Abstract
Alcaligenes denitrificans Y2k-2 was obtained by selective enrichment followed by screening from soil samples, which showed ω-amino acid:pyruvate transaminase activity, to kinetically resolve aliphatic β-amino acid, and the corresponding structural gene (aptA) was cloned. The gene was functionally expressed in Escherichia coli BL21 by using an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pET expression system (9.6 U/mg), and the recombinant AptA was purified to show a specific activity of 77.2 U/mg for l-β-amino-n-butyric acid (l-β-ABA). The enzyme converts various β-amino acids and amines to the corresponding β-keto acids and ketones by using pyruvate as an amine acceptor. The apparent Km and Vmax for l-β-ABA were 56 mM and 500 U/mg, respectively, in the presence of 10 mM pyruvate. In the presence of 10 mM l-β-ABA, the apparent Km and Vmax for pyruvate were 11 mM and 370 U/mg, respectively. The enzyme exhibits high stereoselectivity (E > 80) in the kinetic resolution of 50 mM d,l-β-ABA, producing optically pure d-β-ABA (99% enantiomeric excess) with 53% conversion.
β-Amino acids are emerging as an interesting class of compounds for medicinal chemists. They are commonly found as building blocks in many natural and synthetic drugs, such as antibiotics, enzyme inhibitors, and peptide mimetic drugs with pharmacological properties (4, 13). For example, functionalized β-amino acids are key components of a variety of bioactive molecules, such as taxol (i.e., one of the most active antitumor agents, which contains phenylisoserine as its side chain) (7). Given the significance of the β-amino acids, their efficient synthesis in optically pure form has become an attractive challenge to organic chemists in recent years. Numerous chemical synthetic methodologies have been devised and extensively reviewed (10). However, biocatalytic methods of the synthesis of optically pure β-amino acids have been relatively less explored, because the enzymes commonly used to resolve α-amino acids, such as transaminase, dehydrogenase, and protease, have narrow substrate tolerance to β-amino acids and often exhibit low enantioselectivity (7, 10).
Transaminase reaction has some superior features compared to other enzymatic and chemical methods for the production of chiral amino acids and amines (8, 23, 24). Transaminase shows rapid reaction rates, with no requirement for external cofactor regeneration, and it allows asymmetric synthesis from prochiral ketone compounds, depending upon the properties of target chemical compounds. Although transaminase group II (3, 12), containing ω-transaminase, shows reactivity to specific β-amino acids such as β-alanine, few attempts have been undertaken to resolve various racemic β-amino acids. In this study, we have attempted to screen for microorganisms having transaminase activity on β-amino acids containing aliphatic chains.
To select such microorganisms, soil samples collected from various domestic sites were incubated in 50 ml of Luria-Bertani (LB) broth medium for 1 h. After incubation, 0.1 ml of the culture broth was transferred to 10 ml of minimal medium containing 10 mM d,l-β-amino-n-butyric acid (ABA) as the sole nitrogen source (100 mM glycerol, 50 mM potassium phosphate buffer [pH 7.0], 1.0-g/liter MgSO4 · 7H2O, 0.2 mM CaCl2, 0.1-mg/liter ZnCl2, 0.1-mg/liter MnSO4 · 4H2O, 0.02-mg/liter H3BO3, 0.1-mg/liter CuSO4 · 5H2O, 0.05-mg/liter CoCl2, 0.1-mg/liter NiSO4 · 6H2O, 2.0-mg/liter NaMoO4, 4.0-mg/liter FeSO4 · 7H2O) (18). The enrichment of the microorganism was carried out by dilution (20- to 30-fold) of culture broth with fresh minimal medium at 37°C whenever the optical density of the culture broth reached 0.2 to 0.4. After reinoculation, the diluted culture broth was spread out on the minimal agar plates. Each single colony from the plates was cultivated in 50 ml of LB medium. The harvested cells (2 mg of cell [dry weight]) were subjected to whole-cell reaction in 0.5 ml of 100 mM borate buffer (pH 9.0) containing 10 mM d,l-β-ABA and 10 mM pyruvate for 5 h at 37°C. The remaining β-ABA was analyzed, and the enantiomeric excess (ee) was calculated. Quantitative chiral analysis of d,l-β-ABA was performed with a C18 symmetry column (Waters, Billerica, Mass.) with a Waters high-performance liquid chromatography (HPLC) system after the derivatization of β-ABA with 2,3,4,6-tetra-o-acetyl-α-d-glucopyranosyl isothiocyanate (6). Separation of each enantiomer was achieved with an isocratic elution condition using 10 mM phosphate buffer (pH 2.8) and methanol (80:20 [vol/vol]). The quantification of d,l-β-ABA was carried out by an o-phthaldialdehyde derivatization method (5). A strain (Y2k-2) later identified as Alcaligenes denitrificans by 16S rRNA analysis (Korea Research Institute of Bioscience and Biotechnology, Taejon, Korea) showed the highest reaction rate and L-specific stereoselectivity. When the kinetic resolution of 10 mM d,l-β-ABA was carried out with 4 mg (cell dry weight) of A. denitrificans Y2k-2 in 1 ml of 100 mM borate buffer (pH 9.0) containing 10 mM pyruvate, 56% conversion yield with 99% ee of d-β-ABA was achieved in 5 h.
To investigate whether or not the kinetic resolution of the d,l-β-ABA is really due to a transaminase activity, the enzyme activity was examined in the presence of typical inhibitors of pyridoxal 5′-phosphate (PLP)-dependent enzyme by using the crude extract of A. denitrificans Y2k-2. To prepare the crude extract, the harvested cells (10 g of cell wet weight) of A. denitrificans Y2k-2 were suspended in 20 ml of sonication buffer, which contains 20 μM PLP, 0.2 mM EDTA, 1.0 mM phenylmethylsulfonyl fluoride (PMSF), and 0.01% (wt/wt) 2-mercaptoethanol in 10 mM phosphate buffer (pH 7.0), and disrupted by sonication at 4°C. It was strongly inhibited by 5 mM 3-amino-2,3-dihydrobenzoic acid (gabaculine), hydroxylamine, or aminooxyacetic acid (25), and equivalent amounts of l-alanine were produced according to the consumed pyruvate. The enzyme involved in the kinetic resolution was confirmed to be an ω-amino acid:pyruvate transaminase (ω-APT).
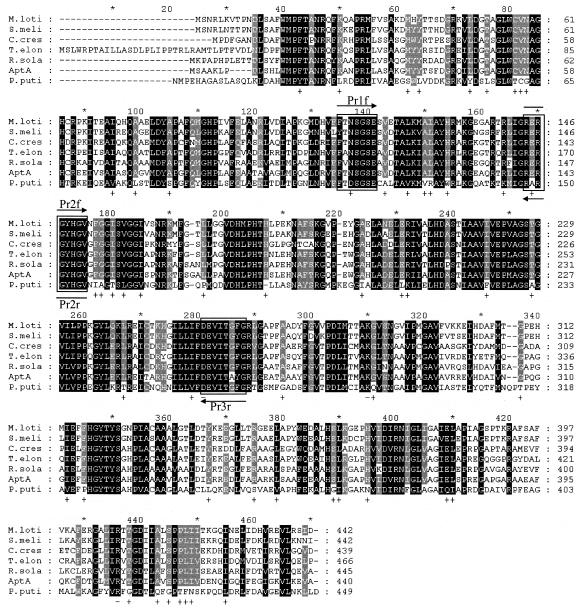
The BLAST search was done with amine:pyruvate transaminase from Vibrio fluvialis JS17 (22). The BLAST search using the GenBank database showed that seven ω-APTs exist in proteobacteria. The consensus amino acid sequences were found through the alignment of these sequences (Fig. 1). Degenerate PCR primers were designed based upon the consensus amino acid sequences to clone the gene (aptA) encoding amine:pyruvate transaminase from A. denitrificans Y2k-2. Pr1f (5′-TTCTTCACCRRYTCIGGITCSGA-3′), Pr2f (5′-CGCGIICGCGG-CTATCACGGSGT-3′), Pr2r (5′-ACSCCGTGATAGCCGCGIICGCG-3′), and Pr3r (5′-CCRAA-RCCGGTGATSACCTCGTC-3′) were synthesized, in which I, R, S, and Y indicate inosine, A or G, C or G, and C or T, respectively. DNA manipulations, including preparation of plasmids, restriction enzyme digestion and ligation, transformation of Escherichia coli, Southern hybridization, and colony hybridization followed the methods of Sambrook et al. (17). PCR was performed using genomic DNA of A. denitrificans Y2k-2 as a template. When Prlf/Pr3r, Pr2f/Pr3r, and Pr1f/Pr2r were used as primers, gene fragments of about 470, 360, and 130 bp were obtained, respectively. The sizes of the amplified fragments are in good agreement with the expected sizes from the consensus sequences (Fig. 1). A 470-bp gene fragment was ligated into pGEM-T vector (Promega), and the DNA sequence was analyzed by ABI 3100 (Perkin-Elmer Biosystems, Foster City, Calif.). The result of BLAST search with a 155-amino-acid sequence deduced from the 465-bp sequence revealed that this partial fragment is highly homologous to the ω-APT from Ralstonia solanacearum (RSp1056; GenBank accession no. NP5222617) with 74% identity (16), and the deduced sequence of 155 amino acids includes the consensus peptide sequences (Fig. 1).
FIG. 1.
Multiple sequence alignment of A. denitrificans Y2k-2 AptA and other related proteins. Highly conserved residues are in black, and less strongly conserved residues are in gray. The boxes indicate the consensus amino acid sequences used to design the degenerate primers. M.loti, β-alanine:pyruvate transaminase from M. loti (mll1632; GenBank accession no. NP103175; 56% identity); S.meli, putative ω-APT from S. meliloti (SMc01534; GenBank accession no. NP386510; 58% identity); C.cres, ω-APT from C. crescentus (cc3143; GenBank accession no. AAK25105; 56% identity); T.elon, ω-APT from T. elongatus BP-1 (tlr0408; GenBank accession no. NP681198; 59% identity); R.sola, probable β-alanine:pyruvate transaminase from R. solanacearum (RSp1056; GenBank accession no. NP5222617; 64% identity); AptA, AptA from A. denitrificans Y2k-2; P.puti, ω-APT from P. putida (45% identity).
The 465-bp PCR product was used as a probe after labeling with [32P]dCTP with a random-labeling kit (Promega). The chromosomal DNA of A. denitrificans Y2k-2 was partially digested with Sau3AI to obtain DNA fragments of 30 to 40 kb. The fragments were ligated into the cosmid vector pOJ446 (1) and digested with BamHI and HpaI. The ligation mixture was packaged in vitro by using a lambda packaging system (Stratagene) and transfected into E. coli XL1-Blue MRF. The colonies were selected on an LB agar plate containing apramycin (100 μg/ml). Several positive clones (13 out of 2,000 clones) were identified by Southern hybridization, and six cosmids were randomly selected. Southern hybridization revealed that five cosmids contained 3.4-kb PstI and 2.8-kb EcoRI fragments hybridizing to the probe. The 3.4-kb PstI fragment and the 2.8-kb EcoRI fragment were each ligated with pGEM-3 (Promega). One complete 1,323-bp open reading frame (ORF) (ATG initiation codon at nucleotide 247 and TAG termination codon at nucleotide 1259) was identified in the sequence; this ORF encodes a polypeptide of 440 amino acid residues with theoretical pI of 6.5 and a calculated molecular mass of 46,843 Da. A putative ribosomal binding site, GGAG, was found 8 bp upstream from the presumptive start codon, ATG. The putative −35 and −10 promoter sequences were observed 64 bp upstream from the start codon. The stop codon (TAG) was followed by a sequence of dyad symmetry (10-nucleotide perfect repeat), which is expected to function as a transcription terminator.
Homology analysis of the deduced amino acid sequence of AptA by the National Center for Biotechnology Information (NCBI) protein database using BLAST searches revealed that the enzyme showed high similarity to RSp1056 from R. solanacearum (16), Tlr0408 from Thermosynechococcus elongatus BP-1 (14), SMc01534 from Sinorhizobium meliloti (2), Mll1632 from Mesorhizobium loti (9), and Cc3143 from Caulobacter crescentus (15), with 56 to 64% identity. These enzymes were found by bacterial genome sequencing projects and annotated as ω-APT. However, none of these proteins has been biochemically characterized yet. AptA had 45% identity to ω-APT from Pseudomonas putida, which is the only well-characterized enzyme (25, 26). The linear alignment of AptA and the six related proteins mentioned above showed that these enzymes are highly homologous to each other (Fig. 1). Some essential key residues of the ω-APT from P. putida, such as D259 (hydrogen bond with PLP), K288 (active-site lysine), and R414 (salt bridge with α-carboxylate group of substrate) are well conserved in all of the ω-APTs (Fig. 1). However, despite the high similarity, the amino acid sequences of the ω-APT from P. putida and the ω-APTs from other strains are quite different. Among the identical residues conserved in the six ω-APTs (except ω-APT from P. putida), 54 amino acid residues were different in the ω-APT from P. putida (Fig. 1), suggesting that biochemical properties of the six enzymes could be different from the ω-APT from P. putida.
To express the enzyme without excessive flanking parts, the coding region of AptA was amplified by PCR using P1 (5′-CATATGAGCGCTGCCAAACTGCCC-3′) and P2 (5′-GGATCCCTAAGCCACTTCCTTGAGCAC-3′) primers. To express C-terminal fusion enzyme with a His-tagged polypeptide, the coding region of AptA was amplified by PCR using P1 and P3 (5′-CTCGAGAGCCACTTCCTTGAGCACCTT-3′) primers. The PCR product was digested with restriction enzymes (NdeI/BamHI or NdeI/XhoI) and inserted into the vector pET24ma (22). The plasmid was introduced into E. coli BL21, and transformants were grown in LB broth containing kanamycin (50 μg/ml) at 37°C. When the optical density reached 0.75, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. After 5 h of induction, the cells were harvested and disrupted by sonication. After centrifugation, the enzyme activity of the cell extract was measured to evaluate expression efficiency. Unless otherwise specified, the activity assay was carried out with 100 mM borate buffer (pH 9.0) at 37°C using the cell extract. One unit of the enzyme activity was defined as the amount of the enzyme that catalyzes the formation of 1 μmol of l-alanine from 10 mM l-β-ABA and 10 mM pyruvate for 1 min.
Although the relative quantities of the recombinant proteins were almost equal in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (data not shown), the specific activity (9.6 U/mg) of the recombinant enzyme without the histidine tag was 10-fold higher than that of the His-tagged AptA (1.0 U/mg) in crude extract and 100-fold higher than that of A. denitrificans Y2k-2. Since the His-tagged AptA showed a low activity, the recombinant AptA without the histidine tag was purified. All purification steps were carried out at 4°C and were monitored by SDS-PAGE. Column chromatography was performed with Biologic HR system (Bio-Rad, Richmond, Calif.). The cell pellet harvested from 1 liter of E. coli culture broth (LB medium) was resuspended in 50 ml of 100 mM potassium phosphate buffer (pH 7.0) containing 20 μM PLP, 0.2 mM EDTA, 1.0 mM PMSF, and 0.01% (vol/vol) 2-mercaptoethanol and then subjected to ultrasonic disruption for 25 min. The supernatant was obtained after centrifugation (17,000 × g, 30 min). Polyethyleneimine (0.8%) was slowly added to the supernatant with gentle stirring. After equilibration for 20 min, the solution was centrifuged again. The supernatant was fractionated by ammonium sulfate precipitation between 20 and 60% saturation. The dissolved ammonium sulfate fraction was loaded in butyl-Sepharose FF column (10 mm by 30 cm; Pharmacia) pre-equilibrated with 50 mM potassium phosphate buffer (pH 7.0). The proteins were eluted with 50 mM potassium phosphate buffer (pH 7.0) by using a 1.0 to 0 M linear gradient of ammonium sulfate. Active fractions were concentrated with an Amicon PM-10 ultrafiltration unit. The sample was applied to a HiPrep 16/60 Sephacryl S-200 HR (Phramacia) pre-equilibrated with 50 mM potassium phosphate buffer (pH 7.0) containing 150 mM NaCl. The proteins were isocratically eluted by using the same buffer at a flow rate of 0.8 ml/min. (The active fractions were dialyzed against 50 mM potassium phosphate buffer [pH 7.0] and concentrated with an Amicon PM-10 ultrafiltration unit.) The recombinant AptA was purified with over 22% yield (Table 1). The purified transaminase gave a single protein band on SDS-PAGE (data not shown).
TABLE 1.
Purification of AptA produced by E. coli BL21
| Purification step | Total protein (mg) | Sp act (U/mg) | Yield (%) | Fold purifi- cation |
|---|---|---|---|---|
| Crude extract | 430 | 9.6 | 100 | 1 |
| 0.8% Polyethyleneimine | 134 | 18.5 | 60 | 1.9 |
| Ammonium sulfate (20∼60%) | 78.4 | 27.9 | 53 | 2.9 |
| Butyl-Sepharose FF | 30.1 | 52.2 | 38 | 5.4 |
| Sephacryl S-200 FF | 11.8 | 77.2 | 22 | 8.0 |
A small amount of the purified enzyme solution (400 μl) was applied to a Superdex 200 preparative-grade column (HR 10/30; Pharmacia) and eluted with 0.1 M Tris-HCl (pH 8.0) containing 0.1 M KCl at a flow rate of 0.7 ml/min. Only a single protein peak was observed, and the corresponding fractions displayed the AptA activity. The column was calibrated with bovine thyroglobulin (Mr, 669,000), catalase (Mr, 232,000), aldolase (Mr, 158,000), bovine serum albumin (Mr, 67,000), chymotrypsinogen A (Mr, 25,000), and RNase A (Mr, 13,700) as reference proteins (gel filtration calibration kit; Pharmacia). The molecular mass of the AptA was estimated to be 180 kDa by gel filtration chromatography. The subunit molecular mass deduced from the DNA sequence is 46.8 kDa, suggesting that the enzyme has a tetrameric structure. Isoelectric focusing was performed with a 5% disk-type polyacrylamide gel (pH gradient ranging from 5 to 7), and the isoelectric point of the enzyme was 6.2. The enzyme activity versus pH was determined within a pH range of 6.0 to 10.0 with 10 mM l-β-ABA and 10 mM pyruvate at 37°C. Using two kinds of reaction buffer, 100 mM potassium phosphate (pH 6.0 to 7.5) and 100 mM boric acid (pH 7.5 to 10.0), the optimum pH of the enzyme was identified as ca. 9.0.
To measure amino donor specificity, the purified AptA was subjected to the reaction assay in 0.2 ml of 100 mM borate buffer (pH 9.0) containing 10 mM pyruvate and 10 mM each amino donor, and the initial reaction rate was measured by analyzing the amount of pyruvate consumed by using an Aminex HPX-87H HPLC column (Bio-Rad) with 5 mM sulfuric acid eluent solution. To measure amine acceptor specificity, the reaction conditions were the same as that for amine donor specificity, except 10 mM (S)-α-methylbenzylamine (MBA) and 10 mM each amino acceptor were used. The acetophenone was quantified with a C18 Symmetry HPLC column (Waters) with isocratic elution of acetonitrile and water (35:65 [vol/vol]) at 1 ml/min.
In terms of the substrate specificity of the purified enzyme, the AptA showed high activity toward l-β-ABA, which was 10-fold higher than that for β-alanine (Table 2). Other aliphatic β-amino acids, such as l-β-leucine, l-β-homoleucine, and l-β-homoisoleucine, also showed considerable reactivity (0.4 U/mg for l-β-homoisoleucine), whereas aromatic β-amino acids (l-3-amino-3-phenylpropanoic acid) were inert. Among the examined amines, α-MBA was the most reactive amino donor, whose reactivity is 1.4-fold higher than that of β-alanine. Nineteen natural amino acids were inert, except for alanine. In the case of alanine, its substrate specificity could not be examined, because pyruvate was used as the amino acceptor. In the case of amino acceptor, butyraldehyde was the most reactive amino acceptor. Among ketoacids, pyruvate and glyoxylate showed good reactivity, whereas oxaloacetate and α-keto-glutarate were inert. The enzyme showed (S)-enantioselectivity toward d,l-β-ABA and α-MBA, and the enantiomeric ratios were above 80. This high enantioselectivity is quite beneficial for the preparation of enantiopure β-amino acids and amines via kinetic resolution (18, 19).
TABLE 2.
Substrate specificity of AptA
| Amino donor | Relative activity (%)a | Amino acceptor | Relative activity (%)b |
|---|---|---|---|
| l-β-Alanine | 10 | Pyruvate | 100 |
| l-β-ABA | 100 | Oxalacetate | NRc |
| l-β-Leucine | 4.3 | α-Ketoglutarate | NR |
| l-β-Homoleucine | 2.6 | Glyoxylate | 13 |
| l-β-Homoisoleucine | 0.5 | Propionaldehyde | 105 |
| l-3-Amino-3-phenylpropanoic acid | NR | Butyraldehyde | 118 |
| (R,S)-sec-Butylamine | 7.3 | Pyruvicaldehyde | 5 |
| (R,S)-1,2-Dimethylpropylamine | 7.5 | Ethylacetoacetate | NR |
| (R,S)-1,3-Dimethylbutylamine | 0.5 | ||
| (R,S)-2-Aminoheptane | 9.1 | ||
| (R,S)-1.5-Dimethylhexylamine | 8.3 | ||
| (S)-α-MBA | 14 | ||
| (R,S)-α-Ethylbenzylamine | 0.3 | ||
| (R,S)-1-Aminoindan | 2.6 |
Pyruvate (10 mM) and 10 mM each amino donor were used, and the initial reaction rate was measured by analyzing the amount of pyruvate consumed. The activity for l-β-ABA, corresponding to 77.2 U/mg, was taken as 100%. For racemic amines, the concentration of amino donor was 20 mM.
(S)-α-MBA (10 mM) and 10 mM each amino acceptor were used, and the initial reaction rate was measured by analyzing the acetophenone produced. The activity for (S)-α-MBA, corresponding to 10.8 U/mg, was taken as 100%.
NR, not reactive.
The transaminase family is divided into four subgroups on the basis of their mutual structural relatedness (3, 12). α-Transaminases belong to subgroups I, III, and IV, whereas ω-transaminases belong to subgroup II, which contains ω-APT (EC 2.6.1.18), ornithine transaminase (EC 2.6.1.11), γ-aminobutyrate transaminase (EC 2.6.1.19), diaminopelargonate transaminase (EC 2.6.1.62), etc. (11, 12). The enzyme AptA cloned in this study showed high homology to the other known ω-APTs (five enzymes are annotated, and one enzyme is characterized) (Fig. 1). The substrate specificity study of AptA (Table 2) revealed that pyruvate is the only reactive amino acceptor among the typical amino acceptors for transaminases, such as oxaloacetate, α-ketoglutarate, and pyruvate. Based on these results, AptA belongs to the ω-APT family (E.C. 2.6.1.18). Generally β-alanine is one of the most reactive amino donors for the ω-APT. However, AptA showed the highest activity for l-β-ABA, which is higher than that for β-alanine by 10-fold. The enzyme can use not only amines but also various aliphatic β-amino acids, such as l-β-leucine, l-β-homoleucine, and l-β-homoisoleucine, as the amine donor. None of these characteristics of ω-APT has been reported before.
Homology analysis revealed that among the identical residues of the six ω-APTs (except ω-APT from P. putida) in Fig. 1, 54 amino acid residues are different from the ω-APT from P. putida, suggesting that the biochemical properties of the six enzymes would be very similar to one another but different from those of the ω-APT from P. putida (Fig. 1). One such gene from M. loti, mll1632 (9), was cloned and overexpressed in E. coli. BL21. The substrate specificity of Mll1632 was quite similar to that of AptA. Similarly, l-β-ABA was the most reactive amino donor for Mll1632, so that it is a more reactive amino donor than β-alanine by 9.9-fold, and Mll1632 aminotransferase can transfer the amine group from various aliphatic β-amino acids, such as l-β-leucine and l-β-homoleucine, to pyruvate (data not shown).
To evaluate the inactivation rate constant of AptA, purified AptA (0.5 mg/ml) was incubated in 1 ml of 100 mM borate buffer (pH 9.0) for a certain period of time (0.1 to 20 h) at desired temperatures (16 to 60°C). Aliquots of the enzyme solution were periodically added to the reaction solution containing 10 mM l-β-ABA and 10 mM pyruvate, and then the initial rate was measured by analyzing the amount of pyruvate consumed. The kinetics of thermal inactivation of the enzyme usually follows a pseudo-first-order enzyme inactivation equation, suggesting that the inactivation rate constant does not change depending on the enzyme concentration. The measured half-lives of AptA at 16, 30, 37, 45, and 60°C were 34, 17, 5.7, 2.9, and 1.2 h, respectively. An Arrhenius plot of the inactivation rate constants (5.7 × 10−6, 1.1 × 10−5, 3.4 × 10−5, 6.6 × 10−5, and 1.5 × 10−4 s−1 at 16, 30, 37, 45, and 60°C, respectively) yielded a deactivation energy of 64 kJ mol−1. To evaluate the substrate concentration and PLP effects on enzyme stability, the enzyme solutions (0.5 mg/ml) in 0.5 ml of 100 mM borate buffer (pH 9.0) containing the predetermined substrate concentrations were incubated at 37°C. By adding 5 mM pyruvate and 1 mM PLP in the enzyme solutions, the half-life of the enzyme dramatically increased from 5.7 h to 23 and 35 h, respectively, suggesting that the addition of pyruvate and PLP greatly prolonged the half-life of the enzyme. In contrast, the addition of 5 mM l-β-ABA only dramatically decreased (ca. fivefold) the half-life of the enzyme down to 1 h. However, the destabilizing effect of l-β-ABA was leveled off by adding 1 mM PLP, resulting in a half-life of 28 h. Because apoenzyme is usually more susceptible to irreversible inactivation than holoenzyme (3) and the apo form of transaminase usually binds more tightly to PLP than to pyridoxamine 5′-phosphate (PMP), it is not surprising to see that PLP shows strong stabilizing effect on AptA. In another study, the dissociation constants of TyrAT with PLP and PMP were found to be 5.6 × 10−9 and 5.3 × 10−7 M, respectively, at pH 7.6 in the presence of 0.1 M chloride (3), supporting the above explanation. Therefore, the instability to l-β-ABA can be explained mechanistically, so that l-β-ABA converts PLP enzyme to PMP enzyme at the initiation of the transaminase reaction, whereas pyruvate is the amino acceptor that converts PMP enzyme to PLP enzyme.
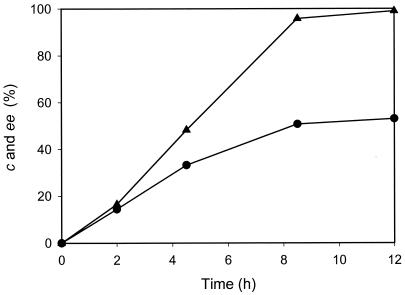
The kinetic resolution of l-β-ABA was attempted to produce d-β-ABA by using AptA. Although propionaldehyde and butyraldehyde were cheaper and more reactive amino acceptors than pyruvate, pyruvate was the best amino acceptor, considering both reactivity and enzyme stability, which enabled us to use pyruvate as the amino acceptor in the kinetic resolution of d,l-β-ABA. Since the reaction rate of ω-APT is greatly influenced by product and substrate concentrations (18-21), the product and substrate inhibitions against AptA were primarily examined. The apparent Km and Vmax for l-β-ABA were 56 mM and 500 U/mg, respectively, in the presence of 10 mM pyruvate. The substrate inhibition by l-β-ABA was not observed at all up to 1 M. In the presence of 10 mM l-β-ABA, the apparent Km and Vmax for pyruvate were 11 mM and 370 U/mg, respectively. The substrate inhibition by pyruvate was not observed up to 50 mM. However, as its concentration exceeded 50 mM (4.5-fold above the apparent Km), the substrate inhibition was more pronounced. The initial rate of AptA at 1 M pyruvate was about 45% of that of AptA at 50 mM pyruvate in the presence of 10 mM l-β-ABA. In terms of the product inhibition, AptA activity was inhibited severely by l-alanine. In the presence of 100 mM l-alanine in the reaction mixture containing 10 mM l-β-ABA and 10 mM pyruvate, the initial rate of AptA decreased by about 90% of that in the absence of alanine. The product inhibition by the β-keto acid could not be evaluated properly, because the β-keto acid is spontaneously decomposed in the reaction mixture. One remarkable thing in the kinetic resolution of the β-amino acid is the spontaneous decomposition of the β-keto acid by a β-elimination mechanism, so that the equilibrium would be rapidly shifted. When the kinetic resolution of 50 mM d,l-β-ABA was carried out with 30 mM pyruvate and 10 mg of the crude cell extract containing AptA in 10 ml of 100 mM borate buffer (pH 9), 53% of the conversion was achieved with 99% ee of d-β-ABA (Fig. 2).
FIG. 2.
Kinetic resolution reaction profile of the reaction mixture with 50 mM dl-β-ABA, 30 mM pyruvate, and 10 mg of a cell extract of recombinant ΑptA in 10 ml of 0.1 M borate buffer (pH 9.0). The conversion (c •) and ee (▴) of l-β-ABA are shown.
In this study, the basic properties (thermal stability, substrate specificity, and kinetic parameter) of a new l-stereospecific ω-APT (i.e., AptA from A. denitrificans Y2k-2) were characterized. In addition, this study successfully demonstrated the production of d-β-ABA using d,l-β-ABA and pyruvate with AptA. The yield of d-β-ABA from 50 mM d,l-β-ABA was high and efficient (23.5 mM d-β-ABA with 99% ee). Perhaps AptA can be further developed as a useful biocatalyst for the production of other aliphatic d-β-amino acids, such as d-β-leucine and homoleucine, from the corresponding racemic β-amino acids. The directed evolution study of this enzyme to broaden its substrate specificity and to improve its reaction rate for various β-amino acids is in progress now.
Nucleotide sequence accession number.
The nucleotide sequence of the aptA gene has been deposited in the GenBank nucleotide sequence database under accession no. AY330220.
Acknowledgments
This paper was partly supported by Korea Energy Management Corporation and 21C Frontier R&D Programs (Microbial Genomics and Applications Center, Korea).
REFERENCES
- 1.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 2.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christen, P., and D. E. Metzler. 1985. Transaminases. Wiley, New York, N.Y.
- 4.Harmat, N. J., C. Di Bugno, M. Criscuoli, R. Giorgi, A. Lippi, A. Martinelli, S. Monti, and A. Subissi. 1998. 1,2-Disubstituted cyclohexane derived tripeptide aldehydes as novel selective thrombin inhibitors. Bioorg. Med. Chem. Lett. 8:1249-1254. [DOI] [PubMed] [Google Scholar]
- 5.Hill, D. W., F. H. Walters, T. D. Wilson, and J. D. Stuart. 1979. High performance liquid chromatographic determination of amino acids in the picomole range. Anal. Chem. 51:1338-1341. [DOI] [PubMed] [Google Scholar]
- 6.Ito, S., A. Ota, K. Yamamoto, and Y. Kawashima. 1992. Resolution of the enantiomers of thiol compounds by reversed-phase liquid chromatography using chiral derivatization with 2,3,4-tetra-O-acetyl-β-d-glucopyranosyl isothiocyanate. J. Chromatogr. 626:187-196. [Google Scholar]
- 7.Juaristi, E. 1997. Enantioselective synthesis of β-amino acid. Wiley-VCH, New York, N.Y.
- 8.Kamphuis, J., E. M. Meijer, W. H. Boesten, T. Sonke, W. J. van den Tweel, and H. E. Schoemaker. 1992. New developments in the synthesis of natural and unnatural amino acids. Ann. N. Y. Acad. Sci. 672:510-527. [PubMed] [Google Scholar]
- 9.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 10.Liu, M., and M. P. Sibi. 2002. Recent advances in the stereoselective synthesis of β-amino acids. Tetrahedron 58:7991-8035. [Google Scholar]
- 11.Mehta, P. K., and P. Christen. 1994. Homology of 1-aminocyclopropane-1-carboxylate synthase, 8-amino-7-oxononanoate synthase, 2-amino-6-caprolactam racemase, 2,2-dialkylglycine decarboxylase, glutamate-1-semialdehyde 2,1-aminomutase and isopenicillin-N-epimerase with aminotransferases. Biochem. Biophys. Res. Commun. 198:138-143. [DOI] [PubMed] [Google Scholar]
- 12.Mehta, P. K., T. I. Hale, and P. Christen. 1993. Aminotransferases: demonstration of homology and division into evolutionary subgroups. Eur. J. Biochem. 214:549-561. [DOI] [PubMed] [Google Scholar]
- 13.Miura, K., T. Sawa, T. Takeuchi, and H. Umezawa. 1986. Effects of enzyme inhibitors in inhibiting the growth and inducing the differentiation of human promyelocytic leukemia cells, HL-60. J. Antibiot. (Tokyo) 39:734-735. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura, Y., T. Kaneko, S. Sato, M. Ikeuchi, H. Katoh, S. Sasamoto, A. Watanabe, M. Iriguchi, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9:123-130. [DOI] [PubMed] [Google Scholar]
- 15.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, C. M. Fraser, and J. Eisen. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Shin, J.-S., and B.-G. Kim. 1997. Kinetic resolution of α-methylbenzylamine with ω-transaminase screened from soil microorganisms: application of a biphasic system to overcome product inhibition. Biotechnol. Bioeng. 55:348-358. [DOI] [PubMed] [Google Scholar]
- 19.Shin, J.-S., and B.-G. Kim. 1998. Kinetic modeling of ω-transamination for enzymatic kinetic resolution of α-methylbenzylamine. Biotechnol. Bioeng. 60:534-540. [DOI] [PubMed] [Google Scholar]
- 20.Shin, J. S., and B. G. Kim. 2001. Comparison of the omega-transaminases from different microorganisms and application to production of chiral amines. Biosci. Biotechnol. Biochem. 65:1782-1788. [DOI] [PubMed] [Google Scholar]
- 21.Shin, J.-S., B.-G. Kim, A. Liese, and C. Wandrey. 2001. Kinetic resolution of chiral amines using enzyme-membrane reactor. Biotechnol. Bioeng. 73:179-187. [DOI] [PubMed] [Google Scholar]
- 22.Shin, J. S., H. Yun, J. W. Jang, I. Park, and B. G. Kim. 2003. Purification, characterization, and molecular cloning of a novel amine:pyruvate transaminase from Vibrio fluvialis JS17. Appl. Microbiol. Biotechnol. 61:463-471. [DOI] [PubMed] [Google Scholar]
- 23.Stewart, J. D. 2001. Dehydrogenases and transaminases in asymmetric synthesis. Curr. Opin. Chem. Biol. 5:120-129. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, P. P., D. P. Pantaleone, R. F. Senkpeil, and I. G. Fotheringham. 1998. Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends Biotechnol. 16:412-418. [DOI] [PubMed] [Google Scholar]
- 25.Yonaha, K., S. Toyama, and K. Soda. 1987. Omega-amino acid-pyruvate aminotransferase. Methods Enzymol. 143:500-504. [DOI] [PubMed] [Google Scholar]
- 26.Yonaha, K., M. Nishie, and S. Aibara. 1992. The primary structure of omega-amino acid:pyruvate aminotransferase. J. Biol. Chem. 267:12506-12510. [PubMed] [Google Scholar]