Abstract
A method has been developed for collecting air-water interface (AWI) microbes and biofilms that enables analysis of the same sample with various combinations of bright-field and fluorescence light microscopy optics, scanning and transmission electron microscopy (TEM), and atomic force microscopy. The identical sample is then subjected to molecular analysis. The sampling tool consists of a microscope slide supporting appropriate substrates, TEM grids, for example, that are removable for the desired protocols. The slide with its substrates is then coated with a collodion polymer membrane to which in situ AWI organisms adhere upon contact. This sampling device effectively separates the captured AWI bacterial community from the bulk water community immediately subtending. Preliminary data indicate that the AWI community differs significantly from the water column community from the same sample site when both are evaluated with microscopy and with 16S ribosomal DNA sequence-based culture-independent comparisons. This microbe collection method can be used at many levels in research and teaching.
The air-water interface (AWI), while being a ubiquitous feature of all surface water ecosystems, is a very specific, perhaps underreported niche for microorganisms. Every open body of water, large or small, natural or man-made, supports an AWI, the interface between the atmosphere and the hydrosphere. Through this interface occurs a dynamic exchange of gases, water, and organic and inorganic compounds. State and energy transformations occur at the AWI; aerosols form here. The very specific molecular properties that exist at a liquid-gas interface differ from those of the same molecules when they are located beneath the surface in bulk. These properties are observable as surface tension in aquatic systems.
The physical and molecular forces generated at water surfaces where surface tension exists, even taken alone, are a considerable challenge to the integrity of many small life forms. In natural settings, however, the water surface habitat is additionally exposed to all the extreme variations in conditions possible in both the atmospheric and the aquatic environments. Great fluctuations can occur on a moment-by-moment basis in these two different, but fluid, matrices. Notable variables include radiation, temperature, gas and salt concentrations, and mechanical perturbation. In addition, gravity is responsible for the accumulation of high concentrations of small particulates that are heavier than air as well as of substances that are less dense than water. All of these floating substances may become incorporated into the surface microlayers (9). Analysis of sea surface microlayer samples shows that significant accumulations of metals and polychlorinated compounds, as well as of proteins, carbohydrates, lipids, fatty acids, and other organic carbon compounds, occur here. Organic films form at the AWI and stabilize this layer, which is reported by various authors to measure 30 to 300 μm thick. This active and enriched zone is also home to a concentration of microorganisms (24).
The terms “bacterioneuston layer” and “surface microlayers,” among others, have been used to describe the AWI habitat for microorganisms in surface waters to depths specified variously by different authors but within the upper micrometer (for recent reviews, see references 15 and 24). The organisms that by definition reside in the surface biofilm layer and contact both the atmospheric milieu and the aquatic one are those considered for this collection and analysis method, along with any subsurface microorganisms attached to these surface tension inhabitants.
This new method is very simple and has been developed for collecting specifically the microorganisms that survive and apparently thrive (see Fig. 3A) at the AWI (17), as opposed to those dwelling somewhere within the top several millimeters of the water, thus providing a culture-independent analytical tool that also characterizes a subcommunity of an aquatic system. A key element of the collection device is a layer of collodion polymer, for which AWI microbes have a marked affinity, just as they do for many sorts of collection vessels, upon or in which they often become trapped. The specific adhesive properties of this transparent polymer have yet to be elucidated, but its hydrophobicity seems to play a part in its strong binding of bacteria. Prepared as described, collodion from a commercially available solution forms a thin (approximately 60-nm) transparent polymer membrane. This membrane applied to a glass microscope slide makes a sampling tool that can be used both to capture and to observe AWI microbes as they were arrayed in life on the water surface. Even without additional types of analysis, microscopic observation of the preserved in situ arrangement of microbes can offer new insights into community structure and interactions within and among species.
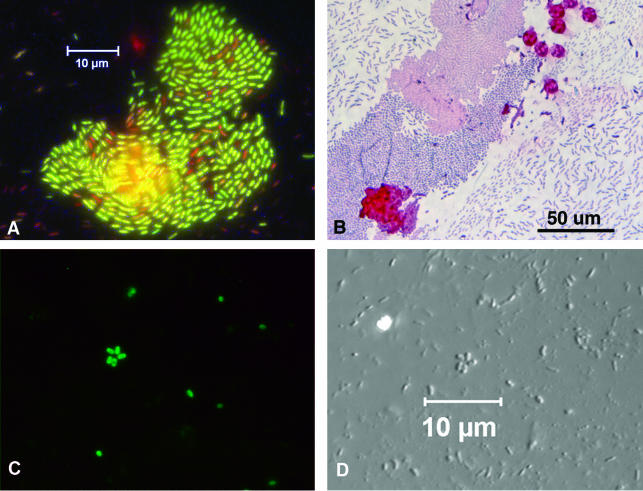
FIG. 3.
Stained AWI collections. (A) Molecular Probes Live/Dead BacLight viability kit staining indicates that most of the bacteria in this freshly collected AWI specimen are intact (green), while some cells are moribund (red). (B) A routine Gram stain does not require the use of a naked flame for heat fixation. Results are easily distinguishable with a ×40 objective. Some algal cells are visible as large red circles. (C and D) Organisms collected from an environmental site viewed with fluorescence optics (C) and with DIC optics (D). The slide was stained with a fluorescein isothiocyanate-conjugated rabbit antiserum.
However, other types of characterization also may be applied correlatively to the same collected sample with the method described here. Additional small substrates for other types of analysis, i.e., supports for transmission electron microscopy (TEM), scanning electron microscopy (SEM), atomic force microscopy (AFM), or paper substrates from which organisms may be detached, are subtended by the slide and held in position by the collodion membrane during the collection process. These can be removed for other purposes, including molecular techniques, either before or after microscopy has been carried out. None of the more than 20 different types of bulk collection devices (for reviews, see references 24 and 15), nor glass slides, nor other microscopic surface collecting devices (7) currently used for sampling surface microlayers simultaneously preserve the in situ arrangement of the AWI microorganisms while allowing for microscopic observation and for correlative molecular analysis. In addition, the commonly used microscopic methods of biofilm collection involve extended periods of immersion or flotation of glass microscope slides or TEM grids, therefore collecting not the existing air-water community considered in this work but a nascent solid-liquid interface biofilm grown on the immersed surface. Although an untreated slide does have a slight affinity for AWI organisms, glass alone does not bind such microbes sufficiently to allow for subsequent treatments, made possible only by the collodion membrane in this work, nor is there the possibility of performing analyses other than light microscopy.
Construction of an AWI collecting slide.
A strainer was constructed by melting several holes in the bottom of a 100- by 15-mm polystyrene disposable petri dish (Sigma Aldrich) with a soldering iron. The culture dish lid was used as a container, into which was placed the strainer, a 70-mm circular sheet of absorbent filter paper, (Whatman no. 1; VWR International), and a clean end-labeled microscope slide, label side up (Super Up-Rite; Richard-Allan Scientific). Fifty milliliters of deionized water was added to completely immerse the slide. Any other desired substrates for processing AWI collections, such as copper TEM grids (Electron Microscopy Sciences), aluminum foil circles for SEM, 10-mm-diameter glass coverslips for AFM, or 10- by 10-mm squares of Whatman no. 1 filter paper tabs for removal of sample to other analyses, were immersed and positioned on the slide surface, leaving an area free for routine light microscopy observation. From a height of about 4 cm, 50 μl of 2% collodion monomer in amyl acetate (Electron Microscopy Sciences) was dropped onto the water surface and allowed to spread and polymerize as a floating membrane. The strainer was slowly lifted straight up out of the water, allowing water to drain out the holes and the collodion membrane to settle on the slide, sandwiching any additional substrates in between. The assembly was left to dry completely for 24 h or more. The collodion-coated slide was scored around the edges to separate it from the filter paper (Fig. 1A). The completed collection slide with its additional substrates (Fig. 1B) was stored in a slide box or other protective container until ready for use. Simple field kits were made by placing two collodion-coated slides back-to-back in 4-oz plastic screw-on lidded specimen cups (Electron Microscopy Sciences) and filling a second cup with water for rinsing the collected AWI biofilm.
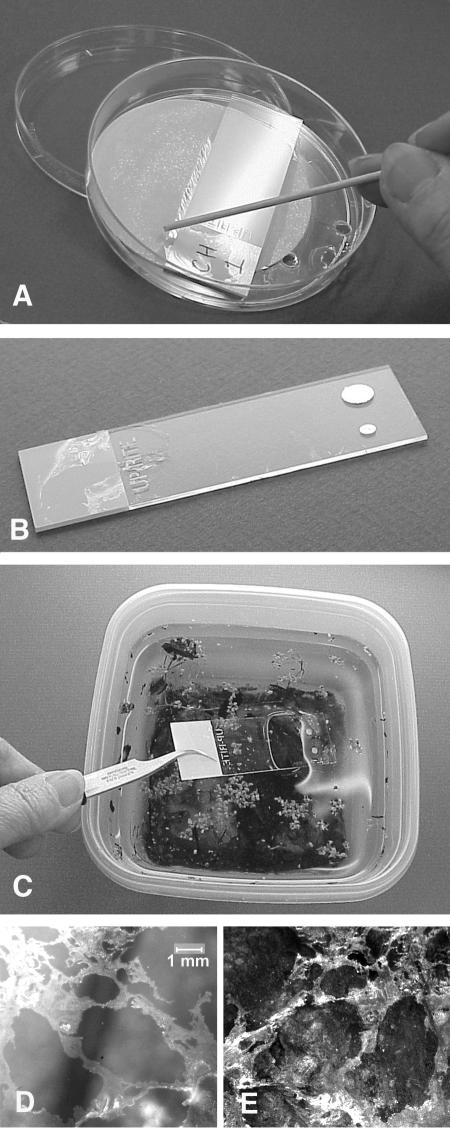
FIG. 1.
Collection kit. (A) Use of an applicator stick to cut the collodion membrane at the edge of the collection slide. The culture dish lid, strainer, and filter paper pad are shown. (B) This collecting slide supports a copper grid for TEM and a paper circle for removal of the collodion polymer and its attached AWI biofilm for molecular biology or other purposes. (C) Preparing to collect an AWI biofilm from the surface of a small ecosystem sample. The collodion surface faces the water surface and will bind AWI microorganisms upon contact. (D) A floating AWI biofilm thick enough to be seen with the naked eye. (E) The same biofilm firmly adhering to the collodion slide upon which it was collected, after the slide was vigorously rinsed in tap water. Note mirror image appearance of collected biofilm.
Collection and analysis of the AWI sample.
The collodion-coated slides were touched horizontally to a desired AWI, immediately and completely immersed, and then immediately withdrawn vertically, all in one motion (Fig. 1C). The slides were rinsed by immersing and withdrawing them vertically one or more times in water (tap, distilled, filtered from source, or as desirable for the particular collection) over a period of several seconds. The AWI biofilm having been effectively transferred from the water surface to the collecting slide (Fig. 1D and E), the back of each slide was wiped dry, and the slides were stored in protective containers. When samples were to be embedded and sectioned for TEM, the paper squares supporting that portion of the sample were immediately removed to a general TEM fixative, where the intact collodion membrane with attached AWI organisms floated free of the filter paper. The membranes were processed by a routine TEM fixation protocol (2) and photographed with a JEOL 100CX TEM. For light microscopy, living organisms were observed (Nikon Microphot with Spot RT digital camera) within 24 h. Dried preparations were observed stained or unstained, and their additional substrates were removed for the various procedures. For fluorescence antibody labeling, dried preparations were stained for 15 min with a 1:100 dilution of fluorescein isothiocyanate-conjugated rabbit antiserum prepared against Vibrio sp. (a gift from the laboratory of R. J. Siebeling) and rinsed and mounted with phosphate-buffered saline. For SEM, aluminum foil circles were attached to SEM specimen stubs, coated with gold-palladium in an Edwards S-150 sputter coater, and observed with a Cambridge 260 SEM. For TEM whole mounts, copper grids were routinely platinum shadowed with a Balzers MED 010 vacuum evaporator or negatively stained on a 50-μl drop of 2% uranyl acetate. For atomic force microscopy, glass coverslips were air dried and observed with a Digital Instruments nanoscope AFM. For molecular analysis, paper squares were immersed in water or buffer to release the collodion film with its attached environmental AWI organisms for isolation of DNA (18). Partial 16S ribosomal DNA (rDNA) sequences were amplified from the total environmental DNA samples by using the primers GM5F-GC-clamp and DS907-reverse described by Teske et al. (23), using touchdown PCR (6). Denaturing gradient gel electrophoresis (DGGE) was performed with 6% polyacrylamide gels using the D-CODE system from Bio-Rad. Material pelleted from a 10-ml sample of bulk water directly subtending the environmental AWI collection site was given treatment identical to that for the collodion-bound organisms.
Adhesive coating.
Collodion, a pyroxylin mixture consisting chiefly of nitrocellulose, is available in solution and has been used for many scientific, commercial, and medical purposes, many of which utilize its ability to form strong but thin membranes. Among its many applications has been its use as a support film for tiny particulates being observed in a TEM. Properties of collodion membranes essential to this technique include its transparency to light and electrons, its intrinsic strength and stability, its various affinities for solid surfaces, and its interactions with water. Its physical properties and chemical structure when dry appear to be particularly attractive to AWI organisms, binding them upon contact, while exhibiting negligible adhesion to subsurface organisms (Fig. 2A). This striking differential affinity emphasizes the special properties of AWI organisms and serves as a means of distinguishing and separating them from water column microbes.
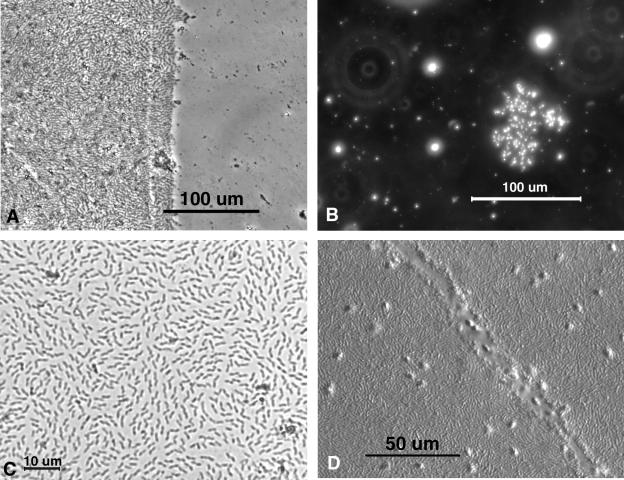
FIG. 2.
Light microscopy of unstained AWI collections. (A) Phase contrast, showing concentration and adhesion differences between AWI biofilm, on the left, and water column organisms, on the right. A temporary aluminum foil cover was removed from the right half of the collection slide after the AWI biofilm had been collected but while it was still immersed in the water sample. A small number of particulates were captured on the immersed collodion on the right as the slide was stirred through the water sample for 30 s. (B) Dark-field image of a small colony that had grown on the surface of tap water that had been boiled 3 days previously. (C) Bright-field image of a monolayer biofilm. Such samples are visible with ×20 objectives without staining. (D) DIC image of a thick biofilm to which living flagellates are adhering. The numerous flagellates, visible as out-of-focus bumps on this thick preparation, move freely about but are not easily removed from the biofilm. The furrow may have been disturbed or grazed by a larger protozoan.
Hundreds of collections performed in the optimization of this method have demonstrated that prokaryotes and other microorganisms are captured in their in situ arrangements upon the water surface (see Fig. 1D and E.) Isolated microcolonies (Fig. 2B) and large monolayer biofilms (Fig. 2C), as well as more complex multitiered biofilms (Fig. 2D), can be stabilized and observed with various modes of microscopy. Light microscopy methods include phase contrast (Fig. 2A), dark field (Fig. 2B), bright field (Fig. 2C), and differential interference contrast (DIC) (Fig. 2D). Since the cells are affixed to the slide, specific fluorescence (Fig. 3A, C, and D) and other staining protocols, such as Gram staining (Fig. 3B), are also possible. When the appropriate specimen support is used with the collection polymer, both TEM (Fig. 4A to C) and SEM (Fig. 4D) can be carried out with their assorted elemental detection devices, and AFM (Fig. 4E) can be utilized for analytical imaging. Several microscopy methods may be utilized in tandem on the same sample collection, and some individual cells or groups of cells may be specifically analyzed by two or more methods (Fig. 5A).
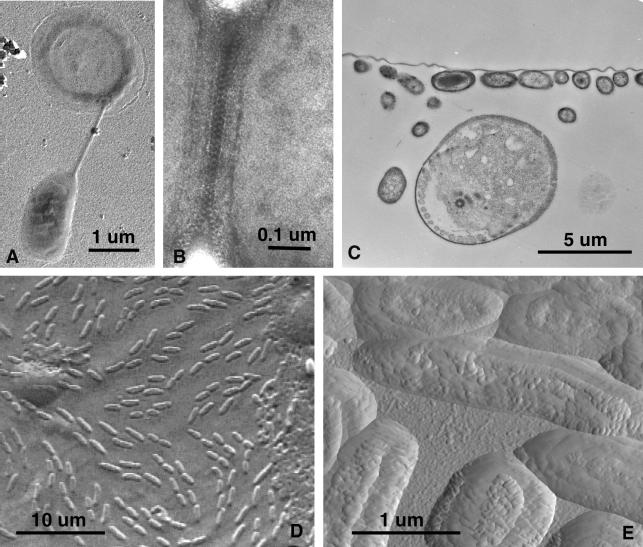
FIG. 4.
TEM, SEM, and AFM of AWI collections. (A) A platinum-shadowed preparation showing a bacterium suspended from a halo-like flotation appendage. (B) A uranyl acetate-negative stain of s-particles at the interface between two prokaryote cells from a tightly packed, raft-like microcolony. (C) The collodion membrane layer with its attached AWI organisms was removed from the slide, fixed, embedded, and sectioned for TEM. The continuous horizontal line is the membrane with attached bacteria, and the large circular object is a section through a protozoan with at least one flagellum. (D) Air-dried SEM sample shows relief image of prokaryotes and various unknown patches in this view of the AWI as seen from beneath the water surface. (E) Atomic force micrograph of the same sample seen in the previous panel.
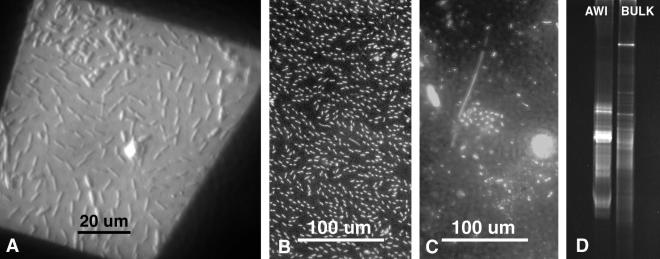
FIG. 5.
Multiple uses of the same AWI collections. (A) A DIC image through the opening in a copper grid that was later observed using TEM. (B) DAPI-stained AWI collection. (C) DAPI-stained organisms filtered from 10 ml of subsurface water directly beneath AWI sample in previous panel. (D) DGGE of rDNA populations collected from AWI and subsurface bulk water organisms from the same samples used in two previous panels. These preliminary results indicate two distinct communities, the AWI probably exhibiting less diversity but higher density of several cell populations.
AWI microorganisms captured on collodion membranes can also be used for molecular analyses. An appropriate release substrate, such as a small piece of filter paper, can underlie the collection membrane. When the paper substrate and its collodion-bound sample are removed from the slide and immersed in an aqueous liquid, the membrane and its tightly bound microbes will float away from the paper, which is then removed (Fig. 4C). The collodion attached to the microbial sample takes up a very small volume and has been shown to have no adverse effect on DNA isolation and subsequent PCR-mediated amplification of 16S rDNA.
The selective affinity of the collection polymer for AWI dwellers makes possible the comparison of organisms from this very specific surface habitat with organisms that inhabit the same body of water beneath the surface. Routine 4′,6′-diamidino-2-phenylindole (DAPI) staining and preliminary correlative DGGE data (Fig. 5B to D) indicate that a freshwater-environment AWI bacterial community was quite distinct from the subsurface community directly beneath it. Other less-specific collection methods for freshwater (11) and marine (16) surface microorganisms have provided qualitative data that have resulted in conclusions similar to those found in the present study but with no correlative microscopic data.
Although the collodion polymer itself provides a considerable affinity for AWI dwellers, its binding properties may be altered or enhanced to provide selective adhesion of other types of organisms as well. The collodion layer of the collection slide can include additional “transparent” adhesive elements, such as antibodies, as previously demonstrated for TEM analysis of collected virus particles (5). Other specific adhesives, such as surface active compounds (14), may also prove to select specific organisms not necessarily affiliated with the AWI in order to perform microscopy and molecular analysis.
Light microscopy applications.
Any routine sort of light microscopy can be done with the collection slides. The polymer membrane does not interfere with observations. The slides should be handled properly, however. The AWI organisms are firmly attached to the slide, but some care should be taken not to scrape off the collodion polymer layer with its attached microbes. If coverslips are applied for observing living organisms or for better optics, the slide should be held collodion side down and brought into contact with a small drop (about 20 μm for a 1- by 1-inch coverslip) of liquid on the coverslip, allowing capillary action to draw the coverslip against the slide. Coverslips can be floated away by immersing the whole slide in a container of water. The microbes remain firmly attached to the collodion layer on the collection slide.
Observation of living samples with light microscopy provides insight into interactions among AWI microorganisms and bulk water dwellers, including grazing activity of small flagellates, amoebae, and even larger invertebrates such as worms, which can sometimes be seen adhering to the collected AWI biofilms. This collection method can be readily applied to grazing studies (e.g., see references 13, 17, and 19).
Staining protocols are facilitated by the adhesion of the sample to the slide; no other matrix is required, and the flame fixation step is unnecessary for bacterial protocols such as Gram staining. The collodion polymer does not interfere with most fluorescence work, and tedious sample adhesion steps can be avoided. Procedures such as autofluorescent chlorophyll determinations, routine DAPI DNA localization, and more complicated determinative staining techniques, such as those involving Molecular Probes Live/Dead BacLight viability kits, are improved. A fluorescently labeled sample can easily be correlated with light microscopy images, since no carbonate filter is present to block substage illumination (Fig. 3C and D), and the fact that the organisms are immobilized facilitates photography, since drifting or Brownian motion are eliminated and longer exposures will not be blurred. The easily obtainable and ideal microscopy samples provided by this method may be able to facilitate many types of bacterial characterization studies (such as those described in references 8, 10, and 14).
This type of microorganism collection also provides an excellent introductory microbiology teaching laboratory opportunity, in that monolayers of bacteria are the ideal microscopy sample; interesting living environmental samples are observed (1); and the time, expense, and hazards of dealing with large concentrations of cultured cells are eliminated while basic protocols, such as Gram staining, are still applied (M. Henk, R. Bridges, J. Enticknap, M. Clements, and F. Rainey, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol., abstr. Q-103, 2002).
Electron microscopy applications.
When this collection device is used, typical sorts of EM processing and observation of AWI samples can be carried out without tedious specimen attachment and dispersal procedures normally required for microbes. When SEM and TEM samples are to be collected, substrates are added to the collection slide under the collodion membrane as described and then removed before or after other analyses as the protocol requires. Samples for SEM and TEM negative staining of whole cells are best removed from the collection slide after the sample has been allowed to dry. Air drying is not destructive for many bacteria, and in SEM preparations the bacteria attached to small aluminum foil substrates are viewed from the under side, since their upper surface is adhered to the collodion membrane, which in turn is attached to the aluminum foil substrate and SEM specimen pin. TEM specimen grids act as substrates for negative staining or metal shadowing, and the collodion membrane and its collected organisms can be easily viewed as whole mounts for observation of many types of extracellular structures as well as for cell-cell connections and interactions. TEM samples that will be fixed, embedded, and sectioned are collected on small paper square substrates under the collodion membrane. These must be removed to a fixative solution immediately after the collection is made, while the AWI organisms are still alive. Vials of fixative can be included in field kits when environmental samples are being taken.
Electron microscopy of all types reveals unusual structural features of AWI microbes and the surrounding surface film, including flotation or attachment appendages and paracrystalline surface-borne particles, as well as other particular characteristics, such as specific affinities for metallic nanoparticles or particular interspecies affiliations (M. C. Henk and B. M. Territo, Abstr. 96th Gen. Meet. Am. Soc. Microbiol., abstr. J-11, 1996). Observations made to date indicate that this collection and analysis method might be applied productively to such varied studies as the transfer of mercury resistance plasmids (e.g., see reference 4) and the elucidation and application of s-layers (e.g., see reference 22) in the growing field of surface chemistry.
AFM applications.
Convenient substrates for AFM protocols can be 10 mm diameter coverslips placed beneath the collodion membrane on the collection slide. These can be removed from the slide and air dried for AFM analysis without any additional treatment. Future AFM analyses may help elucidate the precise nature of physical and molecular properties essential for successful existence at the AWI (e.g., see references 3 and 20). This method also shows promise for application in the burgeoning research fields of water surface dynamics, thin films, and nano-construction, perhaps especially with s-particle subunits (e.g., see reference 22) and surface active compounds (e.g., see reference 14).
Molecular analyses.
This collection method provides the ability to separate subsets of aquatic microbes, i.e., the AWI from the water column organisms. Also demonstrated in this work is the rationale for doing so, in that both the microscopy and the DGGE results indicate the existence of two distinct aquatic bacterial community subsets, the AWI bacteria and the bulk water bacteria, existing within millimeters of each other. This method provides for microscopical analysis of a system subset, e.g., the AWI of an aquatic ecosystem, that may already be under genomic or proteomic investigation. It shows promise in supplying a rich resource of culture-independent information, facilitating studies of environmental gene flow (e.g., see references 10 and 11) and elucidating molecular and other data collected from environmental systems with multiple or unknown variables (e.g., see references 12 and 21).
Both microscopy and molecular analysis provide valuable data for characterization of aquatic microbe populations. When both are used correlatively on the same sample, interpretation of data may be substantially improved. The ultrastructural and biochemical adaptations that enable organisms to survive the harsh and variable conditions existing at the AWI may account for some of the previously unexplained biological or chemical data from surface waters. Although surface water collections may specifically include the AWI, the microbes themselves often have a tendency to adhere firmly to the sides of collection vessels, and depending on the particular collection, subsequent analytical methods may yield puzzling results without representative and correlative microscopy. The present collection and analysis method capitalizes on the “troublesome” adhesive properties of AWI microbes (which often trap them inside or outside of collection containers) to improve characterization of environmental samples.
Development of databases.
Besides providing a useful technique for specific research and teaching applications, the widespread use of this method can be instrumental in the acquisition of images and databases of the organisms in the very specialized AWI niche. The website at http://blc.biolab.udel.edu/, established by Robert C. Hodson at the University of Delaware, included access to a light microscopy image database for AWI collections made by undergraduate microbiology classes. Other useful information may emerge from databases, including elucidation of the role of bacteria in mineral cycling in aquatic ecosystems, development of indicator tests for the existence of other organisms or of specific environmental conditions, patterns of microbial dispersal via aerosols, and the discovery and application of organisms with particular types of useful characteristics.
Acknowledgments
I thank the laboratories of R. J. Siebeling, V. R. Srinivasan, and F. A. Rainey at Louisiana State University for helpful discussions and support. M. Clements prepared DNA extracts, J. Enticknap carried out PCR and DGGE, and R. McCarley and J. Aucoin assisted with AFM.
REFERENCES
- 1.Avant, T. 2002. It's alive. Sci. Teach. 69:42-44.
- 2.Bozzola, J. J., and L. D. Russell. 1999. Electron microscopy: principles and techniques for biologists, 2nd ed., p. 19-22. Jones and Bartlett Publishers, Sudbury, Mass.
- 3.Dahlback, B., M. Hermansson, S. Kjelleberg, and B. Norkrans. 1981. The hydrophobicity of bacteria—an important factor in their initial adhesion at the air-water interface. Arch. Microbiol. 128:267-270. [DOI] [PubMed] [Google Scholar]
- 4.Dahlberg, C., C. Linberg, V. L. Torsvik, and M. Hermansson. 1997. Conjugative plasmids for bacteria in marine environments show various degrees of homology to each other and are not closely related to well-characterized plasmids. Appl. Environ. Microbiol. 63:4692-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrick, K. S., and R. H. Brlansky. 1976. Assay for viruses and mycoplasmas using serologically specific electron microscopy. Phytopathology 66:815-820. [Google Scholar]
- 6.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuerst, J. A., A. McGregor, and M. R. Dickson. 1987. Negative staining of fresh-water bacterioneuston sampled directly with electron-microscope specimen support grids. Microb. Ecol. 13:219-228. [DOI] [PubMed] [Google Scholar]
- 8.Glockner, F. O., H.-D. Babenzein, and R. Amann. 1998. Phylogeny and identification in situ of Nevskia ramosa. Appl. Environ. Microbiol. 64:1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermansson, M. 1990. The dynamics of dissolved and particulate organic material in surface microlayers, p. 145-159. In R. S. Wotton (ed.), The biology of particles in aquatic systems. CRC Press, Boca Raton, Fla.
- 10.Hermansson, M., and Cristel Linberg. 1994. Gene transfer in the marine environment. FEMS Microbiol. Ecol. 15:47-54. [Google Scholar]
- 11.Jones, G. W., L. Baines, and F. J. Genthner. 1991. Heterotrophic bacteria of the freshwater neuston and their ability to act as plasmid recipients under nutrient deprived conditions. Microb. Ecol. 22:15-26. [DOI] [PubMed] [Google Scholar]
- 12.MacGregor, B. J. 1999. Molecular approaches to the study of aquatic microbial communities. Curr. Opin. Biotechnol. 10:220-224. [DOI] [PubMed] [Google Scholar]
- 13.Monger, B. C., M. R. Landry, and S. L. Brown. 1999. Feeding selection of heterotrophic marine nanoflagellates based on the surface hydrophobicity of their picoplankton prey. Limnol. Oceanogr. 44:1917-1927. [Google Scholar]
- 14.Neu, T. 1996. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol. Rev. 60:151-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norkrans, B. 1980. Surface microlayers in aquatic environments, p. 51-85. In M. Alexander (ed.), Advances in microbial ecology, vol. 4. Plenum Press, New York, N.Y. [Google Scholar]
- 16.Plusquellec, A., M. Beucher, C. Lelay, Y. Legal, and J. J. Cleret. 1991. Quantitative and qualitative bacteriology of the marine water-surface microlayer in a sewage-polluted area. Mar. Environ. Res. 31:227-239. [Google Scholar]
- 17.Preston, T. M., H. Richards, and R. S. Wotton. 2001. Locomotion and feeding of Acanthamoeba at the water-air interface of ponds. FEMS Microbiol. Lett. 194:143-147. [DOI] [PubMed] [Google Scholar]
- 18.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 19.Ricci, N., F. Erra, A. Russo, and R. Banchetti. 1991. The air-water-interface—a microhabitat for hypotrictious settlers (Protista, Ciliata). Limnol. Oceanogr. 36:1178-1188. [Google Scholar]
- 20.Schafer, A., H. Harms, and A. J. B. Zehnder. 1998. Bacterial accumulation at the air-water interface. Environ. Sci. Technol. 32:3704-3712. [Google Scholar]
- 21.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sletyr, U. B., M. Sara, D. Pum, and B. Schuster. 2001. Characterization and use of crystalline bacterial cell surface layers. Prog. Surf. Sci. 68:231-278. [Google Scholar]
- 23.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulphate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuev, B. K., V. V. Chudinova, V. V. Kovalenko, and V. V. Yagov. 2001. The conditions of formation of the chemical composition of the sea surface microlayer and techniques for studying organic matter in it. Geochem. Int. 39:702-710. [Google Scholar]