses. Most of the genes that were down regulated in the atcsp3–2 mutant were found to be inducible upon salt and drought stresses, and upregulated in the AtCSP3-overexpressors. Together, our data demonstrates that AtCSP3 is involved in the regulation of salt and drought stress tolerance in Arabidopsis.
Keywords: ABA, Arabidopsis thaliana, Cold shock domain protein, Drought stress, RNA chaperone, Salt stress
Abbreviations: ABA, abscisic acid; CBF, C-repeat binding factors; COR, cold-regulated; CSD, cold shock domain; CSPs, cold shock domain proteins; mRNP, messenger ribonucleoprotein; RNP, ribonucleoprotein
Highlights
-
•
Arabidopsis thaliana COLD SHOCK DOMAIN PROTEIN 3 (AtCSP3) is induced during salt and drought stresses.
-
•
A knockout mutant of AtCSP3 showed lower survival rates after salt and drought stresses.
-
•
AtCSP3-overexpressing plants displayed higher survival rates after salt and drought stresses.
-
•
AtCSP3 is involved in the regulation of salt and drought stress tolerance in Arabidopsis.
1. Introduction
The bacterial cold shock proteins (CSPs) function as RNA chaperones that melt secondary structures of mRNAs and facilitate transcription and translation [1,2]. In Escherichia coli, nine members of the CSP gene family (cspA–cspI) have been identified. Four of them (cspA, cspB, cspG, and cspI) are induced by cold shock, whereas the others are either constitutive or induced by nutritional starvation [3]. CSPs are essential for growth of E. coli under low temperature conditions, and quadruple deletion mutation of cspA, cspB, cspG, and cspE results in deficient growth at low temperature [4]. CSPs bind to single-stranded RNA and DNA without apparent sequence specificity [2]. However, CspA has been shown to target and unwind the secondary structures of partially double-stranded RNA, which may arise during exposure to low temperatures [2]. In prokaryotic transcriptional machinery, RNA secondary structures can lead to premature transcription termination. CspA, CspC, and CspE have been confirmed to possess in vivo and in vitro transcription anti-termination activity [5], and CspA is also thought to enhance translation at low temperature through the elimination of stabilized RNA secondary structures [2]. These reported activities of CSPs have established their function as RNA chaperones in response to cold [2].
Plant cold shock domain (CSD) proteins typically contain an N-terminal CSD and a C-terminal glycine-rich region interspersed with various numbers of retroviral-like CCHC zinc fingers [6]. The first functionally characterized plant CSD protein was wheat (Triticum aestivum) Cold Shock domain Protein 1 (WCSP1), which accumulates in crown tissue during prolonged cold acclimation [7]. WCSP1 mRNA accumulation is not modulated by other environmental stresses such as salt, drought and heat, or treatment with abscisic acid (ABA) [7], which suggests that the function of WCSP1 is specific to cold adaptation. WCSP1 has nucleic acid-binding activity [7,8] and unwinds nucleic acid duplexes in vitro and in vivo [9]. These findings indicate that, similar to bacterial CSPs, WCSP1 functions as an RNA chaperone to destabilize RNA secondary structures during cold acclimation.
In Arabidopsis, four CSD protein genes (AtCSP1–AtCSP4) have been identified in the genome [6,10]. Expressions of AtCSP1 (At4g36020), AtCSP2 (At4g38680), and AtCSP3 (At2g17870) are cold-inducible, and the proteins show RNA chaperone activity [10–14]. AtCSP3 is expressed mainly in meristematic tissues, where the protein accumulates during cold acclimation. A loss-of-function mutant of AtCSP3 (atcsp3–2) is sensitive to freezing as compared to wild-type plants under both non-acclimated and cold-acclimated conditions [14]. Overexpression of AtCSP3 in transgenic plants confers enhanced freezing tolerance. These genetic studies have shown that AtCSP3 plays an important role for freezing tolerance in Arabidopsis [11]. C-repeat binding factors (CBFs) regulate freezing tolerance through activation of cold-regulated (COR) gene expression [15–17]. However, AtCSP3 does not affect expression of CBFs or COR genes, suggesting that AtCSP3 is involved in the regulation of a CBF-independent pathway [11]. AtCSP3 is localized to the nucleus and cytoplasm. In the nucleus, AtCSP3 interacts with several different proteins within the nucleolus and nuclear speckles [18]. Based on this it has been suggested that AtCSP3 is involved in several different classes of ribonucleoprotein (RNP) complexes.
In our previous study, we found that some of the down-regulated genes in the AtCSP3 knockout mutant (atcsp3–2) are related to not only freezing tolerance but also to tolerance of other abiotic stresses such as salt and drought. Here, we present evidence from analysis of transgenic and mutant plants that AtCSP3 is involved in the regulation of salt and drought stress tolerance.
2. Materials and methods
2.1. Plant materials
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used as the wild-type in this study. The atcsp3–2 mutant line (WiscDsLox35G12) was obtained from the ABRC collection [11]. Production of 35S:AtCSP3 transgenic plants has been described previously [11]. Two new transgenic lines (S3–3 and S3–29) were isolated for this study. Homozygous T4 plants were used for salt and drought tolerance tests.
2.2. Immunoblot analysis
Ten-d-old seedlings were homogenized and extracted with a protein extraction buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2 mM EDTA). Protein concentration was determined with the Bio-Rad protein assay kit (Bio-Rad). Total protein (30 μg on SDS–PAGE gels) was transferred to Hybond-C Extra membrane (GE Healthcare). The membranes were probed with custom rabbit polyclonal antibodies against the cold shock domain (CSD) of AtCSP3 (1:5,000 v/v, Hokudo, Hokkaido, Japan) and anti-rabbit IgG peroxidase-linked secondary antibodies (1:10,000 v/v, GE Healthcare). Chemiluminescent detection of the signal was carried out using the ECL kit (GE Healthcare) according to the manufacturer's instructions.
2.3. RT-PCR analysis
For stress treatments, 10-d-old seedlings hydroponically grown with MS medium were transferred to MS medium supplemented with 200 mM NaCl (salt stress), a filter paper (drought stress), or MS medium supplemented with ABA (100 μM). Total RNAs were extracted from the seedlings using the RNeasy Plant Mini Kit (Qiagen). First strand cDNA was synthesized from 2 μg DNase I (Takara, Shiga, Japan)-treated total RNA using the High Capacity RNA-to-cDNA Kit (GE Healthcare). Semi-quantitative RT-PCR was performed with the Expand High Fidelity PCR System (Roche), using gene-specific primers (Supplementary Table S1 and [11]).
2.4. Analysis of salt and drought tolerance
For salt-tolerance tests, 7-d-old seedlings were transferred to MS agar plates supplemented with 200 mM NaCl and grown for either 3 d (atcsp3–2 mutant) or 6 d (35S:AtCSP3 plants). Survival was scored 7 d after transfer back to MS agar plates. Effects of salt on root growth were determined with 7-d-old seedlings. The root length was measured 7 d after transfer to MS agar plates supplemented with 125 mM NaCl.
For drought tolerance tests, 2-week-old seedlings grown on MS agar plates were transferred to soil and further grown for a week. Three weeks after germination, plants were subjected to drought stress by withholding water for 5 d for the comparison of the atcsp3–2 mutant and wild-type plants or 10 d for the comparison of 35S:AtCSP3 and wild-type plants. Surviving plants were counted after 5 d of re-watering.
3. Results
3.1. Expression of AtCSP3 is induced by abiotic stresses and ABA
We previously identified a group of genes that are down-regulated in atcsp3–2 mutant by microarray analysis [11]. An expression database search using the Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) revealed that these down-regulated genes are potentially inducible by abiotic stresses such as salt and drought as well as cold (Supplementary Table S2). This suggested the possibility that AtCSP3 is involved in the regulation of salt and drought stress tolerance in addition to freezing tolerance acquired through cold acclimation. To explore this possibility, we first determined changes in AtCSP3 expression in response to drought and salt by RT-PCR, finding that the transcript levels of AtCSP3 were up-regulated under both salt and drought stress conditions (Fig. 1A and B). AtCSP3 expression levels gradually increased up to 12 h under salt stress (200 mM NaCl), and up to 8 h under drought stress and decreased thereafter (Fig. 1A and B). Expression of AtCSP3 was also up-regulated by exogenous ABA treatment (Fig. 1C). AtCSP3 expression reached a maximum level 12 h after initiation of ABA treatment and was maintained at that level through 48 h of treatment (Fig. 1C). Since the ABA-induced expression of AtCSP3 was detectable only after 8 h, the initial induction of AtCSP3 upon salt and drought stresses is likely not directly regulated by ABA. Together, these data support the idea that AtCSP3 is involved in responses to salt and drought stresses.
Fig. 1.
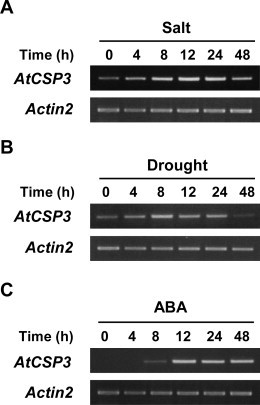
Expression of AtCSP3 in response to salt, drought, and ABA. Semi-quantitative RT-PCR was performed with RNA from 10-d-old seedlings treated with salt (200 mM NaCl) (A), drought (B), or ABA (100 μM) (C). The ACTIN2 gene was amplified as a control.
3.2. The atcsp3–2 mutant is sensitive to high salinity and drought, whereas overexpression of AtCSP3 improves salt and drought tolerance in Arabidopsis
To test for a function of AtCSP3 in salt and drought stress tolerance, a knockout mutant of AtCSP3 (atcsp3–2) [11] was first analyzed. Compared to wild-type, atcsp3–2 had significantly decreased survival rates when grown on MS medium supplemented with 200 mM NaCl (Fig. 2A and B). In addition, drought tolerance of the mutant and wild-type was compared after withholding water for 5 d. Analysis of survival rates indicated that the atcsp3–2 mutant was also less tolerant of drought stress (Fig. 2C and D). These data indicate that AtCSP3 is important in both salt and drought stress tolerance in Arabidopsis.
Fig. 2.
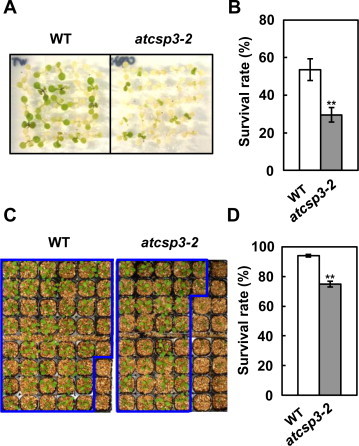
Response of the atcsp3–2 mutant to salt or drought stress. (A) Phenotype of atcsp3–2 on high-salt medium (200 mM NaCl). Seven-d-old wild type and mutant seedlings grown on MS agar medium were transferred to MS medium supplemented with 200 mM NaCl. The photograph was taken after 3 d of salt treatment. (B) Survival rates were calculated from three independent experiments (n = 25) and are expressed as the means ± SE. (C) Phenotype of atcsp3–2 mutants after re-watering subsequent to drought stress induced by withholding water for 5 d. The photographs were taken 1 d after re-watering. Note that pots were rearranged to better show the differences between genotypes. Survivors are surrounded by lines. (D) Survival rates were calculated from three independent experiments (n = 48). Data represent the means ± SE. Asterisks in (B) and (D) indicate significantly lower survival rates than wild-type (WT) plants as determined by Student's t test (**P < 0.01).
To further examine this, the effect of AtCSP3 overexpression on salt and drought stress tolerance was analyzed (Fig. 3). In addition to the previously isolated 35S:AtCSP3 line (S3–31) [11], we isolated two new 35S:AtCSP3 transgenic lines (S3–3 and S3–29) with higher transgene expression levels (Fig. 3A). Accumulation of the AtCSP3 protein in these lines was confirmed by immunoblot analysis (Fig. 3B). All three 35S:AtCSP3 transgenic lines showed higher survival rates than wild-type on 200 mM NaCl-containing plates (Fig. 3C and D), although the levels of AtCSP3 accumulation in the transgenic plants did not correlate with the survival rates. In addition, 35S:AtCSP3 seedlings displayed less inhibition of root growth than wild-type on medium containing 125 mM NaCl (Fig. 3E and F).
Fig. 3.
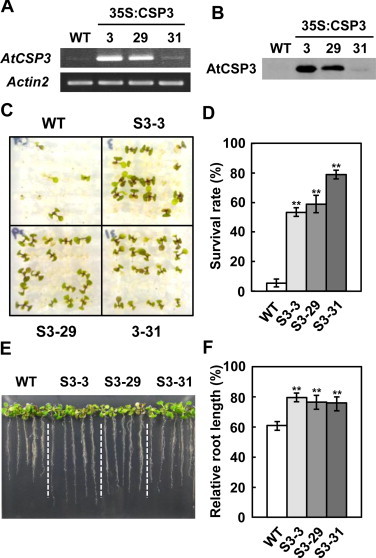
Salt tolerance of AtCSP3 overexpressors. Semi-quantitative RT-PCR (A) and immunoblot analysis (B) of AtCSP3 expression in 10-d-old seedlings. Three independent transgenic lines (S3–3, S3–29, and S3–31) were analyzed. (C) Phenotype of AtCSP3-overexpressing transgenic plants on high-salt medium (200 mM NaCl). The photograph was taken after 6 d of salt treatment. (D) Survival rates were calculated from three independent experiments (n = 25). Data represent the means ± SE. (E) Root growth of AtCSP3 transgenic plants in response to salt treatment (125 mM NaCl). The picture was taken 7 d after transfer to salt-supplemented medium. (F) Relative root length to plants without the salt stress was calculated from three independent experiments (n = 5). The data represent the means ± SE. Asterisks in (D) and (F) indicate significantly higher survival rates than wild-type (WT) plants as determined by Student's t test (**P < 0.01).
We also determined the drought stress tolerance of the 35S:AtCSP3 plants (Fig. 4). Water-deprived 35S:AtCSP3 transgenic plants of lines S3–3 and S3–29 grown in soil exhibited substantially higher survival rates than did wild-type plants after drought stress, while the S3–3 line showed only a small increase in survival rate (Fig. 4B). Thus, overexpression of AtCSP3 confers salt and drought stress tolerance in Arabidopsis, further demonstrating the importance of AtCSP3 in salt and drought stress responses.
Fig. 4.
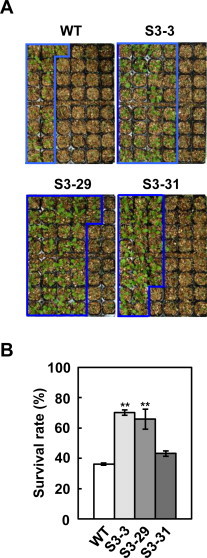
Drought tolerance of AtCSP3-overexpression lines. (A) Phenotype of AtCSP3-overexpressing plants after re-watering. Plants were grown in separate pots (one plant per pot) and drought-stressed by withholding water for 10 d. The photograph was taken 1 d after re-watering. Note that pots were rearranged to better show the differences between genotypes. Survivors are surrounded by lines. (B) Survival rates and standard deviations were calculated from three independent experiments (n = 48). Asterisks indicate significantly higher survival compared to wild-type (WT) as determined by Student's t test (**P < 0.01).
3.3. Overexpression of AtCSP3 results in up-regulation of salt- and drought-responsive genes in Arabidopsis
Our results suggest that AtCSP3 is involved in the regulation of salt and drought stress tolerance. Supplementary Table S2 lists candidate genes whose expression levels are altered by AtCSP3 expression. From these, fifteen candidate genes were selected and further analyzed for their expression pattern under salt and drought stress conditions (Fig. 5A and B). Semi-quantitative RT-PCR analysis revealed that the expressions of all candidate genes were salt-inducible except for that of DOMAIN-CONTAINING PROTEIN 1 (DC1), which was down-regulated by salt (Fig. 5A). Drought stress also induced all of the candidate genes except for DARK INDUCIBLE 2 (DIN2), GLUTATHIONE S-TRANSFERASE 7 (GSTF7), CYTOCHROME P450 FAMILY 81 SUBFAMILY F POLYPEPTIDE F2 (CYP81F2), GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1 (ASN1), and FLOTILLIN 1 (FLOT1) (Fig. 5B). These data suggest that the regulatory mechanism in which AtCSP3 is involved regulates expression of genes for salt and drought stress responses.
Fig. 5.
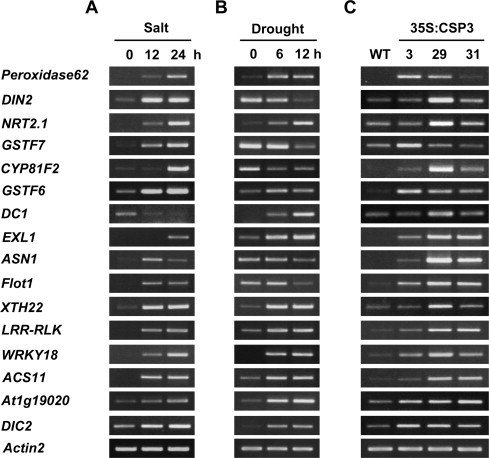
Expressions of salt and drought stress-responsive genes in AtCSP3-overexpression lines. Semi-quantitative RT-PCR was performed with total RNA from 10-d-old seedlings grown on agar plates treated with 200 mM NaCl (A) or subject to drought (B) for 0, 12, and 24 h. (C) Stress-responsive marker gene expression in three independent 35S:CSP3 lines (S3–3, 29, and 31) and wild-type. Gene-specific primers for PEROXIDASE62 (At5g39580), NRT2.1 (NITRATE TRANSPORTER 2; At1g08090), CYP81F2 (CYTOCHROME P450 FAMILY 81 SUBFAMILY F POLYPEPTIDE 2; At5g57220), GSTF6 (GLUTATHIONE S-TRANSFERASE; At1g02930), DC1 (DC1 DOMAIN-CONTAINING PROTEIN 1; At5g40590), and WRKY18 (WRKY TRANSCRIPTIONAL FACTOR 18; At4g31800) were described in Kim et al. [11]. Primers for DIN2 (DARK INDUCIBLE 2; At3g60140), GSTF7 (GLUTATHIONE S-TRANSFERASE 7; At1g02920), EXL1 (EXORDIUM-LIKE 1; At1g35140), ASN1 (GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1; At3g47340), FLOT1 (FLOTILLIN 1; At5g25250), XTH22 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE 22; At5g57560), LRR-RLK (PUTATIVE LEUCINE-RICH REPEAT RECEPTOR-LIKE KINASE; At2g34930), ACS11 (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 11; At4g08040), At1g19020 (unknown protein), and DIC2 (DICARBOXYLATE CARRIER 2; At4g24570) were described in Supplementary Table S1. The ACTIN2 gene was amplified as a control.
Among the fifteen candidate genes, eleven were up-regulated in the three AtCSP3-overexpressing lines (Fig. 5C). NITRATE TRANSPORTER 2.1 (NRT2.1), DC1, DIN2, and XYLOGLUCAN ENDOTRANSGLUCOSYLASE 22 (XTH22) were up-regulated in two of the overexpressing lines. However, GSTF7 was up-regulated only in the S3–3 line, therefore it may not be under the same regulation.
4. Discussion
AtCSP3 has been shown to be involved in freezing tolerance in Arabidopsis [11]. Since plant CSD proteins have been characterized in view of functional conservation with bacterial CSPs, their functions in tolerance of other abiotic stresses have not been fully characterized. Here, we demonstrated that AtCSP3 is inducible upon salt and drought stress and involved in these stress tolerance, likely through up-regulation of stress-related proteins.
WCSP1 was the first plant CSD protein to be functionally characterized [7]. Expression analysis of WCSP1 in seedlings revealed that WCSP1 is highly inducible by cold but not by other stresses such as heat, salt and drought [7]. Therefore, it was assumed that WCSP1 function is specifically related to cold stress response [7]. AtCSP1(CSDP1) [14], AtCSP2 (CSDP2) [12–14], and AtCSP3 [11] were also identified as cold-inducible genes, although the responses of plant CSD proteins to stresses other than cold have not been fully characterized. In fact, it has been reported that AtCSP1 is down-regulated by drought and salt stresses and that AtCSP2 is slightly up-regulated by salt stress but down-regulated by drought stress [19]. In this regard, our findings that AtCSP3 is inducible under both salt and drought stress conditions (Fig. 1) and functions in tolerance of both stresses (Figs. 2–4) widen the scope of known functions of CSD proteins in plants.
Most of the genes that are down-regulated in the atcsp3-2 knockout mutant showed increased expression in AtCSP3-overexpressing plants (Fig. 5). These genes are potentially regulated by AtCSP3 directly or indirectly. These genes showed up-regulation by both salt and drought stresses (Fig. 5), which is in agreement with the up-regulation of AtCSP3 in response to salt and drought stresses (Fig. 1). WRKY18 and At1g19020 are up-regulated in transgenic Arabidopsis plants that overexpress ZAT10 [20], which also demonstrate salt and drought stress tolerance [21]. We found that expression of EXL1, which is up-regulated in a Zat12 overexpressor according to microarray data [22], was elevated during salt and drought stress, as well as in AtCSP3-overexpressing plants (Fig. 5). It is therefore of interest to determine whether there is crosstalk between the regulatory pathways that involve ZAT10/12 and AtCSP3, respectively.
GLUTATHIONE S-TRANSFERASE 6 (GSTF6, GST1/ERD11), GSTF7 (GST11), XTH22 were up-regulated in salt- and drought-stressed plants (Fig. 5A and B), as well as in AtCSP3 overexpressors (Fig. 5C). These genes are also up-regulated in transgenic plants overexpressing the ethylene-responsive factor, AtERF1. AtERF1 is involved in both biotic and abiotic stress responses and overexpression of AtERF1 confers tolerance of salt and drought stresses [23]. Another gene that was upregulated in the AtCSP3 overexpressors was 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 11 (ACS11) (Fig. 5), which is involved in pathogen-induced ethylene production [24]. These data suggest a possible interaction of AtCSP3 with the ethylene signaling pathway.
Park et al. [19] characterized Arabidopsis plants overexpressing either AtCSP1 (CSDP1) or AtCSP2 (CSDP2). The overexpression lines were tested for tolerance of freezing, salt and drought stresses. In contrast to AtCSP3, overexpression of either AtCSP1 or AtCSP2 does not result in enhanced tolerance against these stresses [19]. However, these two genes may be involved in the responses to salt and drought stresses at a specific stage of development, as overexpression of AtCSP1 or AtCSP2 leads to negative or positive effects, respectively, on germination on medium containing 150 mM NaCl or 250 mM mannitol [19].
Our recent research has shown that AtCSP3 localizes to both the nucleus and cytoplasm, and forms a variety of messenger ribonucleoproteins (mRNPs) by interacting with different RNA-associated proteins [18]. For example, AtCSP3 interacts with poly A-binding proteins within the nuclear speckles, suggesting a function in poly A length control. AtCSP3 also interacts with DECAPPING PROTEIN 5 (DCP5) and may regulate the mRNA-decapping process. These data suggest that AtCSP3 is involved in the regulation of mRNA stability or degrqdation. Other interacting proteins have functions in mRNA splicing, mRNA export, or ribosome biogenesis, suggesting that AtCSP3 may also be involved in other steps of post transcriptional regulation.
Castiglioni et al. [25] created transgenic plants that overexpress the bacterial CSD proteins E. coli CspA and Bacillus subtilis CspB and demonstrated that these genes confer multiple abiotic stress tolerance. Transgenic rice plants expressing CspA or CspB showed improved growth under chilling and heat stress conditions [25]. The CspB-expressing rice plants showed improved growth under water deficit as well. In field trials, transgenic maize plants expressing CspA or CspB showed 10.9–30.8% increases in end-of-season grain yield under water-limited conditions [25]. The results suggested that bacterial CspA and CspB confer enhanced tolerance to drought, cold, and heat by protecting and improving vegetative growth, photosynthesis, and reproductive development in plants [25]. Although bacterial and plant CSD proteins share a function as RNA chaperones, it is not known if bacterial CSPs regulate gene expression in plants in a similar manner to plant CSPs such as AtCSP3.
In conclusion, the data presented in this report provide evidence that one of the plant CSD proteins, AtCSP3, can confer salt and drought stress tolerance. The regulatory function of AtCSP3 in salt and drought stress tolerance was demonstrated using gain- and loss-of-function approaches. The multiple stress tolerance conferred by AtCSP3 reported herein suggests that this gene will be useful in molecular crop breeding for high yield.
Acknowledgment
This work was supported by grants from the Japan Society for the Promotion of Science (KAKENHI Scientific Research B nos. 22380063 and 19380063) and the Salt Science Research Foundation (No.1216).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Supplementary material
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fob.2013.10.003.
Appendix. Supplementary materials
Supplementary materials for Cold Shock Domain Protein 3 is involved in salt and drought stress tolerance in Arabidopsis.
References
- 1.Graumann P.L., Marahiel M.A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 2.Jiang W., Hou Y., Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 3.Wang N., Yamanaka K., Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J. Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia B., Ke H., Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- 5.Bae W., Xia B., Inouye M., Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki K., Imai R. Pleiotropic roles of cold shock domain proteins in plants. Front. Plant Sci. 2012;2:116. doi: 10.3389/fpls.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlson D., Nakaminami K., Toyomasu T., Imai R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 2002;277:35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- 8.Nakaminami K., Sasaki K., Kajita S., Takeda H., Karlsona D.T., Ohgi K., Imai R. Heat stable ssDNA/RNA-binding activity of a wheat cold shock domain protein. FEBS Lett. 2005;579:4887–4891. doi: 10.1016/j.febslet.2005.07.074. [DOI] [PubMed] [Google Scholar]
- 9.Nakaminami K., Karlson D.T., Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proc. Natl. Acad. Sci. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlson D., Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiol. 2003;131:12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim M.H., Sasaki K., Imai R. Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. J. Biol. Chem. 2009;284:23454–23460. doi: 10.1074/jbc.M109.025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sasaki K., Kim M.-H., R.Imai Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochem. Biophys. Res. Commun. 2007;364:633–638. doi: 10.1016/j.bbrc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 13.Fusaro A.F., Bocca S.N., Ramos R.L.B., Barrôco R.M., Magioli C., Jorge V.C., Coutinho T.C., Rangel-Lima C.M., Rycke R., Inzé D. AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta. 2007;225:1339–1351. doi: 10.1007/s00425-006-0444-4. [DOI] [PubMed] [Google Scholar]
- 14.Kim J.S., Park S.J., Kwak K.J., Kim Y.O., Kim J.Y., Song J., Jang B., Jung C.H., Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Research. 2007;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- 16.Thomashow M.F. Role of cold-responsive genes in plant freezing tolerance. Plant physiology. 1998;118:1–8. doi: 10.1104/pp.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcriptional factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim M.H., Sonoda Y., Sasaki K., Kaminaka H., Imai R. Interactome analysis reveals versatile functions of Arabidopsis COLD SHOCK DOMAIN PROTEIN 3 in RNA processing within the nucleus and cytoplasm. Cell Stress Chaperones. 2013;18:517–525. doi: 10.1007/s12192-012-0398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S.J., Kwak K.J., Oh T.R., Kim Y.O., Kang H. Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 2009;50:869–878. doi: 10.1093/pcp/pcp037. [DOI] [PubMed] [Google Scholar]
- 20.Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell. 2007;19:4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittler R., Kim Y., Song L., Coutu J., Coutu A., Ciftci-Yilmaz S., Lee H., Stevenson B., Zhu J.K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizhsky L., Davletova S., Liang H., Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J. Biol. Chem. 2004;279:11736–11743. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- 23.Cheng M.C., Liao P.M., Kuo W.W., Lin T.P. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S. Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 2012;8:e1002767. doi: 10.1371/journal.pgen.1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castiglioni P., Warner D., Bensen R.J., Anstrom D.C., Harrison J., Stoecker M., Abad M., Kumar G., Salvador S., D’Ordine R., Navarro S., Back S., Fernandes M., Targolli J., Dasgupta S., Bonin C., Luethy M.H., Heard J.E. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 2008;147:446–455. doi: 10.1104/pp.108.118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials for Cold Shock Domain Protein 3 is involved in salt and drought stress tolerance in Arabidopsis.


