Abstract
Background
Bone marrow plays a key role in bone formation and healing. Although a subset of marrow explants ossifies in vitro without excipient osteoinductive factors, some explants do not undergo ossification. The disparity of outcome suggests a significant heterogeneity in marrow tissue in terms of its capacity to undergo osteogenesis.
Questions/Purposes
We sought to identify: (1) proteins and signaling pathways associated with osteogenesis by contrasting the proteomes of ossified and poorly ossified marrow explants; and (2) temporal changes in proteome and signaling pathways of marrow ossification in the early and late phases of bone formation.
Methods
Explants of marrow were cultured. Media conditioned by ossified (n = 4) and poorly ossified (n = 4) subsets were collected and proteins unique to each group were identified by proteomic analysis. Proteomic data were processed to assess proteins specific to the early phase (Days 1–14) and late phase (Days 15–28) of the culture period. Pathways involved in bone marrow ossification were identified through bioinformatics.
Results
Twenty-eight proteins were unique to ossified samples and eight were unique to poorly ossified ones. Twelve proteins were expressed during the early phase and 15 proteins were specific to the late phase. Several identified pathways corroborated those reported for bone formation in the literature. Immune and inflammatory pathways were specific to ossified samples.
Conclusions
The marrow explant model indicates the inflammatory and immune pathways to be an integral part of the osteogenesis process.
Clinical Relevance
These results align with the clinically reported negative effects of antiinflammatory agents on fracture healing.
Electronic supplementary material
The online version of this article (doi:10.1007/s11999-012-2459-4) contains supplementary material, which is available to authorized users.
Introduction
The dynamics of protein synthesis inside and around the cells constantly change through biochemical interactions with the genome and the cell microenvironment. As a result of the protein diversity created by alternative splicing and posttranslational modifications [48, 78], gene expression-based analyses cannot always be helpful in understanding and characterizing the role and involvement of proteins in complex biological processes.
Proteomics is defined as a large-scale study of the function and structure of the complete set of proteins in a tissue or organism [104]. Therefore, proteomics has become a commonly used research tool for directly assessing protein expression at the posttranslational stage in biological and clinical research [60, 104]. Secretome analysis and quantitative proteomics have enabled the discovery of important signaling dynamics and new bone-related markers, facilitating new therapeutic targets and diagnostic approaches for patients with bone diseases [11, 104]. Proteomic studies have also been used for comparing the protein expression profiles between normal and disease conditions such as bone sarcoma, osteoarthritis, and osteonecrosis [27, 45, 90]. Secretome-based analyses and proteomics were performed on mesenchymal stem cells, osteoblasts, and osteoclasts to understand their roles in the context of bone homeostasis and bone diseases [11, 14]. These studies at the cellular level have been helpful and pivotal in identifying new hormones, growth factors, and cytokines, which regulate the differentiation, growth, maturation, and activity of these cells [9, 10, 17, 20, 32, 61, 87].
Bone homeostasis is a complex process, which involves the interaction of multiple cell types in a complex three-dimensional (3-D) microenvironment. Therefore, a functional in vitro tissue model with a higher degree of complexity is needed to better capture the process and proteome dynamics of the bone formation cascade.
Bone marrow is a rich collection of stem and progenitor cells (hematopoietic and mesenchymal) and other accessory cells derived therefrom [6, 30, 99]. Bone marrow is intimately related with bone homeostasis, bone turnover, and can be associated with bone diseases [28]. Early organ cultures of marrow have demonstrated an inherent osteogenic potential in the absence of osteoinductive factors [30, 31] and in the absence of serum [29]. In a series of articles [29–31], we have demonstrated that the 3-D in vitro cultures of marrow at microliter volumes undergo osteogenesis to form plate-like mineralized structures, which are up to 4 to 5 mm in diameter and up to 150 μm in thickness. These studies demonstrated that bone marrow tissue has a remarkable bone-forming potential in the absence of exogenous osteoinductive factors. Further characterization of extracellular matrix in these ossified marrow explants confirmed bone tissue characteristics (eg, Type I collagen expression, alkaline phosphatase expression, and carbonated apatite crystals) and residency of osteocyte-like cells embedded within the matrix, which stained positive for sclerostin [30], commonly used osteocyte-specific marker. The plate-like bone structures were mechanoresponsive and displayed differences in cytokine expression dynamics in response to mechanical stimulation [31]. Maintaining the spatial integrity of marrow tissue immediately after explantation was critical such that homogenized micropellets of marrow did not display ossification to the same extent as the minimally processed marrow tissue samples [30]. The probability of ossification in this marrow explant model was approximately 50%, in which half of the samples did not undergo ossification at the same level. However, it is unclear which unique proteins and pathways are involved in ossified and poorly ossified marrow samples, and in early and late phases of ossification.
We therefore determined: (1) the comprehensive proteome of marrow explants and the unique proteins in cultures that underwent ossification and that displayed poor ossification; (2) the proteome of marrow ossification in the early phase (Days 1–14) and late phase (Days 15–28); and (3) bone-related pathways and pathway maps based on the list of identified proteins.
Materials and Methods
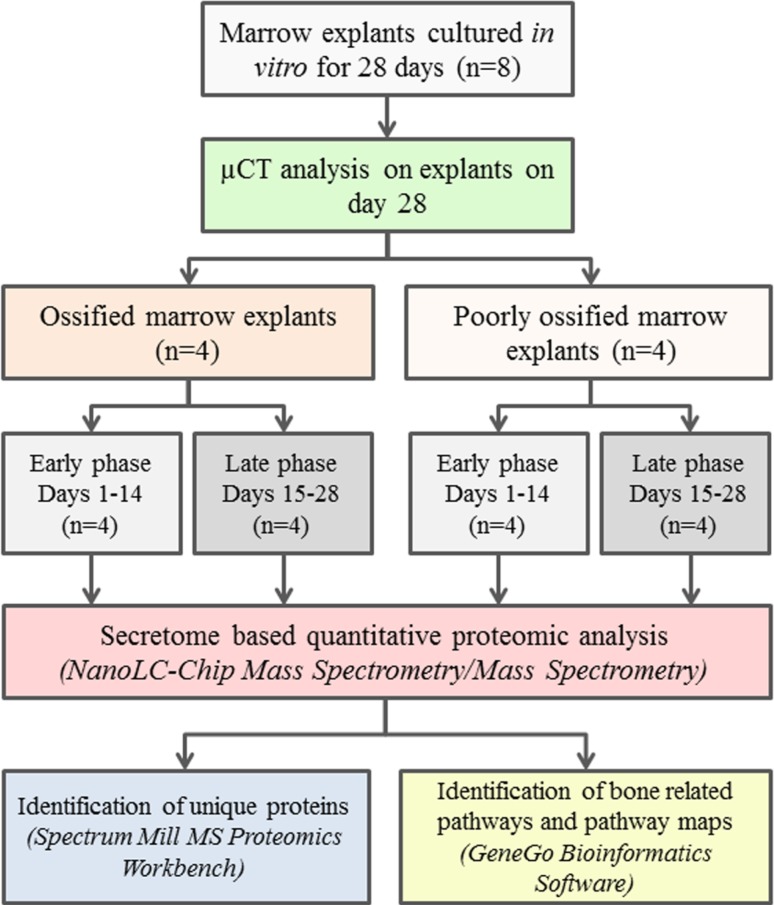
Eight marrow explants from four femurs and four tibiae from two rats were cultured and the conditioned media was collected for up to 28 days. Ossification was confirmed and quantified by microcomputed tomography (μCT) of explants at Day 28 (Fig. 1). Ossification level of the marrow samples indicated that the ossified group had an ossified volume of 0.0556 mm3 (SD: 0.0464, n = 4) and the poorly ossified group had an ossified volume of 0.0029 (SD: 0.0007, n = 4). Based on the ossified volume, samples were grouped as ossified (O) and poorly ossified (PO) (Fig. 1). Conditioned media samples were also classified based on time: Days 1 to 14 were considered early-stage and Days 15 to 28 were considered late-stage groups. The list of proteins involved was identified and presented for the following experimental groups: (1) early-stage media from ossified cultures (O-E); (2) late-stage media from ossified cultures (O-L); (3) early-stage media from poorly ossified cultures (PO-E); (4) late-stage media from poorly ossified cultures (PO-L); (5) media from ossified cultures pooled over all time points (O); (6) media from poorly ossified cultures pooled over all time points (PO); (7) late-stage cultures (including both ossified and poorly ossified samples); and (8) early-stage cultures (including both ossified and poorly ossified samples).
Fig. 1.
Experimental design and data analyses process are presented as a flow chart. Marrow explants were cultured in vitro for 28 days (n = 8). Conditioned culture medium that contained the secreted proteins from the marrow explants were collected (Days 2, 5, 7, 10, 12, 14, 17, 19, 21, 23, 26, and 28) and stored for secretome based quantitative proteomic analysis. At the end of the culture period, ossified and poorly ossified samples were identified and grouped using micro computed tomography (μCT), which quantified the ossified volume. Next, the collected conditioned medium samples were grouped based on early (Days 1–14) and late (Days 15–28) phases. The samples were processed using NanoLC-Chip Mass Spectrometry/Mass Spectrometry to identify the secreted proteins. Unique proteins for ossified and poorly ossified samples and in early and late phases of ossification were identified using Spectrum Mill MS Proteomics Workbench Software. Bone related pathways and pathway maps were identified using Genego Bioinformatics software.
The proteins in the media were digested by trypsin and the resulting peptides were subjected to NanoLC-Chip Mass Spectrometry/Mass Spectrometry (MS/MS) (Agilent Technologies, Santa Clara, CA, USA). MS/MS analysis was followed by targeted MS/MS. We analyzed the resulting spectra acquired on NanoLC-Chip-MS/MS using Spectrum Mill A.03.02.060 software (Agilent Technologies) and searches were performed against the National Institutes of Health National Center for Biotechnology Information protein database. To explore biological functionality, data were analyzed using GeneGo (GeneGo Bioinformatics Software, Inc, St Joseph, MI, USA). GeneGo is a data-mining tool that facilitates analysis of biological pathways for high-throughput data sets.
Whole bone marrow was isolated from both femurs and tibiae of two 80- to 90-day-old male Long-Evans rats (approved by Purdue Animal Care and Use Committee) using a previously described method for bone marrow extraction and explant culturing [29]. Briefly, diaphyseal bone marrow was extracted with a centrifugation-based technique that minimizes manual handling and transfer of bony fragments from the donor bone tissue into the culture environment. Marrow tissue was cultured on inserts with porous PET membranes (0.4-μm pore size Transwell; Corning, Corning, NY, USA) by applying marrow samples at 7 μL (7 mm3) volume using a micropipette. Standardization of the starting volume of samples as such eliminated the need to normalize the final ossified tissue volume to original explant volume. The serum-free growth medium was modified from Lennon et al. [53] and composed of 60% DMEM, 40% MCDB-201 supplemented with 1% ITS + 1 (Sigma, St Louis, MO, USA), 50 μg/mL ascorbic acid, 5 mM Na-β-glycerophosphate, 3.5 mg/mL glucose, 40 U/mL penicillin and 40 μg/mL streptomycin, and 1.5 μg/mL fungizone. No osteoinductive factors (eg, dexamethasone, bone morphogenetic protein-2) were added into the culture media at any point in time. We cultured the explants for 28 days and media was replenished on Days 2, 5, 7, 10, 12, 14, 17, 19, 21, 23, 26, and 28. The medium conditioned by bone marrow explants was collected from each ossifying explant before each fresh media addition and stored in sterile low protein-binding tubes (LoBind, Eppendorf, Germany) separately. The samples were stored at −80°C for secretome-based proteomic analysis, which was performed at the end of the experiment collectively. Repeated freezing and thawing of the collected conditioned media was eliminated with appropriate aliquoting.
At the end of the culture period, marrow explants were fixed with 10% formalin and kept in the fixative. The ossified volume of the marrow explants was quantified by μCT (SCANCO Medical AG, μCT 40, Brüttisellen, Switzerland) with a 16-μm voxel resolution (I = 145 μA, E = 55 kVp, integration time = 200 ms). The data sets were reconstructed and analyzed with commercial software (SCANCO evaluation software) and the segmentation parameters of 0.8 (Sigma), 1 (support), and 100 (threshold) were used [63, 67, 73]. The total ossified volume (BV, mm3) was used to determine the level of ossification and to differentiate between ossified and poorly ossified samples.
The conditioned medium samples (100 μL) were dessicated and resuspended in 20 μL 0.1 M HEPES at pH = 8.0 (Applied Biosystems, Foster City, CA, USA). Samples containing buffer were denatured with 10 μL of 2% SDS (Applied Biosystems), reduced with 50 mM tris-(2-carboxyethyl)phosphine (Applied Biosystems), and alkylated with 1 μL 200 mM methyl methanethiosulfonate (Applied Biosystems). We added trypsin (5 μL of 1 μg/μL 5% w/w; Applied Biosystems) and samples were incubated at 37°C overnight. Finally, 2.5 μL of 10% trifluoroacetic acid was added and samples were incubated at 37°C for 30 minutes and then spun at 13,000 × g for 10 minutes. We removed the supernatant and applied it to a C18 microspin column (Nest Group, Southborough, MA, USA) for buffer exchange followed by a G25 Sephedex column (Nest Group) for desalting. The resulting peptides were dried down and resuspended in 100 μL 0.01% trifluoroacetic acid in water.
Trypsin-digested proteins were separated using a NanoLC-Chip system (1100 Series LC equipped with HPLC Chip interface; Agilent, Santa Clara, CA, USA). After injection of 0.5 μg, the peptides were concentrated on the on-chip 300SB-C18 enrichment column and washed with 5% acetonitrile, 0.01% trifluoroacetic acid at a flow rate of 4 μL/min for 5 minutes. The enrichment column was switched into the nanoflow path and further separated with the on-chip C-18 reversed-phase ZORBAX 300SB-C18 (0.75 μm × 150 mm; Agilent) analytical column coupled to the electrospray ionization source of the ion trap mass spectrometer (XCT Plus; Agilent). The column was eluted with a 55-minute linear gradient from 5% to 35% buffer B (100% acetonitrile, 0.01% trifluoroacetic acid) at a rate of 300 nL/min followed by a 10-minute gradient from 35% to 100% buffer B. The column was reequilibrated with an isocratic flow (5% buffer B) at 300 nL/min. The system was controlled by ChemStation software (Agilent). We acquired NanoLC-MS chromatograms in positive ion mode under the following conditions: a capillary voltage of 1850 V and an end plate offset of 500 V. The dry temperature was set at 300°C. Dry gas flow was maintained at 6 L/min. Acquisition range was 350 to 2000 m/z with 0.15-second maximum accumulation time and scan speed of 8100 m/z per second.
To obtain the comprehensive list of proteins involved in each sample group (Fig. 1), raw MS data were exported to mzXML format using Bruker’s CompassXport program and uploaded to the Purdue’s Proteomics Discovery Pipeline (Purdue University, West Lafayette, IN, USA) bioinformatics infrastructure. The Proteomics Discovery Pipeline is a multistep web-based proteomics analysis software suite. It consists of the individual tools, XMass, XAlign, and XNormalization and various other tools used for statistical analysis. An initial list of peaks was obtained using XMass, a deconvolution tool that helps differentiate genuine peaks from experimental noise. Parameters for XMass included: minimum LC peak width (number of scans): six; retention time range: 5 to 55 minutes; and M/Z variation between isotopes: 0.5. Peaks from the same molecule that were present in multiple samples were aligned using XAlign. Parameters for XAlign were: M/Z variation: 0.8; retention time variation: 0.7; and LC peak width (number of scans): 60. We normalized aligned data using XNormalization in preparation for statistical analysis. It was specified that a peak must be present in 80% of the samples to be included in the statistical tests.
To identify the unique proteins involved in each sample group, five Student’s t-tests were run using Pipeline’s built-in statistical tools. The groups for the five t-tests were: (1) all poorly ossified and ossified; (2) poorly ossified early and poorly ossified late; (3) ossified early and ossified late; (4) poorly ossified early and ossified early; and (5) poorly ossified late and ossified late. Results from the statistical tests were further filtered by removing peaks with a p value of > 0.05 and a fold change of < 2 or > 0.5. For the remaining peaks in each group, we calculated average retention times and m/z values. Finally, the peaks in each test were separated into two groups. The first group consisted of peaks that were present in both groups of the statistical test and the second group consisted of peaks that were present in only one group of the statistical test. The fold changes and the p values of the statistical tests were reported.
Trypsin-digested peptides were separated on a NanoLC-Chip system (1100 Series LC equipped with HPLC Chip interface; Agilent) using the same platform as described previously. We acquired automated MS/MS spectra during the run in the data-dependent acquisition mode with the selection of the three most abundant precursor ions (0.5 minute active exclusion; 2 + ions preferred). If the peptides associated with our peaks of interest (ie, the substantially large peaks in the spectra) were not identified as a result of low abundance, then the samples were rerun using the same parameters except the specific mass associated with the peaks of interest was selected for preferentially or targeted MS/MS analysis.
MS/MS data were analyzed using Spectrum Mill A.03.02.060 software (Agilent Technologies) and searches were performed against the National Institutes of Health National Center for Biotechnology Information (NCBInr_09, www.ncbi.nlm.nih.gov/Entrez) protein database. Raw MS/MS data were extracted into Spectrum Mill with the mass range for precursor ions set to 300 to 4000 Da. To accelerate the search, we selected HUMAN RODENT as the species setting to narrow the searchable species to homo sapiens, mus musculus, and rattus norvegicus. The parameters of the search were as follows: no more than two tryptic miscleavages allowed, cysteine searched as S-methylmethane thiolsulfonate, 1.0 Da peptide mass tolerance, and 0.7 Da MS/MS mass tolerance. Only peptides with a score of 6 or higher and score peak intensity of 60% or higher were considered true-positives.
To link differentially expressed peptides identified in MS-only mode with specific proteins, we matched peaks from the MS findings with data generated by the shotgun MS/MS based on the methods in the literature [56]. The retention time and m/z value were used as unique identifiers of each peak (these values were averaged in the MS data). The matching process consisted of identifying all of the peaks from the MS/MS data that fell within a range of ± 1-minute retention time and ± 0.5 m/z from the MS peaks.
A targeted MS/MS analysis was conducted to increase the number of differentially expressed peptides that matched with named proteins. Eighty peaks were selected from the list of unmatched peptides. These peaks included the twenty most highly expressed ossified and poorly ossified peaks. Peaks with overlapping retention times were omitted to increase the accuracy of our analysis.
To explore biological functionality, we analyzed data using GeneGo (GeneGo Bioinformatics Software, Inc). GeneGo is a data-mining tool that facilitates analysis of biological pathways for high-throughput data sets. The proteins in the data set were split into eight different groups depending on the pattern of upregulation: ossified early, ossified late, poorly ossified early, poorly ossified late, all ossified, all poorly ossified, all early, and all late. These eight groups were uploaded to GeneGo as eight separate files. Ontology enrichment analyses were run to search our data set against the GO Processes database, GeneGo Process Networks, and GeneGo Pathway Maps. Parameters were as follows: p value threshold, 0.05; threshold, 0; and signals, both. A GO process is a set of events or operations with a definite starting and end point relating to the function of cells, tissues, organs, and organisms. A GeneGo pathway map is a prebuilt functional ontology that focuses on consensus multistep canonical pathways. In other words, the GeneGo pathway maps database (www.genego.com/mapsearch.php) is a collection of pathways that was compiled from the literature, including metabolic pathways (eg, glycogen metabolism), regulatory pathways (eg, apoptotic tumor necrosis factor family pathways), or disease process pathways (eg, angiotensin signaling through PYK2). Each step in a pathway map is well defined in the public domain or validated by GeneGo scientists. A GeneGo process network can be defined as a comprehensive biological process with a specific functional theme. A process network is a broad concept that consists of all the pathways associated with a process and all the documented connections between genes/proteins. A process network can consist of several pathway maps. An example process network might be inflammation, whereas the pathway maps might be effects of lipoxin on neutrophil migration. The process network connects all the intracellular processes and is not confined to what happens in a single cell. A network may contain more or less information than a pathway, depending on the parameters used when creating the network.
Results
Proteomic profiling indicated 28 proteins specific to the marrow cultures that ossified, of which 12 were specific to only ossified early group, 15 specific to ossified late group, and one specific to both early and late groups (Table 1). On the other hand, five proteins were specific to the poorly ossified early group, three proteins to the poorly ossified late group, and none specific to both poorly ossified early and late groups (Table 1). Sixty-seven proteins were identified with a fold change of at least ± 2 and p < 0.05, and the list of proteins expressed in each group is included in Table 2. Several of these included apolilipoprotein L, Zinc-finger proteins 17 and 34, tumor necrosis factor receptor superfamily, ADAM metallopeptidase with thrombospondin Type 1 motif, collagen XII, and Complement factor H (Table 2). Among these proteins, 26 of them were directly related to bone homeostasis or bone marrow, supported by studies in the literature (Table 3).
Table 1.
Number of proteins specifically expressed in individual groups
| Sample type | Total | Early only | Late only | Early and late |
|---|---|---|---|---|
| Ossified samples | 28 | 12 | 15 | 1 |
| Poorly ossified samples | 8 | 5 | 3 | 0 |
Table 2.
List of identified proteins with a fold change of at least ± 2 and p < 0.05*
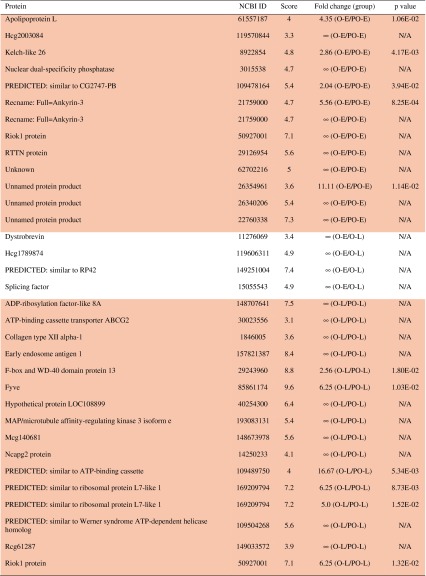
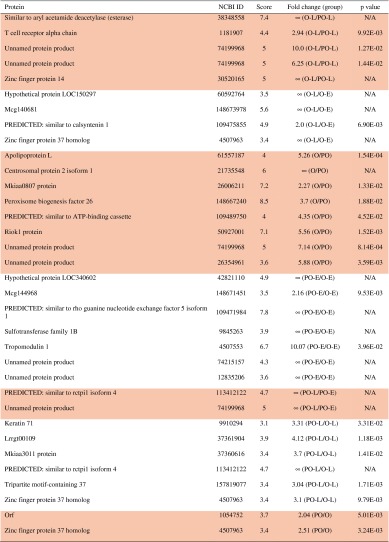
* Peptides with a single protein match. Each row (delineated by shading) represents an individual peptide that matched with a single protein. Proteins were grouped according to expression pattern (indicated by shaded background). The ∞ symbol indicates that a protein was present in the numerator of the group and absent in the denominator, eg, ∞ (O-E/O-L), indicates that a protein was present in ossified early and absent in ossified late); O-E = ossified early; O-L = ossified late; PO-E = poorly ossified early; PO-L = poorly ossified late; O = ossified total; PO = poorly ossified total, in which O signifies the ossified marrow explants, PO signifies the poorly ossified marrow explants, early stands for the samples collected in Days 1–14, and late stands for the samples collected in Days 15–28. O or PO, when used alone, signifies the samples collected for all days (1–28) for the ossified or poorly ossified groups. N/A = not available.
Table 3.
Bone and bone marrow related proteins identified as the result of proteomic analysis performed on the secretome of marrow explants*
| Protein name | NCBI ID | Overexpressed group | References | Function |
|---|---|---|---|---|
| ADAM metallopeptidase with thrombospondin type 1 motif, 13 | 21265043 | O-L | [55] | Identified in cultures of rat osteoblasts treated with molecules known to drive osteoblast differentiation; shown to accumulate around osteoblast extracellular matrix during differentiation |
| Ankyrin-3 | 21759000 | O-E | [37] | Present in bone marrow-derived macrophages |
| Apob protein | 71051035 | O-E | [62] | Has been associated with development of nontraumatic osteonecrosis |
| Apolipoprotein L | 61557187 | O-E, O-L, O | [75] | May play a role in osteoarthritis mechanism |
| ATP-binding cassette transporter ABCG2 | 30023556 | O-L | [81] | Shown to be the sole molecular determinant of the SP phenotype in bone marrow cells |
| Collagen XII | 1846005 | O-L | [72] | Association with Collagen 1 modifies interaction with surrounding matrix; shown to be upregulated in response to mechanical stress |
| Complement factor H | 77861917 | O-E, O-L | [19] | Complexes with bone sialoprotein and osteopontin |
| Dimethylarginine dimethylaminohydrolases 2 | 148694731 | O-E, O-L | [33, 47, 52] | Increases vascular endothelial growth factor in endothelial cells; has been associated with alterations of bone homeostasis |
| Dr1 associated protein 1 (negative cofactor 2 alpha) | 148701178 | O-L | [25, 26] | Expressed in bone and bone marrow; abundantly expressed in CD34+ cells |
| E2F transcription factor 3 | 4503433 | O-E | [66, 91] | Highly expressed in proliferating mesenchymal cells; plays a role in apoptosis |
| Early endosome antigen 1 | 157821387 | O-L | [16, 51, 96, 105] | Effector of Rab5C protein, which is implicated in resorbing osteoclasts |
| Fyve | 85861174 | O-L | [39, 69] | Expressed in bone marrow; involved in leukocyte signaling |
| Low density lipoprotein receptor-related protein 1 | 124494256 | PO | [43, 70, 102] | Plays an important role in the uptake of postprandial lipoproteins and vitamin K1 by osteoblasts; mediates mitogenic effect of lactoferrin in osteoblasts |
| MAP/microtubule affinity-regulating kinase 3 isoform e | 193083131 | O-L | [88] | Associated with bone mineral density |
| mKIAA0807 protein | 26006211 | O | † | Differentially expressed in CD34+ cells |
| Nuclear dual-specificity phosphatase | 3015538 | O-E | [7, 8] | Gene family involved in control of MAP kinase function |
| PREDICTED: calsyntenin-1 | 109475855 | O-L | [21] | Found to be expressed in bone, bone marrow, and connective tissue |
| PREDICTED: similar to heterogeneous nuclear ribonucleoprotein A1 | 88958985 | O-E | [40] | Expressed in bone and bone marrow; shown to play a role in myelopoiesis |
| T cell receptor alpha chain | 1181907 | O-L | [2] | Expressed in mesenchymal cells; implicated in bone loss |
| Toll-like receptor 2 | 38454274 | O-E, PO-L | [4, 98] | Affects osteoclast differentiation and activity; implicated as a negative regulator of osteoclastogenesis |
| Tumor necrosis factor receptor superfamily | 6678383 | O-E, O-L | [3] | Implicated in many diseases of the bone such as familial expansile osteolysis, familial Paget disease of bone, and osteopetrosis |
| Ubiquitin-conjugating enzyme E2B | 149052524 | O-E, PO-L | [84] | Upregulated in tendon tears |
| Unknown protein | 62702216 | O-E | ‡ | Involved in zinc ion binding, proteolysis, and metalloendopeptidase activity |
| Vasodilator-stimulated phosphoprotein | 11414808 | PO-L, PO-E | [103] | Identified as having altered expression in osteogenic induced human mesenchymal stem cells |
| Zinc finger protein 14 | 30520165 | O-L | [23] | Known to be involved in growth of skeletal elements (bone, teeth, and cartilage) |
| Zinc finger protein 37 | 4507963 | O-L, PO-L, PO | [23] | Known to be involved in growth of skeletal elements (bone, teeth, and cartilage) |
* Differentially expressed proteins identified by name and NCBI number; † http://oai.dtic.mil/oai/oai?verb=getRecord&metadataPrefix=html&identifier=ADA446682; ‡ http://www.uniprot.org/uniprot/Q53S40; O-E = ossified early; O-L = ossified late; PO-E = poorly ossified early; PO-L = poorly ossified late; O = ossified total; PO = poorly ossified total, in which O signifies the ossified marrow explants, PO signifies the poorly ossified marrow explants, early stands for the samples collected in Days 1–14, and late stands for the samples collected in Days 15–28. O or PO, when used alone, signifies the samples collected for all days (1–28) for the ossified or poorly ossified groups.
Based on the proteins present, eight bone-related pathways (Table 4) and four bone-related pathway maps (Table 5) were identified by GeneGO analysis. We identified a number of peptides with duplicate protein matches (Supplemental Table 1), which were not included in pathway and network analysis. In addition to the bone-related pathways, and pathway maps presented in Tables 4 and 5, a comprehensive list of other processes, process networks, and pathway maps were identified: GeneGO Processes (Supplemental Table 2), GeneGo Process Networks (Supplemental Table 3), and GeneGo Pathway Maps (Supplemental Table 4).
Table 4.
Bone-related pathways and GeneGO process networks from our data set that have previously been studied in relation to bone and bone formation*
| Pathways and networks | References |
|---|---|
| Apoptosis—apoptosis stimulation by external signals | [54, 79, 106] |
| Cell adhesion—integrin-mediated cell-matrix adhesion | [35] |
| Inflammation—interferon gamma signaling | [24] |
| Development—neurogenesis | [13, 42, 58] |
| Cardiac development—Wnt beta-catenin, Notch, VEGF, IP3, and integrin signaling | [22, 36, 68, 74, 76, 77, 85, 89, 95, 101, 102] |
| Immune response—interleukin-5 signaling | [59, 92] |
| Immune—phagocytosis | [64, 71, 100] |
| Chemotaxis | [15, 83] |
* References to relevant literature are included; VEGF = vascular endothelial growth factor.
Table 5.
Bone-related GeneGo pathway maps*
| GeneGo pathway maps | References |
|---|---|
| Immune response—interleukin-6 signaling pathway | [5, 44, 93, 94] |
| Immune response—Toll-like receptor signaling pathways | [86] |
| Transport—clathrin-coated vesicle cycle | [65] |
| Development—growth hormone signaling through STATs and PLC/IP3 | [46] |
* Pathway maps from our data set that have previously been studied in relation to bone and bone formation. References to relevant literature are included.
Discussion
Bone marrow plays a major role in bone formation, regeneration, and healing and inherently ossifies in vitro without addition of excipient osteoinductive factors. Proteome of marrow ossification would have major implications in understanding the proteomic mechanisms of bone formation, regeneration, and healing. We studied the proteome of marrow ossification using quantitative secretome-based proteomics. We compared the proteome of ossified and poorly ossified marrow samples cultured under equivalent conditions. We further analyzed and compared the expression of secreted proteins from rat marrow explants during ossification in two phases: early phase (Days 1–14) and late phase (Days 15–28). Based on the list of proteins, we identified pathways and pathway maps closely relating to bone biology.
Our study is subject to certain limitations. First, the marrow samples were obtained from rats and therefore may present differences with human marrow. The use of rat marrow samples may have provided a more homogenous composition as a result of the smaller size of the medullary cavity, which is substantially larger in humans. It has been reported that human marrow tissue also displays ossification potential in vitro [57]. However, future studies are needed to explore the inherent ossification potential of human marrow tissue, its characteristics, and proteome. Second, the study analyzed the secreted proteins from the same samples over time. Therefore, the proteins contained within the cells or bound to the matrix synthesized by cells are not generally secreted and were not included in the analyses. Although secretome-based proteomic analyses have been used in studying biological systems [11, 14], to have a complete picture of the marrow ossification process, future studies are needed, which include the proteins in cells.
We have identified unique proteins in ossified and poorly ossified marrow samples (Table 1). We observed the expression of both ADAMTS and APOL1 proteins increases in ossified marrow samples. More specifically, ADAMTS was overexpressed late in the ossified marrow (O-L) samples suggesting active osteogenic differentiation resulting in bone formation. APOL1 was overexpressed in both O-E and O-L samples. ADAMTS is overexpressed during osteoblast differentiation [41]. The overexpression of E2F transcription factor early in ossified marrow samples (O-E) indicates that it may be involved in regulating various processes such as the apoptosis of hematopoietic cells and differentiation of marrow stromal cells (MSCs) toward the bone lineage. E2F transcription factor 3 reportedly regulates the expression of genes involved in proliferation, differentiation, and apoptosis [26]. We have previously reported that although the hematopoietic cells decrease in number, fraction of MSCs is maintained in the marrow explant culture system [30]. We observed that dimethylarginine dimethylaminohydrolases (DDAH 2) was overexpressed both early and late in ossified marrow samples. DDAH 2 reportedly upregulates the expression of vascular endothelial growth factor (VEGF) in endothelial cells [33]. On the other hand, VEGF is not only an angiogenic factor, but it is involved in osteogenesis [38, 82]. The overexpression of DDAH2 together with the evidence of the involvement of VEGF-related pathway suggests that the in vitro marrow explant culture system may be a useful tool to study bone vascularization by using cocultures of ossifying marrow explants with endothelial cells. A coculture model involving bone marrow explants and endothelial cells may further augment VEGF expression and reveal intrinsic details about the complex process of bone vascularization. The key difference between the envisioned in vitro marrow model of vascularization and existing in vitro models [18, 49, 80] is that existing models generally use single populations of cells (generally osteoblasts or stromal cells) with endothelial cells. Although such models provide a greater degree of control, the marrow model ossifies to form bone-like plates and it provides a more heterogeneous population of cells that would reflect the complexity of the in vivo setting in a Petri dish. Such diversity may elicit forms of in vitro vasculogenesis that may not be observed in coculture of two cell populations. On the other hand, the marrow model would still be more complex than culture of two cell types, and interpretation of the results would be more challenging.
The pathway and network analysis helped identify several key bone-related pathways that may be involved in the ossification process of the marrow explants. One of the key pathways we found involved in the ossified marrow samples is the notch signaling pathway (Table 4). Induction of notch signaling in human bone marrow stromal cells (hBMSCs) through transduction of lentiviral vectors containing human notch1 intracellular domain (NICD) enhances the osteogenic differentiation of hBMSCs while inhibiting adipogenesis [24]. It is likely that notch signaling regulated the osteogenic differentiation of MSCs in ossified marrow samples. Moreover, notch signaling is reportedly involved in the maintenance of MSC progenitor population [58] as observed in marrow explant cultures [30]. Wnt signaling pathway involved in ossification of bone marrow explants (Table 4) plays a dominant role in bone development/repair [41], bone metabolism, and regulating osteoblast proliferation, function, and survival [50]. Ossified marrow samples also displayed interleukin-5 signaling (Table 4). It has been widely accepted that proinflammatory factors generally tend to favor bone formation [12, 34], whereas antiinflammatory factors (such as nonsteroidal antiinflammatory drugs [NSAIDs]) impair fracture healing [1, 97]. The association between inflammatory factors and bone formation is believed to stem from the initial presence of inflammatory cells after injury and proinflammatory environment promotes the recruitment of cells for repair and revascularization. IL-5, as identified in the current study, has not been reported extensively within the context of osteogenesis. However, there is one study that investigated a transgenic mouse model overexpressing IL-5 and reported heterotopic ossification in the spleen as well as increased bone formation in long bones [59]. This is in agreement with our observation of the presence of an IL-5-associated pathway in ossified marrow population. Analysis of pathways specific to bone-forming cultures indicated apoptosis stimulation by external signals, phagocytosis, neurogenesis (axonal guidance), cardiac development (Wnt beta-catenin), Notch, VEGF, IP3 and integrin signaling, cell adhesion (integrin-mediated cell-matrix adhesion), immune response (IL-5 signaling), inflammation (interferon-gamma signaling), and development (neurogenesis) in general (Table 4). At the pathway map level, bone-forming marrow cultures displayed immune response (interleukin-6 signaling pathway, TLR signaling pathways), transport (Clathrin-coated vesicle cycle), and development (growth hormone signaling through STATs and PLC/IP3) (Table 5). On the other hand, there were no pathways identified in the ossified late group, which suggests that the pathway database is not comprehensive for bone formation-related pathways.
The ossification process in 3-D marrow cultures is a highly complex process that involves en masse apoptosis of the hematopoietic population and a surge in the mesenchymal compartment. The most important finding of our study was that those marrow cultures that ossified displayed immune and inflammatory pathway maps more specifically than those that did not ossify. Because the model involved disruption of the status quo (ie, isolation of marrow sitting in the confines of bone and disruption of vascular networks), it becomes clear that groundwork for osteogenesis is set by immune and inflammatory cells. The model could be particularly relevant to investigating the underlying mechanisms of bone formation after trauma or surgery. The model and the platform developed here can also be used to assess the osteogenic potential of bone marrow samples obtained from patients to evaluate the risk of bone diseases such as osteoporosis. Future studies will use immune and inflammatory blockers to assess their effect on the level of marrow ossification.
Electronic supplementary material
Acknowledgments
We thank David VanSickle PhD, DVM for insightful discussions on bone marrow ossification model and Pamela Lachik for help with the μCT system.
Footnotes
This study was funded by a grant from the Musculoskeletal Transplant Foundation (OA).
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research editors and board members are on file with the publication and can be viewed on request.
Contributor Information
Umut Atakan Gurkan, Email: ugurkan@rics.bwh.harvard.edu.
Ozan Akkus, Email: ozan.akkus@case.edu.
References
- 1.Aspenberg P. Drugs and fracture repair. Acta Orthop. 2005;76:741–748. doi: 10.1080/17453670510045318. [DOI] [PubMed] [Google Scholar]
- 2.Baker PJ, Garneau J, Howe L, Roopenian DC. T-cell contributions to alveolar bone loss in response to oral infection with Porphyromonas gingivalis. Acta Odontol Scand. 2001;59:222–225. doi: 10.1080/00016350152509247. [DOI] [PubMed] [Google Scholar]
- 3.Bao B-Y, Lin VC, Huang S-H, Pao J-B, Chang T-Y, Lu T-L, Lan Y-H, Chen L-M, Ting W-C, Yang W-H, Hsieh C-J, Huang S-P. Clinical significance of tumor necrosis factor receptor superfamily member 11b polymorphism in prostate cancer. Ann Surg Oncol. 2010;17:1675–1681. doi: 10.1245/s10434-010-0994-3. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Shavit Z. Taking a toll on the bones: regulation of bone metabolism by innate immune regulators. Autoimmunity. 2008;41:195–203. doi: 10.1080/08916930701694469. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard F, Duplomb L, Baud’huin M, Brounais B. The dual role of IL-6-type cytokines on bone remodeling and bone tumors. Cytokine Growth Factor Rev. 2009;20:19–28. doi: 10.1016/j.cytogfr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Bruder SP, Fink DJ, Caplan AI. Mesenchymal stem-cells in bone-development, bone repair, and skeletal regeneration therapy. J Cell Biochem. 1994;56:283–294. doi: 10.1002/jcb.240560303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camps M, Nichols A, Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 2000;14:6–16. [PubMed] [Google Scholar]
- 8.Carlson J, Cui W, Zhang Q, Xu X, Mercan F, Bennett AM, Vignery A. Role of MKP-1 in osteoclasts and bone homeostasis. Am J Pathol. 2009;175:1564–1573. doi: 10.2353/ajpath.2009.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celebi B, Elcin AE, Elcin YM. Proteome analysis of rat bone marrow mesenchymal stem cell differentiation. J Proteome Res. 2010;9:5217–5227. doi: 10.1021/pr100506u. [DOI] [PubMed] [Google Scholar]
- 10.Celebi B, Elcin YM. Proteome analysis of rat bone marrow mesenchymal stem cell subcultures. J Proteome Res. 2009;8:2164–2172. doi: 10.1021/pr800590g. [DOI] [PubMed] [Google Scholar]
- 11.Chiellini C, Cochet O, Negroni L, Samson M, Poggi M, Ailhaud G, Alessi MC, Dani C, Amri EZ. Characterization of human mesenchymal stem cell secretome at early steps of adipocyte and osteoblast differentiation. BMC Mol. Biol. 2008;9. [DOI] [PMC free article] [PubMed]
- 12.Cho HH, Shin KK, Kim YJ, Song JS, Kim JM, Bae YC, Kim CD, Jung JS. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J Cell Physiol. 2010;223:168–177. doi: 10.1002/jcp.22024. [DOI] [PubMed] [Google Scholar]
- 13.Cho MH, Lee JH, Ahn HH, Lee JY, Kim ES, Kang YM, Min BH, Kim JH, Lee HB, Kim MS. Induction of neurogenesis in rat bone marrow mesenchymal stem cells using purine structure-based compounds. Mol Biosyst. 2009;5:609–611. doi: 10.1039/b905598n. [DOI] [PubMed] [Google Scholar]
- 14.Choi YA, Lim J, Kim KM, Acharya B, Cho JY, Bae YC, Shin HI, Kim SY, Park EK. Secretome analysis of human BMSCs and identification of SMOC1 as an important ECM protein in osteoblast differentiation. J. Proteome Res. 2010;9:2946–2956. doi: 10.1021/pr901110q. [DOI] [PubMed] [Google Scholar]
- 15.Cooper CR, Sikes RA, Nicholson BE, Sun Y-X, Pienta KJ, Taichman RS. Cancer cells homing to bone: the significance of chemotaxis and cell adhesion. Cancer Treat Res. 2004;118:291–309. doi: 10.1007/978-1-4419-9129-4_12. [DOI] [PubMed] [Google Scholar]
- 16.Coxon FP, Taylor A. Vesicular trafficking in osteoclasts. Semin Cell Dev Biol. 2008;19:424–433. doi: 10.1016/j.semcdb.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Czupalla C, Mansukoski H, Riedl T, Thiel D, Krause E, Hoflack B. Proteomic analysis of lysosomal acid hydrolases secreted by osteoclasts—implications for lytic enzyme transport and bone metabolism. Mol Cell Proteomics. 2006;5:134–143. doi: 10.1074/mcp.M500291-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Dohle E, Fuchs S, Kolbe M, Hofmann A, Schmidt H, Kirkpatrick CJ. Comparative study assessing effects of sonic hedgehog and VEGF in a human co-culture model for bone vascularisation strategies. Eur Cell Mater. 2011;21:144–156. doi: 10.22203/ecm.v021a12. [DOI] [PubMed] [Google Scholar]
- 19.Fedarko NS, Fohr B, Robey PG, Young MF, Fisher LW. Factor H binding to bone sialoprotein and osteopontin enables tumor cell evasion of complement-mediated attack. J Biol Chem. 2000;275:16666–16672. doi: 10.1074/jbc.M001123200. [DOI] [PubMed] [Google Scholar]
- 20.Foster LJ, Zeemann PA, Li C, Mann M, Jensen ON, Kassem M. Differential expression profiling of membrane proteins by quantitative proteomics in a human mesenchymal stem cell line undergoing osteoblast differentiation. Stem Cells. 2005;23:1367–1377. doi: 10.1634/stemcells.2004-0372. [DOI] [PubMed] [Google Scholar]
- 21.Fung S, Wang F, Chase M, Godt D, Hartenstein V. Expression profile of the cadherin family in the developing Drosophila brain. J Comp Neurol. 2008;506:469–488. doi: 10.1002/cne.21539. [DOI] [PubMed] [Google Scholar]
- 22.Galli C, Passeri G, Ravanetti F, Elezi E, Pedrazzoni M, Macaluso GM. Rough surface topography enhances the activation of Wnt/beta-catenin signaling in mesenchymal cells. J Biomed Mater Res A. 2010;95:682–690. doi: 10.1002/jbm.a.32887. [DOI] [PubMed] [Google Scholar]
- 23.Ganss B, Jheon A. Zinc finger transcription factors in skeletal development. Crit Rev Oral Biol Med. 2004;15:282–297. doi: 10.1177/154411130401500504. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y, Grassi F, Ryan MR, Terauchi M, Page K, Yang X, Weitzmann MN, Pacifici R. IFN-gamma stimulates osteoclast formation and bone loss in vivo via antigen-driven T cell activation. J Clin Invest. 2007;117:122–132. doi: 10.1172/JCI30074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes I, Sharma TT, Mahmud N, Kapp JD, Edassery S, Fulton N, Liang J, Hoffman R, Westbrook CA. Highly abundant genes in the transcriptosome of human and baboon CD34 antigen-positive bone marrow cells. Blood. 2001;98:93–99. doi: 10.1182/blood.V98.1.93. [DOI] [PubMed] [Google Scholar]
- 26.Gu J, Zhang QH, Huang QH, Ren SX, Wu XY, Ye M, Huang CH, Fu G, Zhou J, Niu C, Han ZG, Chen SJ, Chen Z. Gene expression in CD34(+) cells from normal bone marrow and leukemic origins. Hematol J. 2000;1:206–217. doi: 10.1038/sj.thj.6200020. [DOI] [PubMed] [Google Scholar]
- 27.Guo DM, Tan WF, Wang F, Lv Z, Hu J, Lv TR, Chen Q, Gu XY, Wan B, Zhang ZN. Proteomic analysis of human articular cartilage: identification of differentially expressed proteins in knee osteoarthritis. Joint Bone Spine. 2008;75:439–444. doi: 10.1016/j.jbspin.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Gurkan UA, Akkus O. The mechanical environment of bone marrow: a review. Ann. Biomed. Eng. 2008;36:1978–1991. doi: 10.1007/s10439-008-9577-x. [DOI] [PubMed] [Google Scholar]
- 29.Gurkan UA, Gargac J, Akkus O. The sequential production profiles of growth factors and their relations to bone volume in ossifying bone marrow explants. Tissue Eng Part A. 2010;16:2295–2306. doi: 10.1089/ten.tea.2009.0565. [DOI] [PubMed] [Google Scholar]
- 30.Gurkan UA, Kishore V, Condon KW, Bellido TM, Akkus O. A scaffold-free multicellular three-dimensional in vitro model of osteogenesis. Calcif Tissue Int. 2011;88:388–401. doi: 10.1007/s00223-011-9467-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gurkan UA, Krueger A, Akkus O. Ossifying bone marrow explant culture as a three-dimensional mechanoresponsive in vitro model of osteogenesis. Tissue Eng Part A. 2011;17:417–428. doi: 10.1089/ten.tea.2010.0193. [DOI] [PubMed] [Google Scholar]
- 32.Ha BG, Hong JM, Park J-Y, Ha M-H, Kim T-H, Choi J-Y, Ryoo H-M, Choi J-Y, Shi H-I, Chun SY, Kim S-Y, Park EK. Proteomic profile of osteoclast membrane proteins: identification of Na +/H + exchanger domain containing 2 and its role in osteoclast fusion. Proteomics. 2008;8:2625–2639. doi: 10.1002/pmic.200701192. [DOI] [PubMed] [Google Scholar]
- 33.Hasegawa K, Wakino S, Tanaka T, Kimoto M, Tatematsu S, Kanda T, Yoshioka K, Homma K, Sugano N, Kurabayashi M, Saruta T, Hayashi K. Dimethylarginine dimethylaminohydrolase 2 increases vascular endothelial growth factor expression through Sp1 transcription factor in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:1488–1494. doi: 10.1161/01.ATV.0000219615.88323.b4. [DOI] [PubMed] [Google Scholar]
- 34.Hess K, Ushmorov A, Fiedler J, Brenner RE, Wirth T. TNFalpha promotes osteogenic differentiation of human mesenchymal stem cells by triggering the NF-kappaB signaling pathway. Bone. 2009;45:367–376. doi: 10.1016/j.bone.2009.04.252. [DOI] [PubMed] [Google Scholar]
- 35.Hidalgo-Bastida LA, Cartmell SH. Mesenchymal stem cells, osteoblasts and extracellular matrix proteins: enhancing cell adhesion and differentiation for bone tissue engineering. Tissue Eng Part B Rev. 2010;16:405–412. doi: 10.1089/ten.teb.2009.0714. [DOI] [PubMed] [Google Scholar]
- 36.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoock TC, Peters LL, Lux SE. Isoforms of ankyrin-3 that lack the NH2-terminal repeats associate with mouse macrophage lysosomes. J Cell Biol. 1997;136:1059–1070. doi: 10.1083/jcb.136.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Z, Nelson ER, Smith RL, Goodman SB. The sequential expression profiles of growth factors from osteoprogenitors [correction of osteroprogenitors] to osteoblasts in vitro. Tissue Eng. 2007;13:2311–2320. doi: 10.1089/ten.2006.0423. [DOI] [PubMed] [Google Scholar]
- 39.Huber C, Martensson A, Bokoch GM, Nemazee D, Gavin AL. FGD2, a CDC42-specific exchange factor expressed by antigen-presenting cells, localizes to early endosomes and active membrane ruffles. J Biol Chem. 2008;283:34002–34012. doi: 10.1074/jbc.M803957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iervolino A, Santilli G, Trotta R, Guerzoni C, Cesi V, Bergamaschi A, Gambacorti-Passerini C, Calabretta B, Perrotti D. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol Cell Biol. 2002;22:2255–2266. doi: 10.1128/MCB.22.7.2255-2266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson ML, Kamel MA. The Wnt signaling pathway and bone metabolism. Curr Opin Rheumatol. 2007;19:376–382. doi: 10.1097/BOR.0b013e32816e06f9. [DOI] [PubMed] [Google Scholar]
- 42.Jones CM, Lyons KM, Hogan BLM. Involvement of bone morphogenetic protein-4 (BMP-4) and VGR-1 in morphogenesis and neurogenesis in the mouse. Development. 1991;111:531. doi: 10.1242/dev.111.2.531. [DOI] [PubMed] [Google Scholar]
- 43.Jones KS, Bluck LJC, Wang LY, Stephen AM, Prynne CJ, Coward WA. The effect of different meals on the absorption of stable isotope-labelled phylloquinone. Br J Nutr. 2009;102:1195–1202. doi: 10.1017/S0007114509371688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kastelan D, Kastelan M, Massari LP, Korsic M. Possible association of psoriasis and reduced bone mineral density due to increased TNF-alpha and IL-6 concentrations. Med Hypotheses. 2006;67:1403–1405. doi: 10.1016/j.mehy.2006.04.069. [DOI] [PubMed] [Google Scholar]
- 45.Kawai A, Kondo T, Suehara Y, Kikuta K, Hirohashi S. Global protein-expression analysis of bone and soft tissue sarcomas. Clin Orthop Relat Res. 2008;466:2099–2106. doi: 10.1007/s11999-008-0330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly PA, Finidori J, Moulin S, Kedzia C, Binart N. Growth hormone receptor signalling and actions in bone growth. Horm Res. 2001;55:14–17. doi: 10.1159/000063467. [DOI] [PubMed] [Google Scholar]
- 47.Kielstein JT, Zoccali C. Asymmetric dimethylarginine: a cardiovascular risk factor and a uremic toxin coming of age? Am J Kidney Dis. 2005;46:186–202. doi: 10.1053/j.ajkd.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 48.Kiernan UA. Quantitation of target proteins and post-translational modifications in affinity-based proteomics approaches. Expert Rev Proteomics. 2007;4:421–428. doi: 10.1586/14789450.4.3.421. [DOI] [PubMed] [Google Scholar]
- 49.Kirkpatrick CJ, Fuchs S, Unger RE. Co-culture systems for vascularization—learning from nature. Adv Drug Deliv Rev. 2011;63:291–299. doi: 10.1016/j.addr.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Kubota T, Michigami T, Ozono K. Wnt signaling in bone metabolism. J Bone Miner Metab. 2009;27:265–271. doi: 10.1007/s00774-009-0064-8. [DOI] [PubMed] [Google Scholar]
- 51.Lawe DC, Sitouah N, Hayes S, Chawla A, Virbasius JV, Tuft R, Fogarty K, Lifshitz L, Lambright D, Corvera S. Essential role of Ca2 +/calmodulin in early endosome antigen-1 localization. Mol Biol Cell. 2003;14:2935–2945. doi: 10.1091/mbc.E02-09-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leiper JM, Maria JS, Chubb A, MacAllister RJ, Charles IG, Whitley GSJ, Vallance P. Identification of two human dimethylarginine dimethylaminohydrolases with distinct tissue distributions and homology with microbial arginine deiminases. Biochem J. 1999;343:209–214. doi: 10.1042/0264-6021:3430209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lennon DP, Haynesworth SE, Young RG, Dennis JE, Caplan AI. A chemically-defined medium supports in-vitro proliferation and maintains the osteochondral potential of rat marrow-derived mesenchymal stem-cells. Exp Cell Res. 1995;219:211–222. doi: 10.1006/excr.1995.1221. [DOI] [PubMed] [Google Scholar]
- 54.Li G, Dickson GR, Marsh DR, Simpson H. Rapid new bone tissue remodeling during distraction osteogenesis is associated with apoptosis. J Orthop Res. 2003;21:28–35. doi: 10.1016/S0736-0266(02)00097-9. [DOI] [PubMed] [Google Scholar]
- 55.Lind T, McKie N, Wendel M, Racey SN, Birch MA. The hyalectan degrading ADAMTS-1 enzyme is expressed by osteoblasts and up-regulated at regions of new bone formation. Bone. 2005;36:408–417. doi: 10.1016/j.bone.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 56.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 57.Luria EA, Owen M, Friedenstein AJ, Morris J, Kuznetsow SA, Joyner C. Bone formation in organ culture of marrow pieces. Cell Tissue Res. 1986;7:313. doi: 10.1007/BF00218212. [DOI] [PubMed] [Google Scholar]
- 58.Machold RP, Kittell DJ, Fishell GJ. Antagonism between Notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2007;2:5. doi: 10.1186/1749-8104-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macias MP, Fitzpatrick LA, Brenneise I, McGarry MP, Lee JJ, Lee NA. Expression of IL-5 alters bone metabolism and induces ossification of the spleen in transgenic mice. J Clin Invest. 2001;107:949–959. doi: 10.1172/JCI11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 61.Mareddy S, Broadbent J, Crawford R, Xiao Y. Proteomic profiling of distinct clonal populations of bone marrow mesenchymal stem cells. J Cell Biochem. 2009;106:776–786. doi: 10.1002/jcb.22088. [DOI] [PubMed] [Google Scholar]
- 62.Miyanishi K, Yamamoto T, Irisa T, Noguchi Y, Sugioka Y, Iwamoto Y. Increased level of apolipoprotein B/apolipoprotein A1 ratio as a potential risk for osteonecrosis. Ann Rheum Dis. 1999;58:514–516. doi: 10.1136/ard.58.8.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morgan EF, Mason ZD, Chien KB, Pfeiffer AJ, Barnes GL, Einhorn TA, Gerstenfeld LC. Micro-computed tomography assessment of fracture healing: relationships among callus structure, composition, and mechanical function. Bone. 2009;44:335–344. doi: 10.1016/j.bone.2008.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mukae H, Hogg JC, English D, Vincent R, Van Eeden SF. Phagocytosis of particulate air pollutants by human alveolar macrophages stimulates the bone marrow. Am J Physiol Lung Cell Mol Physiol. 2000;279:L924–L931. doi: 10.1152/ajplung.2000.279.5.L924. [DOI] [PubMed] [Google Scholar]
- 65.Mukherjee A, Rotwein P. Akt promotes BMP2-mediated osteoblast differentiation and bone development. J Cell Sci. 2009;122:716–726. doi: 10.1242/jcs.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner JD, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller R, Van Campenhout H, Van Damme B, Van Der Perre G, Dequeker J, Hildebrand T, Ruegsegger P. Morphometric analysis of human bone biopsies: a quantitative structural comparison of histological sections and micro-computed tomography. Bone. 1998;23:59–66. doi: 10.1016/S8756-3282(98)00068-4. [DOI] [PubMed] [Google Scholar]
- 68.Nagayama M, Iwamoto M, Hargett A, Kamiya N, Tamamura Y, Young B, Morrison T, Takeuchi H, Pacifici M, Enomoto-Iwamoto M, Koyama E. Wnt/beta-catenin signaling regulates cranial base development and growth. J Dent Res. 2008;87:244–249. doi: 10.1177/154405910808700309. [DOI] [PubMed] [Google Scholar]
- 69.Nakanishi H, Takai Y. Frabin and other related Cdc42-specific guanine nucleotide exchange factors couple the actin cytoskeleton with the plasma membrane. J Cell Mol Med. 2008;12:1169–1176. doi: 10.1111/j.1582-4934.2008.00345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naot D, Grey A, Reid IR, Cornish J. Lactoferrin—a novel bone growth factor. Clin Med Res. 2005;3:93–101. doi: 10.3121/cmr.3.2.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neale SD, Haynes DR, Howie DW, Murray DW, Athanasou NA. The effect of particle phagocytosis and metallic wear particles on osteoclast formation and bone resorption in vitro. J Arthroplasty. 2000;15:654–662. doi: 10.1054/arth.2000.6622. [DOI] [PubMed] [Google Scholar]
- 72.Nemoto T, Kajiya H, Tsuzuki T, Takahashi Y, Okabe K. Differential induction of collagens by mechanical stress in human periodontal ligament cells. Arch Oral Biol. 2010;55:981–987. doi: 10.1016/j.archoralbio.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 73.Oest ME, Dupont KM, Kong HJ, Mooney DJ, Guldberg RE. Quantitative assessment of scaffold and growth factor-mediated repair of critically sized bone defects. J Orthop Res. 2007;25:941–950. doi: 10.1002/jor.20372. [DOI] [PubMed] [Google Scholar]
- 74.Oh I-H. Microenvironmental targeting of Wnt/beta-catenin signals for hematopoietic stem cell regulation. Expert Opin Biol Ther. 2010;10:1315–1329. doi: 10.1517/14712598.2010.504705. [DOI] [PubMed] [Google Scholar]
- 75.Okabe T, Ohmori Y, Tanigami A, Hishigaki H, Suzuki Y, Sugano S, Kawaguchi A, Nakaya H, Wakitani S. Detection of gene expression in synovium of patients with osteoarthritis using a random sequencing method. Acta Orthop. 2007;78:687–692. doi: 10.1080/17453670710014400. [DOI] [PubMed] [Google Scholar]
- 76.Oldershaw RA, Hardingham TE. Notch signaling during chondrogenesis of human bone marrow stem cells. Bone. 2010;46:286–293. doi: 10.1016/j.bone.2009.04.242. [DOI] [PubMed] [Google Scholar]
- 77.Oldershaw RA, Tew SR, Russell AM, Meade K, Hawkins R, McKay TR, Brennan KR, Hardingham TE. Notch signaling through jagged-1 is necessary to initiate chondrogenesis in human bone marrow stromal cells but must be switched off to complete chondrogenesis. Stem Cells. 2008;26:666–674. doi: 10.1634/stemcells.2007-0806. [DOI] [PubMed] [Google Scholar]
- 78.Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, Sugano S. Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol Cell Proteomics. 2007;6:1000–1006. doi: 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- 79.Palumbo C, Ferretti M, De Pol A. Apoptosis during intramernbranous ossification. J Anat. 2003;203:589–598. doi: 10.1046/j.1469-7580.2003.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Papadimitropoulos A, Scherberich A, Guven S, Theilgaard N, Crooijmans HJ, Santini F, Scheffler K, Zallone A, Martin I. A 3D in vitro bone organ model using human progenitor cells. Eur Cell Mater. 2011;21:445–458. doi: 10.22203/ecm.v021a33. [DOI] [PubMed] [Google Scholar]
- 81.Pfister O, Oikonomopoulos A, Sereti K-I, Sohn RL, Cullen D, Fine GC, Mouquet F, Westerman K, Liao R. Role of the ATP-binding cassette transporter Abcg2 in the phenotype and function of cardiac side population cells. Circ Res. 2008;103:825-U110. [DOI] [PMC free article] [PubMed]
- 82.Pufe T, Wildemann B, Petersen W, Mentlein R, Raschke M, Schmidmaier G. Quantitative measurement of the splice variants 120 and 164 of the angiogenic peptide vascular endothelial growth factor in the time flow of fracture healing: a study in the rat. Cell Tissue Res. 2002;309:387–392. doi: 10.1007/s00441-002-0605-0. [DOI] [PubMed] [Google Scholar]
- 83.Rabinovitch M, Destefano MJ. In vitro chemotaxis of mouse bone marrow neutrophils. Proc Soc Exp Biol Med. 1978;158:170–173. doi: 10.3181/00379727-158-40164. [DOI] [PubMed] [Google Scholar]
- 84.Schmutz S, Fuchs T, Regenfelder F, Steinmann P, Zumstein M, Fuchs B. Expression of atrophy mRNA relates to tendon tear size in supraspinatus muscle. Clin Orthop Relat Res. 2009;467:457–464. doi: 10.1007/s11999-008-0565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Silkstone D, Hong H, Alman BA. beta-Catenin in the race to fracture repair: in it to Wnt. Nat Clin Pract Rheumatol. 2008;4:413–419. doi: 10.1038/ncprheum0838. [DOI] [PubMed] [Google Scholar]
- 86.Sioud M, Floisand Y. TLR agonists induce the differentiation of human bone marrow CD34(+) progenitors into CD11c(+) CD80/86(+) DC capable of inducing a Th1-type response. Eur J Immunol. 2007;37:2834–2846. doi: 10.1002/eji.200737112. [DOI] [PubMed] [Google Scholar]
- 87.Spreafico A, Frediani B, Capperucci C, Chellini F, Paffetti A, D’Ambrosio C, Bernardini G, Mini R, Collodel G, Scaloni A, Marcolongo R, Santucci A. A proteomic study on human osteoblastic cells proliferation and differentiation. Proteomics. 2006;6:3520–3532. doi: 10.1002/pmic.200500858. [DOI] [PubMed] [Google Scholar]
- 88.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Snorradottir S, Center JR, Nguyen TV, Alexandersen P, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 89.Takahashi-Yanaga F, Sasaguri T. The Wnt/beta-catenin signaling pathway as a target in drug discovery. J Pharmacol Sci. 2007;104:293–302. doi: 10.1254/jphs.CR0070024. [DOI] [PubMed] [Google Scholar]
- 90.Tan XY, Cai DZ, Wu YL, Liu B, Rong LM, Chen ZS, Zhao QC. Comparative analysis of serum proteomes: discovery of proteins associated with osteonecrosis of the femoral head. Transl Res. 2006;148:114–119. doi: 10.1016/j.trsl.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 91.Towers M, Fisunov G, Tickle C. Expression of E2F transcription factor family genes during chick wing development. Gene Expr Patterns. 2009;9:528–531. doi: 10.1016/j.gep.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 92.Toyosaki-Maeda T, Takano H, Tomita T, Tsuruta Y, Maeda-Tanimura M, Shimaoka Y, Takahashi T, Itoh T, Suzuki R, Ochi T. Differentiation of monocytes into multinucleated giant bone-resorbing cells: two-step differentiation induced by nurse-like cells and cytokines. Arthritis Res. 2001;3:306–310. doi: 10.1186/ar320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tsangari H, Findlay DM, Kuliwaba JS, Atkins GJ, Fazzalari NL. Increased expression of IL-6 and RANK mRNA in human trabecular bone from fragility fracture of the femoral neck. Bone. 2004;35:334–342. doi: 10.1016/j.bone.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 94.Tsangari H, Findlay DM, Zannettino ACW, Pan B, Kuliwaba JS, Fazzalari NL. Evidence for reduced bone formation surface relative to bone resorption surface in female femoral fragility fracture patients. Bone. 2006;39:1226–1235. doi: 10.1016/j.bone.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 95.Ugarte F, Ryser M, Thieme S, Fierro FA, Navratiel K, Bornhaeuser M, Brenner S. Notch signaling enhances osteogenic differentiation while inhibiting adipogenesis in primary human bone marrow stromal cells. Exp Hematol. 2009;37:867–875. doi: 10.1016/j.exphem.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 96.Vaaraniemi J, Halleen JM, Kaarlonen K, Ylipahkala H, Alatalo SL, Andersson G, Kaija H, Vihko P, Vaananen HK. Intracellular machinery for matrix degradation in bone-resorbing osteoclasts. J Bone Miner Res. 2004;19:1432–1440. doi: 10.1359/JBMR.040603. [DOI] [PubMed] [Google Scholar]
- 97.Vuolteenaho K, Moilanen T, Moilanen E. Non-steroidal anti-inflammatory drugs, cyclooxygenase-2 and the bone healing process. Basic Clin Pharmacol Toxicol. 2008;102:10–14. doi: 10.1111/j.1742-7843.2007.00149.x. [DOI] [PubMed] [Google Scholar]
- 98.Walsh MC, Kim N, Kadono Y, Rho J, Lee SY, Lorenzo J, Choi Y. Osteoimmunology: interplay between the immune system and bone metabolism. Ann Rev Immunol. 2006;24:33–63. doi: 10.1146/annurev.immunol.24.021605.090646. [DOI] [PubMed] [Google Scholar]
- 99.Wan C, He Q, McCaigue M, Marsh D, Li G. Nonadherent cell population of human marrow culture is a complementary source of mesenchymal stem cells (MSCs) J Orthop Res. 2006;24:21–28. doi: 10.1002/jor.20023. [DOI] [PubMed] [Google Scholar]
- 100.Wang W, Ferguson DJP, Quinn JMW, Simpson A, Athanasou NA. Osteoclasts are capable of particle phagocytosis and bone resorption. J Pathol. 1997;182:92–98. doi: 10.1002/(SICI)1096-9896(199705)182:1<92::AID-PATH813>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 101.Weber JM, Calvi LM. Notch signaling and the bone marrow hematopoietic stem cell niche. Bone. 2010;46:281–285. doi: 10.1016/j.bone.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 103.Zhang AX, Yu WH, Ma BF, Yu XB, Mao FF, Liu W, Zhang JQ, Zhang XM, Li SN, Li MT, Lahn BT, Xiang AP. Proteomic identification of differently expressed proteins responsible for osteoblast differentiation from human mesenchymal stem cells. Mol Cell Biochem. 2007;304:167–179. doi: 10.1007/s11010-007-9497-3. [DOI] [PubMed] [Google Scholar]
- 104.Zhang HW, Recker R, Lee WNP, Xiao GGS. Proteomics in bone research. Expert Rev Proteomics. 2010;7:103–111. doi: 10.1586/epr.09.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao HB, Ettala O, Vaananen HK. Intracellular membrane trafficking pathways in bone-resorbing osteoclasts revealed by cloning and subcellular localization studies of small GTP-binding rab proteins. Biochem Biophys Res Commun. 2002;293:1060–1065. doi: 10.1016/S0006-291X(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 106.Zhu H, Miosge N, Schulz J, Schliephake H. Regulation of multilineage gene expression and apoptosis during in vitro expansion of human bone marrow stromal cells with different cell culture media. Cells Tissues Organs. 2010;192:211–220. doi: 10.1159/000313417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.