Abstract
A PCR-restriction fragment length polymorphism (RFLP) method was developed in order to screen a large number of strains for impaired adhesion to epithelial cells due to expression of truncated InlA. inlA polymorphism was analyzed by PCR-RFLP in order to correlate inlA PCR-RFLP profiles and production of truncated InlA. Thirty-seven Listeria monocytogenes strains isolated from various sources, including five noninvasive and two invasive reference strains, were screened. Two endonucleases (AluI and Tsp509I) were used, and they generated five composite profiles. Thirteen L. monocytogenes isolates were characterized by two specific PCR-RFLP profiles similar to PCR-RFLP profiles of noninvasive reference strains previously described as strains that produce truncated InlA. Ten of the 13 isolates showed low abilities to invade human epithelial Caco-2 cells. However, 4 of the 13 isolates were able to invade Caco-2 cells like reference strains containing complete InlA. Sequencing of inlA and Western blot analysis confirmed that truncated InlA was expressed in the 10 L. monocytogenes strains which were isolated from food. This PCR-RFLP method allowed us to identify 10 new strains expressing a truncated internalin. Based on the results obtained in this study, the PCR-RFLP method seems to be an interesting method for rapidly screening L. monocytogenes strains deficient in the ability to invade Caco-2 cells when a sizeable number of strains are studied.
Listeria monocytogenes, a facultative intracellular pathogen, is widespread in the environment (1, 24, 25). This bacterium can contaminate processed foods, and several studies have established that listeriosis is an important food-borne infection (14, 21). Moreover, L. monocytogenes can be isolated from the gastrointestinal tracts of healthy persons (9, 23). It has been estimated that between 1 to 6% of the general population carry this bacterium (4, 11, 13, 22). Recently, variable capacities of human carriage isolates of L. monocytogenes to invade human cell cultures were observed. In fact, Jonquières et al. (10) reported that L. monocytogenes human carriage isolate LO28 entered a fibroblast line expressing L-CAM (the chicken homolog of E-cadherin) poorly and produced a truncated form of the protein InlA (63 kDa), an internalin implicated in entry into host cells, while virulent and invasive strains of L. monocytogenes produced an 80-kDa InlA. In the same study, Jonquières et al. (10) also described one clinical and three food L. monocytogenes isolates expressing a truncated InlA. In a recent study, Olier et al. (18, 19) reported that several L. monocytogenes human carriage isolates were attenuated for virulence, were affected in the ability to invade Caco-2 cells, and also produced truncated InlA (47 kDa). Sequence analysis of inlA revealed that point mutations were responsible of production of the truncated InlA and that there were polymorphisms in inlA (10, 19, 20). In this paper, we describe a PCR-restriction fragment length polymorphism (RFLP) method based on inlA polymorphism for rapidly screening potentially noninvasive L. monocytogenes strains when a sizeable number of strains are examined. Furthermore, we present evidence concerning the occurrence of potentially noninvasive L. monocytogenes.
MATERIALS AND METHODS
Bacterial strains.
The L. monocytogenes isolates used are listed in Table 1. Nine L. monocytogenes human fecal carriage isolates (H2, H6, H11, H12, H27, H28, H31, H35, and H38), three isolates from sporadic human listeriosis (H4, H21, and H22), three isolates from food-processing facilities (1E, 3E, and 6E), an isolate from compost (C9), six food isolates from brine (1S, 2S and 3S) and cheese (1F, 3F, and 7F), and four rook fecal carriage isolates (23, 38, 81, and 97) were obtained from the strain collection of Laboratoire de Microbiologie UMR 1232, Dijon, France. Four isolates from meat (NV4, NV5, NV7, and NV8) were provided by the Laboratoire départemental de la Haute Vienne, Limoges, France. Strain Scott A was obtained from the collection of Institut Pasteur, Paris, France, and strains LO28 and EGD-e were kindly provided by P. Cossart, Institut Pasteur, Paris, France. Strains Scott A and EGD-e were used as virulent reference strains for comparative analysis. Human fecal carriage isolates H1, H17, H32, and H34 (18, 19), as well as LO28 (10), were recently described as strains that produce a truncated InlA; thus, the virulence potential of these strains was affected. These five isolates were used as noninvasive reference strains for development of the PCR-RFLP method.
TABLE 1.
PCR-RFLP profiles of L. monocytogenes strains and entry percentages with Caco-2 cells
| Strain | Origin | Serotype | AluI profilea | Tsp509I profilea | Composite profileb | Internalin theorical molecular mass (kDa) | Entry % | Reference |
|---|---|---|---|---|---|---|---|---|
| H2 | Healthy 3-year-old child (Beaune, France, 1992) | 4b | 2 | 1 | B | 80 | 9.74 ± 1.52 | 18 |
| H4 | Sporadic patient isolate, blood culture (Beaune, France, 1992) | NDc | 3 | 2 | C | 80 | 17.54 ± 0.75 | This study |
| H6 | Healthy 30-year-old woman (Beaune, France, 1991) | 3 (immobile) | 2 | 1 | B | 80 | 6.73 ± 0.86 | 19 |
| H11 | Healthy 35-year-old man (Beaune, France, 1992) | 1/2a | 3 | 2 | C | 80 | 16.70 ± 1.69 | 19 |
| H12 | Healthy 39-year-old man, oral isolate (Beaune, France, 1992) | 4b | 2 | 1 | B | 80 | 14.29 ± 1.08 | 19 |
| H21 | Sporadic patient isolate, blood culture (Strasbourg, France, 1992) | ND | 2 | 1 | B | 80 | 20.18 ± 1.38 | 19 |
| H22 | Sporadic patient isolate, blood culture (Strasbourg, France, 1992) | 4b | 2 | 1 | B | 80 | ND | 19 |
| H27 | Healthy 2-year-old child (Beaune, France, 1994) | 4b | 2 | 1 | B | 80 | 12.54 ± 4.27 | 19 |
| H28 | Healthy 7-year-old child (Beaune, France, 1994) | 1/2b | 2 | 1 | B | 80 | 10.86 ± 0.47 | 19 |
| H31 | Healthy 55-year-old man (Beaune, France, 1995) | 1/2a | 3 | 2 | C | 80 | 10.41 ± 2.75 | 19 |
| H35 | Healthy 11-year-old child (Beaune, France, 1997) | 1/2b | 2 | 1 | B | 80 | 10.80 ± 2.53 | 19 |
| H38 | Healthy carrier (Beaune, France, date not communicated) | 1/2a | 1 | 3 | A | 80 | 16.55 ± 0.86 | This study |
| NV4d | Minced beef | 1/2a | 4 | 2 | D | 67.5 | 0.124 ± 0.007 | This study |
| NV5d | Minced beef | 1/2c | 4 | 2 | D | 68 | 0.131 ± 0.006 | This study |
| NV7d | Bovine carcass | 1/2c | 4 | 2 | D | 50 | 0.32 ± 0.03 | This study |
| NV8d | Bovine carcass | 1/2a | 4 | 2 | D | 43 | 0.48 ± 0.03 | This study |
| 1Ed | Machine at an industrial cheese-making plant (Dijon, France, 1990) | 1/2b | 2 | 1 | B | 80 | 6.62 ± 1.44 | This study |
| 3Ed | Sink at an industrial cheese-making plant (Dijon, France, 1990) | 1/2b | 2 | 1 | B | 80 | 6.83 ± 2.90 | This study |
| 6Ed | Wall at an industrial cheese-making plant (Dijon, France, 1990) | 1/2a | 5 | 3 | E | 80 | 24.21 ± 2.06 | This study |
| 1S | Brine (Dijon, France, 1990) | 1/2a | 1 | 3 | A | 47 | 0.36 ± 0.09 | This study |
| 2Sd | Brine (Dijon, France, 1990) | 1/2a | 1 | 3 | A | 47 | 0.48 ± 0.38 | This study |
| 3S | Brine (Dijon, France, 1990) | 1/2a | 1 | 3 | A | 47 | 0.31 ± 0.01 | This study |
| 1Fd | Cheese (Dijon, France, 1990) | 1/2a | 1 | 3 | A | 47 | 0.93 ± 0.19 | This study |
| 2Fd | Cheese (Dijon, France, 1990) | 1/2a | 1 | 3 | A | 47 | 0.38 ± 0.12 | This study |
| 7Fd | Cheese (Dijon, France, 1990) | 3b | 1 | 3 | A | 47 | 0.51 ± 0.16 | This study |
| C9 | Compost (Dijon, France, 2002) | ND | 2 | 1 | B | 80 | 17.22 ± 1.99 | This study |
| 23d | Rook feces (Besançon, France, 1995) | ND | 2 | 1 | B | 80 | 24.86 ± 0.81 | 1 |
| 38d | Rook feces (Besançon, France, 1995) | ND | 1 | 3 | A | 80 | 14.54 ± 1.32 | 1 |
| 81d | Rook feces (Besançon, France, 1995) | ND | 4 | 2 | D | 80 | 15.81 ± 0.53 | 1 |
| 97d | Rook feces (Besançon, France, 1995) | ND | 3 | 2 | C | 80 | 8.71 ± 0.54 | 1 |
| Reference strains | ||||||||
| Scott A | Massachusetts milk outbreak (1983) | 4b | 2 | 1 | B | 80 | 9.55 ± 1.70 | 5 |
| EGD-e | Laboratory strain | 1/2a | 4 | 2 | D | 80 | 22.86 ± 1.60 | 16 |
| LO28 | Healthy pregnant carrier (Spain) | 1/2c | 4 | 2 | D | 63 | 0.89 ± 0.28 | 10 |
| H1 | Healthy pregnant carrier (Beaune, France, 1991) | 1/2a | 1 | 3 | A | 47 | 0.95 ± 0.42 | 18 |
| H17 | Healthy 10-year-old child (Beaune, France, 1992) | 3a | 1 | 3 | A | 47 | 0.024 ± 0.006 | 19 |
| H32 | Healthy 19-year-old women (Beaune, France, 1996) | 1/2a | 1 | 3 | A | 47 | 0.36 ± 0.15 | 19 |
| H34 | Healthy 8-year-old child (Beaune, France, 1997) | 1/2a | 1 | 3 | A | 47 | 0.39 ± 0.04 | 19 |
Each different set of banding profiles for each restriction endonuclase was given an arbitrary number.
Reflects the total differences or similarities in the banding patterns with the two restriction endonucleases.
ND, not determined.
Isolates from the same source whose lysotypes were different.
Restriction analysis (PCR-RFLP) of inlA.
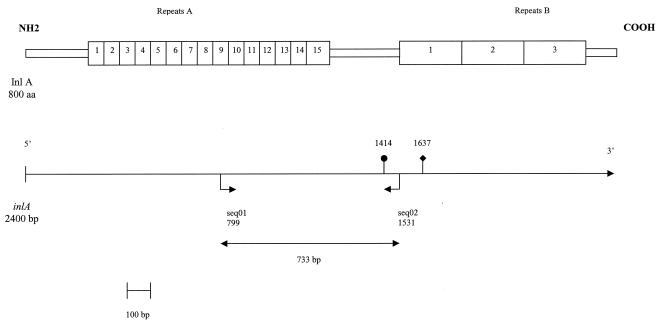
A 733-bp inlA fragment was amplified with primers seq01 (5′-AATCTAGCACCACTGTCGGG-3′) and seq02 (5′-TGTGACCTTCTTTTACGGGC-3′). This fragment encodes a region between repeat A10 and part of repeat B1 of InlA (Fig. 1). This inlA fragment was selected for the polymorphism study because of its genetic heterogeneity due to point mutations (18, 19, 20). PCRs were performed with 50-μl (total volume) reaction mixtures containing 5 μl of 10× Taq polymerase buffer (Appligene-Oncor, Illkirch, France), each deoxynucleoside triphosphate at a concentration of 200 μM, 1.5 mM MgCl2, each primer at a concentration of 0.5 μM, 1.25 U of Taq DNA polymerase (Appligene-Oncor), and 25 ng of DNA template. The following program was used: denaturation for 4 min at 94°C, followed by 30 cycles consisting of 94°C for 30 s, 52°C for 1 min, and 72°C for 2.5 min, and a final extension step consisting of 72°C for 7 min.
FIG. 1.
Structural organization of internalin and partial map of inlA (20). The horizontal arrows indicate the positions of primers seq01 and seq02 used to amplify an inlA fragment with genetic heterogeneity. The solid circle indicates the position of the mutation which created a nonsense codon in inlA of noninvasive reference strains H1, H17, H32, and H34. The solid diamond indicates the position of the deletion which created a nonsense codon in inlA of noninvasive reference strain LO28.
Restriction endonucleases AluI and Tsp509I were selected on the basis of a partial sequence analysis of inlA of invasive reference strain EGD-e (accession number LMO00433), noninvasive reference strains H1, H17, H32, H34, and LO28 (accession numbers AF468816, AY126441, AY126442, AY126443, and AY166686, respectively), and invasive reference strain Scott A (accession number AY166685). Ten (EGD-e and LO28), nine (H1, H17, H32, and H34), and seven (Scott A) AluI (AG/CT) restriction sites were detected. Ten (Scott A), seven (EGD-e and LO28), and six (H1, H17, H32, and H34) Tsp509I restriction sites were detected. These two restriction endonucleases were used independently. PCR-RFLP fragments were separated by electrophoresis on a 3.5% agarose gel (type 05 DNA grade; Euromedex, Mundolsheim, France). Gels were stained with ethidium bromide and were recorded with Bio-Rad gel doc 2000.
Plaque formation in Caco-2 cells.
The capacity of L. monocytogenes isolates to invade and disseminate in Caco-2 cells was evaluated by a plaque formation assay. The human colon carcinoma cell line Caco-2, obtained from the European Collection of Cell Cultures (ECACC no. 86010202), was used between passages 43 and 50. Cells were routinely grown in 25-cm2 plastic tissue culture flasks (Greiner) at 37°C in a humidified atmosphere containing 5% (vol/vol) CO2 in air. The culture medium used for growth of the cell line was Dulbecco's modified Eagle's minimum essential medium supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 1% (vol/vol) nonessential amino acids, and antibiotics (100 U of penicillin ml−1, 100 μg of streptomycin ml−1). All reagents were purchased from Invitrogen (Life Technologies).
Early confluent cell monolayers were prepared in six-well tissue culture plates. After overnight growth at 37°C in brain heart infusion broth (Biomerieux), listerial cells were centrifuged (6,000 × g for 10 min at room temperature) and serially diluted in phosphate-buffered saline (Dulbecco's phosphate-buffered saline with 1 mg of glucose ml−1 and 36 mg of sodium pyruvate ml−1; Life Technologies). Cells were counted by plating suitable dilutions on brain heart infusion medium. The plates were incubated for 24 to 48 h at 37°C. Caco-2 cells were infected with 0.1-ml portions of the 10−4 and 10−5 dilutions. Three replicate wells were used per dilution. After 2 h of contact at 37°C, nonadherent bacteria were removed from the monolayers by washing them three times with phosphate-buffered saline and by overlaying them with 2 ml of Dulbecco's modified Eagle's minimum essential medium containing 0.8% agarose, 20% fetal calf serum, and 2 mM l-glutamine. To kill extracellular adherent bacteria, 10 μg of gentamicin ml−1 was added. After 24 h of incubation at 37°C, 100 μl of trypan blue (0.4%), which stained dead cells, was added to each well, and plaque formation was observed 24 h later. The initial entry was determined by determining the ratio of the number of plaques observed to the initial number of bacteria added, expressed as a percentage. Two independent assays were performed in triplicate for each isolate on separate days with the Caco-2 cell line between passages 43 and 50 (n = 6). A statistical analysis (t test) was performed.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
Bacterial surface proteins were extracted by the method of Kochs et al. (12). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting were done as previously described (18). Membrane hybridization was performed with mouse antibodies directed against InlA (L7.7) (15).
DNA sequencing.
Sequencing was performed with DNA fragments generated by PCR performed with primers seq01 and seq02. Each PCR product was sequenced in both orientations by Genome Express.
Nucleotide sequence accession numbers.
The nucleotide sequences have been deposited in the GenBank/EMBL database, and the accession numbers of partial inlA sequences of the isolates are AJ564715 (1S), AJ564716 (2S), AJ564717 (3S), AJ564712 (1F), AJ564713 (2F), AJ564714 (7F), AJ608706 (NV4), AJ578445 (NV5), AJ564719 (NV7), and AJ564720 (NV8).
RESULTS
Restriction analysis (PCR-RFLP) of inlA.
The PCR-RFLP method was developed to study polymorphism of an inlA region in 37 L. monocytogenes isolates (Table 1), which allowed us to screen isolates that produced truncated InlA and thus were deficient in the ability to invade Caco-2 cells.
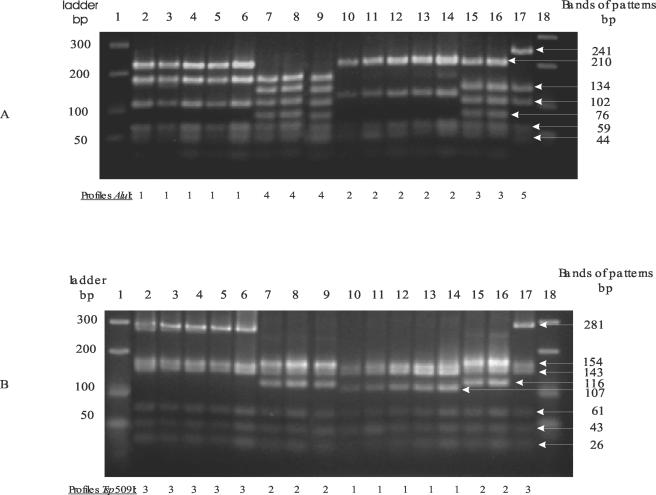
With primers seq01 and seq02, single DNA fragments of the expected size were obtained for all L. monocytogenes isolates. PCR-RFLP profiles of some of the isolates tested are shown Fig. 2.
FIG. 2.
PCR-RFLP profiles of different strains obtained after restriction with endonucleases AluI (A) and Tsp509I (B). Lanes 1 and 18, molecular weight ladder; lane 2, noninvasive reference strain H32; lane 3, food isolate 1S; lane 4, food isolate 2S; lane 5, food isolate 1F; lane 6, food isolate 2F; lane 7, food isolate NV4; lane 8, noninvasive reference strain LO28; lane 9, invasive reference strain EGD-e; lane 10, human fecal carriage isolate H2; lane 11, sporadic patient isolate H21; lane 12, isolate 1E from food-processing facilities; lane 13, compost isolate C9; lane 14, invasive reference strain Scott A; lane 15, sporadic patient isolate H4; lane 16, rook fecal isolate 97; lane 17, isolate 6E from food-processing facilities.
Restriction of the amplified inlA fragment with endonuclease AluI (Fig. 2A and Table 1) generated five different profiles with 8 to 11 bands ranging in size from 15 to 241 bp. Most of the 30 isolates were associated with PCR-RFLP profile 1 or 2. Eight isolates were grouped in PCR-RFLP profile 1 with noninvasive reference strains H1, H17, H32, and H34. Twelve isolates were grouped in PCR-RFLP profile 2 with invasive reference strain Scott A. Four L. monocytogenes isolates were associated with PCR-RFLP profile 3, and five isolates were grouped in PCR-RFLP profile 4 with invasive reference strain EGD-e and noninvasive reference strain LO28. Only one isolate (6E) had PCR-RFLP profile 5.
Restriction endonuclease Tsp509I generated three profiles (Fig. 2B and Table 1) with 7 to 11 bands ranging in size from 8 to 281 bp. Twelve L. monocytogenes isolates were grouped in PCR-RFLP profile 1 with invasive reference strain Scott A, and nine isolates were grouped in PCR-RFLP profile 2 with noninvasive reference strain LO28 and invasive reference strain EGD-e. Noninvasive reference strains H1, H17, H32, and H34 and nine L. monocytogenes isolates were associated with PCR-RFLP profile 3.
Five composite profiles were generated from the PCR-RFLP profiles obtained with the two restriction endonucleases (for example, composite profile A described L. monocytogenes isolates characterized by PCR-RFLP profile 1 obtained with AluI and by PCR-RFLP profile 3 obtained with Tsp509I) (3) (Table 1). There was no correlation between composite profiles and isolate origin. Eight L. monocytogenes isolates from brine (1S, 2S, and 3S), cheese (1F, 2F, and 3F), a human fecal carrier (H38), and a rook (38) were grouped in composite profile A with noninvasive reference strains H1, H17, H32, and H34. L. monocytogenes isolates from sporadic listeriosis patients (H21 and H22), compost (C9), food-processing facilities (1E and 3E), a rook (23), and human fecal carriers (H2, H6, H12, H27, H28, and H35) were grouped in composite profile B with invasive reference strain Scott A. Only four L. monocytogenes isolates were grouped in composite profile C; these isolates were isolates from a sporadic listeriosis patient (H4), human fecal carriers (H11 and H31), and a rook (97). Isolates from meat (NV4, NV5, NV7, and NV8) and rook isolate 81 were grouped in composite profile D with invasive reference strain EGD-e and noninvasive reference strain LO28. Composite profile E was exhibited by only one isolate, strain 6E from food-processing facilities.
Uncharacterized isolates with composite profiles A and D (H38, 1S, 2S, 3S, 1F, 2F, 7F, 38, NV4, NV5, NV7, NV8, and 81) were tested to determine their abilities to invade by using a plaque formation assay with Caco-2 cells.
Analysis of plaque formation in Caco-2 cells.
Ten of 13 L. monocytogenes isolates, 1F, 2F, 7F, 1S, 2S, and 3S with composite profile A and NV4, NV5, NV7, and NV8 with composite profile D, showed a weak ability (entry percentages, less than 1%) to invade Caco-2 cells compared to the abilities of the other strains (entry percentages, 6 to 24%) (Table 1). The differences in the entry percentages are statically significant. Western blot analyses showed that the InlA expressed by these 10 noninvasive isolates had a molecular mass that was less than 80 kDa. However, the entry percentages for L. monocytogenes isolates H38, 38, and 81 were more than 14%, and an 80-kDa InlA was detected by Western blotting (data not shown).
DNA sequencing.
inlA sequences of L. monocytogenes isolates 1S, 2S, 3S, 1F, 2F, 7F, NV4, NV5, NV7, and NV8 were analyzed to validate the PCR-RFLP method and to determine the reasons for production of the truncated InlA and thus the deficiency in the ability to invade Caco-2 cells.
DNA sequencing of inlA from strains 1S (accession number AJ564715), 2S (accession number AJ564716), 3S (accession number AJ564717), 1F (accession number AJ564712), 2F (accession number AJ564713), and 7F (accession number AJ564714) with composite profile A showed that there were silent mutations. One single-point mutation consisting of substitution of a cytosine for a thymidine at position 1414 (position based on the first translated codon) was detected. This point mutation created a nonsense codon (TAG) in the coding sequence that led to production of a protein with a theoretical molecular masses of 47 kDa (Table 1). These six isolates produced a truncated InlA (47 kDa) similar to the InlA produced by noninvasive reference strains H1, H17, H32, and H34.
DNA sequencing of inlA from NV4 (accession number AJ608706), NV5 (accession number AJ578445), NV7 (accession number AJ564719), and NV8 (accession number AJ564720) with composite profile D also revealed point mutations. For NV4, insertion of a thymidine at position 1901 was observed. This frameshift mutation led to creation of a nonsense codon, TGA, for position 1969, which generated an open reading frame encoding a theoretical 67.5-kDa InlA. For NV5, substitution of a guanine for an adenine at position 1994 (position based on the first translated codon) created a nonsense codon (TAG) in the coding sequence, which led to production of a protein with a theoretical molecular mass of 68 kDa (Table 1). For NV7, deletion of a guanine at position 1480 was observed. This frameshift mutation led to creation of a nonsense codon, TGA, at position 1496, which generated an open reading frame encoding a theoretical 50-kDa InlA. For NV8, substitution of a guanine for an adenine at position 1320 (position based on the first translated codon) created a nonsense codon (TGA) in the coding sequence that led to production of a protein with a theoretical molecular masse of 43 kDa (Table 1).
These results are in agreement with InlA molecular masses determined by Western blot analysis.
DISCUSSION
In this work, a PCR-RFLP method was developed for rapidly screening less invasive L. monocytogenes strains expressing truncated InlA proteins. Five PCR-RFLP profiles of the inlA region were observed, suggesting that inlA genetic heterogeneity was likely due to point mutations, particularly in a region between repeats A10 and B1 of InlA (Fig. 1), as previously described (5, 8, 20, 26). Most of these inlA polymorphism studies were carried out with L. monocytogenes isolates from food, animals, plants, and the environment (5, 8, 26), and little attention has been paid to clinical isolates (10, 22).
Restriction endonucleases AluI and Tsp509I were good discriminating enzymes for this polymorphism analysis. In other studies workers have also reported using restriction endonuclease AluI to study virulence gene polymorphism (5, 8, 20). In this study, composite profile A was obtained for eight uncharacterized L. monocytogenes isolates (H38, 1S, 2S, 3S, 1F, 2F, 7F, and 38) and four noninvasive L. monocytogenes reference strains (H1, H17, H32, and H34). Like noninvasive L. monocytogenes reference strains H1, H17, H32, and H34, six isolates (1S, 2S, 3S, 1F, 2F, and 7F) produced a truncated InlA. Moreover, five L. monocytogenes isolates (NV4, NV5, NV7, NV8, and 81), noninvasive reference strain LO28, and invasive reference strain EGE-e were characterized by composite profile D. Four of these strains (NV4, NV5, NV7, and NV8) produced a truncated InlA. Altogether, 15 L. monocytogenes isolates (strains 1S, 2S, 3S, 1F, 2F, 7F, NV4, NV5, NV7, and NV8 and noninvasive reference strains H1, H17, H32, H34, and LO28) that produced a truncated InlA were characterized by two specific composite profiles (profiles A and D), and 10 new L. monocytogenes strains expressing truncated forms of InlA were identified by using this method. Production of a truncated InlA was not correlated with the serotype of the isolates. Although all strains that produced a truncated InlA were characterized by these two specific composite profiles, isolates H38, 38, and 81 and reference strain EGD-e, which had a complete internalin, had profiles A and D. Sequence analysis of an inlA fragment showed that point mutations responsible for the production of truncated InlA were located at inlA position 1302 (NV8), position 1414 (H1, H17, H32, H34, 1S, 2S, 3S, 1F, 2F, and 7F), position 1496 (NV7), position 1637 (reference strain LO28), position 1901 (NV4), and position 1994 (NV5). These point mutations were not detected as they did not correspond to restriction sites (point mutations at inlA positions 1302, 1414, and 1496) for endonucleases AluI and Tsp509I or were outside the amplified inlA gene fragment (point mutations at inlA position 1637 of reference strain LO28, at inlA position 1901 of NV4, and at inlA position 1994 of NV5). To optimize screening of strains expressing truncated InlA by this rapid method, it would be useful to amplify a longer inlA fragment that might include a higher number of point mutations. We could also increase polymorphism analysis by using other restriction endonucleases to detect numerous point mutations responsible for production of different forms of internalin and to obtain more information about the genetic heterogeneity of inlA.
Despite these limitations, this PCR-RFLP method is a useful tool for screening numerous L. monocytogenes strains that are deficient in the ability to invade in large strain collections. It permitted us to identify 10 new L. monocytogenes strains that produce truncated InlA.
In this study and previous studies (10, 18, 19), 19 strains that produce truncated internalin have been described; it appears that expression of truncated internalin may not be a rare event and not specific to human carriage. In this study, two-thirds of the L. monocytogenes strains with a truncated InlA were isolated from food (brine, cheese, or meat). Similarly, in a previous report, Jonquières et al. (10) reported that three of five L. monocytogenes isolates that produced truncated internalin were isolated from meat, dairy products, and fish. Epidemiological studies have shown that although the rate of exposure to L. monocytogenes is rather high, the probability of contracting listeriosis is low (7, 17). Several hypotheses to explain this finding have been suggested. Most L. monocytogenes strains that occur in food may not be responsible for development of the disease because the levels of strains with attenuated virulence may be higher than the levels previously described. If the high occurrence of food isolates with truncated InlA was confirmed, it could partly explain the low occurrence of food-borne listeriosis. Occasional ingestion of food contaminated with attenuated virulence strains could increase the level of human resistance to Listeria infections (2). Production of truncated InlA may partially explain human carriage. However, several factors may be involved in asymptomatic carriage. In fact, Olier et al. (18, 19) described L. monocytogenes with complete InlA isolated from healthy carriers. It would be interesting to investigate possible epidemiological links between L. monocytogenes strains expressing truncated InlA isolated from healthy carriers and attenuated virulence strains producing truncated InlA isolated from food. It should be interesting to use molecular typing methods, such as pulsed-field gel electrophoresis or random amplification of polymorphic DNA, to address this hypothesis.
Acknowledgments
We gratefully acknowledge P. Cossart (Institut Pasteur, Paris, France) for kindly providing antibodies directed against InlA.
This work was supported by the Ministère de la Recherche et de l'Enseignement and the Conseil Regional de Bourgogne.
REFERENCES
- 1.Bouttefroy, A., J. P. Lemaitre, and A. Rousset. 1997. Prevalence of Listeria sp. in droppings from urban rooks (Corvus frugilegus). J. Appl. Microbiol. 82:641-647. [DOI] [PubMed] [Google Scholar]
- 2.Chakraborty, T., F. Ebel, J. Wehland, J. Dufrenne, and S. Notermans. 1994. Naturally occurring virulence-attenuated isolates of Listeria monocytogenes capable of inducing long term protection against infection by virulent strains of homologous an heterologous serotypes. FEMS Immunol. Med. Microbiol. 10:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Delgado da Silva, M. C., M. T. Destro, E. Hofer, and A. Tibana. 2001. Characterization and evaluation of some virulence markers of Listeria monocytogenes strains isolated from Brazilian cheeses using molecular, biochemical and serotyping techniques. Int. J. Food Microbiol. 63:275-280. [DOI] [PubMed] [Google Scholar]
- 4.Durst, J., and M. Zimanyi. 1976. The Listeria monocytogenes carrier state of the staff of maternity centres in non epidemic periods. Zentralbl. Bakteriol. 234:281-283. [PubMed] [Google Scholar]
- 5.Ericsson, H., P. Stalhandske, M. L. Danielsson-Tham, E. Bannerman, J. Bille, C. Jacquet, J. Rocourt, and W. Tham. 1995. Division of Listeria monocytogenes serovar 4b strains into two groups by PCR and restriction enzyme analysis. Appl. Environ. Microbiol. 61:3872-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleming, D. W. Cochi, S. L. MacDonald K. L. Brondum, J. Hayes, P. S. Plikaytis B. D. Holmes, M. B. Audurier, A. Broome, C. V., and A. L. Reingold. 1985. Pasteurized milk as a vehicle of infection in an outbreak of listeriosis. N. Engl. J. Med. 3120:404-407. [DOI] [PubMed] [Google Scholar]
- 7.Gellin, B. C., and C. V. Broome. 1989. Listeriosis. JAMA 261:1313-1320. [PubMed] [Google Scholar]
- 8.Giovannacci, I., C. Ragimbeau, S. Queguiner, G. Salvat, J. L. Vendeuvre, V. Carlier, and G. Ermel. 1999. Listeria monocytogenes in pork slaughtering and cutting plants: use of RAPD, PFGE and PCR-REA for tracing and molecular epidemiology. Int. J. Food Microbiol. 53:127-140. [DOI] [PubMed] [Google Scholar]
- 9.Grif, K., I. Hein, M. Wagner, E. Bandl, O. Mpamugo, J. MacLauchlin, M. P. Dierich, and F. Allerberger. 2001. Prevalence and characterization of Listeria monocytogenes in the faeces of healthy Austrians. Wien Klin. Wochenschr. 113:737-742. [PubMed] [Google Scholar]
- 10.Jonquières, R., H. Bierne, J. Mengaud, and P. Cossart. 1998. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect. Immun. 66:3420-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kampelmacher, E. H., and L. M. van Noorle Jansen. 1972. Further studies on the isolation of L. monocytogenes in clinically healthy individuals. Zentralbl. Bakteriol. 221:70-77. [PubMed] [Google Scholar]
- 12.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. moncytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 13.Macgowan, A. P., R. J. Marshall, I. M. Mackay, and D. S. Reeves. 1991. Listeria faecal carriage by renal transplant recipients, haemodialysis patients and patients in general practice: its relation to season, drug therapy, foreign travel, animal exposure and diet. Epidemiol. Infect. 106:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLauchlin, J., N. A. Saunders, A. M. Ridley, and A. G. Taylor. 1988. Listeriosis and food-borne transmission. Lancet i:177-178. [DOI] [PubMed] [Google Scholar]
- 15.Mengaud, J., M. Lecuit, M. Lebrun, F. Nato, J. C. Mazie, and P. Cossart. 1996. Antibodies to the leucine-rich repeat region of internalin block entry of Listeria monocytogenes into cells expressing E-cadherin. Infect. Immun. 64:5430-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, E. G. D. Webb, R. A., and M. B. R. Swann. 1926. A disease of rabbits characterized by large mononuclear leucocytosis caused by a hitherto undescribed bacillus Bacterium monocytogenes (n. sp.) J. Pathol. Bacteriol. 29:407-439. [Google Scholar]
- 17.Notermans, S., J. Dufrenne, P. Teunis, and T. Chackraborty. 1998. Studies on the risk assessment of Listeria monocytogenes. J. Food Prot. 61:244-248. [DOI] [PubMed] [Google Scholar]
- 18.Olier, M., F. Pierre, J. P. Lemaître, C. Divies, A. Rousset, and J. Guzzo. 2002. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology 148:1855-1862. [DOI] [PubMed] [Google Scholar]
- 19.Olier, M., F. Pierre, S. Rousseaux, J. P. Lemaître, A. Rousset, P. Piveteau, and J. Guzzo. 2003. Expression of truncated internalin A is involved in impaired internalization of some Listeria monocytogenes human carriage isolates. Infect. Immun. 71:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poyart, C., P. Trieu-Cuot, and P. Berche. 1996. The inlA gene required for cell invasion is conserved and specific to Listeria monocytogenes. Microbiology 142:173-180. [DOI] [PubMed] [Google Scholar]
- 21.Schlech, W. F., 3rd, P. M. Lavigne, R. A. Bortolussi, A. C. Allen, E. V. Haldane, A. J. Wort, A. W. Hightower, S. E. Johnson, S. H. King, E. S. Nicholls, and C. V. Broome. 1983. Epidemic listeriosis—evidence for transmission by food. N. Engl. J. Med. 308:203-206. [DOI] [PubMed] [Google Scholar]
- 22.Schuchat, A., K. Deaver, P. S. Hayes, L. Graves, L. Mascola, and J. D. Wenger. 1993. Gastrointestinal carriage of Listeria monocytogenes in household contacts of patients with listeriosis. J. Infect. Dis. 167:1261-1262. [DOI] [PubMed] [Google Scholar]
- 23.Slutsker, L., and A. Schuchat. 1999. Listeriosis in animals, p. 75-95. In Listeria, listeriosis and food safety. E. T. Ryser and E. H. Marth (ed.), Marcel Dekker, Inc., New York, N.Y.
- 24.Watkins, J., and K. P. Sleath. 1981. Isolation and enumeration of Listeria monocytogenes from sewage, sewage sludge and river water. J. Appl. Bacteriol. 50:1-9. [DOI] [PubMed] [Google Scholar]
- 25.Weis, J., and H. P. Seeliger. 1975. Incidence of Listeria monocytogenes in nature. Appl. Microbiol. 30:29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiedmann, M., J. L. Bruce, C. Keating, A. E. Johnson, P. L. McDonough, and C. A. Batt. 1997. Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65:2707-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]